Abstract
The promise of “personalized medicine” guided by an understanding of each individual’s genome has been fostered by increasingly powerful and economical methods to acquire clinically relevant features. We describe operational implementation of prospective genotyping linked to an advanced clinical decision support system to guide individualized healthcare in a large academic health center. This approach to personalized medicine includes patient and healthcare provider engagement, identifying relevant genetic variation for implementation, assay reliability, point-of-care decision support, and necessary institutional investments. In one year, approximately 3,000 patients, most scheduled for cardiac catheterization, were genotyped on a multiplexed platform including CYP2C19 variants that modulate response to the widely-used antiplatelet drug clopidogrel. These data are deposited into the Electronic Medical Record and point-of-care decision support is deployed when clopidogrel is prescribed for those with variant genotypes. The establishment of programs such as this is a first step toward implementing and evaluating strategies for personalized medicine.
Keywords: Drug-Drug Interactions, Personalized Medicine, Pharmacogenetics, Translational Medicine, Adverse Drug Reactions
INTRODUCTION
An increasingly robust body of knowledge indicates that human genetic variation modulates disease susceptibility and drug responses. Compelling arguments have been put forward supporting the use of genetic variation information to choose among medications or dose 1,2. The US Food and Drug Administration (FDA) now includes pharmacogenomic data in drug labels 3, some of which have acquired “Black Box” status. While incorporating information about individual genomic variability can improve health care 1, there are challenges to implementing this vision so that the fundamental idea remains largely untested.
The conventional approach to using genetic information to guide prescribing is reactive and often labor intensive: for instance, a practitioner must recognize the potential utility of knowing a patient’s genetic variant status when considering a therapeutic, order the test, receive and interpret the result, and re-contact the patient to relay the treatment decision or alter a prescription if already dispensed. Unfortunately, as knowledge relating genomic data to healthcare expands, the systems to deliver information have not, and the current approach therefore is becoming increasingly impractical even for a limited number of drugs.
An alternate strategy is to deposit genomic information in patient records preemptively – that is, prior to its being needed in care 1,4. In this scenario, when a drug is considered for a patient with known genetic variants modulating response, electronic decision support would alert the practitioner to potential decreased efficacy or adverse effect risks, and would recommend alternate therapies as appropriate. Implicit in this idea is that genetic information is stored and advice is provided in an advanced health information technology environment: no healthcare provider can reasonably be expected to have a personal fund of knowledge large enough to take appropriate action in an era of genomically-enabled personalized medicine without automated clinical decision support 5,6. The preemptive approach presents substantial challenges, such as selecting the genetic variants for prospective testing and identifying which patients to test. Test results must be aligned with synthesized evidence, formatted to be acted upon by decision support algorithms and rules, and presented as clearly actionable guidance to prescribers.
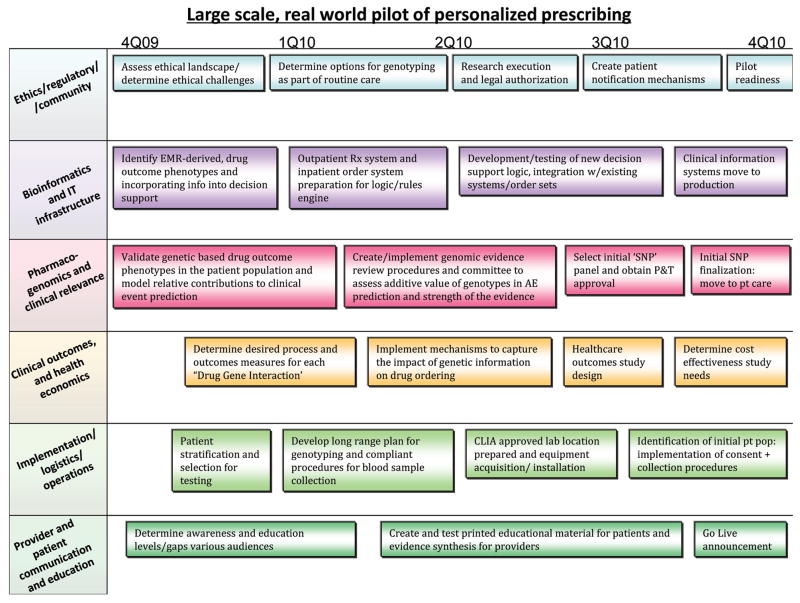
Addressing these challenges is the key goal of a pharmacogenomics implementation project at Vanderbilt University Medical Center (VUMC) launched in September 2010. We describe the elements of this program, a 1-year report of initial implementation focused first on antiplatelet therapy following placement of cardiovascular stents, and its potential generalizability. The present implementation focuses on prospective assessment of genomic variants that have relevance for drug prescribing and is designated the Pharmacogenomic Resource for Enhanced Decisions in Care and Treatment (PREDICT). The long-term goal of the PREDICT initiative is to establish a framework and comprehensive infrastructure for preemptive incorporation of genomic and other high dimensional patient-specific data into the VUMC Electronic Medical Record (EMR). The PREDICT planning team was formed in September 2009 with the goal of developing and begin implementing the program within a year. The key elements of the planning process are shown in Figure 1 and the implementation steps and components are described here.
Figure 1.
