Abstract
Background & Aims
Hepatocyte nuclear factor 4 alpha (HNF4α) is a transcription factor that has been shown to be required for hepatocyte differentiation and development of the liver. It has also been implicated in regulating expression of genes that act in the epithelium of the lower gastrointestinal tract. This implied that HNF4α might be required for development of the gut.
Methods
Mouse embryos were generated in which Hnf4a was ablated in the epithelial cells of the fetal colon by using Cre-loxP technology. Embryos were examined by using a combination of histology, immunohistochemistry, DNA microarray, reverse-transcription polymerase chain reaction, electrophoretic mobility shift assays, and chromatin immunoprecipitation analyses to define the consequences of loss of HNF4α on colon development.
Results
Embryos were recovered at E18.5 that lacked HNF4α in their colons. Although early stages of colonic development occurred, HNF4α-null colons failed to form normal crypts. In addition, goblet-cell maturation was perturbed and expression of an array of genes that encode proteins with diverse roles in colon function was disrupted. Several genes whose expression in the colon was dependent on HNF4α contained HNF4α-binding sites within putative transcriptional regulatory regions and a subset of these sites were occupied by HNF4α in vivo.
Conclusions
HNF4α is a transcription factor that is essential for development of the mammalian colon, regulates goblet-cell maturation, and is required for expression of genes that control normal colon function and epithelial cell differentiation.
The primary functions of the gastrointestinal tract are to process ingested food, absorb nutrients and water, and regulate energy homeostasis.1–4 The gut tube consists of tissue layers called tunics that contribute to specific aspects of gut function. The innermost tunic is the mucosa, which contains an epithelial layer that interfaces with the lumen of the gut and is derived from the definitive endoderm that forms during gastrulation.5,6 At around embryonic day (E) 8.5 in mouse development, the endoderm invaginates to form 2 portals. An anterior portal generates the foregut, and the hindgut is formed from the caudal portal.5 Initially, the gut endoderm forms a simple tube consisting of a psuedostratified epithelium surrounded by gut mesenchyme.5,7 At around E14.5, the epithelium transitions to a simple columnar epithelium and the topology of the mucosa changes to include villi and crypts within the small intestine and only crypts within the colon.5 The architecture and tissue patterning of the gut is established during embryonic development and involves cross-talk between the fetal gut endoderm and mesenchyme.2,8 Signaling pathways that control patterning of the gut epithelium have been intensely studied and include wingless-related MMTV integration site proteins, Hedgehog, fibroblast growth factor, bone morphogenic protein, Notch, and platelet-derived growth factor.2,7,9 In addition, several transcription factors have been identified that are required for epithelial cell differentiation. Atoh1 (also called Math1) controls the commitment of cellular differentiation toward the secretory cell lineage, a process that is antagonized by Hes110,11; Klf4 and Ngn3 are required for establishing goblet and enteroendocrine cells,12–14 respectively; and Elf3 is essential for the terminal differentiation of both columnar and goblet cells in the small intestine.15 Analyses of transcriptional regulatory elements within genomic DNA have suggested that hepatocyte nuclear factor 4 alpha (HNF4α) also regulates expression of many genes throughout the lower gastrointestinal tract.16–23 In addition, studies using the Caco2 intestinal epithelial tumor cell line have shown that HNF4α occupies the promoter of the α1-antitrypsin (Serpina1) gene coincident with the onset of expression of α1-antitrypsin during Caco2 cell differentiation.24 Further support for HNF4α being an important developmental regulator comes from studies using knockout mice. Most relevant is the finding that ablation of Hnf4a in fetal livers prevents the hepatic parenchyma from establishing an epithelium and blocks hepatocyte differentiation.25–27
To directly address the role of HNF4α in development of the colon, mice were derived in which the Hnf4a gene was ablated in the colonic epithelial cells by using the Cre-loxP system.28 Examination of these embryos revealed that loss of HNF4α disrupts formation of crypt topology in fetal colons by E18.5. Loss of crypts was accompanied by reduced goblet cell maturation and proliferation of epithelial cells. Gene expression and HNF4α DNA–binding analyses revealed that loss of HNF4α is required for expression of several target genes that are central to colon function.
Materials and Methods
Animals
The derivation of Hnf4a+/− (Hnf4atm1Dnl), Hnf4aloxP/loxP (Hnf4atm1Sad), and Foxa3Cre (Tg[Foxa3-cre]1Khk) mice has been described previously.28–30 Mating of Hnf4a+/− and Hnf4aloxP/loxP with Foxa3Cre mice produced Hnf4a+/−Foxa3Cre and Hnf4aloxP/+ Foxa3Cre male mice that were used as studs. These males were mated with Hnf4aloxP/loxP female mice to generate control Hnf4aloxP/+Foxa3Cre or experimental Hnf4aloxP loxPFoxa3Cre and Hnf4aloxP/−Foxa3Cre embryos. Analysis of HNF4α loss of function was restricted to the colon because deletion of HNF4α was inefficient in the small intestine. Noon on the day of the appearance of a vaginal plug was considered as 0.5 days postcoitum, and the genotype of all embryos was determined by polymerase chain reaction (PCR) analysis of genomic DNA. The Medical College of Wisconsin’s IACUC committee approved all animal experiments and procedures.
Histology and Immunohistochemistry
Five millimeters of colon lying 5 mm distal to the cecum was dissected from E18.5 embryos in phosphate-buffered saline, fixed overnight with 4% paraformaldehyde (Sigma, St Louis, MO) in phosphate-buffered saline, and embedded in paraffin. Five-micrometer paraffin sections were used in all staining procedures. H&E, alcian blue, and β-galactosidase staining were performed by using standard histological protocols.31 Immunohistochemistry was performed by using antigen retrieval in citrate buffer as described previously,26 except for PECAM1 (platelet/endothelial cell adhesion molecule 1) staining in which sections were treated with 0.25% trypsin for 20 minutes at room temperature. Primary antibodies recognized HNF4α (sc-6556, 1:500; Santa Cruz, Santa Cruz, CA), E-cadherin (#610181, 1:16,000; BD Transduction Laboratories, San Jose, CA), Ki-67 (sc-7846, 1:500; Santa Cruz), phosphoHistone H3 (#06-570, 1:1500; Upstate, Charlottesville, VA), acetylated tubulin (T6793, 1:1000; Sigma, St Louis, MO), α smooth-muscle actin (A2547, 1:1600; Sigma), laminin (L9393, 1:500; Sigma), and PECAM1 (#553370, 1:50; Pharmingen, San Jose, CA). Micrographs were collected by using a SPOT digital camera (Diagnostic Instruments, Sterling Heights, MI) and images from control and experimental samples were processed identically using Adobe Photoshop 7.0 (Adobe, San Jose, CA). Cell counts were taken from digital images of 3 sections from each embryo analyzed. Thickness of the circular muscle was measured using Metamorph software (Molecular Devices, Sunnyvale, CA). Data were subjected to statistical analyses by using analysis of variance (ANOVA) (StatView 5.0.1; Adept Scientific Inc, Acton, MA) and were considered significant at P < .001.
DNA Microarrays and Reverse-Transcriptase PCR
Affymetrix GeneChip Mouse Expression 430A Arrays were used for the analyses. Total RNA was extracted from fetal colons using Qiagen-RNeasy kit (Qiagen, Valencia, CA). Biotin-labeled complementary RNA probes were generated from 5 µg of total RNA by using a bioarray transcript labeling kit (Affymetrix). After hybridization, array data were analyzed by using DNA-Chip Analyzer (dChip) software (http://www.dchip.org). The microarray data (Supplementary Table 1; see supplemental material online at www.gastrojournal.org) presented in this manuscript have been deposited in National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through Gene Expression Omnibus series accession number GSE3116. Reverse-transcriptase PCR (RT-PCR) was performed as described previously21 by using oligonucleotide sequences shown in Supplementary Table 2 (see supplemental material online at www.gastrojournal.org). Amplification of Hprt was used to standardize loading and reactions lacking reverse transcriptase were included to confirm the absence of contaminating DNA.
