Abstract
The ubiquitously expressed Polycomb Group protein Yin-Yang1 (YY1) is believed to regulate gene expression through direct binding to DNA elements found in promoters or enhancers of target loci. Additionally, YY1 contains diverse domains that enable a plethora of protein–protein interactions, including association with the Oct4/Sox2 pluripotency complex and Polycomb Group silencing complexes. To elucidate the in vivo role of YY1 during gastrulation, we generated embryos with an epiblast specific deletion of Yy1. Yy1 conditional knockout (cKO) embryos initiate gastrulation, but both primitive streak formation and ingression through the streak is severely impaired. These streak descendants fail to repress E-Cadherin and are unable to undergo an appropriate epithelial to mesenchymal transition (EMT). Intriguingly, overexpression of Nodal and concomitant reduction of Lefty2 are observed in Yy1 cKO embryos, suggesting that YY1 is normally required for proper Nodal regulation during gastrulation. Furthermore, definitive endoderm is specified but fails to properly integrate into the outer layer. Although anterior neuroectoderm is specified, mesoderm production is severely restricted. We show that YY1 directly binds to the Lefty2 locus in E7.5 embryos and that pharmacological inhibition of Nodal signaling partially restores mesoderm production in Yy1 cKO mutant embryos. Our results reveal critical requirements for YY1 during several important developmental processes, including EMT and regulation of Nodal signaling. These results are the first to elucidate the diverse role of YY1 during gastrulation in vivo.
Keywords: Yy1, Nodal, Lefty2, Gastrulation, Endoderm, EMT
Introduction
Yin-Yang1 (YY1, also called delta, NF-E1, and UCRBP) is a multifunctional protein that is thought to alter gene expression through a variety of mechanisms. YY1 contains a GLI-Krüppel zinc finger domain in the C-terminus allowing for direct binding at target loci, which are found throughout the genome often within 1 kb of transcription start sites (Galvin and Shi, 1997; Shi et al., 1991). Functional YY1 binding sequences have also been demonstrated at distant enhancers of many genes including Snail and Otx2 (Palmer et al., 2009; Peinado et al., 2004). YY1 binding sites are highly conserved across the genomes of many species, second only to conservation of SP1, which is itself a binding partner of YY1 in certain cell types (Lee et al., 1993). Despite numerous efforts to functionally define YY1, its endogenous activity remains enigmatic.
YY1 is believed to influence gene activity by inhibiting or strengthening interactions between target loci and other transcriptional activators, such as SP1 and E2F (Dong and Pfister, 1999; Lee et al., 1993; Schlisio et al., 2002; Shi et al., 1997; Yakovleva et al., 2004). It is also clear that YY1 participates in epigenetic regulation of gene expression through interactions with chromatin modifying complexes such as PRMT1 and members of the Polycomb Repressive Complexes, PRC1 and PRC2 (Garcia et al., 1999; Rezai-Zadeh et al., 2003). Because the known members of PRC2 lack DNA binding capability and the single Drosophila ortholog of YY1, Pho, has been shown to perform this function, it has been postulated that YY1 is responsible for PRC2 targeting (Brown et al., 1998; Mohd-Sarip et al., 2002). However, it remains unclear if mammalian YY1 does perform this function in vivo.
Yy1 has been knocked-out or knocked-down in several species in order to functionally define this dynamic protein. Pho homozygous null pupae exhibit impaired pattern formation and aberrant nervous system development (Fritsch et al., 1999; Girton and Jeon, 1994). Pho directs PRC2 to target loci, and expression of mammalian Yy1 partially rescues the Pho mutant phenotype (Atchison et al., 2003), indicating that the ability to target PRC2 is at least partially conserved in mammalian YY1. The YY1 paralogues in Xenopus are required for gastrulation and neural crest epithelial to mesenchymal transition (EMT), presumably governed at least in part by YY1's role in activation of the Slug promoter (Morgan et al., 2004).
