Abstract
Four actinomycete strains previously isolated from Brazilian soils were tested for their antimicrobial activity against Bacillus pumilus LF-4 and Desulfovibrio alaskensis NCIMB 13491, bacteria that are well known to be involved in biofilm formation and biocorrosion. Strain 235, belonging to the species Streptomyces lunalinharesii, inhibited the growth of both bacteria. The antimicrobial activity was seen over a wide range of pH, and after treatment with several chemicals and heat but not with proteinase K and trypsin. The antimicrobial substances present in the concentrated supernatant from growth media were partially characterized by SDS-PAGE and extracellular polypeptides were seen. Bands in the size range of 12 to 14.4 kDa caused antimicrobial activity. Transmission electron microscopy of D. alaskensis cells treated with the concentrated supernatant containing the antimicrobial substances revealed the formation of prominent bubbles, the spherical double-layered structures on the cell membrane, and the periplasmic space completely filled with electron-dense material. This is the first report on the production of antimicrobial substances by actinomycetes against bacteria involved in biocorrosion processes, and these findings may be of great relevance as an alternative source of biocides to those currently employed in the petroleum industry.
1. Introduction
Corrosion is a leading cause of pipe failure and high maintenance costs in gas pipelines [1]. Biocorrosion is defined as a corrosive damage initiated or aggravated by the direct or indirect activities of microorganisms [2]. A wide range of bacteria are present in most if not all areas of oil production and have been described from water injection plants, drilling mud, and live reservoir cores [3–6]. Among the aerobic and anaerobic bacteria related to biocorrosion in oil industries, the sulfate-reducing bacteria (SRB) have been extensively studied because as well as forming corrosive biofilms they also produce hydrogen sulfide [3], which can result in health risks to workers [7]. A variety of Bacillus species have been shown to form biofilms on metal surfaces and produce elaborated multicellular communities that display conspicuous architectural features [8]. Jack et al. [9] demonstrated a significant increase in corrosion rates of carbon steel in a continuously flowing freshwater reactor where a biofilm of Bacillus and SRB had been observed.
The most common method for controlling microbial growth in industrial water systems is the use of biocides [10]. Oxidizing (chlorine, ozone) or nonoxidizing compounds (quaternary ammonium salts, aldehydes, and tetrakis (hydroxymethyl)phosphonium sulfate—THPS) are commonly applied [11, 12]. However, the environmental impact and cost of adding large quantities of these compounds must also be considered when such biocides are used [13]. So, the use of biocides of a microbial nature and origin offers another option. Indeed, the production of antimicrobial substances (AMSs) able to inhibit SRB growth has already been shown as an attractive alternative to the use of biocides [5, 13].
Actinomycetes are well known as potent producers of a variety of secondary metabolites with distinct biological activities [14, 15], including AMSs active against both pathogenic [16–18] and phytopathogenic microorganisms [18–20]. However, the production of AMSs active against bacteria involved in biocorrosion process has been poorly described thus far. The exploration of soils and other habitats for microbes of biotechnological interest has led to the isolation of novel actinomycete strains [21]. Brazilian soils offer great potential for bioprospection for novel strains and new bioactive compounds. In previous studies, we have isolated several actinomycete strains from these soils. Therefore, the search among these strains for those with antimicrobial properties against microorganisms that negatively impact on the oil industry was likely to reveal a strain with biotechnology potential.
In this study four different strains of actinomycetes, previously isolated from Brazilian tropical soils and previously selected as promising for AMS production, had their ability to produce AMSs against the Bacillus pumilus LF-4 and the Desulfovibrio alaskensis NCIMB 13491 evaluated. In this paper, one strain, identified as belonging to the Streptomyces lunalinharesii species, is reported and discussed. A preliminary characterization of the AMS obtained from the culture supernatant and its effect on SRB cells are also described. The antimicrobial substance produced by S. lunalinharesii 235 might be an important alternative biocide for use against undesirable bacteria that colonize pipe and flow lines used in the production of oil and gas.
2. Material and Methods
2.1. Bacterial Strains and Growth Conditions
The actinomycete strains used in this study were originally isolated from Brazilian tropical soils prior to 2001. The genus and the origin of these bacterial strains are presented in Table 1. They were previously selected as promising for the production of AMSs against different microbial strains [22–25] and now were used to test their capability to produce AMSs with antimicrobial activity against aerobic and anaerobic bacteria associated with biocorrosion processes. Unless otherwise stated, these strains were grown in yeast extract-malt extract-agar (YMA) [26] under aerobic conditions at 28°C for 7 days.
Table 1.
Genus and origin of the actinomycete strains.
| Strain | Genus | Origin |
|---|---|---|
| 221 | Streptomyces [22] |
Soil of the Atlantic Forest (Vista Chinesa—RJ, Brazil) [23] |
| 224 | nda | |
| 235 | Streptomyces [22] | |
| 606 | Streptomyces [22] | Soil of the Atlantic Forest (Mendanha—RJ, Brazil) [22] |
and: not determined.
The Bacillus pumilus LF-4 and the SRB Desulfovibrio alaskensis NCIMB 13491 were used as indicator strains for the production of AMSs. B. pumilus LF-4 was originally isolated from an oil reservoir in Brazil located in a deep-water production basin at an off-shore platform in Rio de Janeiro [27]. It was grown under aerobic conditions for 24 h at 30°C, in the Luria-Bertani broth (LB) [28]. The SRB strain was isolated from a soured oil reservoir [29] and grown in Postgate C medium [30] at 30°C for 3 days, in anaerobic conditions using sealed serum bottles (10 mL). The bottles were purged with a N2 flux to achieve anaerobiosis. All the bacterial strains were maintained in a long-term storage by freezing the cells at −20°C in 20% glycerol.
2.2. Antimicrobial Activity Assay
For testing the actinomycete strains, the overlay method was performed, as described by Rosado and Seldin [31]. All strains were spot inoculated (5 μL) on the surface of YMA plates, and after incubation at 28°C for 7 days, they were killed by exposure to chloroform vapors for 15 min. The plates were then flooded with either B. pumilus LF-4 or D. alaskensis and incubated at 30°C for 24 h or 5 days, respectively. Manipulation of D. alaskensis was performed in an anaerobic chamber (Plas Labs, Lansing, MI, USA). Clear inhibition zones around the spot inoculum indicated antimicrobial production. The diameters of the inhibition zones were scored as follows: (−) no inhibition, (+) weak inhibition with clear zones <7 mm, (++) moderate inhibition with clear zones between 7 and 12 mm, and (+++) strong inhibition with clear zones >12 mm [32].