Electrophoretic Mobility Shift Assays
Radiolabeled probes were incubated at room temperature for 20 minutes with liver or Cos-7 cell nuclear extracts in the presence or absence of HNF4α antiserum or a negative control antibody (anti-Pes132). For competition analyses, double-strand oligonucleotide sequence representing the HNF4α-binding site (H4.2133) from the ApoCIII promoter34 was labeled by using [P32]-deoxyadenosine triphosphate and incubated with 1 µg of nuclear extracts from Cos-7 cells or Cos-7 cells expressing exogenous HNF4α in the presence or absence of 150-fold molar excess of each test binding site as described previously.35 Alternatively, radiolabeled binding sites were incubated with mouse liver nuclear extract prepared as described previously.34 Complexes were resolved by electrophoresis in 4% polyacrylamide gels in 0.25X Tris-borate- EDTA buffer at 400 V for 1 hour. The oligonucleotide sequences representing putative HNF4α-binding sites used in electrophoretic mobility shift assay (EMSA) reactions are provided in Supplementary Table 2.
Chromatin Immunoprecipitation
Brains and colons were isolated from wild-type CD1 mice, fixed with 1% formaldehyde, and cells dissociated by using a BD Medimachine (BD Biosciences, #340587). Chromatin immunoprecipitation (ChIP) assays were performed using the Upstate ChIP Assay Kit (Upstate #17-295) following the manufacturer’s instructions with anti-HNF4α (Santa Cruz sc-8987) or anti-Pes132 antibodies. Oligonucleotide primers used for PCR amplification of precipitated genomic DNA are given in Supplementary Table 2.
Results
HNF4α Protein Expression Is Restricted to the Epithelial Cells of the Developing Lower Gastrointestinal Tract
Hnf4a mRNA has been previously localized to the fetal gastrointestinal tract.36,37 However, to facilitate cell-specific disruption of the Hnf4a gene using the Cre-loxP system required clarification of which specific cell types in the developing gut expressed HNF4α protein.Figure 1 shows the result of immunohistochemistry staining analyses of mouse embryos using an antibody (Santa Cruz, anti-HNF4α C19) that specifically recognizes the carboxyl end of all major isoforms of HNF4α.26 At E9.5, HNF4α protein was undetectable in the foregut but could be identified in the developing hepatic rudiment as well as in the epithelium of the midgut and hindgut (Figure 1A and B). By E11.5, staining became more pronounced in these tissues presumably reflecting an increase in the abundance of HNF4α (Figure 1C and D). At this developmental time, HNF4α protein was detected throughout the epithelium of the lower gastrointestinal tract with a rostral boundary of expression occurring within the developing stomach (Figure 1C). HNF4α expression was maintained in all epithelial cells during the conversion of the gut epithelium from psuedostratified to simple columnar (Figure 1E–H). The restriction of HNF4α protein to the epithelium of both small and large intestine continued throughout development into adulthood (not shown) with expression found in both absorptive and secretory cell lineages of the intestinal epithelium.
Figure 1.
HNF4α is expressed in the epithelium of the lower gastrointestinal tract during embryogenesis. Embryos were collected at E9.5 (A and B), E11.5 (C and D), E14.5 (E), E15.5 (F), and E17.5 (G and H) and transverse sections processed for immunohistochemistry using an anti-HNF4α antibody. At E9.5, HNF4α (brown nuclear staining) was identified in the hepatoblasts (A, arrowhead) as well as in the epithelium of the hindgut and midgut (B, arrow) but not in the foregut (A, *). There was a sharp rostral boundary of expression of HNF4α within the stomach (C, ^). Caudal to this boundary, HNF4α was restricted to the epithelium (C–H, arrows) of the gut throughout the remainder of development, including that of the small intestine (D–G) and colon (H).
HNF4α-Null Colons Were Recovered From Both Hnf4aloxP/loxPFoxa3Cre and Hnf4aloxP/−Foxa3Cre E18.5 Embryos
Development of Hnf4a knockout embryos arrests during gastrulation because of defects in visceral endoderm function.30,36 This has prevented the use of Hnf4a−/− embryos to study the role of HNF4α in gut development. The Cre-loxP system was therefore used to circumvent this early embryonic lethality and to generate embryos lacking HNF4α in the fetal colonic epithelium. The production of mice (Hnf4aloxP/loxP) that contain an allele of Hnf4a in which exon2 is flanked by loxP elements has been described previously.26,28 Mice that are homozygous for this allele are healthy and fertile. However, in the presence of Cre, recombination between loxP sites generates a null allele.26,28 A line of transgenic mice (Foxa3Cre) in which Cre recombinase is expressed from a yeast artificial chromosome encompassing all Foxa3 transcriptional regulatory elements has also been previously described.29 The tissue distribution of Cre activity in transgenic mice can be determined by breeding Cre mice to a LacZ reporter mouse (Gt[ROSA]26Sortm1Sho).38 Using this approach confirmed that Cre recombinase activity was present from E8.5 onwards in the definitive endoderm and its derivatives including the epithelial cells of the colon (Figure 2A and B).29 Foxa3Cre mice were therefore chosen to delete Hnf4a from the colon because Cre was active at the very onset of colon development in these mice. To disrupt HNF4α in the gastrointestinal tract, Hnf4aloxP/loxP mice or Hnf4α+/− mice were mated to Foxa3Cre mice. Double-heterozygous offspring were then bred to Hnf4aloxP/loxP mice to generate either Hnf4aloxP/loxPFoxa3Cre and Hnf4aloxP/−Foxa3Cre experimental embryos or Hnf4aloxP/+Foxa3Cre control embryos. In contrast to control embryos, colons could be recovered from Hnf4aloxP/loxPFoxa3Cre embryos at E18.5 in which HNF4α protein was undetectable by immunohistochemistry (Figures 2C and D). E18.5 was the latest stage at which HNF4α-null embryos could be recovered because Hnf4aloxP/loxPFoxa3Cre newborn mice also lacked HNF4α in the liver, which resulted in neonatal lethality as previously reported.26 The efficiency of ablating HNF4α in the colon of Hnf4aloxP/loxPFoxa3Cre embryos was variable, with <10% of the mice showing complete absence of HNF4α and the remaining colons exhibiting varying levels of chimerism (Figure 2E). The observed variation in loss of HNF4α in different Hnf4aloxP/loxPFoxa3Cre embryos most likely reflects variation in recombination efficiency. In an attempt to increase the efficacy of recovering HNF4α-null colons, Hnf4aloxP/−Foxa3Cre embryos were derived in which 40% had undetectable levels of HNF4α in the colon (Figure 2F). The continued presence of HNF4α in a subset of fetal colons could potentially confound any phenotypic analyses and so only embryos in which either HNF4α protein was judged absent by immunohistochemistry or Hnf4a mRNA was undetected by using RT-PCR were studied further. In addition, Hnf4aloxP/loxPFoxa3Cre and Hnf4aloxP/−Foxa3Cre embryos were used interchangeably because they displayed indistinguishable phenotypes.
Figure 2.