Program components overview and timeline, with key milestones reflected
RESULTS
Overview
The PREDICT team established the overall goal of the PREDICT program to prospectively genotype patients for “high-value” genetic variants that could improve drug selection and dosing to decrease medication related adverse events. The evidence review process identified the relationship between variant CYP2C19 genotypes and reduced clopidogrel efficacy 7–11, included in the FDA’s March 2010 relabeling of the drug 12, as the first PREDICT implementation interaction. The evidence for a pharmacogenomic contribution to clopidogrel response is sufficiently strong that our initial implementation was executed as a quality improvement initiative. This report therefore describes the multiplexed genotyping we have undertaken and includes a major focus on the process for moving CYP2C19 genotype information into routine healthcare processes at VUMC.
Patient attitude pilot survey
An initial step was to take advantage of the patient portal oMyHealthAtVanderbilt.com, currently used by >140,000 patients at our medical center, to survey patient attitudes. An optional patient survey link was placed on the portal and available over 4 days in May 2010, and a total of 644 patients responded. Patient demographic characteristics are shown in Table 1. While 84% percent of respondents found prospective genotyping for use in future care acceptable, they also felt that the practice is not yet routine relative to other common laboratory tests (Table 2): 87% identified cholesterol levels as routine, while only 20% felt similarly about genetic tests conducted to avoid adverse drug outcomes (p<0.0001 via Chi-square). “Routine” had no pre-specified definition provided to participants, and the goal was to elicit relative perspectives.
Table 1.
Demographics of patient pilot survey respondents
| DEMOGRAPHIC - AGE | # OF RESPONDENTS |
|---|---|
| Age 20–39 | 202 |
| Age 40–59 | 292 |
| Age 60–79 | 143 |
| Age 80+ | 8 |
| DEMOGRAPHIC - RACE | # OF RESPONDENTS |
| Caucasian | 521 |
| African-American | 44 |
| Asian | 8 |
| American Indian | 1 |
| Unknown | 71 |
| DEMOGRAPHIC - GENDER | # OF RESPONDENTS |
| Male | 224 |
| Female | 421 |
Table 2.
Responses as to whether a specific test was viewed as routine
| Test | Routine | Not Routine | Not Sure | No Response |
|---|---|---|---|---|
| Cholesterol levels | 87% | 12% | 1% | 1% |
| Blood counts used to detect anemia or infection | 73% | 24% | 3% | 1% |
| Hemoglobin A1c (for diabetes management) | 59% | 32% | 10% | <1% |
| Urine test for drug abuse | 18% | 78% | 5% | 1% |
| Testing for HIV/AIDS | 24% | 71% | 5% | 2% |
| Information in genes to avoid bad side effects from medicines | 20% | 69% | 12% | 1% |
| Information in genes to test risk for specific diseases | 24% | 70% | 7% | 1% |
Patient focus groups
Focus group discussions generated a wide range of findings related to clinical pharmacogenomics 13. Patients expressed preference for brief verbal notification of testing from their provider rather than a process requiring them to sign a formal patient consent document. Patients expressed a wide-range of preferences about whether they would want to be informed of their personal ancillary findings related to genetic susceptibility to disease, but came to a consensus that (1) patients should have a choice about how much and which types of ancillary findings they would receive, (2) providers should not be provided with test results that are not communicated to patients, and (3) work to implement PREDICT should not be delayed by issues related to ancillary findings.
Patient notification of test ordering
To raise awareness of the program, information related to PREDICT has been included in the standard Consent to Treatment forms all patients sign upon registration. Electronic prompts within the patient chart remind providers and other members of the care team to discuss the testing program verbally, document the conversation, record preferences, and provide the brochure. In this way, patients are notified of the PREDICT program and have the opportunity to have questions addressed. [A copy of the brochure is available in Supplementary Figure 1.]
Drug-gene interaction evidence synthesis and review
In 2009, clopidogrel was the third most commonly prescribed drug in the United States, and the primary indication for its use is in patients with coronary artery disease and in particular in the prevention of stent thrombosis in patients treated with drug-eluting stents. While clopidogrel was marketed in 1997, it was only in 2006 that bioactivation by CYP2C19 was identified as a major factor in its anti-platelet efficacy 14. In 2009, multiple reports from single centers reported that individuals homozygous for CYP2C19*2, a loss of function allele, displayed increased cardiovascular event rates during clopidogrel prescribed for coronary stenting 7–9,15. More recently, meta-analyses have confirmed this risk and have extended it to include individuals who are heterozygous (termed CYP2C19*1/*2) for this variant, and suggested variants in the intestinal transporter encoded by ABCB1 may also contribute 10,11,16,17. CYP2C19*17 is a “gain-of-function” allele that has been associated with increased bleeding during clopidogrel treatment 18. However, the evidence for this outcome is not as strong as for the *2 allele, and the FDA label does not recommend acting on this genotype. Accordingly, the initial implementation described here does not consider different management in subjects carrying this variant. The development of an implementation plan started with review of these data by a subcommittee of the standing Pharmacy and Therapeutics (P&T) committee described further in Methods, and that review agreed with the FDA label and consensus statements from professional societies 12 that there is substantial individual variability in response to clopidogrel, and that individuals with decreased CYP2C19 activity display an increased incidence of stent thrombosis and other cardiovascular events. In addition, a retrospective case-control validation study using data from BioVU, a resource that links DNA extracted from discarded blood samples to deidentified medical records 19, found a statistically significant increase in coronary events in patients with variant CYP2C19 genotypes treated with clopidogrel after coronary stenting (hazard ratio 1.54, 95% CI 1.16–2.06, p=0.003) 20. At VUMC, approximately 4000 patients underwent coronary angiography in 2008, and clopidogrel was ultimately prescribed in 1735 (42.5%). Accordingly, the initial group of patients targeted for preemptive genotyping in PREDICT were those scheduled for coronary arteriography, prior to any decision to prescribe clopidogrel.
