Abstract
Background
The 3 most commonly encountered bacteria in acute otitis media (AOM) are Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis. Conventional culture methods detect these pathogens in only 60% to 70% of cases of AOM. Alloiococcus otitidis, another potential pathogen, has often been ignored.
Methods
Tympanocentesis was performed in 97 children with AOM presenting with a bulging tympanic membrane (TM) producing 170 middle ear fluids (MEFs). S. pneumoniae, H. influenzae, M. catarrhalis, and A. otitidis were isolated in 21%, 32%, 8%, and 0% of MEFs, respectively; no otopathogen was isolated in 29% of MEFs. In nasopharyngeal cultures at the time of AOM diagnosis, 34%, 36%, 17%, and 0% and in oropharyngeal cultures, 7%, 31%, 11%, and 0% grew S. pneumoniae, H. influenzae, M. catarrhalis, and A. otitidis, respectively. No otopathogen was isolated in 23% of nasopharyngeal and 20% of oropharyngeal cultures. Multiplex polymerase chain reaction (PCR) was used to detect DNA of the 4 bacterial species in culture negative samples.
Results
All culture-positive MEF, nasopharyngeal and oropharyngeal samples tested were also multiplex-PCR positive, indicating the reliability of the method. Culture-negative samples of MEF from children with a bulging TM yielded S. pneumoniae, H. influenzae, M. catarrhalis, and A. otitidis DNA in 51%, 35%, 14%, and 32% of MEF, in 45%, 31%, 10%, and 9% of nasopharyngeal and in 31%, 23%, 0%, and 3% of oropharyngeal, respectively. In 9% of the cases A. otitidis DNA was found without detection of a second organism in MEF.
Conclusions
Conventional culture detected otopathogens in MEF of children with a bulging TM in 71%; using multiplex-PCR, otopathogens were detected in 88% of MEF (P < 0.01). Similar improved detection of otopathogens was noted with nasopharyngeal and oropharyngeal cultures.
Keywords: acute otitis media, Alloiococcus otitidis, multiplex-PCR, otopathogen, Streptococcus pneumoniae
Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis are the bacterial pathogens most commonly cultured from the middle ear fluid (MEF) samples of children with acute otitis media (AOM).1–4 Approximately 35% of samples are sterile by culture.5,6 However, these previous studies did not state whether a bulging TM was required for the diagnosis of AOM and a bulging TM is now recognized as the most important diagnostic finding of AOM.7 Polymerase chain reaction (PCR) methods can detect bacterial DNA of otopathogens in MEF and can also detect fastidious organisms,8–11 Therefore, PCR has the potential to improve identification of otopathogens in culture negative MEF, as well as in nasopharyngeal and oropharyngeal samples.
Alloiococcus otitidis, a largely overlooked organism, was first found by Faden and Dryja in 1989 in MEF from children with otitis media with effusion (OME).12 This bacterium is not identified by conventional culture because it requires about 5 days of growth. Since its discovery A. otitidis has been detected by culture in only a few studies,13–15 and mostly by PCR from OME patients.10,16–21 A. otitidis has only been described in 2 studies in AOM patients.10,22
The objectives of this study were (1) to assess the value of adding a molecular diagnosis assay, multiplex-PCR (M-PCR), for better etiologic diagnosis of AOM and for identification of otopathogens colonizing the nasopharyngeal and oropharyngeal when cultures were negative (2); to determine the frequency of detection of A. otitidis in MEF of children with AOM and in nasopharyngeal and oropharyngeal samples; and (3) to evaluate whether the M-PCR signal found in culture-negative samples represented fossilized bacterial DNA or viable bacteria, by use of RT-PCR targeting mRNA of the identified bacteria.
MATERIALS AND METHODS
Sample Collection
A total of 165 children were enrolled in a prospective study of AOM, July 2006 to July 2008. The children ranged from 6 months to 36 months of age. Written consent was obtained from all the parents or guardians before sample collection. Tympanocenteses were performed in 97 children with AOM producing 170 MEFs. Children with recurrent AOM underwent repeat tympanocentesis. MEF samples varied in quantity of material obtained from 50 to 250 μL; the entire sample was added to 1 mL of phosphate buffered saline. Otitis media with effusion patient samples were not collected in the study. S. pneumoniae, H. influenzae, and M. catarrhalis were identified using CLSI standard culture methods. In addition, nasopharyngeal and oropharyngeal samples were obtained from the 97 children with AOM at the time of the AOM visit and again 3 weeks later (194 child visits). Nasopharyngeal samples were obtained with a dacron-tipped wire swab inserted midway into the nasal canal below the inferior turbinate. Oropharyngeal cultures were obtained by rubbing both tonsils and the posterior pharynx. The details of the sample processing are given elsewhere.23 After immediate culture plating, the remainder of the MEF, nasopharyngeal, and oropharyngeal samples were stored at −80°C for later analysis in batches.