Loss of HNF4α in the epithelial cells disrupts development of the colon. (A and B) Foxa3cre mice were bred to a line of transgenic mice, Gt(ROSA)26SortmlSho, that expresses β-galactosidase only after Cre-mediated deletion of loxP-flanked DNA sequences and resulting E9.5 embryos were stained for β-galactosidase expression. Expression of β-galactosidase was detected in the colon (arrowhead) of whole embryos (A) and sections (B) through these embryos revealed the expression to be restricted to the epithelial cells of the colon (arrowhead). (C–F) Micrographs of immunohistochemistry experiments using an anti-HNF4α antibody. HNF4α was identified in the nuclei (open arrowheads) of control (C, Hnf4aloxP/+Foxa3Cre) but was undetectable in a subset of experimental (D, Hnf4aloxP/loxPFoxa3Cre; F, Hnf4aloxP/−Foxa3Cre) E18.5 colons. Some Hnf4aloxP/loxPFoxa3Cre colons displayed a chimeric expression of HNF4α (E). (G and H) H&E–stained sections of E18.5 embryos revealed the formation of crypts (G, arrow) and the presence of mature goblets cells (G, arrowhead) in control E18.5 colons, whereas crypt formation and goblet cell maturation was disrupted in colons lacking HNF4α (H). Boxed areas are shown at a higher resolution in the inset.
HNF4α Is Essential for Normal Development of the Colon
Examination of E18.5 HNF4α-null colons by histochemistry uncovered abnormalities in lower gut topology and tissue organization. H&E-stained sections of control colons revealed that the epithelium contained characteristic crypts extending from a relatively compact lumen (Figure 2G). In contrast, HNF4α-null colons showed no obvious sign of crypt formation with the mucosa arranged as a simple band surrounding a greatly expanded lumen (Figure 2H).
To more accurately define the consequence of the absence of HNF4α on the development of the colon, the structure of various tunics was examined by using immunohistochemistry. Although H&E staining revealed that crypt formation was severely affected by loss of HNF4α, it was also apparent from these analyses that the epithelium had successfully transitioned from pseudostratified to simple columnar in both control and HNF4α-null E18.5 colons (Figure 2G and H). In support of the formation of a normal epithelium, the cell-adhesion protein, E-cadherin, could be readily detected between apposing epithelial cells regardless of the presence of HNF4α (Figure 3A and B). It has been previously shown that loss of HNF4α in the fetal liver results in reduced expression of E-cadherin.26 Nevertheless, in contrast to HNF4α-null livers in which E-cadherin (Cdh1) mRNA is barely detectable,26 RT-PCR analyses revealed that the level of Cdh1 mRNA in HNF4α-null colons was reduced by a moderate 2-fold compared with controls (Figure 3K).
Figure 3.
HNF4α is necessary for multiple aspects of colon development. (A–J) Micrographs showing results of immunohistochemistry (brown staining) performed on sections of control (A, C, E, G, I; Hnf4aloxP/+Foxa3Cre) and mutant (B, D, F, H, J; Hnf4aloxP/loxPFoxa3Cre) E18.5 embryos using antibodies that recognize E-cadherin (A and B), PECAM1 (C and D), laminin (E and F), neural acetylated tubulin (G and H,) and α smooth-muscle actin (I and J). (K) RT-PCR analyses of mRNA isolated from Hnf4aloxP/+Foxa3Cre (control, lanes 2–4) and Hnf4aloxP/loxPFoxa3Cre (null, lanes 5–7) E18.5 colons using oligonucleotide pairs that amplified Hnf4a, Cdh1, Bmp4, Foxl1, Tcf21, Myh11, and Ihh mRNAs. Amplification of Hprt mRNA was used as a loading control and reactions lacking input DNA (0 DNA, lane 1) confirmed that amplicons originated from input RNA. Levels of amplicons were determined by phosphoimager analyses and were presented as an average fold difference (fold change) between control and mutant samples after normalizing to levels of the Hprt amplicon.
Although HNF4α expression is restricted to epithelial cells, signaling between the mesenchymal and endodermal compartments coordinate development of all layers of the gastrointestinal tract.7 It was therefore important to determine whether development of the enteric nervous system, lamina propria, and intestinal smooth muscle was affected in HNF4α-null colons. First, the integrity of the capillary network within the lamina propria of the colon was investigated by using antibodies to identify the endothelial cell marker PECAM1. In control colons, PECAM1-positive endothelial cells could be identified extending through the lamina propria to the top of the crypts (Figure 3C). In contrast, this network of capillaries was disrupted in colons lacking HNF4α (Figure 3D). The lamina propria, in which the intestinal capillary network is housed, consists of loose connective tissue that secretes extracellular matrix proteins including laminin. It has been proposed that the extracellular matrix defines an environment that supports cell-cell communication, thereby contributing to the control of cell differentiation.39,40 Figure 3E shows that laminin extends throughout the mucosa occupying the spaces between the crypts of control colons. In HNF4α-null colons, laminin was still clearly detectable but instead was organized as a lining at the base of the epithelium (Figure 3F). The observation that laminin is present in HNF4α-null colons suggests that the disorganization of laminin localization is a consequence of defective crypt formation rather than complete absence of the lamina propria per se. Consistent with this proposal, RT-PCR analyses of colons lacking HNF4α revealed no significant change in the levels of mRNAs encoding Bmp4, Tcf21, and Foxl1 (Figure 3K), all of which are expressed in the lamina propria.41–43
The development of the enteric nervous system and the smooth-muscle layers of HNF4α-null colons were examined next. Immunohistochemical staining for neural acetylated tubulin, whose expression is restricted to the enteric neurons, identifies the myenteric plexus as indistinguishable between control and mutant colons (Figure 3G and H). The muscularis layers were detected by using antibodies against α smooth-muscle actin (Figure 3I and J). Although the muscularis mucosa and muscularis externa were identifiable in both control and HNF4α-null colons, the musclaris externa appeared thinner in mutant colons compared with controls. The apparent difference in muscle volume was confirmed by morphometric analyses using Metamorph software, which calculated a 58% reduction (ANOVA, P < .0001) in the volume of the external smooth-muscle layer of HNF4α-null colons compared with controls. In support of a reduction in overall muscle mass of the colon, RT-PCR analyses revealed that smooth muscle myosin heavy chain (Myh11) mRNA, whose expression is restricted to the muscle cells of the colon, was reduced by 2-fold in HNF4α-null colons (Figure 3K).
The phenotype associated with loss of HNF4α was similar in some aspects to that associated with loss of the signaling molecule Indian hedgehog (IHH). Like HNF4α-null colons, Ihh−/− embryos have a severely dilated intestine and display disrupted crypt formation.44 Examination of Ihh−/− embryos revealed a reduction in intestinal smooth-muscle content and loss of enteric neurons, and it was proposed that this contributed to the abnormal dilation of the colon.44 Although loss of smooth-muscle volume was also identified in HNF4α-null colons (Figure 3J), the observation that these colons contained neural cells (Figure 3H) suggested that IHH signaling was unlikely to be affected by loss of HNF4α. This was confirmed by RT-PCR analyses (Figure 3K) that identified similar levels of Ihh mRNA in both control and HNF4α-mutant colons. Therefore, we conclude that HNF4α has an integral role in controlling multiple aspects of colon development and that this regulation is not mediated through control of IHH expression.