Assay Performance
The VeraCode ADME Core Panel that includes 184 variants relevant for drug responses was selected as the initial genotyping platform for the program. To maximize reporting efficiency, patient call rates were established at 97.30%. Average observed call rates for controls is 98.6%. After implementation of the assay for patient testing, repeat testing was performed on 150 patients to measure the concordance of results. The loci showing the highest discordance was GSTT1 CNV and DPYD*9B, SLC22A6 and CYP2D6*9. However, CYP2C19 genotype results demonstrated 100% concordance in all patients. Seven subsequent monthly QC plates have shown similar findings with GSTT1 CNV and DPYD*9B showing the highest discordant results and indicating that patient management utilizing these results will not be instituted. Likewise, on these QC plates, no discordant results have been observed for CYP2C19 on an additional 200 patient specimens tested in duplicate. Importantly, CYP2C19 allele frequencies for *2/*2 and *17/*17 homozygotes and *2 and *17 heterozygotes are as expected when compared to the NCBI dbSNP (Table 3). Ten loci are considered poor performing markers with locus call rates <95% and include CYP1A2*1C (94.47%); ABCC2 – I1324I (94.4%); SLC22A2-M165I (94.19%); DPYD*9B (92.97%); UGT2B17-CNV (91.68%); CYP1A2*3 (91.65%); UGT2B15*2 (91.43%); TPMT*4 (88.65%); GSTM1*B (50.42%) and GSTT1 (46.51%).
Table 3.
Genotype data and dosing recommendation
| Category | Genotypes | Recommendation | Total |
|---|---|---|---|
| Poor metabolizer | CYP2C19*2/*2 | Alternative therapy | 82 (2.6%) |
| Rapid metabolizer | CYP2C19*17/*17 | Usual care | 154 (4.9%) |
| Intermediate metabolizer | CYP2C19*2 Heterozygote, CYP2C19*3 Heterozygote | Alternative therapy | 601 (19.1%) |
| Normal metabolizer | CYP2C19*1/*1, CYP2C19*17 Heterozygote, CYP2C19*4 Heterozygote | Usual care (e.g. clopidogrel 75mg) | 2,059 (65.5%) |
| Indeterminant | CYP2C19*5 Heterozygote, CYP2C19*6 Heterozygote, CYP2C19*8 Homozygote, CYP2C19*8 Heterozygote, CYP2C19*12 Heterozygote | Usual care | 249 (7.9%) |
Clinical decision support systems/architecture
After genotype results are generated, they are stored in a database that is separate from the electronic medical record. Data is archived to a privately owned path on the secure PREDICT application server to protect patient confidentiality. Genetic data that has not been approved for use is stored in a sequestered Oracle database which resides at the VUMC data center behind their firewall. This data is not accessible by patients or providers but is linked to the patient. The data will be stored long term and will not be released until appropriate, i.e. a new genotype is deemed actionable. Patient genotyping data is also protected through The Genetic Information Nondiscrimination Act of 2008 (GINA), the Federal law that prohibits discrimination in health coverage and employment based on genetic information.
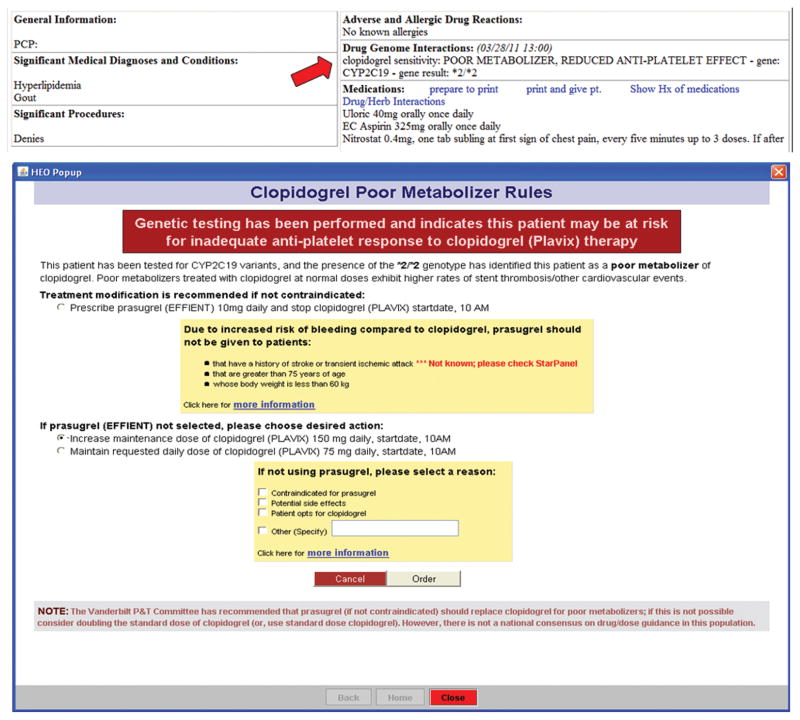
Genotypes that have been validated by the established quality control metrics, and reviewed and approved for clinical implementation by the P&T process above are deemed “actionable”. After an actionable genotype is recorded, it is converted into a standard notation and interpretation (e.g., CYP2C19*2, “Poor Metabolizer: Reduced anti-platelet effect”), stored in the EMR as a molecular diagnostic lab result, and displayed within a “Drug-Genome Interaction” (DGI) section of the patient summary page of the EMR (Figure 2A). Decision support modules were developed in collaboration with informaticists, medical geneticists, clinical pharmacologists, clinicians practicing in relevant fields, and the P&T committee. Current decision support modules are integrated with inpatient computerized provider order entry (CPOE) and the outpatient electronic prescribing application.