Genomic DNA and RNA Preparation for PCR Analysis
Nasopharyngeal and oropharyngeal swabs were taken from the freezer and swirled and mixed into 1 mL of phosphate buffered saline. One hundred microliters of MEF, nasopharyngeal, and oropharyngeal samples were then dissolved in Tris-EDTA buffer (pH 8.0) for genomic DNA extraction. After adding 10% SDS and proteinase K enzyme, the mixture was incubated for 1.0 hour at 37°C. An equal volume of phenol/chloroform was added and the aqueous phase was extracted after centrifugation, and 3.0M sodium acetate (pH 5.2) and isopropanol were added in the aqueous phase to precipitate the DNA. After centrifugation, DNA precipitates were washed with 70% ethanol and dissolved in water. DNA from different samples was stored at −20°C for later PCR study. DNA of each species was also obtained from pure culture. The control strain of A. otitidis was provided by Dr. Howard Faden, who discovered this bacterium.12
For RNA work, samples collected in the first 1.5 years were not preserved to allow study, so only those samples that were processed in a manner suitable for RNA work are described in the study. RNase-free components and plastic ware were used in the dilution and processing of the MEF and other samples. For the mRNA isolation, diluted MEF samples from AOM patients were centrifuged and the pellet dissolved in Trizol reagent, and RNA isolation was performed according to the manufacturer’s instructions (Molecular Research Center Inc.).24 To remove any DNA contamination, each sample was treated with DNAse in a digestion step using the Turbo DNA-free kit (Ambion Inc.) before RT-PCR amplification.
PCR Protocol
A multiplex PCR procedure was used for the simultaneous detection of S. pneumoniae, H. influenzae, M. catarrhalis, and A. otitidis using primers as described with some modifications.9 The detailed protocols of M-PCR and RT-PCR are provided in Appendix, Supplemental Digital Content 1, http://links.lww.com/INF/A428.
Sequencing of PCR Products
To determine the specificity of each amplified product, 2 to 3 bands from each PCR product were randomly picked and purified using a gel purification kit (QIAGEN). The sequencing reactions were performed on these purified products using 0.3 μM of common reverse primer on an Applied Biosystems Prism 377 automated sequencer using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). The sequenced data was compared with the 16S rRNA gene sequence entries in the GenBank database for verification of the PCR products.
RESULTS
Culture Results
As shown in Table 1, cultures of 170 MEF samples obtained from children with bulging TMs yielded S. pneumoniae, H. influenzae, and M. catarrhalis in 21%, 32%, and 8% of cases, respectively. No bacteria were detected by conventional culture in 29% of cases. No S. pyogenes was found but S. aureus was detected in 1% of the MEFs. The culture results of nasopharyngeal and oropharyngeal samples from children with AOM are also shown in Table 1. From AOM children, 23% of nasopharyngeal and 20% of oropharyngeal samples were culture negative for the 3 major otopathogens. S. aureus was detected in 4% of nasopharyngeal and oropharyngeal samples from AOM children. A. otitidis was not found using standard microbial culture methods (Because we did not anticipate the M-PCR detection of A. otitidis, the cultures were not retained for 5 days to identify this slow-growing organism).
TABLE 1.
Frequency of Major Otopathogens Identified by Standard Culture Method in MEF, Nasopharyngeal and Oropharyngeal Cultures from Children With AOM
| Pathogen | MEF (N = 170) (%) | Nasopharyngeal (N = 194) (%) | Oropharyngeal (N = 194) (%) |
|---|---|---|---|
| S. pneumoniae | 35 (20.6) | 66 (34.0) | 13 (6.7) |
| H. influenzae | 54 (31.8) | 70 (36.0) | 60 (30.9) |
| M. catarrhalis | 13 (7.6) | 33 (17.0) | 21 (10.8) |
| A. otitidis | 0 (0) | 0 (0) | 0 (0) |
| S. aureus | 2 (1.2) | 7 (3.6) | 4 (2.1) |
| No otopathagen | 49 (28.8) | 44 (22.6) | 38 (19.6) |
N is the total number of samples.