Loss of HNF4α Results in a Reduction in Epithelial Cell Numbers in the Developing Colon
The observation that the colonic mucosa seemed thinner in the absence of HNF4α could result from a reduction in the number of epithelial cells present within the mucosa. Epithelial cells were therefore counted in three independent H&E-stained sections per colon in either control or mutant embryos. The total number of epithelial cells counted per section was reduced by 40% (ANOVA, P < .0001) in HNF4α-null colons compared with controls (Figure 4A). The reduction in HNF4α-null epithelial cells did not appear to be caused by increased apoptotic cell death because immunohistochemistry using anti-cleaved Caspase 3 antibodies revealed few apoptotic cells in colons from either control or experimental embryos (not shown). To test whether cell proliferation was affected by loss of HNF4α, sections were processed for immunohistochemistry to detect Ki-67 protein, which acts as marker of cells that are actively proliferating. Figure 4B shows that, as expected, immunohistochemistry detected Ki-67–positive cells located in the lower portion of the crypts in control colons. In contrast, in HNF4α-null colons Ki-67 was identified in epithelial cells that were present as intermittent clusters throughout the epithelium (Figure 4C). This analysis shows that although crypt architecture is disrupted by loss of HNF4α, proliferating cell compartments were still present, suggesting that some level of crypt to surface epithelium patterning was maintained in the absence of HNF4α. Although proliferating cells were detected in HNF4α-null colons, they appeared to be less numerous compared with control colons (Figure 4B and C). Cell counting revealed that the number of Ki-67–positive cells was reduced by 35% (ANOVA, P = .0002) in colons lacking HNF4α compared with controls (Figure 4D). Similar results were also found by using antiphosphohistone H3 antibodies to detect mitotic cells (data not shown). Expression of transcription factors c-Myc and Foxm1 has been shown to closely correlate with cell proliferation in the gut.45–47 RT-PCR analyses (Figure 4E) showed that the levels of both Myc and Foxm1 mRNAs were reduced by 2-fold in colons lacking HNF4α, which is consistent with the reduction in proliferating cells observed using immunohistochemistry. These data imply that the observed reduction in the total number of epithelial cells associated with the absence of HNF4α is at least in part a consequence of a reduction in epithelial cell proliferation.
Figure 4.
Epithelial cell proliferation is reduced in colons lacking HNF4α. (A) Epithelial cells were counted in 3 H&E-stained colon sections from 2 Hnf4aloxP/+Foxa3Cre (Con.1 and Con. 2, shaded box) or 2 Hnf4aloxP/loxPFoxa3Cre (Null 1, Null 2, open box) E18.5 embryos, and results are presented graphically. Statistical significance (**) was calculated by using ANOVA. (B and C) Proliferating cells were identified by using immunohistochemistry with anti–Ki-67 antibodies (brown staining, arrowhead) on control (B) and mutant (C) E18.5 colons. A high-resolution image of the boxed area is contained within inset. (D) Counts of proliferating cells were collected from 3 independent sections and are presented as in A. (E) RT-PCR analyses of mRNA isolated from Hnf4aloxP/+Foxa3Cre (control, lanes 2–4) and Hnf4aloxP/loxPFoxa3Cre (null, lanes 5–7) E18.5 colons using oligonucleotide pairs that amplified Hprt as a loading control, Myc and Foxm1 mRNAs. 0 DNA (lane 1) was included to exclude the possibility of contaminating DNA. Average fold changes in amplicon levels between control and mutant colons are shown.
Goblet-Cell Maturation in the Colon Is Dependent on HNF4α
H&E staining indicated a reduction in the number of goblet cells present in HNF4α-null colons (Figure 2G and H). Alcian blue histochemistry, which identifies the presence of acidic mucins, was therefore used to differentiate between goblet cells and the rest of the epithelial cells of the colon. In control colons, alcian blue–positive mature goblet cells and accumulation of mucin within the gut lumen could easily be detected (Figure 5A). A pregoblet cell can be distinguished from a mature goblet cell by the grouping of a limited number of mucous granules into a small theca. As the pregoblet cell matures, the number of mucus granules increases and the theca expands to occupy a large portion of the cytoplasm, which gives the mature goblet cell its characteristic morphology.48 Examination of control colons revealed that the majority of the alcian blue–positive cells were mature goblet cells (Figure 5A). However, few mature goblet cells could be identified in HNF4α-null colons, which instead had an increase in the abundance of epithelial cells containing small clusters of mucin (Figure 5B). It seems most likely that these cells are pregoblet cells that have failed to mature, although it is formally possible that they represent colonocytes that ectopically express mucins. In support of the view that cells containing small clusters of alcian blue material are pregoblet cells, cell counting revealed that the total number of alcian blue–positive cells was similar but that the ratio of presumptive immature to mature goblet cells was reversed between control and HNF4α-null colons (Figure 5C). The apparent block in goblet cell maturation was reminiscent of the phenotype associated with loss of the zinc finger transcription factor Klf4.14 However, RT-PCR analyses revealed that Klf4 mRNA levels remained unchanged in HNF4α-null colons (Figure 5D). In addition, levels of Trefoil factor 3 mRNA (Ttf3, also called Itf), which is considered a marker of differentiated goblet cells,49 was unchanged in colons lacking HNF4α (data not shown).
Figure 5.
Goblet-cell maturation is blocked by the loss of HNF4α. (A and B) Alcian blue histochemistry identified goblet cells in both control (Hnf4aloxP/+Foxa3Cre) and null (Hnf4aloxP/loxPFoxa3Cre) E18.5 colons (blue-stained cells). Insets contain high-resolution image of boxed regions showing that mature goblet cells (arrow) predominate in control colons, whereas immature goblet cells (arrowhead) are most abundant in HNF4α-null colons. (C) The number of mature or immature goblet cells were counted in 3 sections from each of 2 Hnf4aloxP/+Foxa3Cre (control 1, control 2; shaded boxes) and Hnf4aloxP/loxPFoxa3Cre (null 1, null 2; open boxes) E18.5 colons and data are presented as a bar graph. Statistical significance (**) was calculated by using ANOVA. (D) RT-PCR analyses of mRNA isolated from Hnf4aloxP/+Foxa3Cre (control, lanes 2–4) and Hnf4aloxP/loxPFoxa3Cre (null, lanes 5–7) E18.5 colons using oligonucleotide pairs that amplified Hprt as a loading control; mucins-1, -2, -3, and -4; and Klf4. 0 DNA (lane 1) showed the absence of contaminating DNA, and average fold changes in amplicon levels between control and mutant colons are shown.
The finding that HNF4α-null goblet cells failed to accumulate normal amounts of mucin (Figure 5B) raised the possibility that the expression of genes encoding mucin core proteins could be disrupted in HNF4α-null colons. The major mucins found in the intestine include Muc1, Muc2, Muc3, and Muc4.50 The steady-state levels of mRNA encoding these proteins in control and HNF4α-null colons were therefore compared by RT-PCR. The level of Muc3 mRNA was reduced by 18-fold in HNF4α-null livers compared with controls (Figure 5D). However, somewhat surprisingly, expression of Muc2 was only minimally reduced (2-fold), and Muc1 and Muc4 were unaffected in the mutant colons (Figure 5D). Muc2 is the major gel-forming mucin in goblet cells and mucin production is undetectable by alcian blue staining in Muc2−/− colons; in contrast, Muc1, 3, and 4 are primarily transmembrane glycoproteins.49,51 The fact that Muc2 mRNA is detected in reasonable abundance in HNF4α-null colons implies that the observed loss of goblet cell morphology likely reflects a block in maturation of mucous granules as opposed to a loss of expression of secretory mucin core protein genes.