Figure 2.
Lab result presentation of genetic results in EMR and clinical decision support guidance within order entry system.
The data implicating CYP2C19*2 as a modulator of response to clopidogrel did not translate unambiguously into standardized clinical recommendations 21,22. However, the approach that our program ultimately adopted was to recommend use of prasugrel for patients with genotypes associated with decreased clopidogrel effectiveness. (The option of recommending a higher dose of clopidogrel was not supported by any data at the time, or since 23.) Use of prasugrel in patients with acute coronary syndrome has been associated with a 20% reduction in adverse cardiovascular events at 12 months compared to clopidogrel but at the expense of an increased risk of major bleeding 24. In patient subgroups with elevated rates of bleeding events (e.g., those aged >75 years), prasugrel is contraindicated and so in PREDICT, the drug is not recommended in these patients. Figure 2B shows the point of care decision support guidance that is triggered when a prescription order is initiated for a patient with a variant CYP2C19 genotype. The guidance implemented in September 2010 focused on CYP2C19*2/*2 homozygotes only; with the availability of data from subsequent meta-analyses 10,11, the guidance was later extended to CYP2C19*1/*2 heterozygotes in 2011. Provider behavior in response to genotypes and outcomes of individuals are being followed for Quality Improvement program evaluation at a later date.
Initial uptake and CYP2C19 variant assay results
The program was launched on September 15, 2010. As of August 1, 2011, 3449 patients had undergone left heart catheterization, and genotyping was ordered in 2165 (63%). The results of CYP2C19 genotyping are shown in Table 3; as expected of a largely Caucasian patient population, 19% were heterozygous for the *2 allele, and 3% were homozygous. Genotypes are classified as “Normal Metabolizer” (two copies of the CYP2C19*1 allele), “Intermediate Metabolizer” (one copy of either the *2,*3, or *4 allele), “Poor Metabolizer” (two copies of either the *2, *3, or *4 allele), or “Rapid Metabolizer” (two copies of the *17 allele). “Indeterminate” is used to note variants that have insufficient evidence to be clinically actionable or variants that could not be called due to low genotyping performance or low signal.
DISCUSSION
We have gleaned lessons that might be beneficial for other sites considering such a program, listed below:
Commitments across multiple disciplines as well as by institutional leadership are necessary: the requisite disciplines include clinicians, geneticists, informaticists, user interface experts, pharmacists, pharmacologists, clinical pathologists, and program managers. Major institutional commitments are critical for salary support for faculty and staff effort; purchase of new equipment for clinical genotyping; and funding for genotyping for the initial set of patients. The estimated total is ~$5 million in the first two years. Seeking payer reimbursement for genetic screening to guide drug therapy as specified in approved FDA labels is a next phase of the program. With this in place, we expect the program to ultimately generate cost savings to payers and patients through reduced adverse outcomes associated with lack of efficacy and toxicity.
Collaboration with interventional cardiology was essential. This involved specific domain expertise as well as buy-in to the program by users. We anticipate that each new drug-genotype rollout will require expertise in terms of pharmacogenomic content as well as domain expertise from physicians who work within the specific targeted practice settings. The initial testing rate was approximately 75%, and it is possible that the mechanics of ordering a new test or uncertainties over how to interpret the result may have resulted in some clinicians’ omitting the test order from the pre-catheterization protocol.
A key initial step for implementation was to establish attitudes in our patient population. To accomplish this goal we leveraged the MyHealthAtVanderbilt.com portal and conducted patient focus groups. We recognize that this group of patients may differ in some ways than the general VUMC population; for example, survey participants are likely to be more familiar with diagnostic testing since receipt of lab results is a common reason to access the portal. However, this population represents 27% of the current VUMC population and is growing25. These methods provided strong support for using genetic information to guide choice of drugs or dosage, and provided invaluable input for implementation of PREDICT.
The procedure for developing and refining decision support rules is time consuming; it required careful review of the literature and input from multiple constituencies, including clinicians, pharmacy, informatics, and genomics, and repetitive iterations of the formats in which it would be provided (Figure 1). There were advantages to starting the program with clopidogrel: the drug is widely-used; there was good evidence that failure of drug efficacy confers risk of substantial morbidity; there is a single common risk allele in subjects of European descent; and the assay is reliable. The drug prescribing information also has a “Black Box” warning from the FDA. However, even this “simple” example highlighted the nuances and complexities of an implementation program. For example, there is an evolving set of data relating genotypes to outcomes: since initiating PREDICT in September 2010, there have been over 60 manuscripts published that might influence the approach to clopidogrel and CYP2C19. These include clinical outcomes in poor metabolizers11,26, interaction with proton pump inhibitors27,28, evaluations of other antiplatelet therapies29, dose adjustments based on genotype23,30,31, comparison to ticagrelor32, further characterization of rare 2C19 alleles33, use of platelet aggregometry testing 34 and platelet inhibition response11,26–33,35. Indeed, our initial implementation focused on patients with the *2/*2 genotype, and was extended to those with the *1/*2 genotype in March 2011, after publication of a large meta-analysis describing poorer outcome in both homozygotes and heterozygotes11. Maintaining ongoing awareness of evolving pharmacogenetic evidence for purposes of translation to practice is a daunting task, and with the ever-increasing number of genetic tests and medications approved for pre-treatment genetic tests, additional program staff may be necessary to keep up with the field. An increase in institutional funding may be necessary to do so.