To test the antimicrobial activity of the supernatants, 20 μL aliquots were spotted on solid LB media containing a growth lawn of B. pumilus LF-4. Plates were maintained at 30°C/24 h and then inhibition zones were observed and their size recorded.
2.3. Influence of Growth Conditions on the AMS Production
The overlay method was also performed using the actinomycete strains grown in two chemically defined agar media [32], containing a mineral salt solution and either glucose or glycerol as a carbon source. After incubation in aerobic conditions at 28°C for 7 days, the antimicrobial activity of strains was tested against B. pumilus LF-4.
The influence of pH and aeration on AMS production was tested in liquid medium using the chemically defined medium containing glucose [32]. Four different pH values (5.0, 6.0, 7.0, and 8.0) and two aeration conditions (stationary or shaking, 200 rpm) were tested. After incubation at 28°C for 7 days, each culture was filtered with filter Whatman paper no. 1, and the supernatants were lyophilized for further analysis. The lyophilized supernatants were concentrated 100-fold and 300-fold. The antimicrobial activity of each concentrated supernatant was tested as described above.
2.4. Molecular Identification of Streptomyces sp. 235
After growth of Streptomyces sp. 235 in YMA for four days at 28°C with agitation (200 rpm), genomic DNA was extracted as described in earlier reports [33]. PCR amplification of the rrs gene was performed using a GoTaq Green Master Mix Kit (Promega Corporation) according to the manufacturer's instructions. The amplification was carried out using the pair of universal primers 27F [34] and 1541R [35]. The PCR amplification conditions were 35 cycles of 95°C (30 s), 55°C (30 s), and 72°C (50 s) in a thermal cycler model Gene Amp PCR System 9700 (Applied Biosystems). A hot start (5 min at 95°C) was applied to avoid initial mispriming and enhance primer specificity. A final extension step was run for 7 min at 72°C and the reaction tubes were then cooled to 4°C. Amplified fragments were purified using the Illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare), which was used according to the manufacturer's instructions. A negative control (without DNA) was run in all amplifications. DNA preparation and PCR products were visualized after electrophoresis in 1x TBE buffer on a 1.2% agarose gel [36]. The purification product was sequenced by the Center for Human Genome Studies at the University of São Paulo, Brazil. All sequences were identified using BLAST [37] at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast). Sequences retrieved were aligned with the most similar-type strains obtained using CLUSTAL X [38]. BioEdit v. 7.0.0 (http://www.mbio.ncsu.edu/Bioedit/bioedit.html) was used for manual editing of the sequences and a phylogenetic tree with 1000 bootstrap replicates was constructed using MEGA 4 software (http://www.megasoftware.net/). The sequence obtained was deposited in the GenBank database under accession number GU126551.
Genomic DNA-DNA hybridization experiments were performed to compare strain 235 with its closest-type strain relative as determined by 16S rRNA gene sequences. Levels of genomic DNA-DNA similarity were determined at the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ). Cells were disrupted by using a French pressure cell (Thermo Spectronic) and the DNA in the crude lysate was purified by chromatography on hydroxyapatite [39]. DNA-DNA hybridization was carried out as described by De Ley et al. [40] under consideration of the modifications described by Huss et al. [41] using a model Cary 100 Bio UV/VIS spectrophotometer equipped with a Peltier-thermostatted 6X6 multicell changer and a temperature controller with in situ temperature probe (Varian).
2.5. Partial Characterization of AMSs Produced by Strain 235
Partial characterization was performed using a concentrated supernatant containing the AMSs, obtained from growth of the strain 235 in the liquid medium containing glucose at pH 7.0 [32]. After incubation at 28°C for 7 days, the culture was filtered with Whatman paper no. 1, and the supernatant lyophilized and 300-fold concentrated in the same chemically defined medium.
To estimate the molecular mass of AMSs the concentrated supernatant (500 μL) was submitted to centrifugation at 4°C and 3 000 rpm for 1 h in an ultrafiltration membrane (Millipore Corp., USA) with a 10 000 Da molecular mass cut off. Both fractions, the concentrated one (MM > 10 000 Da) and the excluded one (MM < 10 000 Da), were tested for antimicrobial activity against B. pumilus LF-4 in LB plates.
The effect of organic solvents, chemicals, proteolytic enzymes, heat and pH on activity of AMSs was tested (Table 2) using 20 μL of the concentrated supernatant. Systems containing the concentrated supernatant and equal volume of each solvent, chemical, or enzyme were incubated for 2 hours at room temperature (for organic solvents and chemicals) or at 37°C (for enzymes). For the heat treatment, the concentrated supernatant was incubated during 45 or 60 min, at 40, 60, 80, and 100°C, or autoclaved (121°C for 20 min). For pH stability, the concentrated supernatants were mixed to the same volume of citric acid-sodium citrate buffer to achieve different pH values lower than 6.0 and with Tris-HCl buffer for pH higher than 7.0. Antimicrobial activities against B. pumilus LF-4 were determined before and after all treatments.
Table 2.
Properties of the antimicrobial substances produced by Streptomyces lunalinharesii 235.
| Responses to | Sensitiveb |
|---|---|
| Enzymesa | |
| Pronase E | − |
| Proteinase K | + |
| Trypsin | + |
| Solvents (10% and 50%) | |
| Methanol | − |
| Ethanol | − |
| Acetone | − |
| Chloroform | − |
| Chemicals | |
| Urea 6 M | − |
| NaOH (0.2 M and 1 M) | − |
| HCl 0.2 M | − |
| Heat treatment | |
| 40°C for 20, 45 and 60 min | − |
| 60°C for 20, 45 and 60 min | − |
| 80°C for 20, 45 and 60 min | − |
| 100°C for 20, 45 and 60 min | − |
| Autoclavation (121°C for 20 min) | + |
| pHc | |
| 3.0–6.0 | − |
| 7.0–9.0 | − |
aAll enzymes were used at final concentration of 1 mg/mL, and tested with 300-fold concentrated supernatant containing the AMS.
b(−) inhibition zones similar to those observed in control without treatment, (+) no inhibition zone observed.
cpH of supernatants was adjusted from 3.0 to 9.0 varying 1.0 unit before testing their activity.