In the mouse, perimplantation lethality of Yy1 null embryos precludes analysis of later embryonic functions (Donohoe et al., 1999). Affar et al. demonstrated a dose-dependent necessity for YY1 during mid-gestation, and defined a requirement for YY1 in cytokinesis and cell cycle progression (Affar el et al., 2006). Conditional deletion studies have also revealed that YY1 is required for VDJ recombination during pro-B-cell differentiation, as well as oligodendrocyte differentiation and myelination (He and Casaccia-Bonnefil, 2008; He et al., 2010; Liu et al., 2007). A role for YY1 has also been documented in male germ cells, where YY1 is critical for double strand break repair and heterochromatin formation (Wu et al., 2009), and deletion of Yy1 in growing oocytes results in a failure of follicle expansion and paracrine signaling defects in the ovary (Griffith et al., 2011). While these studies have elucidated requirements for mammalian Yy1 in committed cell types, little is known about its role during gastrulation in vivo.
Here we report the developmental and molecular consequences of epiblast-specific deletion of Yin-Yang1, demonstrating a critical role for YY1 in appropriate primitive streak (PS) formation and E-Cadherin repression in cells exiting the streak. Although both mesoderm and endoderm are specified, neither cell type is able to properly migrate. We also observe impaired embryonic to extraembryonic signaling when YY1 is removed from the epiblast, resulting in aberrant morphogenetic movements during gastrulation. Additionally, we show that YY1 is required for negative regulation of Nodal, possibly through direct activation of Lefty2 during gastrulation in vivo.
Materials and methods
Embryo production and genotyping
Timing of embryonic development was determined by presence of a vaginal plug the morning after mating (E0.5). Embryos for analysis were generated by mating female mice homozygous for a Yy1 conditional allele, Yy1flox/flox (Affar el et al., 2006), to Yy1Δ/WT; Sox2-Cre+/+ males (Hayashi et al., 2002). For visualization of Yy1 null cells by B-galactosidase staining, females homozygous for both the Yy1 conditional allele and the R26 reporter (Soriano, 1999) were mated to Yy1Δ/WT; Sox2-Cre+/+ males. At the time of embryo dissection, ectoplacental cone (EPC) DNA extraction was used for PCR genotyping both the Yy1 (Affar el et al., 2006) and Sox2-Cre alleles (Hayashi et al., 2002). Embryos were sexed by Sry specific PCR using the primers: 5′TTTATGGTGTGGTCCCGTGG3′ and 5″CCAGTCTTGCCTGTATGTGAT3′. During gastrulation, all genotypes were recovered at expected Mendelian ratios. Mutant embryos are of the genotype Yy1flox/Δ, Sox2-Cre+/−, hereafter referred to as “mutants” or cKO embryos for simplicity. All other genotypes are referred to as “control” or “wild-type”.
RNA extraction and RT-PCR
RNA extraction was performed with Roche High Pure RNA Isolation Kit (Roche 11828665001). cDNA was synthesized with both random hexamers and oligo-dT primers as described previously (Griffith et al., 2011). RT-PCR was performed with 3 μl (1/16 of each embryo) as template for 36 cycles of 30 s at 60 °C, 72 °C and 94 °C with the following gene specific primer pairs (given 5′ to 3′): ActB (GGCCCAGAGCAAGAGAGGTATCC and ACGCACGATTTCCCTCTCAGC); Drap1 (GAAATGCCAAAACCATGACC and TTGTCTTTGCCT TTGCTTCC); Eomes (CCAGGGTTCTCCGCTCTAC and GTCACTTCCACGATGTGCAG); Fgf4 (CTTGCCCTAGTTCCTTGCTG and GGACTGATGGGAATGATTGG,); Fgf8 (TGTTGCACTTGCTGGTTCTC and ACTCGGACTCTGCTTCCAAA); Foxa2 (TGAGGTGGGTAGCCAGAAAG and GCTCAGACTCGGACT-CAGGT); Lefty2 (AACTTTTCAGGGCACTTTTAGGGAC and GGACAAGCTCACTGAGAATACATCTG); Nodal (CGCATCCTTCTTCTTCAAGC and GCCTGGTGGAAAATGTCAAT); Snail (CTTGTGTCTGCACGACCTGT and CTTCACATCCGAGTGGGTTT); Tbx6 (GGGACTCAGATCCAGAGCAG and ACTTCAATGCGGATGCTACC); Twist1 (ACGAGCTGGACTCCAAGATG and CCTCTGGGAATCTCTGTCCA); T (CATGTACTCTTTCTTGCTGGG and GGTCTCGGGAAAGCAGTGGC); Wnt3 (CGCTCAGCTATGAACAAGCA and GGTGTTTCTCCACCACCATC). Quantitative RT-PCR assays were performed using Taqman Gene Expression Assays (Snail MM00441533_g1 and Eomes MM01351985_m1 multiplexed with ActB 4352341E) and PerfeCTa® qPCR SuperMix, Low ROX™ (Quanta Biosciences # 95052-02K) and run on a Stratagene 3001 mx Q-PCR machine using Quanta's recommended cycling conditions.