In some of the samples, more than one organism was isolated. The specific combinations of multiple pathogens are described elsewhere.10
M-PCR Results
Multiplex-PCR was used for the detection of bacterial DNA of H. influenzae, S. pneumoniae, M. catarrhalis, and A. otitidis in culture negative MEF, nasopharyngeal, and oropharyngeal samples. An example of an agarose gel run is shown in Figure 1.
FIGURE 1.
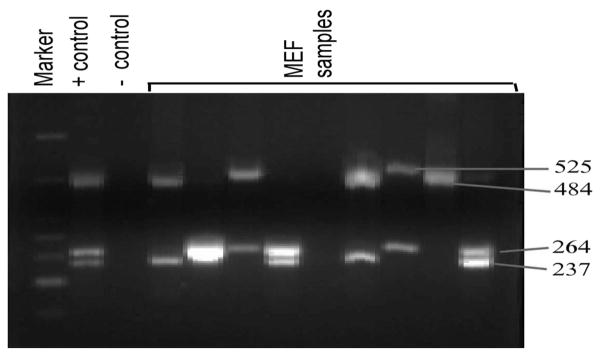
Multiplex-PCR detection of 4 bacterial pathogens of MEF samples from children with AOM. In every set of reactions positive and negative controls are included. The size of the 4 PCR products (525, 484, 264, and 237 base pairs) are also shown on the side of the figure, which corresponds to H. influenzae, S. pneumoniae, A. otitidis, and M. catarrhalis, respectively.
Sensitivity of the M-PCR method was determined for all 4 organisms based on dilutional experiments. The method reliably detected the bacterial genome for all 4 organisms when 10 organisms per reaction were present, which is consistent with previous studies.9 To confirm that PCR products were specific to each species, 3 of the PCR products corresponding to each species were randomly picked and verified by sequencing. The sequences showed 100% correspondence, which confirms the presence of genomic DNA of that species in the samples.
To test the reliability of the PCR method, 48 culture positive MEF samples and 52 culture positive nasopharyngeal samples for H. influenzae, S. pneumoniae, and M. catarrhalis were studied. All the culture positive samples for H. influenzae, S. pneumoniae, and M. catarrhalis were also PCR positive for that species. Alloiococcus otitidis was also detected in 21% of MEF and 12% of nasopharyngeal samples from children who were culture positive for H. influenzae, S. pneumoniae, and M. catarrhalis.
Multiplex-PCR was then performed on 49, 43, and 38 culture negative MEF, nasopharyngeal and oropharyngeal samples, respectively. Culture negative MEF samples yielded S. pneumoniae, H. influenzae, M. catarrhalis, and A. otitidis in 51%, 35%, 14%, and 32% of cases, respectively. In 9% of the cases, A. otitidis was detected as the sole AOM pathogen.
Culture negative samples that are positive for bacterial DNA by PCR can be explained in some cases by recent or current antibiotic therapy received by the host. Table 2 summarizes all the PCR data from culture negative MEF, nasopharyngeal, and oropharyngeal samples of children with AOM whether they were on an antibiotic or not. About 50% of the AOM patients who had culture negative samples were receiving an antibiotic at the time of operation, just before sample collection.
TABLE 2.
Frequency of Detection of Major Otopathogens Using Multiplex-PCR in Culture-Negative MEF, Nasopharyngeal, and Oropharyngeal Cultures from Children With AOM
| Pathogen | MEF (N = 49), Antibiotic*
|
Nasopharyngeal (N = 43), Antibiotic*
|
Oropharyngeal (N = 38), Antibiotic*
|
|||
|---|---|---|---|---|---|---|
| Yes (n = 26 ) (%) | No (n = 23) (%) | Yes (n = 23) (%) | No (n = 20) (%) | Yes (n = 20) (%) | No (n = 18) (%) | |
| S. pneumoniae | 57.6 | 43.4 | 34.7 | 55.0 | 35.0 | 27.7 |
| H. influenzae | 30.7 | 39.1 | 30.4 | 35.0 | 30.0 | 16.6 |
| M. catarrhalis | 15.3 | 13.0 | 8.7 | 10.0 | 0 | 0 |
| A. otitidis | 38.4 | 26.0 | 13.0 | 5.0 | 5.0 | 0 |
N = total number of samples.