2.6. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
Ten microliters of the same 300-fold concentrated supernatant described above were treated with equal volume of SDS-PAGE sample buffer (125 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 2% mercaptoethanol, 2% bromophenol blue). Proteins were analyzed in 20–5% gradient SDS-PAGE by the method described by Laemmli [42]. Following electrophoresis conducted at 100 V at 4°C for 140 min, each lane of the gel was cut vertically. One lane was stained with 0.025% Comassie brilliant blue R-250 in methanol-acetic acid-water (40 : 7 : 53) and destained in the same solvent, to determine the secretory protein profile. The apparent molecular masses of the polypeptides were calculated by comparison with the mobility of Full-Range Rainbow Molecular Weight Markers (GE Healthcare, Buckinghamshire, UK). To localize the in situ antimicrobial activity, a bioautography was performed: the other lane was prewashed with 10 volumes of 1% Triton X-100 in water for 1 h at room temperature, under agitation, to remove the SDS. Then, the gel strips were washed three times (20 min each) with double-distilled water, transferred to a LB agar plate, and overlaid with 4 mL semisolid (0.6%) LB containing 0.4 mL of the indicator strain (B. pumilus LF-4). Plates were then incubated in aerobic conditions at 30°C for 24 h and examined for the presence of an inhibitory zone.
2.7. Inhibitory Effect of AMSs on D. alaskensis
The inhibitory effect of AMSs on D. alaskensis was assessed by the determination of the minimal inhibitory concentration (MIC) and/or the minimal bactericide concentration (MBC). The MIC was performed using a microdilution method. The SRB were grown in liquid Postgate C medium for about 24–48 h at 30°C under anaerobic conditions. Experiments were performed using a 96-well plate, where each well contained 100 μL of SRB suspension (107 cells/mL), and suspensions were exposed to different amounts of AMSs (from 0.1 to 0.003 g protein/mL) diluted in Postgate C medium, in order to obtain successive dilutions of AMSs (from 1/1, 1/2, 1/4, 1/8, 1/16, until 1/2032) and a final volume of 200 μL. Experimental controls included a medium without AMSs or a medium plus cell suspensions without AMSs. After incubation at 30°C for 7 days, growth of D. alaskensis was confirmed visually, by the observation of a blackish precipitate (FeS, from the reaction of H2S, produced by the SRB cells, and Fe, present in the medium), and then measured spectrophotometrically at 630 nm. The MIC was defined as the lowest amount of AMSs required to ensure that SRB growth was absent.
To perform the MBC test, aliquots of 10 μL from the wells where growth was absent in the MIC test were used to inoculate fresh Postgate C medium (90 μL). The same controls were used, and after incubation for 7 days at 30°C, the MBC was determined as the lowest concentration where growth was absent. All the inoculation procedures and incubations in MIC and MBC tests were performed in an anaerobic chamber (Plas Labs, Lansing, MI, USA) and the experiments conducted five times. Protein determination was according to Bradford [43].
2.8. Transmission Electron Microscopy (TEM)
TEM was performed to examine the ultrastructural changes in the D. alaskensis cells treated with MIC, sub-MIC, and supra-MIC of AMSs during 7 days. Postgate C medium was also tested as a negative control. The mixtures were harvested by centrifugation at 4 000 ×g for 15 min and the pellet washed three times in a reducing solution (0.0124% sodium thioglycollate, 0.01% ascorbic acid, and 4 mL of a solution of 0.025% resazurin). Then cells were fixed for 2 h, at room temperature, with 2.5% glutaraldehyde and 4% paraformaldehyde in 0.1 M cacodylate buffer, pH 7.2. Postfixation was carried out in 1 % osmium tetroxide in 0.1 M cacodylate buffer for 1 h. Cells were further dehydrated in an acetone series and embedded in PolyBed 812. Ultrathin sections were obtained using a Leica ultramicrotome, stained with uranyl acetate and lead citrate, and observed in a FEI Morgagni TEM at 80 kV.
3. Results
3.1. Antimicrobial Activity Assay
All four actinomycete strains (Table 1) were able to inhibit aerobic B. pumilus LF-4 and anaerobic D. alaskensis when tested using the overlay method in YMA medium. Figure 1 shows the inhibition of D. alaskensis by strain 235, where a 12 mm inhibition zone was observed.
Figure 1.
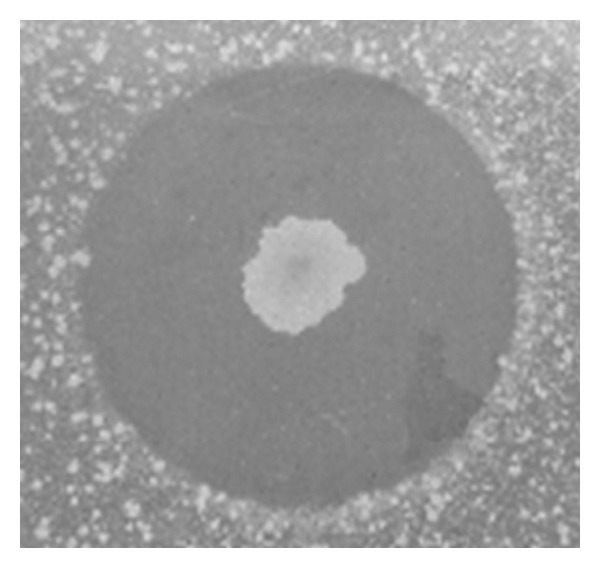
Inhibition zone of D. alaskensis NCIMB 13491 by S. lunalinharesii 235 on agar plate.
3.2. Influence of Growth Conditions on the AMS Production and Strain Selection
All four strains were also able to inhibit B. pumilus LF-4 grown on solid media with either glucose or glycerol as a carbon source. Strains 221 and 235 were more antagonistic on solid media and were selected for further studies in liquid media. Strain 221 supernatants failed to inhibit B. pumilus LF-4 at less than a 400-fold concentration. Given that the 300-fold concentrated supernatant from strain 235 did show inhibitory activity, strain 235 was chosen for identification and further studies.