β-Galactosidase staining
Embryos were fixed for 30 min at room temperature and processed as previously described (Tremblay et al., 2000).
Fixation, embedding, sectioning
Embryos were prepared for histology by fixation in 4% paraformaldehyde (PFA) for 2 h at room temp or overnight at 4 °C. Embryos were dehydrated through a series of methanol washes; 20 min each in 25%, 50%, 75% methanol diluted in phosphate buffered saline/0.01% tween20 (PBT), followed by two 100% methanol washes. Embryos were embedded and sectioned as described (Griffith et al., 2011).
Immunohistochemistry
Sections were deparafinized with three 10-min xylene washes and rehydrated with three 5-min washes in 100% ethanol, followed by successive 1-min washes in 90%, 80%, 70% ethanol and water. Antigen retrieval was performed by boiling for 5 min in 0.01 M Tris Base pH 10.0 with 0.05% Tween20. After slides cooled to room temperature they were washed twice in PBT for 2 min and blocked with 0.5% milk in PBT for 2 h at room temperature in a humidified chamber. Primary antibody was applied in 0.05% milk/PBT overnight at 4 °C in a humid chamber. Three 15-min PBT washes preceded a 1-h secondary treatment in 0.05% milk/PBT in a humid chamber at room temperature. Slides were washed in PBT for 15 min twice and then in PBS for 15 min. Nuclei were countered stained with Dapi (Roche or Molecular Probes) in PBS (1:10,000) for 2 min and then rinsed with PBS. Slides were sealed and coverslipped with Prolong Gold (Invitrogen). Primary antibodies were used at the following concentrations: YY1 [Santa Cruz, sc-1703 (1:100)], CDH1 [Abcam, ab53033 (1:500)], HNF4α [Santa Cruz sc6556 (1:200)]. Secondary antibodies were diluted 1:500 and included Alexa Fluor 488 donkey-anti-rabbit [Molecular Probes (A-21206)] and Alexa Fluor 546 donkey-anti-goat [Molecular Probes (A-11056)].
Whole-mount in situ hybridization
Embryos were fixed in 4%PFA/PBS overnight at 4C with agitation, dehydrated in a series of methanol washes and stored at − 20 °C E7.5 embryos were treated with proteinase K for 10 min and embryos collected at E7.0 were treated with proteinase K for 8 min. In situ hybridization probe synthesis and protocols were performed as previously described (Rivera-Perez and Magnuson, 2005). WISH stained embryos were imaged in PBT. Probes used: Brachyury (Wilkinson et al., 1990), Lefty2 (Meno et al., 1997), Fgf8 (Crossley and Martin, 1995), Bmp4 (probe produced from RT-PCR product using primers 5′AGGAGGAGGAGGAAGAGCAG3′ and 5′TGTGATGAGGTGTCCAGGAA3′), Snail, 5′ACACTGGGTGAGAAGCCATT3′ and 5′GAAGGAGTCCTGGCAGTGAG3′; Fgf4, 5′TTGCGTCCCTATTTGCTCTC3′ and 5′CGGAGGGTCACAGTCTAGGA3′; Eomes (Russ et al., 2000), Nodal (Conlon et al., 1994), Foxa2 (Sasaki and Hogan, 1996), Cer1 (Thomas et al., 1997), Hex (Bedford et al., 1993), Sox2 (Conlon et al., 1994), Otx2 (gift from James Li), Oct4 (Rosner et al., 1990), Shh (gift from James Li). A minimum of 3 mutants were analyzed at each stage for each in situ presented.