In some of the culture negative samples more than one otopathogen was detected.
Whether children were on antibiotic or not prior to sample acquisition.
Evaluation of M-PCR Detection of Viable Bacteria
Given the sensitivity of the PCR technique, fossilized bacterial DNA may be detected in biologic samples and so PCR detection does not establish whether viable organisms are present. To determine how fast DNA of bacteria cleared from the MEF, we compared the M-PCR data between MEF samples collected at different time intervals ranging from 1 week to 3 months from 15 children having recurrent AOM and undergoing a repeat tympanocentesis. Among these 15 children DNA of 4 bacterial species disappeared between 7 days and 21 days except in one case where H. influenzae DNA was detected 20 days after the first tympanocentesis.
RT-PCR Results
As bacterial RNA has a half life of only seconds to minutes and requires intact organisms for its synthesis,25 RT-PCR was performed on 3 of the culture negative but S. pneumoniae M-PCR positive MEF samples to detect RNA and 2 were positive for mRNA; likewise we tested 2 M-PCR positive sample for M. catarrhalis and 1 was positive for mRNA.
DISCUSSION
This is the first study, to our knowledge, where all AOM clinical diagnoses were made by validated otoscopists using a bulging TM as a necessary and sufficient criterion, followed by microbiology and molecular diagnostics to detect the bacterial pathogens. Our main findings were as follows: (1) Bacterial cultures were positive for the 3 main otopathogens Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in 71% of MEF samples and a M-PCR assay improved detection of the 3 otopathogens to 88%. (2) We detected the 3 major otopathogens colonizing the nasopharyngeal in AOM children in 77% of samples by culture but with M-PCR we detected the presence of these otopathogens in 93% of children. (3) Alloiococcus otitidis was detected by M-PCR in 21% of MEF of children with AOM (and in 12% of nasopharyngeal and oropharyngeal samples). (4) When we tested MEF from closely spaced repeat tympanocentesis, we showed bacterial DNA usually clears from the MEF in 1 to 3 weeks and by use of RT-PCR, mRNA of the targeted species was identified in culture negative samples. As a result, the M-PCR results from our study identified viable bacteria and not fossilized bacterial DNA.
Previous publications of microbiologic results from our otitis media research center found positive cultures for the 3 major otopathogens in 53%, 56%, and 56% of tympanocenteses in years 1997, 1998–2000, and 2001–2003, respectively, in patients with recurrent AOM or AOM antibiotic treatment failure.6,26–28 More recently, when our group began to focus and rely on a bulging TM for the diagnosis of AOM, otopathogens were isolated from MEF in patients with recurrent AOM in approximately 75% of cases, similar to the findings of the current report.29,30 However, the results of typmanocenteses will differ in recurrent AOM or AOM antibiotic treatment failure patients as shown in our most recent article when we evaluated culture positive rates in patients with their 1st or 2nd AOM episode and a bulging TM without prior antibiotic treatment and found otopathogens in 87% of AOM episodes.23 In comparison, Kilpi et al,31 in Finland found a culture positive rate of 83% in patients with their first few AOM episodes and in other centers otopathogen isolation rates in the MEF of recurrent AOM patients ranged from 68% to 90%,32,33 and 50% in patients with AOM treatment failure.34 Thus, the results of the current study are in line with previous reports from our center and others. In untreated AOM, culture positivity rates are generally around 85%, among those with recurrent AOM rates are generally around 70% and for AOM treatment failures undergoing tympanocentesis while receiving antibiotics, the isolation rate is around 50%. The addition of a molecular technique such as M-PCR can improve detection of otopathogens in all types of AOM diagnosis situations by 10% to 30%, with higher rates of improved detection occurring in MEFs collected from children who recently received antibiotics or who undergo tympanocentesis while on antibiotics.
Detection of bacterial DNA from culture negative samples by PCR can be explained in some cases by recent or current antibiotic therapy received by the host, lack of sustained viability of the bacteria during transport, and/or low quantities of organisms. We did not systematically evaluate all these variables. We found that almost 50% of the AOM children were receiving antibiotics at the time the MEF sample was collected. Thus, antibiotic therapy could explain half of the culture negative results but was not the sole explanation for culture negative M-PCR positive results. Therefore, it is reasonable to conclude that the PCR is of value in detection of bacterial otopathogens.