3.3. Molecular Identification of Streptomyces sp. 235
The identification of strain 235 was carried out by PCR amplification of rrs gene (1498 bp), its sequencing, and then genomic DNA-DNA whole-genome hybridization homology. According to the phylogenetic tree (Figure 2), the most closely related bacterial type strain was Streptomyces lunalinharesii RCQ1071 (accession number DSM 41876T) [44], which shares 99.0% similarity within its 16S rDNA gene sequence. Genomic DNA homology between the Streptomyces lunalinharesii type strain and 235, analyzed by DNA-DNA hybridization (DSMZ service), demonstrated that these strains belong to the same species.
Figure 2.
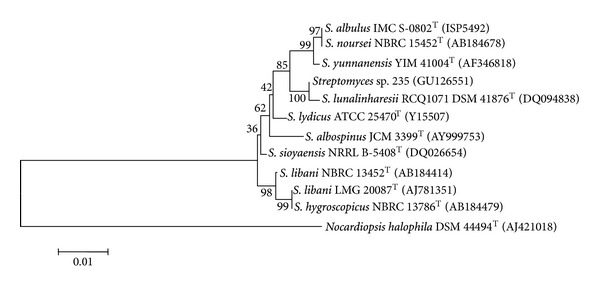
Phylogenetic tree obtained by the neighbor-joining method, based on the alignment of 16S rDNA of strain 235 and other Streptomyces species. Nocardiopsis halophila was used as an out-group. Bootstrap analyses were performed with 1000 repetitions. The scale bar corresponds to 0.01 substitutions per nucleotide position.
3.4. Partial Characterization of AMSs Produced by Streptomyces sp. 235
The AMS that was retained by the ultrafiltration membrane (10 000 Da cutoff) and its characterization can be seen in Table 2. The antimicrobial activity was resistant to pronase E, different chemicals (urea 6 M, NaOH 0.2 and 1 M, and HCl 0.2 M) and organic solvents (acetone, ethanol, methanol, and chloroform at 10 and 50%) but sensitive to proteinase K and trypsin. It was heat stable after incubation at 100°C for 1 h but did not maintain its activity after being autoclaved at 121°C for 20 min. It was active in the range of pH values between 3.0 and 9.0.
3.5. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
Streptomyces sp. 235 was able to secrete polypeptides to the extracellular environment during its growth in a chemically defined medium for 7 days, as demonstrated by SDS-PAGE analysis (Figure 3(a)). The strain released a large amount of extracellular polypeptides with molecular masses varying from 12 to 111 kDa, including one region of the gel (from 12 to 14.4 kDa), which exhibited inhibitory activity when overlaid with the indicator strain B. pumilus LF-4 (Figure 3(b)).
Figure 3.
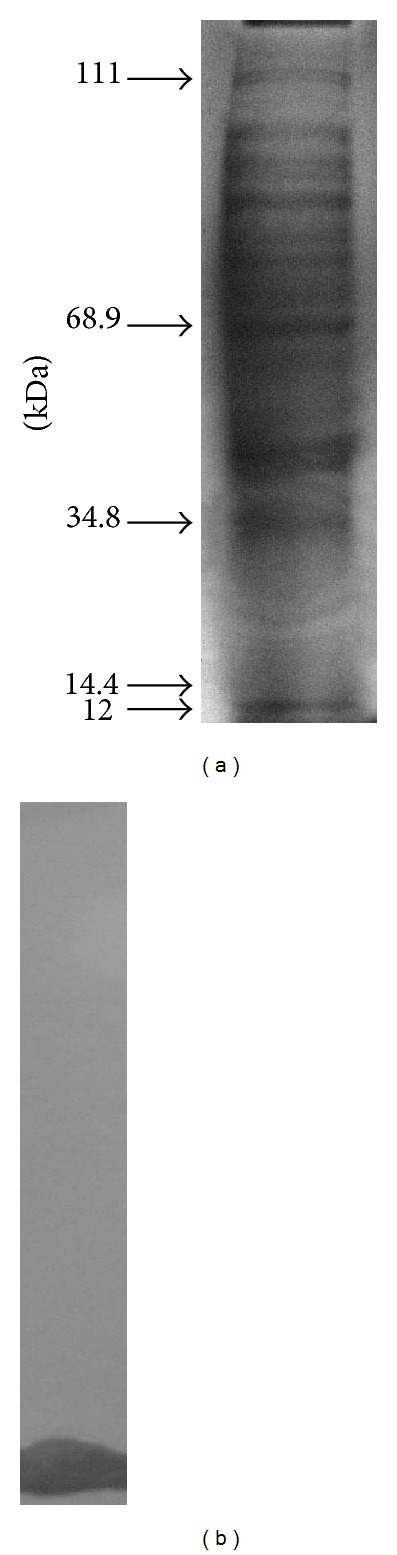
Antimicrobial activity profile presented by strain 235 in SDS-PAGE. The gel strips containing 300-fold concentrated supernatant fluids were stained with Coomassie brilliant blue to reveal the secretory protein profile (a) or overlaid with semisolid LB agar containing the indicator strain (LF-4) to show the inhibition zones (b). For details see text. Numbers on the left indicate relative molecular mass markers (in kDa).
3.6. Inhibitory Effect of AMSs against D. alaskensis
Experiments concerning the type of activity of AMSs against D. alaskensis cells have shown a bacteriostatic effect at a 0.03 g protein/mL (MIC), which corresponded to a 1/4 dilution of the 300-fold concentrated supernatant, whereas a bactericidal effect was observed at a 0.05 g protein/mL (MBC), which corresponded to the 1/2 dilution.
3.7. Transmission Electron Microscopy (TEM)
Untreated D. alaskensis cells in Postgate C medium showed an undamaged structure of the inner membrane and an intact, slightly waved outer membrane, and the periplasmic space was thin with a uniform appearance (Figure 4(a)). D. alaskensis cells treated with sub-MIC (0.01 g protein/mL) of the concentrated supernatant of AMSs presented spherical double-layered structures between inner and outer membranes and out of the cell (black arrows in Figure 4(b)). After incubation with a MIC (0.03 g protein/mL) of AMSs, numerous spherical double-layered structures could be observed (black arrow in Figures 4(c)-4(d)). The periplasmic space became completely filled with electron-dense material (Figures 4(c)-4(d)). For a supra-MIC (0.05 g protein/mL) of AMSs, the formation of prominent bubbles emerging from the cell wall surface was observed (black arrows in Figures 4(e)-4(f)), whereas an electron-dense material was still visible in the periplasmic space (Figures 4(e)-4(f)).
Figure 4.