Imaging
Digital images of whole mount embryos were captured on a Nikon SMZ-1500 stereomicroscope equipped with a Spot Idea Digital Camera and Spot software (v4.6). Digital images of sectioned embryos were taken with a Nikon Eclipse TE2000-S inverted fluorescence microscope and QImaging Retiga Exi Fast 1394 camera fitted with a color-slider for use with brightfield images. All slides were imaged with NIS-Elements BR Software.
Chromatin immunoprecipitation
Putative YY1 binding sites were identified using MacVector software and searching for the YY1 consensus binding sequence as described previously by other groups (5′-(C/g/a)(G/t)(C/t/a)CATN(T/a)(T/g/c)-3′ where the upper case letters represent the preferred bases) (Hyde-DeRuyscher et al., 1995; Shi et al., 1997). E7.5 embryos were collected in PBS/PVP and immediately processed with the MagnaChipA Kit (Millipore MAGNA0001) according to the manufacturer's instructions. Briefly, the embryos were treated with 1% formaldehyde at RT for 10 min, squelched with glycine and washed with cold PBS three times. E7.5 embryos were processed in groups of ten. Samples were incubated in Protease Inhibitor Cocktail with Cell Lysis Buffer on ice for 15 min, pelleted, and resuspended in 500 μl of Protease Inhibitor Cocktail with Nuclear Lysis Buffer. Samples were kept on ice and sonicated twice for 20s with the Heat Systems Sonicator/Ultrasonic Processor (output setting 3) generating chromatin fragments between 200–1000 bp. 1% of each sample was removed for Input control. Immunoprecipitation was performed using 20 μl of Magnetic Protein A Beads and 50 μl anti-YY1 H414 antibody (Santa Cruz, sc-1703) or normal rabbit serum with overnight incubation at 4C followed by 2 h at RT with over-end mixing. After antibody incubation samples were sequentially washed with low salt, high salt, LiCl and TE solutions, washed and collected by spin column as described in the manufacturer's directions and eluted in 50 μl. Quantitative PCR was performed using PerfeCTa® qPCR SybrMix, Low ROX™ (Quanta Biosciences) and run on a Stratagene 3001mx Q-PCR machine using Quanta's recommended cycling conditions. Primers used are shown in Sup. Fig. 3.
Whole embryo culture
E6.5 embryos were carefully dissected in pre-warmed and incubated dissection media (DMEM with 7.5% FBS) on a 37 °C warming plate under a dissecting microscope. After removal of Reichert's membrane, whole litters of embryos were cultured for 20 h in rotating tubes in 5% CO2 37 °C incubator in 75% rat serum/25% DMEM with or without 25 μM SB505124 (Sigma S4696). After culture, embryos were genotyped and processed for IHC/IF or RT-PCR. At least 3 mutant embryos were examined for each data set (IHC/RT-PCR).