HNF4α Is Required for Expression of Several Genes That Contribute to Colon Function
Although at E18.5 the colonic epithelium is not fully mature, it does express gene products related to the function of the adult colon. This implies that any analyses of HNF4α-null E18.5 colons should offer insight into the potential contribution of HNF4α in regulating both colon development and adult colon function. Microarray expression profiling was therefore used to comprehensively define the requirement for HNF4α in controlling colon gene expression. Analyses were performed independently on 2 control and 3 HNF4α-null colons and results were analyzed for fold changes by dChip. Genes whose expression exhibited a mean 2-fold or more differential expression between control and null colons are listed in Supplementary Table 1. By using these criteria, expression of 20 genes was predicted to have increased and 85 to have decreased. However, only 2 mRNAs had a statistically significant (Student t test, P ≤ .05) increase of >3-fold in the mutant colons suggesting that, like in the liver, HNF4α is predominantly an activator of gene expression. Analyses of the gene ontology distribution of genes whose expression was predicted to be reduced revealed that a wide range in physiological and cell biological processes was likely affected by loss of HNF4α in the colon (Supplementary Figure 1; see supplemental material online at www.gastrojournal.org). These included genes encoding proteins with roles in various aspects of cell and physiological metabolism, transport, and localization, all of which are crucial functions of the colon. A statistical comparison of the distribution of down-regulated genes to the occurrence of gene categories in the genome using GOTree Machine52 found few gene ontology categories to be overrepresented (Supplementary Figure 1). This implies that HNF4α action is not restricted to regulating a specific process but rather is required for widespread control of colonic gene expression.
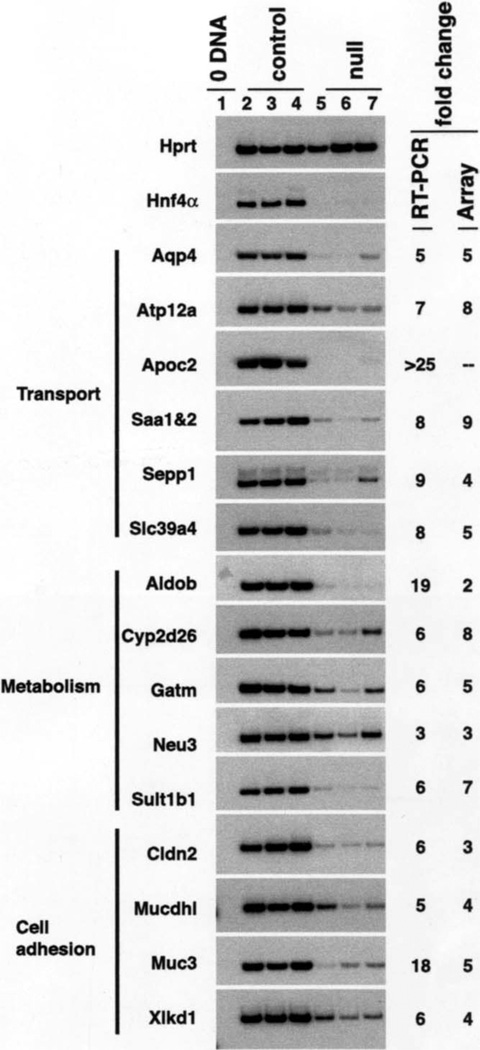
Known genes whose expression was predicted to be reduced ≥3-fold by array analyses were considered further because, based on past experience, such changes are most often verified by RT-PCR. Figure 6 shows that of the genes analyzed 16 were confirmed to express reduced steady-state mRNA levels in mutant colons. In control colons, aquaporin 4 (Aqp4); ATPase H+/K+ transporting nongastric alpha polypeptide (Atp12a); apolipoprotein C-II (Apoc2); serum amyloid 1 and 2 (Saa1&2); selenoprotein P, plasma, 1 (Sepp1); solute carrier family 39 (zinc transporter); member 4 (Slc39a4); aldolase B (Aldob); cytochrome P450, family 2, subfamily d, polypeptide 26 (Cyp2d26); glycine amidinotransferase (Gatm); neuraminidase3 (Neu3); sulfotransferase family 1B, member1 (Sult1b1); claudin2 (Cldn2); mucin and cadherin like (Mucdhl); mucin3 (Muc3); and extracellular link domain-containing 1 (Xlkd1) mRNAs were easily detectable. However, in fetal colons lacking HNF4α, the level of each of these mRNAs was reproducibly reduced. In general, the fold differences in mRNA levels predicted by analyses of the microarray data closely correlated with the changes determined by phosphoimager quantification of the RT-PCR products. Several of the genes with reduced expression encode proteins that have crucial roles in gut function, including ion and water transporters, metabolic proteins, and cell-adhesion molecules. The dependence of gene expression on HNF4α did not appear to be the consequence of disrupting expression of colonic epithelial transcription factors in general because expression of Klf4 (Figure 5), Atoh1, Hes1, Elf3, Cdx1, and Cdx2 (data not shown) was unaffected. These data show that HNF4α has an integral role in controlling the developmental expression of genes that define a functioning colonic epithelium.
Figure 6.
Loss of HNF4α disrupts expression of multiple genes encoding proteins that contribute to colon function. RT-PCR analyses of mRNA isolated from Hnf4aloxP/+Foxa3Cre (control, lanes 2–4) and Hnf4aloxP/loxPFoxa3Cre (null, lanes 5–7) E18.5 colons using oligonucleotide pairs that amplified Hprt as a loading control; Aqp4, Atp12a, Apoc2, Saa1 and 2, Sepp1, and Slc39a4, which encode proteins with a variety roles in transport; Aldob, Cyp2d26, Gatm, Neu3, and Sult1b1 that encode proteins having diverse roles in metabolism; and Cldn2, Mucdhl, Muc3, and Xlkd1 that encode proteins with various contributions to cell adhesion. 0 DNA (lane 1) confirmed the absence of contaminating DNA and average fold changes in mRNA levels between control and mutant colons calculated from RT-PCR analysis or Affymetrix array analysis are shown.
HNF4α Directly Interacts With Multiple Genes Whose Expression Is Reduced in HNF4α-Null Colons
To determine whether the 16 genes whose expression was reduced in HNF4α-null colons were direct targets of HNF4α, genomic sequences lying −10Kb to +10Kb relative to the transcriptional start site (+1) of each gene were searched for the presence of any of 169 known HNF4α-binding-sites33 by using a Knuth-Morris-Pratt exact match algorithm.53 Of the 16 genes studied, 8 were found to contain nucleotide sequences that matched known HNF4α-binding sites (Supplementary Figure 2; see supplemental material online at www.gastrojournal.org). These sites were positioned both upstream and downstream of the transcriptional start site and included intronic sequences (Supplementary Figure 2). All sequences were confirmed to interact with HNF4α by EMSA (Supplementary Figure 2). Although these data showed that the majority of the 16 down-regulated genes contained HNF4α-binding sites, it also seemed possible that previously uncharacterized HNF4α-binding sites could be present within the genes as well. Therefore, we used a permutated Markov model that was recently developed to predict any potential HNF4α-binding sites54 to search the same −10Kb to +10Kb of genomic sequence encompassing the 16 potential target genes. By using this approach, 35 binding sites were identified in 14 genes with several genes containing multiple sites (Supplementary Table 2). Eleven of 14 genes contained potential HNF4α-binding sites within their putative promoter regions (Figure 7A). Further studies were restricted to these 11 genes because they seemed most likely to be responsible for regulating expression by HNF4α. The ability of HNF4α to interact with the predicted binding sites was assessed by EMSA. Nuclear extracts from Cos-7 cells expressing exogenous HNF4α yielded a single specific shift complex when incubated with a 32P-labeled oligonucleotide containing a well-characterized HNF4α-binding site (Figure 7B, compare lanes 1 and 2). As expected, a 150-fold molar excess of the same unlabeled oligonucleotide specifically competed the complex (Figure 7B, compare lanes 2 and 3), whereas control oligonucleotides lacking a predicted HNF4α-binding site did not (Figure 7B, lane 17). Of the 14 putative HNF4α-binding sites analyzed in a similar fashion, 13 were shown to compete the known site and therefore interact with HNF4α. Together with the identification of the previously characterized sites, these analyses show that 13 of the 16 genes whose expression in the colon was dependent on HNF4α-contained sequences that could interact with HNF4α in vitro.