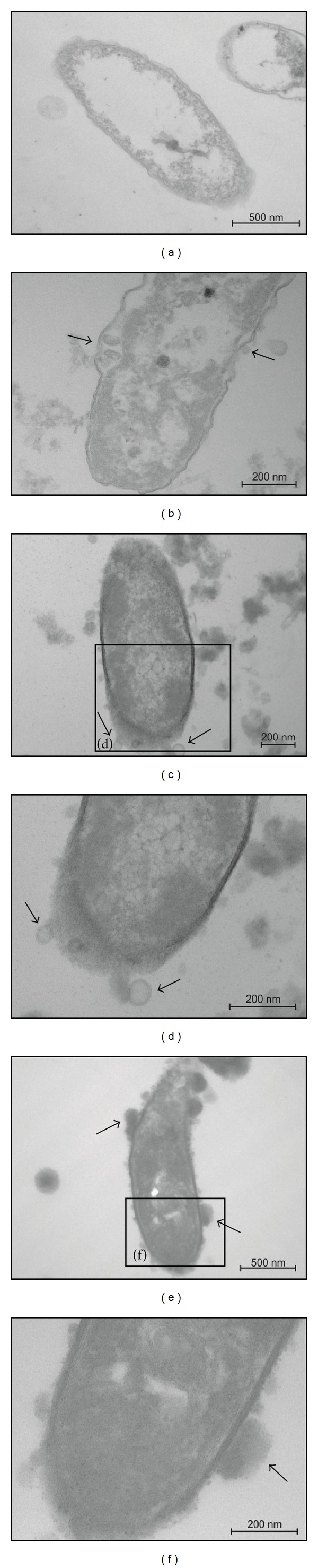
Transmission electron microscopy micrographs of D. alaskensis cells. In untreated cells, the inner and outer membranes are visible as continuous and intact structures (a). After treatment with a sub-MIC of AMSs, spherical double-layered structures inside and outside the cell can be observed ((b), black arrow), and the periplasmic space filled with electron-dense material. MIC of AMSs causes the formation of numerous spherical double-layered structures ((c)-(d), black arrows) and the periplasmic space to be filled with electron-dense material. After treatment with a supra-MIC of AMSs, the periplasmic space is still filled with electron-dense material and prominent bubbles are emerging from the cell surface ((e)-(f), black arrows).
4. Discussion
Although the four actinomycete strains isolated from the Brazilian soils were able to effectively inhibit the growth of B. pumilus LF-4 and D. alaskensis NCIMB 13491 in vitro, strain 235 was the only one able to inhibit them on solid and in liquid media. This strain was originally isolated from soil of the Atlantic Forest, Vista Chinesa, RJ [23], and characterized as belonging to the Streptomyces genus [22]. The molecular characterization by PCR amplification of the rrs gene had 99.0% identity with rrs gene sequences of Streptomyces lunalinharesii RCQ1071. Within the Streptomyces genus, the rrs gene sequences are highly conserved and sequence identities are high and can be more than 99%. As a result, it becomes very difficult to identify streptomycete species based solely on 16S rDNA sequencing, even when 99.0% similarity to a type strain is observed [45]. Dastager et al. [46] described Streptomyces deccanensis as a new Streptomyces species even though its rrs gene is 99.4% similar to its most closely related type strain. Whole-genome DNA-DNA hybridization remains the “gold standard” to distinguish bacterial species and is necessary to decide whether a strain belongs to a species. When DNA-DNA homology is less than 70%, then the two strains being compared do not belong to the same species. Strain 235 shared around 100% homology with S. lunalinharesii RCQ1071, which is far above the 70% threshold recommended for the recognition of separate genomic species [47]. These results have clearly demonstrated that strain 235 indeed belongs to this same species.
S. lunalinharesii-type strain RCQ1071T was described recently by our group [44]. It was isolated from another Brazilian soil, in the Central Plateau, under cerrado vegetation [48]. A preliminary study has shown it has the ability to produce antimicrobial substances against human pathogenic microorganisms [22]. Its activity against some phytopathogenic fungi was assessed and the strain was characterized as an excellent chitinase producer [48]. In another study by our group [49], strain 80 from this same species has been shown to be a promising strain for the biological control of Sclerotinia sclerotiorum. Here we report on a previously undescribed strain of this species which produces bioactive compounds able to inhibit bacteria involved in the microbial colonization and corrosion of pipe systems in the oil industry.
S. lunalinharesii strain 235 was grown in a mineral salt solution containing glucose, pH 7.0, at 28°C for 7 days without shaking. These growth conditions for AMS production are very interesting if we think about future biotechnological application: the use of glucose, a simple, cheap, and easy carbon source along with a mineral salt solution would be very appropriate for an industrial scale production; the use of a neutral pH and fermentation without agitation would be also be very favorable. Most of the methods for antibiotic production described in the literature for Streptomyces [50], other bacteria [5], and fungi [51] require similar conditions of carbon source, pH, and temperature. However, they require growth under agitation. Von Der Weid et al. [32] also observed maximum antimicrobial activity when Paenibacillus peoriae was cultivated in a chemically defined medium containing glucose at pH 7.0 and incubation at 30°C in stationary conditions.
Peptidic antibiotics, an abundant class of special metabolites, are commonly produced by many microbial species including Streptomyces. The preliminary tests for the characterization of AMSs produced by S. lunalinharesii 235 have shown that its antimicrobial activity was sensitive to trypsin and proteinase K, indicating its proteic nature. The SDS-PAGE analysis and the presence of an inhibitory zone between 12 and 14.4 kDa have confirmed this finding. This apparent molecular mass range is in accordance with the ultrafiltration membrane approach. However, the AMS was resistant to pronase E suggesting that it is resistant to some types of proteolytic cleavage. The resistance to proteolytic enzymes may indicate the presence of unusual amino acids in the AMS structure. Moreover, the AMS could also present a compact structure, or the lack of cleavage recognition sites, making it resistant to proteolytic enzymes. Cyclic peptides can be resistant to hydrolysis by proteases because their cyclic structure renders them relatively inflexible, which may make cleavage sites inaccessible because of steric hindrance [52].