Results
Characterization of embryos with epiblast-specific YY1 deletion
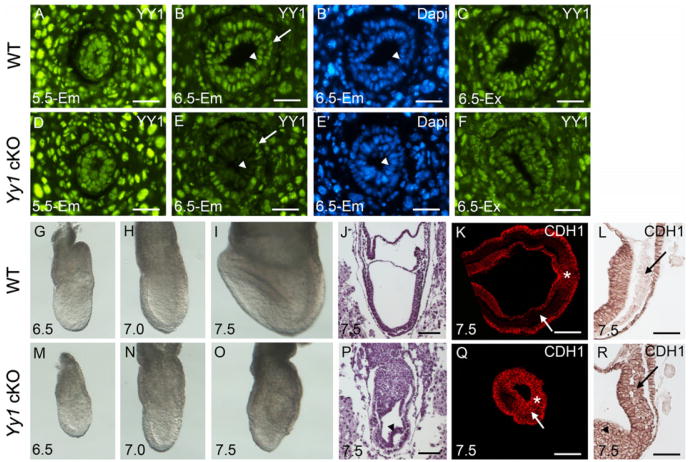
YY1 is ubiquitously expressed in embryonic and extraembryonic tissues throughout gastrulation (Fig. 1A–C). Using a paternally inherited Sox2-Cre transgene (Hayashi et al., 2002), Yy1 was deleted in the epiblast. Although YY1 is present throughout wild type (WT) and cKO embryos at E5.5 (Fig. 1A and D), it is absent from the cKO epiblast at the onset of gastrulation at E6.5 (arrowhead in Fig. 1E and E′). As expected, YY1 expression is maintained in the cKO extraembryonic tissues (Fig. 1F), including the visceral endoderm (VE, arrow in Fig. 1E, E′). Also as predicted, YY1 is absent in the mutant epiblast and epiblast-derived tissues at E7.5 (Sup. Fig. 1).
Fig. 1.
YY1 is required in the epiblast for developmental EMT. YY1 (green) localization in WT (A–C) and mutant (D–F) sectioned embryos. At E5.5 (A,D), YY1 is present throughout both mutant and WT embryos. At E6.5 YY1 is expressed in the WT epiblast (arrowhead in B and B′), VE (arrow in B and B') and extraembryonic tissues (C). YY1 is absent specifically from the mutant epiblast (arrowhead in E and E') starting at E6.5. YY1 expression is unaffected in the mutant visceral endoderm (arrow in E and E') and other extraembryonic tissues (F). B′ and E′ are DAPI fluorescence of the same sections in B and E, respectively. Bright field images of WT (G–I) and mutant (M–O) images during gastrulation. Haematoxylin and eosin staining of sagital E7.5 WT (J) and mutant (P) sections. Arrowhead in P and R indicates abnormal primitive streak derivatives. E-Cadherin (CDH1) localization on transverse (K and Q) and sagital (L and R) E7.5 WT and mutant embryos as indicated. While E-Cadherin is normally present throughout the cells of the PS (asterisk, K and L), it is repressed in the streak derivatives in the WT (arrows, K and L) and remains on in the mutant PS derivatives (arrows, Q and R). Scale bars in A–F represent 25 μm. Scale bars in J–L and P–R represent 75 μm.
Yy1 cKO mutant embryos are smaller than control littermates at E6.5 and E7.0 (compare Fig. 1G–M and H–N) and are easily distinguished by E7.5 due to the absence of a midline, node and head-folds as well as abnormal extraembryonic region (Fig. 1I and O). While the primitive streak (PS) is apparent in mutants, there is a distinct accumulation of cells proximal to the streak (arrowheads, Fig. 1P and R), suggesting defects in EMT.
YY1 is required in the epiblast for EMT and PS elongation
Repression of E-Cadherin (CDH1) is an essential step of EMT, permitting movement of PS cells and their derivatives [reviewed in (Thiery et al., 2009)]. An excess of CDH1 in mutant streak derivatives (compare Fig. 1K–L with Q–R) indicates that although YY1 deficient cells are capable of delaminating from the epiblast and can ingress through the streak, they do not properly repress E-Cadherin. The retention of CDH1 in streak derivatives likely contributes to the posterior accumulation of mutant cells that fail to migrate properly and remain tightly associated.