Figure 7.
HNF4α-binding sites are present in genes whose expression is dependent on HNF4α. (A) Table showing the position and sequence of putative HNF4α-binding sites predicted to be present within genes whose expression is HNF4α dependent. HNF4α-binding site numbers (H4 sites) were assigned by using the criteria established by Yang et al.33 (B) EMSA was performed by using an HNF4α-binding site from the Apoc3 promoter and extracts from Cos-7 cells transfected with a control plasmid (mock, lane 1) or a plasmid expressing exogenous HNF4α (lanes 2–17). A specific shift caused by binding of HNF4α to the Apoc3 HNF4α-binding site is indicated with an arrowhead. Inclusion of 150-fold molar excess of unlabeled oligonucleotides corresponding to HNF4α-binding sites in the either the Apoc3 promoter (H4.21, lane 3) or binding sites predicted through computer modeling in Aldob (lane 4), Apoc2 (lanes 6–7), Aqp4 (lane 8), Cldn2 (lane 9), Gatm (lane 10), Muc3 (lane 11), Mucdh1 (lane 12), Neu3 (lane 13), Saa1 and 2 (lane 14), and Slc39a4 (lanes 15 and 16) all competed for binding of HNF4α to the labeled Apoc3 HNF4α-binding site. One binding site (lane 5) in the Apoc2 gene failed to compete, as did a negative control (N.C.) binding site for the transcription factor Foxa (lane 17).
If any of the sites identified by EMSA (Supplementary Figure 2 and Figure 7) facilitated HNF4α-controlled transcription of the associated gene, then it could be predicted that these sites would be occupied by HNF4α in the colon. To test this prediction, ChIP assays were performed on individual colons isolated from fasted mice. Antibody specific to HNF4α precipitated colonic chromatin encompassing the HNF4α-binding sites in 7 of 13 genes tested but not from a control gene (Hprt) that had no HNF4α-binding site (Figure 8). Only those reactions that yielded a positive result, with the exception of Muc3 BS2, which serves as an example of a negative result, are shown. The positive precipitations were specific for the anti-HNF4α antibody because an unrelated antibody (anti-Pescadillo) failed to precipitate genomic DNA from any samples. Moreover, the anti-HNF4α antibody did not precipitate any genomic sequences from samples of brain tissue that does not express HNF4α. These studies show that in the colon HNF4α interacts with several genes whose expression is dependent on HNF4α implying that they are direct targets of HNF4α transcriptional control.
Figure 8.
Genomic sequences within several genes whose expression is dependent on HNF4α are occupied by HNF4α within the colon. Chromatin immunoprecipitation was performed on 2 independent colons and a brain sample, a negative control tissue that does not express HNF4α, by using antibodies that precipitated either HNF4α or an unrelated protein, PES1. DNA sequences that coprecipitated with these proteins were identified by PCR by using primers that flanked the HNF4α-binding sites within Aldob, Apoc2, Muc3, Saa1, Slc39a4, Sult1b, and Mucdhl. PCR of the Hprt gene or the HNF4α-binding site 2 of the Muc3 gene (Muc3 H4.77) failed to show any enrichment in the colon, confirming that DNA sequences that were precipitated with anti-HNF4α were specific and reflected a bona fide interaction with HNF4α in the colon.
Discussion
The results presented here show that the nuclear receptor HNF4α makes a crucial contribution toward establishing a mature epithelium within the large intestine during embryonic development. Loss of HNF4α in the epithelial tissue of the colon results in a block to formation of normal crypt topology, thinning of muscle cell layers, loss of vascular tissue, a reduction in the number of epithelial cells, and diminished maturation of goblet cells. However, although these disruptions to gut tissue morphology and cell differentiation are quite striking, many aspects of colon development occur normally including specification of all cell types, generation of all tissue tunics, and the conversion of the primitive gut tube from a psuedostratified epithelium to a simple columnar epithelium. The observation that most early processes in colon development occur normally suggests that the predominant requirement for HNF4α is to maintain a differentiated gene expression profile rather than to control early patterning events or cell-fate choices. Molecular studies have revealed that HNF4α regulates expression of several transcription factors in the liver, and disruption to this transcription factor network in HNF4α-null livers results in a widespread dysregulation of hepatic gene expression.25,26 However, in contrast to the liver, our studies using expression profiling and RT-PCR uncovered only minor alterations in the expression of gut transcription factors in HNF4α-deficient colons. Transcription factors whose expression is unaffected by loss of HNF4α include several with well-defined roles in controlling differentiation of intestinal epithelial cells such as those encoded by Klf4, Atoh1, Elf3, and Cdx2.11,14,15,55,56 Although our studies show that the transcription factor profile in the colon is relatively unaffected by loss of HNF4α, we identified a down regulation in expression of many genes that encode proteins with important roles in colon and epithelial cell function. Importantly, EMSA and ChIP analyses showed that many of these genes are direct targets of HNF4α. Cumulatively, these studies suggest that HNF4α acts to control terminal differentiation of the colonic epithelial cells by controlling gene expression downstream of other transcription factors. In this regard, it will be interesting to determine whether other developmental gut transcription factors such as Klf4, Atoh1, and Elf3 control expression of HNF4α in the gut epithelium.
Of the genes analyzed whose expression in the colon is dependent on HNF4α, we have identified 7 that contained HNF4α-binding sites, are bound by HNF4α in vivo, and are therefore likely to be direct targets of HNF4α. However, it seems likely that this is an underestimation of the total number of HNF4α target genes in the colon resulting from the relatively stringent criteria that were applied. For example, we limited our binding-site search to genomic sequences that lay within 10 kb of the gene. Of the novel HNF4α-binding sites identified, we chose to study only those that were in putative promoter regions and those with a sequence close to the established HNF4α–binding-site consensus.33,54 This means that genes with unconventional HNF4α-binding sites or sites downstream of +1, of which there are many (Supplementary Table 2), were excluded by our analyses. Even with these limitations, however, our data show that HNF4α is required for expression of a significant number of genes whose products contribute to gut function. This set of genes includes those encoding proteins involved in a diverse array of physiological and cellular functions implying that HNF4α is not only essential for colon development but will also make an important contribution toward maintaining adult gut physiology.