Another interesting characteristic shown by the AMS in this study was its resistance to organic solvents, indicating that its structure should not contain a lipid portion. Also this AMS was able to resist different chemicals and continued to inhibit growth over a range of pH values and high temperatures. Similar characteristics to those of the AMSs produced by S. lunalinharesii have been observed for the AMSs produced by other bacterial genera, such as Bacillus [5, 53–56], Paenibacillus [32, 57], Lactobacillus [58], and Enterococcus [59]. AMS from Paenibacillus peoriae, for instance, presented a high stability after treatments with a broad pH range (3.2–9.6), heat (100°C for 1 h or 121°C for 10 min), proteolytic enzymes and organic solvents, among other tests [32]. Bacillus subtilis, B. firmus, and B. licheniformis, isolated from an oil reservoir in Brazil, have also produced AMSs that are stable at 100°C for 1 h as well as in presence of other chemicals. In those studies, however, the AMSs were resistant to different proteolytic enzymes and sensitive to several organic solvents, indicating a different chemical nature [5]. Lactobacillus paracasei strains have produced an AMS stable at 100°C for only 3 min, and its inhibitory activity was also totally lost after treatment with different proteolytic enzymes [58]. Enterococcus faecium was able to produce a bacteriocin which has shown stability at 30°C for 1 h and at 100°C for 30 min in the pH range 2.0–7.0 and 2.0–5.0, respectively, being also sensitive to different proteolytic enzymes [59]. The stability at high temperatures and different pH values, in particular, may be very useful in oil reservoirs, where temperatures above 60°C and pH values ranging between 3.0 and 7.0 are usually found [4].
Cellular alterations caused by AMSs in D. alaskensis cells observed by TEM were basically related to the appearance of an electron-dense material in the periplasmic space and to membranes, where spherical double-layered structures appeared and increased from sub-MIC to MIC of AMSs, and prominent bubbles emerge from the cell wall surface at a supra-MIC concentration. Some of these alterations have been observed in other bacterial strains treated with antimicrobial peptides. For instance, Meincken et al. [60] treated Escherichia coli cells with the synthetic peptide peptidyl-glycyl-leucine-carboxamide (PGLa) and observed numerous regularly distributed protrusions from cell surface. Latter, Hartmann et al. [61] have treated E. coli and Staphylococcus aureus cells with gramicidin S, extracted from Aneurinibacillus migulanus, or with PGLa, and observed additional intracellular membranous structures and the periplasmic space completely filled with an electron-dense material. They have also observed numerous bubbles which protruded from E. coli cell surface.
5. Conclusions
Here, for the first time, we report on an antimicrobial substance (AMS) produced by a streptomycete strain, with activity against a sulfate-reducing bacterium D. alaskensis NCIMB 13491 and an aerobic bacterium B. pumilus LF-4, known to be of importance in corrosion. Strain 235 was identified as belonging to S. lunalinharesii species cluster, was originally isolated from a Brazilian soil, and was already known as producer of bioactive compounds against phytopathogenic bacteria and fungi. The AMS, of proteic nature, has shown to be promising for use in oil production plants, given its stability in the presence of several chemicals and solvents, and over a wide range of pH and temperature values. According to TEM, its mechanism of action is probably related to cell membrane alteration and appearance of an electron-dense material in the periplasmic space. The results obtained in this study stimulate further detailed biochemical, molecular, chromatographic, and spectrometric analyses. Having now identified the strain and with the elucidation of the primary structure of the antimicrobial substance, we may have an AMS control agent for sulfate-reducing bacteria commonly found in biofilm and corrosion in the petroleum industry.
Acknowledgments
This work was supported by grants from PETROBRAS, Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ).
References
- 1.Zhu XY, Lubeck J, Kilbane JJ. Characterization of microbial communities in gas industry pipelines. Applied and Environmental Microbiology. 2003;69(9):5354–5363. doi: 10.1128/AEM.69.9.5354-5363.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zuo R. Biofilms: strategies for metal corrosion inhibition employing microorganisms. Applied Microbiology and Biotechnology. 2007;76(6):1245–1253. doi: 10.1007/s00253-007-1130-6. [DOI] [PubMed] [Google Scholar]
- 3.Feio MJ, Rainha V, Reis MA, Lino AR, Fonseca ITE. The influence of the Desulfovibrio desulfuricans 14 ATCC 27774 on the corrosion of mild steel. Materials and Corrosion—Werkstoffe und Korrosion. 2000;51(10):691–697. [Google Scholar]
- 4.Magot M, Ollivier B, Patel BKC. Microbiology of petroleum reservoirs. Antonie van Leeuwenhoek. 2000;77(2):103–116. doi: 10.1023/a:1002434330514. [DOI] [PubMed] [Google Scholar]
- 5.Korenblum E, Von Der Weid I, Santos ALS, et al. Production of antimicrobial substances by Bacillus subtilis LFE-1, B. firmus H2O-1 and B. licheniformis T6-5 isolated from an oil reservoir in Brazil. Journal of Applied Microbiology. 2005;98(3):667–675. doi: 10.1111/j.1365-2672.2004.02518.x. [DOI] [PubMed] [Google Scholar]
- 6.Von Der Weid I, Korenblum E, Jurelevicius D, et al. Molecular diversity of bacterial communities from subseafloor rock samples in a deep-water production basin in Brazil. Journal of Microbiology and Biotechnology. 2008;18(1):5–14. [PubMed] [Google Scholar]
- 7.Gardner LR, Stewart PS. Action of glutaraldehyde and nitrite against sulfate-reducing bacterial biofilms. Journal of Industrial Microbiology and Biotechnology. 2002;29(6):354–360. doi: 10.1038/sj.jim.7000284. [DOI] [PubMed] [Google Scholar]