While the evidence for a genetic predictor to clopidogrel response is strong, there is controversy over the utility of genotyping 22. The best approach to managing patients carrying allele(s) indicating increased risk for drug failure and yet who are not eligible for alternate therapy (such as prasugrel) is unclear. The role of genomic variants in other settings in which clopidogrel is used, such as neurovascular disease, is unknown.
There is a need to perform and continuously monitor assay performance in a CLIA setting. The initial platform that we chose performed well in determining CYP2C19*2 phenotype, but as described above, there were a number of assays that did not meet clinical performance expectations. Additionally, with rapidly emerging genotyping technologies, the future use of DNA samples for additional testing is a possibility, as leftover DNA is currently stored, but this has not been explored in detail by PREDICT implementation teams and warrants further examination.
We are in the process of adding additional drug-gene pairs, including warfarin/CYP2C9-VKORC1 and simvastatin/SLCO1B1 to PREDICT. Each new drug-gene pair presents issues analogous to those we encountered with clopidogrel: an evolving set of evidence and SNPs, variability across ancestries, newer drugs in the same therapeutic class, and changing regulatory advice. Nevertheless, the principles we describe above represent important starting points for any program that proposes to implement genotype data into clinical workflow. The fundamental design principle that we embrace is that genotype data are best used in such an effort when they are available in the EMR prior to drug prescription, “pre-prescription genotyping”.
MATERIALS AND METHODS
Drug-gene variant evidence synthesis and review
The selection of drug-gene variants pairs to be implemented started with consideration of available published evidence, followed by formulation of an initial implementation plan. The criteria for including a drug-genome interaction (DGI) in the program included: an established body of evidence in the biomedical literature linking DGI to patient outcomes, therapeutic guidance from the FDA 36,37, risk allele frequency, and the severity and costs of adverse events that could be averted by genome tailored prescribing. The decision for initial implementation was also guided by practical considerations, such as whether the genotyping platform selected for initial implementation contained the genotypes of interest; the potential complexity of decision support rules; the number of providers involved; and availability of faculty with content expertise. Our initial implementation plan also included local validation of drug-genome associations in BioVU 19, the resource that links DNA samples to de-identified clinical data from the VUMC EMR, so that we had confidence that the conditions of interest did exist in the patient population that would be affected by the intervention.
The original implementation plan was developed by a multidisciplinary team that included individuals with expertise in pharmacy operations, clinical laboratory operations, pharmacogenomics, biomedical informatics, and ethics, as well as clinicians with content expertise. In the case of clopidogrel, interventional cardiologists were involved with the design and implementation of the program. The review process was facilitated by a CTSA studio 38 and final approval was given by the Pharmacy and Therapeutics (P&T) Committee. A Therapeutics Subcommittee of P&T composed of a cross-section of VUMC clinicians, pharmacogeneticists, and pharmacists was organized to perform a preliminary review for any proposed drug-genotype variant. For each actionable genotype, the entire process of genotype validation, review and approval for clinical implementation took approximately one year.
Clinician communication
Since the primary initial PREDICT clinical site was the catheterization lab, the primary providers were the 13 interventional cardiologists performing the procedures and responsible for post procedure anti-platelet therapy. Four prelaunch sessions were held among this group. These involved review of the published and local data supporting the intervention, discussion of the proposed format of the guidance as provided by VUMC’s clinical decision support systems, and the mechanisms for providing a testing prompt within Vanderbilt’s EMR to guide clinicians to order the genotyping test.
Patient attitudes – pilot survey
With Institutional Review Board approval, a pilot patient survey was conducted to gauge attitudes toward aspects of genetic testing. The survey was conducted by soliciting users of the MyHealthAtVanderbilt.com (MHAV) patient portal, a resource that provides general health information, messages healthcare providers, and displays to patients their laboratory results 25. The optional survey link was made available on the MHAV patient portal home page after a user had logged into their account. If the individual elected to take the survey and opened the link, they were directed to the structured survey developed using REDCap Survey (http://redcap.vanderbilt.edu/consortium/index.php). The domains captured in this pilot survey were patient perspectives on medical testing, whether different types of diagnostic and genetic tests were considered routine, the ways health care institutions use and store their health information, and the importance of use of their genetic information in healthcare decisions.
Patient attitudes – focus groups
Previous studies to evaluate patient perceptions of pharmacogenetics have focused on general concerns related to privacy and management of ancillary findings, but have not attended closely to the practical issues of implementing clinical pharmacogenomics 39–45. We conducted ten focus group sessions, including two conducted in Spanish, with Vanderbilt patients to elicit input the design and implementation of PREDICT 13. Discussions addressed a number of issues, including preferences on how patients would like to be informed of PREDICT, how they would like to provide their consent for pharmacogenomic testing, what they would need to learn from their healthcare provider, and how they would like ancillary findings to be managed.