The phenotype associated with loss of HNF4α function in the colon is clearly complex. Although HNF4α expression is restricted to the epithelial cells of the gut, analyses of Hnf4aloxP/loxPFoxa3Cre embryos revealed, in addition to defects in development of the epithelium, abnormal formation of the colon’s muscle and vascular tissues. The phenotype associated with nonepithelial cells most likely reflects a secondary consequence of disrupting HNF4α in the epithelium. Cross-talk between the mesenchymal and epithelial layers of the gut has been well documented and signaling between mesenchymal and epithelial layers is required for differentiation, cell proliferation, and patterning of the gut tube.2,7,9 A large array of growth factors secreted from one intestinal tissue type have been shown to assert their effects on the other including Indian and Sonic hedgehog, FGFs, BMPs, Wnts, Notch, and PDGF.2,7,9 Indeed, Ihh−/− embryos share characteristics of the phenotype associated with loss of HNF4α in the colon including a distended gut and reduced thickness of the muscularis layers.44 However, no changes in expression of specific growth factors in the absence of HNF4α were detected, including expression of Ihh (Figure 3K). Nevertheless, we did find that embryos in which HNF4α was deleted in a subset of epithelial cells displayed an apparently normal morphology, including the generation of crypts (Figure 2E). The formation of morphologically normal crypts and tissue tunics in such chimeric embryos is consistent with at least some aspects of the phenotype being non cell-autonomous. Analyses of the genes whose expression is most significantly affected by loss of HNF4α does not give any clear indication of a single specific gene product that would result in the pleiotropic effects of losing HNF4α in the colon. Mice lacking Aqp4, Atp12a, or Sepp1 have been described, and although some have phenotypes that could be associated with colon dysfunction, none display characteristics that are similar to those associated with Hnf4aloxP/loxPFoxa3Cre embryos.57–59 Although it remains possible that the phenotype associated with loss of HNF4α could be caused by reduced expression of a specific target gene, it seems more likely that the cumulative loss of multiple proteins whose expression is regulated by HNF4α in the epithelial cells results in an environment that is unable to sustain normal colon development. Nevertheless, as more gene knockouts in HNF4α targets become available, it is likely that subsets of defects caused by loss of HNF4α in the colon will be identified.
In summary, we have shown that HNF4α is a critical regulator of gene expression in the epithelial cells of the colon. Ablation of the Hnf4a gene solely in the epithelial cells disrupts many aspects of normal colon development affecting multiple tissue layers. Analyses of expression of other transcription factors with known roles in colon development in Hnf4aloxP/loxPFoxa3Cre embryos suggest that HNF4α is an endpoint mediator of colon epithelial cell gene expression. This notion is supported by the identification of several genes that are new targets of HNF4α regulation in the colon. Based on this, we conclude that HNF4α is not only necessary for colon development but is also likely to act as a crucial regulator of colon physiology.
Supplementary Material
Acknowledgments
Supported by United States National Institutes of Health grants to S.A.D., F.M.S., K.H.K. and M.A.B., and American Heart Association Predoctoral Fellowship to W.D.G.
Abbreviations used in this paper
- ANOVA
analysis of variance
- E
embryonic day
- HNF4α
hepatocyte nuclear factor 4 alpha
- RT-PCR
reverse-transcription polymerase chain reaction
Footnotes
Appendix
Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1053/j.gastro.2006.01.003.
References
- 1.Radtke F, Clevers H. Self-renewal and cancer of the gut: two sides of a coin. Science. 2005;307:1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- 2.Stainier DY. No organ left behind: tales of gut development and evolution. Science. 2005;307:1902–1904. doi: 10.1126/science.1108709. [DOI] [PubMed] [Google Scholar]
- 3.Badman MK, Flier JS. The gut and energy balance: visceral allies in the obesity wars. Science. 2005;307:1909–1914. doi: 10.1126/science.1109951. [DOI] [PubMed] [Google Scholar]
- 4.Pennisi E. The dynamic gut. Science. 2005;307:1896–1899. doi: 10.1126/science.307.5717.1896. [DOI] [PubMed] [Google Scholar]
- 5.Sanderson IR, Walker WA. Development of the gastrointestinal tract. Hamilton, Ontario, Canada: B.C. Decker; 2000. [Google Scholar]
- 6.Wells JM, Melton DA. Vertebrate endoderm development. Annu Rev Cell Dev Biol. 1999;15:393–410. doi: 10.1146/annurev.cellbio.15.1.393. [DOI] [PubMed] [Google Scholar]
- 7.Sancho E, Batlle E, Clevers H. Signaling pathways in intestinal development and cancer. Annu Rev Cell Dev Biol. 2004;20:695–723. doi: 10.1146/annurev.cellbio.20.010403.092805. [DOI] [PubMed] [Google Scholar]
- 8.Roberts DJ. Molecular mechanisms of development of the gastrointestinal tract. Dev Dyn. 2000;219:109–120. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1047>3.3.co;2-y. [DOI] [PubMed] [Google Scholar]
- 9.Wells JM, Melton DA. Early mouse endoderm is patterned by soluble factors from adjacent germ layers. Development. 2000;127:1563–1572. doi: 10.1242/dev.127.8.1563. [DOI] [PubMed] [Google Scholar]
- 10.Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- 11.Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 12.Lee CS, Perreault N, Brestelli JE, Kaestner KH. Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes Dev. 2002;16:1488–1497. doi: 10.1101/gad.985002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenny M, Uhl C, Roche C, Duluc I, Guillermin V, Guillemot F, Jensen J, Kedinger M, Gradwohl G. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. EMBO J. 2002;21:6338–6347. doi: 10.1093/emboj/cdf649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, Yang VW, Kaestner KH. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng AY, Waring P, Ristevski S, Wang C, Wilson T, Pritchard M, Hertzog P, Kola I. Inactivation of the transcription factor Elf3 in mice results in dysmorphogenesis and altered differentiation of intestinal epithelium. Gastroenterology. 2002;122:1455–1466. doi: 10.1053/gast.2002.32990. [DOI] [PubMed] [Google Scholar]
- 16.Walsh A, Azrolan N, Wang K, Marcigliano A, O’Connell A, Breslow JL. Intestinal expression of the human apoA-I gene in transgenic mice is controlled by a DNA region 3′ to the gene in the promoter of the adjacent convergently transcribed apoC-III gene. J Lipid Res. 1993;34:617–623. [PubMed] [Google Scholar]
- 17.Ktistaki E, Lacorte JM, Katrakili N, Zannis VI, Talianidis I. Transcriptional regulation of the apolipoprotein A-IV gene involves synergism between a proximal orphan receptor response element and a distant enhancer located in the upstream promoter region of the apolipoprotein C-III gene. Nucleic Acids Res. 1994;22:4689–4696. doi: 10.1093/nar/22.22.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bisaha JG, Simon TC, Gordon JI, Breslow JL. Characterization of an enhancer element in the human apolipoprotein C-III gene that regulates human apolipoprotein A-I gene expression in the intestinal epithelium. J Biol Chem. 1995;270:19979–19988. doi: 10.1074/jbc.270.34.19979. [DOI] [PubMed] [Google Scholar]
- 19.Ginsburg GS, Ozer J, Karathanasis SK. Intestinal apolipoprotein AI gene transcription is regulated by multiple distinct DNA elements and is synergistically activated by the orphan nuclear receptor, hepatocyte nuclear factor 4. J Clin Invest. 1995;96:528–538. doi: 10.1172/JCI118065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu C, Perlmutter DH. Regulation of alpha1-antitrypsin gene expression in human intestinal epithelial cell line caco-2 by HNF-1alpha and HNF-4. Am J Physiol. 1999;276:G1181–G1194. doi: 10.1152/ajpgi.1999.276.5.G1181. [DOI] [PubMed] [Google Scholar]