- 8.Branda SS, González-Pastor JE, Dervyn E, Ehrlich SD, Losick R, Kolter R. Genes involved in formation of structured multicellular communities by Bacillus subtilis. Journal of Bacteriology. 2004;186(12):3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jack RF, Ringelberg DB, White DC. Differential corrosion rates of carbon steel by combinations of Bacillus sp., Hafnia alvei and Desulfovibrio gigas established by phospholipid analysis of electrode biofilm. Corrosion Science. 1992;33(12):1843–1853. [Google Scholar]
- 10.Videla HA, Herrera LK. Microbiologically influenced corrosion: looking to the future. International Microbiology. 2005;8(3):169–180. [PubMed] [Google Scholar]
- 11.Larsen J, Sanders PF, Talbot RE. Experience with the use of tetrakis hydroxymethyl phosphonium sulfate (THPS) for the control of downhole hydrogen sulfide. Proceedings of the Corrosion Conference; March 2000; NACE; [Google Scholar]
- 12.Videla HA. Prevention and control of biocorrosion. International Biodeterioration and Biodegradation. 2002;49(4):259–270. [Google Scholar]
- 13.Zuo R, Örnek D, Syrett BC, et al. Inhibiting mild steel corrosion from sulfate-reducing bacteria using antimicrobial-producing biofilms in Three-Mile-Island process water. Applied Microbiology and Biotechnology. 2004;64(2):275–283. doi: 10.1007/s00253-003-1403-7. [DOI] [PubMed] [Google Scholar]
- 14.Berdy J. Bioactive microbial metabolites. Journal of Antibiotics (Tokyo) 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 15.Ben Ameur Mehdi R, Sioud S, Fourati Ben Fguira L, Bejar S, Mellouli L. Purification and structure determination of four bioactive molecules from a newly isolated Streptomyces sp. TN97 strain. Process Biochemistry. 2006;41(7):1506–1513. [Google Scholar]
- 16.Sun P, Maloney KN, Nam SJ, et al. Fijimycins A-C, three antibacterial etamycin-class depsipeptides from a marine-derived Streptomyces sp. Bioorganic & Medicinal Chemistry. 2011;19:6557–6562. doi: 10.1016/j.bmc.2011.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trenozhnikova LP, Khasenova AK, Balgimbaeva AS, et al. Characterization of the antibiotic compound no. 70 produced by Streptomyces sp. IMV-70. The Scientific World Journal. 2012;2012:8 pages. doi: 10.1100/2012/594231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiong ZQ, Zhang ZP, Li JH, Wei SJ, Tua GQ. Characterization of Streptomyces padanus JAU4234, a producer of actinomycin X2, fungichromin, and a new polyene macrolide antibiotic. Applied and Environmental Microbiology. 2012;78(2):589–592. doi: 10.1128/AEM.06561-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain PK, Jain PC. Isolation, characterization and antifungal activity of Streptomyces sampsonii GS 1322. Indian Journal of Experimental Biology. 2007;45(2):203–206. [PubMed] [Google Scholar]
- 20.Prapagdee B, Kuekulvong C, Mongkolsuk S. Antifungal potential of extracellular metabolites produced by Streptomyces hygroscopicus against phytopathogenic fungi. International Journal of Biological Sciences. 2008;4(5):330–337. doi: 10.7150/ijbs.4.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ouhdouch Y, Barakate M, Finance C. Actinomycetes of Moroccan habitats: isolation and screening for antifungal activities. European Journal of Soil Biology. 2001;37(2):69–74. [Google Scholar]
- 22.Semêdo LTAS, Linhares AA, Gomes RC, et al. Isolation and characterization of actinomycetes from Brazilian tropical soils. Microbiological Research. 2001;155(4):291–299. doi: 10.1016/S0944-5013(01)80007-0. [DOI] [PubMed] [Google Scholar]
- 23.Coelho RRR, Lopes A, Semêdo LTAS, Cruz FS. Culture filtrates of actinomycetes isolated from tropical soils inhibit Trypanosoma cruzi replication in vitro. Revista de Microbiologia. 1995;26(4):307–313. [Google Scholar]
- 24.Reis SA, Costa LV, Cavalcanti EDC, et al. Protein synthesis inhibitory activity in culture filtrates from new strains of Streptomyces isolated from Brazilian tropical soils. Letters in Applied Microbiology. 2003;37(2):138–143. doi: 10.1046/j.1472-765x.2003.01363.x. [DOI] [PubMed] [Google Scholar]
- 25.Sacramento DR, Coelho RRR, Wigg MD, et al. Antimicrobial and antiviral activities of an actinomycete (Streptomyces sp.) isolated from a Brazilian tropical forest soil. World Journal of Microbiology and Biotechnology. 2004;20(3):225–229. [Google Scholar]
- 26.Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. International Journal of Systematic Bacteriology. 1966;16(3):313–340. [Google Scholar]
- 27.Sebastián GV. Avaliação da população bacteriana presente em um reservatório de petróleo situado em águas profundas brasileiras, com ênfase no isolamento e caracterização de estirpes de Bacillus [dissertação] Universidade Federal do Rio de Janeiro; 1999. [Google Scholar]
- 28.Schleif RF, Wensink PC. Practical Methods in Molecular Biology. New York, NY, USA: Springer; 1981. [Google Scholar]
- 29.Feio MJ, Zinkevich V, Beech IB, et al. Desulfovibrio alaskensis sp. nov., a sulphate-reducing bacterium from a soured oil reservoir. International Journal of Systematic and Evolutionary Microbiology. 2004;54(5):1747–1752. doi: 10.1099/ijs.0.63118-0. [DOI] [PubMed] [Google Scholar]
- 30.Postgate JR. The Sulfate-Reducing Bacteria. Cambridge, UK: Cambridge University Press; 1984. [Google Scholar]
- 31.Rosado AS, Seldin L. Production of a potentially novel anti-microbial substance by Bacillus polymyxa . World Journal of Microbiology & Biotechnology. 1993;9(5):521–528. doi: 10.1007/BF00386287. [DOI] [PubMed] [Google Scholar]
- 32.Von Der Weid I, Alviano DS, Santos ALS, Soares RMA, Alviano CS, Seldin L. Antimicrobial activity of Paenibacillus peoriae strain NRRL BD-62 against a broad spectrum of phytopathogenic bacteria and fungi. Journal of Applied Microbiology. 2003;95(5):1143–1151. doi: 10.1046/j.1365-2672.2003.02097.x. [DOI] [PubMed] [Google Scholar]
- 33.Valente P, Gouveia FC, Lemos GA, Pimentel D, Mendonça-Hagler LC, Hagler AN. PCR-amplified ITS length variation within the yeast genus Metschnikowia . Journal of General and Applied Microbiology. 1997;43(3):179–181. doi: 10.2323/jgam.43.179. [DOI] [PubMed] [Google Scholar]