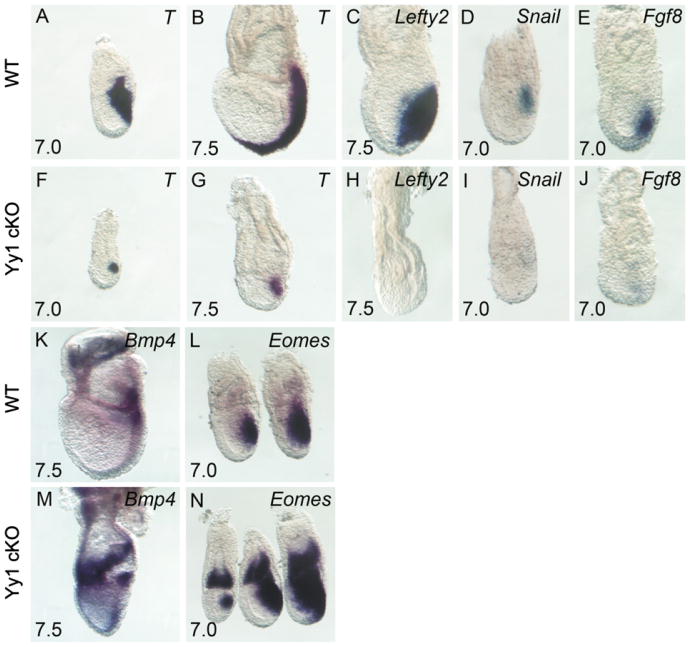
Because of the evident morphological PS defects, we examined expression of genes known to be critical for this transient embryonic structure. In control embryos, brachyury(T) is confined to the PS and nascent mesoderm at E7.0 [Fig. 2A, (Wilkinson et al., 1990)]. At E7.5 T expression continues to mark the streak as it expands distally in WT embryos (Fig. 2B), a requirement for anterior-posterior patterning and proper development (Beddington et al., 1992). At E7.0, the expression domain of T in Yy1 cKO embryos is reduced compared to littermate controls (compare Figs. 2A and F), and at E7.5, T expression has failed to extend distally (Fig. 2G). Despite the drastic reduction in overall streak size, analysis of sectioned mutant embryos reveals a population of T negative mesenchyme surrounding the streak, suggesting that a limited amount of cell migration through the PS has occurred (data not shown).
Fig. 2.
YY1 is required in the epiblast for PS function and paracrine regulation. Whole mount in situ hybridization (WISH) of T (A–B, and F–G), Lefty2 (C and H), Snail (D and I), Fgf8 (E and J), Bmp4 (K and M) and Eomes (L and N). Comparison of WT (A–E and K–L) and mutant embryos (F–J and M–N) reveals compromised PS formation marked by reduced T expression (F and G), loss of the mesoderm markers Lefty2 (H), Snail (I) and Fgf8 (J) and excess extraembryonic and embryonic Bmp4 (M) and Eomes (N) in mutant embryos.
To further examine the PS in Yy1 cKO mutants, we examined other markers of nascent mesoderm, including Lefty2, Snail, Fgf8 and Fgf4 (Carver et al., 2001; Meno et al., 1999; Sun et al., 1999). Lefty2 and Fgf4 were completely absent in mutants at all stages examined (compare Fig. 2C–D with H–I, Fgf4 not shown), and Snail and Fgf8 expression is greatly reduced in cKO mutants, with weak areas of expression confined to the distal-most mesoderm (compare Fig. 2E and J). Because these nascent mesoderm markers are also important for EMT, loss of expression of these genes likely contributes to the EMT defects noted above [reviewed in (Arnold et al., 2008)].
Expression of Bmp4 and Eomes in the extending PS is preceded by their expression in distal extraembryonic ectoderm (EXE). Concomitant with their expression in the newly formed streak, Bmp4 and Eomes are normally down-regulated in EXE (Ciruna and Rossant, 1999; Fujiwara et al., 2002; Russ et al., 2000). In Yy1 mutant embryos Bmp4 and Eomes show appropriate tissue specific localization. However, both genes are overexpressed compared with WT embryos of the same stage (Fig. 2K–N), suggesting a failure of appropriate repression of these genes in both embryonic and extraembryonic tissues of Yy1 cKO embryos. Disruption of gene expression in the extraembryonic region of the conceptus is unanticipated in Yy1 mutants, as the Yy1 locus is not deleted in EXE (Fig. 1E). However, our results are consistent with reports that signaling from embryonic ectoderm is important for proper regulation of Bmp4 and Eomes in EXE [reviewed in (Arnold and Robertson, 2009)], and suggest that this paracrine epiblast activity is YY1 dependent.