Ethics review and patient notification of test ordering
The initial deployment of the PREDICT program was undertaken as a healthcare quality improvement (QI) initiative based on the program objective to implement FDA regulatory guidance for prescribing 36,37; thus, there is no research informed consent process as there would be for a study involving human subjects. However, given the seminal nature of the program, the Medical Center Ethics Committee reviewed the overall program plan prior to implementation to provide guidance. Pilot survey, focus group findings, and committee recommendations were used to develop consent procedures, brochure and other patient notification approaches, and ancillary finding policies. Brochures were subsequently developed with a 7th grade reading level based on the Fry Readability measurement.
Genotyping Assay
The VeraCode ADME Core Panel (Illumina, Inc. San Diego, CA) was chosen for genotyping studies. The assay targets 184 variants in 34 genes involved in drug absorption, distribution, metabolism and excretion (ADME) 46. Due to the highly polymorphic nature of many of the regions assayed, a patient sample is analyzed in 3 separate reactions to optimize the assays and prevent competition from adjacent polymorphic regions within the same gene. Thus, a 96 well plate enables analysis of 32 specimens; the VUMC implementation includes 30 patient specimens as well as a control DNA specimen (with a known genotype at the 184 variants) and a negative control reaction tube with all reagents but no template DNA. Polymorphisms in CYP2C19 analyzed in this panel include both common (*2, *17) and rarer (*3, *4, *5, *6, *7, *8, *12) variants.
Implementation within the institution’s CLIA high complexity molecular diagnostics laboratory required additional institutional investments, including space for designated pre- and post-PCR work stations; personnel; laboratory informatics system work stations; specific instrumentation required for the assay, and an automated DNA extractor. To accommodate the anticipated workflow, 4 medical technologists were trained to perform the assay, with other laboratory staff providing daily assistance as needed for DNA extraction. General workflow was designed to process samples and return results within 3 business days from the sample draw.
Assay validation
Prior to implementation for patient testing, the assay was validated by the laboratory by comparing the observed genotypes for all 184 variants with the previously reported genotypes for these variants in 54 control DNA cell line samples repeatedly tested on training plates and by several technologists. The average concordance was 99.58% (SD=0.4844%) from 23 cell lines (ParagonDx, Jacksonville, WY) and 98.36% (SD=1.91 %) from 31 additional cell line samples (Coriell, Camden, NJ).
Assay quality control
In addition to control parameters included by the manufacturer to monitor the performance of each run, a previously validated control specimen is included on each plate to measure the performance and reproducibility of each variant on each plate. Other quality control indicators established and monitored include the: locus call rate; patient and control call rates; variant allele frequencies; and monthly QC plates containing specimens previously tested and reported in the prior month. The locus call rate represents the percentage of patients for which a result is obtained at a given marker and indicates the overall performance of an individual variant. A “no call” result can either indicate complete failure of the assay at that site or potentially reduced stringency within a particular run and the inability of the assay to discriminate among possible variants. The patient and control call rates indicate the percentage of variants for which a result was obtained for a given patient. The variant allele frequencies generated are compared to those listed in NCBI dbSNP and the monthly QC plates assess the reproducibility of the assay. Due to the amount of data collected, reports were generated using bioinformatic tools and submitted to laboratory personnel for review.
Quality assurance of implementation
To monitor the initial clinician response to PREDICT and ensure the timely transmission of information, we deployed a multi-user web-based application to allow a team of nurses and pharmacists to view which eligible patients had an actionable genotype result. Prescribing providers who had not yet received clinical decision support or reacted to a variant genotype (for example, if the patient’s genetic result was returned after they were discharged) were notified with an electronic clinical message via the EMR, which is a standard means of clinical communication within the institution. The QA mechanism enabled the follow-up of genetic results that returned following discharge and prior to a follow-up visit with a Vanderbilt provider.