The detection of A. otitidis in MEF and nasopharyngeal samples was of interest. A. otitidis has been isolated in the past almost exclusively from patients with OME. We could find only 2 prior studies (from Finland and Japan) where A. otitidis was detected in any patients with AOM.22,35 This is the first report to our knowledge of detection of A. otitidis in the MEF of children with AOM from the United States. It may be that this slow growing bacterium has not been identified in other studies of AOM because only standard culture methods were used. Because A. otitidis is susceptible to aminopenicillins,13 its presence may not have been identified due to antibiotic treatment failures. In 91% of our samples A. otitidis was detected with another bacterium. We are now studying whether A. otitidis acts as a mediator/enhancer of OME following AOM when it is present in a polymicrobial infection with other otopathogens. We performed the M-PCR assays months after all samples had been processed using standard microbiology techniques, so we are unable to report on the correlation between culture and M-PCR detection rates; this is a limitation of our study. When the frequency of A. otitidis detection was compared between MEF versus nasopharyngeal samples, we found nasopharyngeal samples were several fold lower, similar to previous reports.10,19 Previous studies concluded that A. otitidis was more frequently detected in older children (>2 years of age) compared with younger children.16,22 In our study almost all the MEF samples were from children <2 years old.
The detection of bacterial DNA from culture negative samples by PCR does not confirm whether viable organisms are present or not. In a study from children with OME and recurrent AOM, Hall-Stoodley36 found direct detection of biofilms on middle ear mucosa biopsy specimens and showed a correlation of culture negative conditions with biofilm. In a study by Post et al,37 in the AOM chinchilla model it was shown that bacterial DNA does not persist in MEFs in an amplifiable form for greater than 3 days. In our MEF samples from children with recurrent AOM, we observed that DNA clears from MEF in 1 to 3 weeks. The RT-PCR results indicate the presence of intact, viable organisms in some of the culture negative MEF samples, which is consistent with the previous study by Rayner et al.38
In conclusion, using a bulging TM as an important clinical diagnostic criterion, we found a 71% culture positivity rate for bacterial otopathogens. Adding an M-PCR assay allowed the additional detection of bacterial otopathogens in 17% of MEF and 16% of nasopharyngeal/oropharyngeal samples. Multiplex-PCR also effectively detects A. otitidis in about 20% of children with AOM, almost always concurrently with another more common otopathogens. Using M-PCR and RT-PCR, we found that bacterial DNA clears the MEF in 1–3 weeks and the bacterial DNA detected is from viable bacteria.
Acknowledgments
Supported by the National Institute on Deafness and Other Communication Disorders research grant R01DC008671, the Thrasher Research Fund research award 02823–2, and an investigator initiated grant from Wyeth Vaccines, all to Dr. Pichichero as principal investigator.
The authors thank the staff of Legacy, Lewis, Long Pond, and Sunrise Pediatrics for cooperation in sample collection.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.pidj.com).
References
- 1.Rosenfeld RM, Lous J, Bluestone CD, et al. Recent advances in otitis media. 8. Treatment. Ann Otol Rhinol Laryngol Suppl. 2005;194:114–139. [PubMed] [Google Scholar]
- 2.Teele DW, Klein JO, Rosner B. Epidemiology of otitis media during the first seven years of life in children in greater Boston: a prospective, cohort study. J Infect Dis. 1989;160:83–94. doi: 10.1093/infdis/160.1.83. [DOI] [PubMed] [Google Scholar]
- 3.Teele DW, Klein JO, Rosner BA. Epidemiology of otitis media in children. Ann Otol Rhinol Laryngol Suppl. 1980;89:5–6. doi: 10.1177/00034894800890s304. [DOI] [PubMed] [Google Scholar]