- 21.Duncan SA, Nagy A, Chan W. Murine gastrulation requires HNF-4 regulated gene expression in the visceral endoderm: tetraploid rescue of HNF-4−/− embryos. Development. 1997;124:279–287. doi: 10.1242/dev.124.2.279. [DOI] [PubMed] [Google Scholar]
- 22.Stoffel M, Duncan SA. The maturity-onset diabetes of the young (MODY1) transcription factor HNF4alpha regulates expression of genes required for glucose transport and metabolism. Proc Natl Acad Sci U S A. 1997;94:13209–13214. doi: 10.1073/pnas.94.24.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Archer A, Sauvaget D, Chauffeton V, Bouchet PE, Chambaz J, Pincon-Raymond M, Cardot P, Ribeiro A, Lacasa M. Intestinal apolipoprotein A-IV gene transcription is controlled by two hormone-responsive elements: a role for hepatic nuclear factor-4 isoforms. Mol Endocrinol. 2005;19:2320–2334. doi: 10.1210/me.2004-0462. [DOI] [PubMed] [Google Scholar]
- 24.Soutoglou E, Talianidis I. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science. 2002;295:1901–1904. doi: 10.1126/science.1068356. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Ning G, Duncan SA. Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- 26.Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- 27.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parviz F, Li J, Kaestner KH, Duncan SA. Generation of a conditionally null allele of hnf4alpha. Genesis. 2002;32:130–133. doi: 10.1002/gene.10058. [DOI] [PubMed] [Google Scholar]
- 29.Lee CS, Sund NJ, Behr R, Herrera PL, Kaestner KH. Foxa2 is required for the differentiation of pancreatic alpha-cells. Dev Biol. 2005;278:484–495. doi: 10.1016/j.ydbio.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, Bachvarova RF, Darnell JE., Jr Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- 31.Bancroft JD, Gamble M. Theory and practice of histological techniques. London, UK: Elsevier Science; 2002. [Google Scholar]
- 32.Lerch-Gaggl A, Haque J, Li J, Ning G, Traktman P, Duncan SA. Pescadillo is essential for nucleolar assembly, ribosome biogenesis, and mammalian cell proliferation. J Biol Chem. 2002;277:45347–45355. doi: 10.1074/jbc.M208338200. [DOI] [PubMed] [Google Scholar]
- 33.Yang C, Liao H, Bolotin E, Evans J, Ellrott K, Jiang T, Sladek FM. Differences in HNF4a cis-regulatory elements in the human and mouse genomes. (in press) [Google Scholar]
- 34.Sladek FM, Zhong W, Lai E, Darnell JE., Jr Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 35.Jiang G, Nepomuceno L, Hopkins K, Sladek FM. Exclusive homodimerization of the orphan receptor hepatocyte nuclear factor 4 defines a new subclass of nuclear receptors. Mol Cell Biol. 1995;15:5131–5143. doi: 10.1128/mcb.15.9.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duncan SA, Manova K, Chen WS, Hoodless P, Weinstein DC, Bachvarova RF, Darnell JE., Jr Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc Natl Acad Sci U S A. 1994;91:7598–7602. doi: 10.1073/pnas.91.16.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taraviras S, Monaghan AP, Schutz G, Kelsey G. Characterization of the mouse HNF-4 gene and its expression during mouse embryogenesis. Mech Dev. 1994;48:67–79. doi: 10.1016/0925-4773(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 38.Mao X, Fujiwara Y, Orkin SH. Improved reporter strain for monitoring Cre recombinase-mediated DNA excisions in mice. Proc Natl Acad Sci U S A. 1999;96:5037–5042. doi: 10.1073/pnas.96.9.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams JC, Watt FM. Regulation of development and differentiation by the extracellular matrix. Development. 1993;117:1183–1198. doi: 10.1242/dev.117.4.1183. [DOI] [PubMed] [Google Scholar]
- 40.Beaulieu JF. Extracellular matrix components and integrins in relationship to human intestinal epithelial cell differentiation. Prog Histochem Cytochem. 1997;31:1–78. doi: 10.1016/s0079-6336(97)80001-0. [DOI] [PubMed] [Google Scholar]
- 41.Perreault N, Katz JP, Sackett SD, Kaestner KH. Foxl1 controls the Wnt/beta-catenin pathway by modulating the expression of proteoglycans in the gut. J Biol Chem. 2001;276:43328–43333. doi: 10.1074/jbc.M104366200. [DOI] [PubMed] [Google Scholar]
- 42.Kaestner KH, Silberg DG, Traber PG, Schutz G. The mesenchymal winged helix transcription factor Fkh6 is required for the control of gastrointestinal proliferation and differentiation. Genes Dev. 1997;11:1583–1595. doi: 10.1101/gad.11.12.1583. [DOI] [PubMed] [Google Scholar]
- 43.Haramis AP, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus GJ, Clevers H. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science. 2004;303:1684–1686. doi: 10.1126/science.1093587. [DOI] [PubMed] [Google Scholar]
- 44.Ramalho-Santos M, Melton DA, McMahon AP. Hedgehog signals regulate multiple aspects of gastrointestinal development. Development. 2000;127:2763–2772. doi: 10.1242/dev.127.12.2763. [DOI] [PubMed] [Google Scholar]
- 45.Ye H, Kelly TF, Samadani U, Lim L, Rubio S, Overdier DG, Roebuck KA, Costa RH. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol. 1997;17:1626–1641. doi: 10.1128/mcb.17.3.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, Tjon-Pon-Fong M, Moerer P, van den Born M, Soete G, Pals S, Eilers M, Medema R, Clevers H. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 47.Pinto D, Gregorieff A, Begthel H, Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karam SM. Lineage commitment and maturation of epithelial cells in the gut. Front Biosci. 1999;4:D286–D298. doi: 10.2741/karam. [DOI] [PubMed] [Google Scholar]
- 49.Velcich A, Yang W, Heyer J, Fragale A, Nicholas C, Viani S, Kucherlapati R, Lipkin M, Yang K, Augenlicht L. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295:1726–1729. doi: 10.1126/science.1069094. [DOI] [PubMed] [Google Scholar]
- 50.Wang HH, Afdhal NH, Gendler SJ, Wang DQ. Lack of the intestinal Muc1 mucin impairs cholesterol uptake and absorption but not fatty acid uptake in Muc1−/− mice. Am J Physiol Gastrointest Liver Physiol. 2004;287:G547–G554. doi: 10.1152/ajpgi.00097.2004. [DOI] [PubMed] [Google Scholar]
- 51.Jass JR, Walsh MD. Altered mucin expression in the gastrointestinal tract: a review. J Cell Mol Med. 2001;5:327–351. doi: 10.1111/j.1582-4934.2001.tb00169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang B, Schmoyer D, Kirov S, Snoddy J. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;5:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Knuth DE, Morris JJH, Pratt VR. Fast pattern matching in strings. SIAM Journal on Computing. 1977;6:323–350. [Google Scholar]
- 54.Ellrott K, Yang C, Sladek FM, Jiang T. Identifying transcription factor binding sites through Markov chain optimization. Bioinformatics. 2002;18(Suppl 2):S100–S109. doi: 10.1093/bioinformatics/18.suppl_2.s100. [DOI] [PubMed] [Google Scholar]
- 55.Chawengsaksophak K, James R, Hammond VE, Kontgen F, Beck F. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature. 1997;386:84–87. doi: 10.1038/386084a0. [DOI] [PubMed] [Google Scholar]
- 56.Tamai Y, Nakajima R, Ishikawa T, Takaku K, Seldin MF, Taketo MM. Colonic hamartoma development by anomalous duplication in Cdx2 knockout mice. Cancer Res. 1999;59:2965–2970. [PubMed] [Google Scholar]
- 57.Schomburg L, Schweizer U, Holtmann B, Flohe L, Sendtner M, Kohrle J. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem J. 2003;370:397–402. doi: 10.1042/BJ20021853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meneton P, Schultheis PJ, Greeb J, Nieman ML, Liu LH, Clarke LL, Duffy JJ, Doetschman T, Lorenz JN, Shull GE. Increased sensitivity to K+ deprivation in colonic H,K-ATPase-deficient mice. J Clin Invest. 1998;101:536–542. doi: 10.1172/JCI1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma T, Yang B, Gillespie A, Carlson EJ, Epstein CJ, Verkman AS. Generation and phenotype of a transgenic knockout mouse lacking the mercurial-insensitive water channel aquaporin-4. J Clin Invest. 1997;100:957–962. doi: 10.1172/JCI231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.