Acknowledgments
We would like to acknowledge Vanderbilt’s Vice Chancellor and Dean of Medicine for his support, the members of the Medical Center Ethics Committee, the members of the Pharmacy and Therapeutics Committee, and the Vanderbilt Informatics Center. The datasets used for some of the analyses described were obtained from Vanderbilt University Medical Center’s BioVU, which is supported by institutional funding and by the Vanderbilt CTSA grant UL1 RR024975-01 from NCRR/NIH. Components of this effort were also supported in part by RC2 GM092618
Footnotes
Author Contributions:
| JMP | JCD | JFP | GRB | CVJ | AHR | JTD | EB | KB | KJ | DCC | JS | DRM | HHD | RAW | EWC | ES | ML | JM | JNJ | DMR | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| substantial contributions to conception and design | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X |
| substantial contributions acquisition of data | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | |||||
| substantial contributions to analysis and interpretation of data | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | |||||
| drafting the article | X | X | X | X | X | X | X | X | |||||||||||||
| revising it critically for important intellectual content | X | X | X | X | X | X | X | ||||||||||||||
| final approval of the version to be published | X | X | X |
References
- 1.Collins F. Opportunities and challenges for the NIH--an interview with Francis Collins. Interview by Robert Steinbrook. N Engl J Med. 2009;361:1321–3. doi: 10.1056/NEJMp0905046. [DOI] [PubMed] [Google Scholar]
- 2.Green ED, Guyer MS. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–13. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 3.Frueh FW, et al. Pharmacogenomic biomarker information in drug labels approved by the United States food and drug administration: prevalence of related drug use. Pharmacotherapy. 2008;28:992–8. doi: 10.1592/phco.28.8.992. [DOI] [PubMed] [Google Scholar]
- 4.Hamburg MA, Collins FS. The path to personalized medicine. N Engl J Med. 2010;363:301–4. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 5.Masys DR. Effects of current and future information technologies on the health care workforce. Health Aff (Millwood) 2002;21:33–41. doi: 10.1377/hlthaff.21.5.33. [DOI] [PubMed] [Google Scholar]
- 6.Stead WW, Searle JR, Fessler HE, Smith JW, Shortliffe EH. Biomedical informatics: changing what physicians need to know and how they learn. Acad Med. 2011;86:429–34. doi: 10.1097/ACM.0b013e3181f41e8c. [DOI] [PubMed] [Google Scholar]
- 7.Collet JP, et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet. 2009;373:309–17. doi: 10.1016/S0140-6736(08)61845-0. [DOI] [PubMed] [Google Scholar]
- 8.Mega JL, et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360:354–62. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 9.Simon T, et al. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360:363–75. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 10.Hulot JS, et al. Cardiovascular risk in clopidogrel-treated patients according to cytochrome P450 2C19*2 loss-of-function allele or proton pump inhibitor coadministration: a systematic meta-analysis. J Am Coll Cardiol. 2010;56:134–43. doi: 10.1016/j.jacc.2009.12.071. [DOI] [PubMed] [Google Scholar]
- 11.Mega JL, et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304:1821–30. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes DR, et al. ACCF/AHA Clopidogrel clinical alert: approaches to the FDA “boxed warning”: a report of the American College of Cardiology Foundation Task Force on Clinical Expert Consensus Documents and the American Heart Association. Circulation. 2010;122:537–57. doi: 10.1161/CIR.0b013e3181ee08ed. [DOI] [PubMed] [Google Scholar]
- 13.Wright MF, Wright Clayton E, Pulley J, Brothers KB. paper presented at the Congress of the Ethical, Legal and Social Implications Research Program; Chapel Hill, NC. 12 to 14 April 2011. [Google Scholar]
- 14.Hulot JS, et al. Cytochrome P450 2C19 loss-of-function polymorphism is a major determinant of clopidogrel responsiveness in healthy subjects. Blood. 2006;108:2244–7. doi: 10.1182/blood-2006-04-013052. [DOI] [PubMed] [Google Scholar]
- 15.Shuldiner AR, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–57. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mega JL, et al. Genetic variants in ABCB1 and CYP2C19 and cardiovascular outcomes after treatment with clopidogrel and prasugrel in the TRITON-TIMI 38 trial: a pharmacogenetic analysis. Lancet. 2010;376:1312–9. doi: 10.1016/S0140-6736(10)61273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delaney JT, et al. Predicting clopidogrel response using DNA samples linked to an electronic health record. Clinical Pharmacology & Therapeutics. 2011 doi: 10.1038/clpt.2011.221. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sibbing D, et al. Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation. 2010;121:512–8. doi: 10.1161/CIRCULATIONAHA.109.885194. [DOI] [PubMed] [Google Scholar]
- 19.Roden DM, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clinical pharmacology and therapeutics. 2008;84:362–9. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delaney JT, et al. Identifying Genomic predictors of outcome during clopidogrel therapy in an Electronic Medical Record System. Clin Pharmacol Ther. 2011 In press. [Google Scholar]
- 21.Damani SB, Topol EJ. The case for routine genotyping in dual-antiplatelet therapy. J Am Coll Cardiol. 2010;56:109–11. doi: 10.1016/j.jacc.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 22.Fuster V, Sweeny JM. Clopidogrel and the reduced-function CYP2C19 genetic variant: a limited piece of the overall therapeutic puzzle. JAMA. 2010;304:1839–40. doi: 10.1001/jama.2010.1566. [DOI] [PubMed] [Google Scholar]