- 4.Klein JOBC. Otitis media. In: Feigin RD, editor. Textbook of Pediatric Infectious Diseases. Philadelphia: Saunders; 2004. pp. 215–235. [Google Scholar]
- 5.Jacobs MR, Dagan R, Appelbaum PC, et al. Prevalence of antimicrobial-resistant pathogens in middle ear fluid: multinational study of 917 children with acute otitis media. Antimicrob Agents Chemother. 1998;42:589–595. doi: 10.1128/aac.42.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casey JR, Pichichero ME. Changes in frequency and pathogens causing acute otitis media in 1995–2003. Pediatr Infect Dis J. 2004;23:824–828. doi: 10.1097/01.inf.0000136871.51792.19. [DOI] [PubMed] [Google Scholar]
- 7.Pelton SI. Otoscopy for the diagnosis of otitis media. Pediatr Infect Dis J. 1998;17:540–543. doi: 10.1097/00006454-199806000-00032. [DOI] [PubMed] [Google Scholar]
- 8.Ueyama T, Kurono Y, Shirabe K, et al. High incidence of Haemophilus influenzae in nasopharyngeal secretions and middle ear effusions as detected by PCR. J Clin Microbiol. 1995;33:1835–1838. doi: 10.1128/jcm.33.7.1835-1838.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendolin PH, Markkanen A, Ylikoski J, et al. Use of multiplex PCR for simultaneous detection of four bacterial species in middle ear effusions. J Clin Microbiol. 1997;35:2854–2858. doi: 10.1128/jcm.35.11.2854-2858.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harimaya A, Takada R, Somekawa Y, et al. High frequency of Alloiococcus otitidis in the nasopharynx and in the middle ear cavity of otitis-prone children. Int J Pediatr Otorhinolaryngol. 2006;70:1009–1014. doi: 10.1016/j.ijporl.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Post JC, White GJ, Aul JJ, et al. Development and validation of a multiplex PCR-based assay for the upper respiratory tract bacterial pathogens Haemophilus influenzae, Streptococcus pneumoniae, and Moraxella catarrhalis. Mol Diagn. 1996;1:29–39. doi: 10.1054/MODI00100029. [DOI] [PubMed] [Google Scholar]
- 12.Faden H, Dryja D. Recovery of a unique bacterial organism in human middle ear fluid and its possible role in chronic otitis media. J Clin Microbiol. 1989;27:2488–2491. doi: 10.1128/jcm.27.11.2488-2491.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashhurst-Smith C, Hall ST, Walker P, et al. Isolation of Alloiococcus otitidis from indigenous and nonindigenous Australian children with chronic otitis media with effusion. FEMS Immunol Med Microbiol. 2007;51:163–170. doi: 10.1111/j.1574-695X.2007.00297.x. [DOI] [PubMed] [Google Scholar]
- 14.de Miguel Martínez I, Macias AR. Serous otitis media in children: implication of Alloiococcus otitidis. Otol Neurotol. 2008;29:526–530. doi: 10.1097/MAO.0b013e318170b5f8. [DOI] [PubMed] [Google Scholar]
- 15.de Miguel Martínez I, Ramos Macías A, Masgoret Palau E. Bacterial implication in otitis media with effusion in the childhood. Acta Otorrinolaringol Esp. 2007;58:408–412. [PubMed] [Google Scholar]
- 16.Leskinen K, Hendolin P, Virolainen-Julkunen A, et al. The clinical role of Alloiococcus otitidis in otitis media with effusion. Int J Pediatr Otorhinolaryngol. 2002;66:41–48. doi: 10.1016/s0165-5876(02)00186-6. [DOI] [PubMed] [Google Scholar]
- 17.Hendolin PH, Karkkainen U, Himi T, et al. High incidence of Alloiococcus otitis in otitis media with effusion. Pediatr Infect Dis J. 1999;18:860–865. doi: 10.1097/00006454-199910000-00005. [DOI] [PubMed] [Google Scholar]
- 18.Kalcioglu MT, Oncel S, Durmaz R, et al. Bacterial etiology of otitis media with effusion: focusing on the high positivity of Alloiococcus otitidis. New Microbiol. 2002;25:31–35. [PubMed] [Google Scholar]
- 19.Durmaz R, Ozerol IH, Kalcioglu MT, et al. Detection of Alloiococcus otitidis in the nasopharynx and in the outer ear canal. New Microbiol. 2002;25:265–268. [PubMed] [Google Scholar]
- 20.Beswick AJ, Lawley B, Fraise AP, et al. Detection of Alloiococcus otitis in mixed bacterial populations from middle-ear effusions of patients with otitis media. Lancet. 1999;354:386–389. doi: 10.1016/S0140-6736(98)09295-2. [DOI] [PubMed] [Google Scholar]