- 34.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology. 1991;173(2):697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Löffler FE, Sun Q, Li J, Tiedje JM. 16S rRNA gene-based detection of tetrachloroethene-dechlorinating Desulfuromonas and Dehalococcoides species. Applied and Environmental Microbiology. 2000;66(4):1369–1374. doi: 10.1128/aem.66.4.1369-1374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. New York, NY, USA: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 38.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cashion P, Holder Franklin MA, McCully J, Franklin M. A rapid method for the base ratio determination of bacterial DNA. Analytical Biochemistry. 1977;81(2):461–466. doi: 10.1016/0003-2697(77)90720-5. [DOI] [PubMed] [Google Scholar]
- 40.De Ley J, Cattoir H, Reynaerts A. The quantitative measurement of DNA hybridization from renaturation rates. European Journal of Biochemistry. 1970;12(1):133–142. doi: 10.1111/j.1432-1033.1970.tb00830.x. [DOI] [PubMed] [Google Scholar]
- 41.Huss VAR, Festl H, Schleifer KH. Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Systematic and Applied Microbiology. 1983;4(2):184–192. doi: 10.1016/S0723-2020(83)80048-4. [DOI] [PubMed] [Google Scholar]
- 42.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 43.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Analytical Biochemistry. 1976;72(1-2):248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 44.de Souza RF, Coelho RRR, Macrae A, et al. Streptomyces lunalinharesii sp. nov., a chitinolytic streptomycete isolated from cerrado soil in Brazil. International Journal of Systematic and Evolutionary Microbiology. 2008;58(12):2774–2778. doi: 10.1099/ijs.0.65768-0. [DOI] [PubMed] [Google Scholar]
- 45.Lanoot B, Vancanneyt M, Hoste B, et al. Grouping of streptomycetes using 16S-ITS RFLP fingerprinting. Research in Microbiology. 2005;156(5-6):755–762. doi: 10.1016/j.resmic.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 46.Dastager SG, Kim CJ, Lee JC, Agasar D, Park DJ, Li WJ. Streptomyces deccanensis sp. nov., an alkaliphilic species isolated from soil. International Journal of Systematic and Evolutionary Microbiology. 2008;58(5):1089–1093. doi: 10.1099/ijs.0.65525-0. [DOI] [PubMed] [Google Scholar]
- 47.Wayne LG, Brenner DJ, Colwell RR, et al. Report of the Ad Hoc committee on reconciliation of approaches to bacterial systematics. International Journal of Systematic Bacteriology. 1987;37(4):463–464. [Google Scholar]
- 48.Gomes RC, Sêmedo LTAS, Soares RMA, et al. Purification of a thermostable endochitinase from Streptomyces RC1071 isolated from a cerrado soil and its antagonism against phytopathogenic fungi. Journal of Applied Microbiology. 2001;90(4):653–661. doi: 10.1046/j.1365-2672.2001.01294.x. [DOI] [PubMed] [Google Scholar]
- 49.Fróes A, Macrae A, Rosa J, et al. Selection of a Streptomyces strain able to produce cell wall degrading enzymes and active against Sclerotinina sclerotiorum . Journal of Microbiology. 2012;50(5):798–806. doi: 10.1007/s12275-012-2060-2. [DOI] [PubMed] [Google Scholar]
- 50.Thakur D, Bora TC, Bordoloi GN, Mazumdar S. Influence of nutrition and culturing conditions for optimum growth and antimicrobial metabolite production by Streptomyces sp. 201. Journal de Mycologie Medicale. 2009;19(3):161–167. [Google Scholar]
- 51.Yu J, Liu Q, Liu Q, et al. Effect of liquid culture requirements on antifungal antibiotic production by Streptomyces rimosus MY02. Bioresource Technology. 2008;99(6):2087–2091. doi: 10.1016/j.biortech.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 52.Eckart K. Mass spectrometry of cyclic peptides. Mass Spectrometry Reviews. 1994;13(1):23–55. [Google Scholar]
- 53.Cherif A, Chehimi S, Limem F, et al. Detection and characterization of the novel bacteriocin entomocin 9, and safety evaluation of its producer, Bacillus thuringiensis ssp. entomocidus HD9. Journal of Applied Microbiology. 2003;95(5):990–1000. doi: 10.1046/j.1365-2672.2003.02089.x. [DOI] [PubMed] [Google Scholar]
- 54.Cladera-Olivera F, Caron GR, Brandelli A. Bacteriocin-like substance production by Bacillus licheniformis strain P40. Letters in Applied Microbiology. 2004;38(4):251–256. doi: 10.1111/j.1472-765x.2004.01478.x. [DOI] [PubMed] [Google Scholar]
- 55.Kavitha S, Senthilkumar S, Gnanamanickam S, Inayathullah M, Jayakumar R. Isolation and partial characterization of antifungal protein from Bacillus polymyxa strain VLB16. Process Biochemistry. 2005;40(10):3236–3243. [Google Scholar]
- 56.He L, Chen WL, Liu Y. Production and partial characterization of bacteriocin-like pepitdes by Bacillus licheniformis ZJU12. Microbiological Research. 2006;161(4):321–326. doi: 10.1016/j.micres.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 57.Guo Y, Huang E, Yuan C, Zhang L, Yousef AE. Isolation of a Paenibacillussp. strain and structural elucidation of its broad-spectrum lipopeptide antibiotic. Applied and Environmental Microbiology. 2012;78(9):3156–3165. doi: 10.1128/AEM.07782-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Atanassova M, Choiset Y, Dalgalarrondo M, et al. Isolation and partial biochemical characterization of a proteinaceous anti-bacteria and anti-yeast compound produced by Lactobacillus paracasei subsp. paracasei strain M3. International Journal of Food Microbiology. 2003;87(1-2):63–73. doi: 10.1016/s0168-1605(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 59.Ohmomo S, Murata S, Katayama N, et al. Purification and some characteristics of enterocin ON-157, a bacteriocin produced by Enterococcus faecium NIAI 157. Journal of Applied Microbiology. 2000;88(1):81–89. doi: 10.1046/j.1365-2672.2000.00866.x. [DOI] [PubMed] [Google Scholar]
- 60.Meincken M, Holroyd DL, Rautenbach M. Atomic force microscopy study of the effect of antimicrobial peptides on the cell envelope of Escherichia coli . Antimicrobial Agents and Chemotherapy. 2005;49(10):4085–4092. doi: 10.1128/AAC.49.10.4085-4092.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hartmann M, Berditsch M, Hawecker J, Ardakani MF, Gerthsen D, Ulrich AS. Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy. Antimicrobial Agents and Chemotherapy. 2010;54(8):3132–3142. doi: 10.1128/AAC.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]