- 23.Price MJ, et al. Standard- vs high-dose clopidogrel based on platelet function testing after percutaneous coronary intervention: the GRAVITAS randomized trial. JAMA. 2011;305:1097–105. doi: 10.1001/jama.2011.290. [DOI] [PubMed] [Google Scholar]
- 24.Wiviott SD, et al. Prasugrel versus clopidogrel in patients with acute coronary syndromes. N Engl J Med. 2007;357:2001–15. doi: 10.1056/NEJMoa0706482. [DOI] [PubMed] [Google Scholar]
- 25.Osborn CY, et al. MyHealthAtVanderbilt: policies and procedures governing patient portal functionality. J Am Med Inform Assoc. 2011 doi: 10.1136/amiajnl-2011-000184. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paré G, et al. Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. N Engl J Med. 2010;363:1704–14. doi: 10.1056/NEJMoa1008410. [DOI] [PubMed] [Google Scholar]
- 27.Simon T, et al. Clinical events as a function of proton pump inhibitor use, clopidogrel use, and cytochrome P450 2C19 genotype in a large nationwide cohort of acute myocardial infarction: results from the French Registry of Acute ST-Elevation and Non-ST-Elevation Myocard. Circulation. 2011;123:474–82. doi: 10.1161/CIRCULATIONAHA.110.965640. [DOI] [PubMed] [Google Scholar]
- 28.Tran M, Tafreshi J, Pai RG. Review article: combination of clopidogrel and proton pump inhibitors: implications for clinicians. J Cardiovasc Pharmacol Ther. 2010;15:326–37. doi: 10.1177/1074248410369109. [DOI] [PubMed] [Google Scholar]
- 29.Mackenzie IS, Coughtrie MWH, MacDonald TM, Wei L. Antiplatelet drug interactions. J Intern Med. 2010;268:516–29. doi: 10.1111/j.1365-2796.2010.02299.x. [DOI] [PubMed] [Google Scholar]
- 30.Bonello L, et al. Clopidogrel loading dose adjustment according to platelet reactivity monitoring in patients carrying the 2C19*2 loss of function polymorphism. J Am Coll Cardiol. 2010;56:1630–6. doi: 10.1016/j.jacc.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 31.Collet JP, et al. High doses of clopidogrel to overcome genetic resistance: the randomized crossover CLOVIS-2 (Clopidogrel and Response Variability Investigation Study 2) JACC Cardiovasc Interv. 2011;4:392–402. doi: 10.1016/j.jcin.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 32.Tantry US, et al. First analysis of the relation between CYP2C19 genotype and pharmacodynamics in patients treated with ticagrelor versus clopidogrel: the ONSET/OFFSET and RESPOND genotype studies. Circ Cardiovasc Genet. 2010;3:556–66. doi: 10.1161/CIRCGENETICS.110.958561. [DOI] [PubMed] [Google Scholar]
- 33.Harmsze AM, et al. CYP2C19*2 and CYP2C9*3 alleles are associated with stent thrombosis: a case-control study. Eur Heart J. 2010;31:3046–53. doi: 10.1093/eurheartj/ehq321. [DOI] [PubMed] [Google Scholar]
- 34.Alexopoulos D, et al. Prasugrel overcomes high on-clopidogrel platelet reactivity post-stenting more effectively than high-dose (150-mg) clopidogrel: the importance of CYP2C19*2 genotyping. JACC Cardiovasc Interv. 2011;4:403–10. doi: 10.1016/j.jcin.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 35.Kim IS, et al. Platelet inhibition by adjunctive cilostazol versus high maintenance-dose clopidogrel in patients with acute myocardial infarction according to cytochrome P450 2C19 genotype. JACC Cardiovasc Interv. 2011;4:381–91. doi: 10.1016/j.jcin.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 36.Woodcock J, Lesko LJ. Pharmacogenetics--tailoring treatment for the outliers. N Engl J Med. 2009;360:811–3. doi: 10.1056/NEJMe0810630. [DOI] [PubMed] [Google Scholar]
- 37.Lesko LJ, Zineh I. DNA, drugs and chariots: on a decade of pharmacogenomics at the US FDA. Pharmacogenomics. 2010;11:507–12. doi: 10.2217/pgs.10.16. [DOI] [PubMed] [Google Scholar]
- 38.Byrne DW, et al. Clinical and translational research studios: A multidisciplinary internal support program. Academic Medicine. 2011 doi: 10.1097/ACM.0b013e31825d29d4. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rogausch A, Prause D, Schallenberg A, Brockmöller J, Himmel W. Patients’ and physicians’ perspectives on pharmacogenetic testing. Pharmacogenomics. 2006;7:49–59. doi: 10.2217/14622416.7.1.49. [DOI] [PubMed] [Google Scholar]
- 40.Haddy CA, Ward HM, Angley MT, McKinnon RA. Consumers’ views of pharmacogenetics--A qualitative study. Research in social & administrative pharmacy_: RSAP. 2010;6:221–31. doi: 10.1016/j.sapharm.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 41.Almarsdóttir AB, Björnsdóttir I, Traulsen JM. A lay prescription for tailor-made drugs--focus group reflections on pharmacogenomics. Health policy (Amsterdam, Netherlands) 2005;71:233–41. doi: 10.1016/j.healthpol.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 42.Fargher EA, et al. Patients’ and healthcare professionals’ views on pharmacogenetic testing and its future delivery in the NHS. Pharmacogenomics. 2007;8:1511–9. doi: 10.2217/14622416.8.11.1511. [DOI] [PubMed] [Google Scholar]
- 43.Issa AM, Tufail W, Hutchinson J, Tenorio J, Baliga MP. Assessing patient readiness for the clinical adoption of personalized medicine. Public health genomics. 2009;12:163–9. doi: 10.1159/000189629. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen LF, Moldrup C. Lay perspective on pharmacogenomics: a literature review. Personalized Medicine. 2006;3:311–316. doi: 10.2217/17410541.3.3.311. [DOI] [PubMed] [Google Scholar]
- 45.Traulsen JM, Bjornsdóttir I, Almarsdóttir AB. “I”m Happy if I Can Help’. Public views on future medicines and gene-based therapy in Iceland. Community genetics. 2008;11:2–10. doi: 10.1159/000111634. [DOI] [PubMed] [Google Scholar]
- 46.Lin CH, Yeakley JM, McDaniel TK, Shen R. Medium- to high-throughput SNP genotyping using VeraCode microbeads. Methods Mol Bio. 2009;496:129–42. doi: 10.1007/978-1-59745-553-4_10. [DOI] [PubMed] [Google Scholar]