- 21.Frank DN, Spiegelman GB, Davis W, et al. Culture-independent molecular analysis of microbial constituents of the healthy human outer ear. J Clin Microbiol. 2003;41:295–303. doi: 10.1128/JCM.41.1.295-303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leskinen K, Hendolin P, Virolainen-Julkunen A, et al. Alloiococcus otitidis in acute otitis media. Int J Pediatr Otorhinolaryngol. 2004;68:51–56. doi: 10.1016/j.ijporl.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Casey JR, Adlowitz DG, Pichichero ME. New patterns in the otopathogens causing acute otitis media six to eight years after introduction of pneumococcal conjugate vaccine. Pediatr Infect Dis J. 2009 doi: 10.1097/INF.0b013e3181c1bc48. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques. 1993;15:532–537. [PubMed] [Google Scholar]
- 25.Lewin BM. Gene Expression. Vol. 1. London: John Wiley; 1974. pp. 285–403. [Google Scholar]
- 26.Pichichero ME, Pichichero CL. Persistent acute otitis media: I. Causative pathogens. Pediatr Infect Dis J. 1995;14:178–183. doi: 10.1097/00006454-199503000-00002. [DOI] [PubMed] [Google Scholar]
- 27.Pichichero ME, Pichichero CL. Persistent acute otitis media. II. Antimicrobial treatment. Pediatr Infect Dis J. 1995;14:183–188. doi: 10.1097/00006454-199503000-00003. [DOI] [PubMed] [Google Scholar]
- 28.Pichichero ME, McLinn S, Aronovitz G, et al. Cefprozil treatment of persistent and recurrent acute otitis media. Pediatr Infect Dis J. 1997;16:471–478. doi: 10.1097/00006454-199705000-00003. [DOI] [PubMed] [Google Scholar]
- 29.Pichichero ME, Casey JR. Emergence of a multiresistant serotype 19A pneumococcal strain not included in the 7-valent conjugate vaccine as an otopathogen in children. JAMA. 2007;298:1772–1778. doi: 10.1001/jama.298.15.1772. [DOI] [PubMed] [Google Scholar]
- 30.Pichichero ME, Casey JR, Hoberman A, et al. Pathogens causing recurrent and difficult-to-treat acute otitis media, 2003–2006. Clin Pediatr (Phila) 2008;47:901–906. doi: 10.1177/0009922808319966. [DOI] [PubMed] [Google Scholar]
- 31.Kilpi T, Herva E, Kaijalainen T, et al. Bacteriology of acute otitis media in a cohort of Finnish children followed for the first two years of life. Pediatr Infect Dis J. 2001;20:654–662. doi: 10.1097/00006454-200107000-00004. [DOI] [PubMed] [Google Scholar]
- 32.Leibovitz E, Raiz S, Piglansky L, et al. Resistance pattern of middle ear fluid isolates in acute otitis media recently treated with antibiotics. Pediatr Infect Dis J. 1998;17:463–469. doi: 10.1097/00006454-199806000-00005. [DOI] [PubMed] [Google Scholar]
- 33.Del Beccaro MA, Mendelman PM, Inglis AF, et al. Bacteriology of acute otitis media: a new perspective. J Pediatr. 1992;120:81–84. doi: 10.1016/s0022-3476(05)80605-5. [DOI] [PubMed] [Google Scholar]
- 34.Cohen R, de la Rocque F, Boucherat M, et al. Treatment failure in otitis media: an analysis. J Chemother. 1994;6(suppl 4):17–22. [PubMed] [Google Scholar]
- 35.Harimaya A, Takada R, Hendolin PH, et al. High incidence of Alloiococcus otitidis in children with otitis media, despite treatment with antibiotics. J Clin Microbiol. 2006;44:946–949. doi: 10.1128/JCM.44.3.946-949.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall-Stoodley L, Hu FZ, Gieseke A, et al. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA. 2006;296:202–211. doi: 10.1001/jama.296.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Post JC, Aul JJ, White GJ, et al. PCR-based detection of bacterial DNA after antimicrobial treatment is indicative of persistent viable bacteria in the chinchilla model of otitis media. Am J Otolaryngol. 1996;17:106–111. doi: 10.1016/s0196-0709(96)90005-8. [DOI] [PubMed] [Google Scholar]
- 38.Rayner MG, Zhang Y, Gorry MC, et al. Evidence of bacterial metabolic activity in culture-negative otitis media with effusion. JAMA. 1998;279:296–299. doi: 10.1001/jama.279.4.296. [DOI] [PubMed] [Google Scholar]