Organizer formation and function in Yy1 cKO embryos
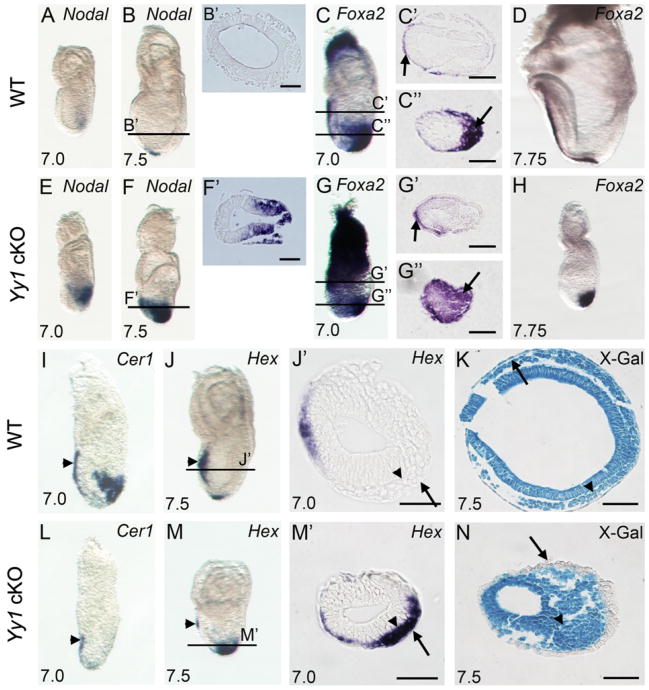
Intact Nodal signaling is critical for a variety of developmental processes including the onset of gastrulation, anterior/posterior patterning, PS formation as well as both endoderm and mesoderm induction [reviewed in (Arnold et al., 2008; Lu et al., 2001; Schier, 2003)]. In accordance with multiple critical roles during development, early Nodal expression is dynamic and tightly regulated. Nodal is normally expressed throughout the epiblast prior to gastrulation and gradually becomes restricted to the PS where its expression is further confined to the anterior streak at E7.0 (Fig. 3A). By E7.5 Nodal is restricted to the node in WT embryos (Fig. 3B). In contrast, Nodal is ectopically expressed throughout the posterior half of the mutant epiblast at E7.0 (Fig. 3E), and throughout the posterior epiblast and streak derivates by E7.5 (Fig. 3F).
Fig. 3.
Defective gastrulation in Yy1 cKO mutants. WISH of Nodal (A–B and E–F) showing failure of spatiotemporal repression in Yy1 mutants. B′ and F′ are transverse sections corresponding to the lines in B and F. WISH of Foxa2 (C–D and G–H). C, C″, G′ and G″ are transverse sections corresponding to the lines in C and G. WISH of Cer1 (I and L); Hex (J and M) marking properly migrating AVE cells in WT (arrowhead in J) and mutant embryos (arrowhead in M). In WT embryos, Hex positive DE cells have intercalated into VE and have migrated anteriorly. Mutant Hex positive cells (arrowhead in M′) fail to intercalate into VE (arrow in M′). J′ and M′ are transverse sections corresponding to the lines in J and M. Arrows in J′ and M′ indicate VE. Transverse sections of X-gal stained R26R WT (K) and mutant (N) embryos. Arrowhead in K and N indicate epiblast derivatives. Arrows in K and N indicate cells in the outer layer of the conceptus, showing β-gal positive epiblast derived DE in WT embryos and β-gal negative extraembryonic derived VE in mutants. Scale bars represent 75 mm.
High levels of Nodal signaling are required to specify the definitive endoderm (DE) and axial midline structures including the node and notochord (Dunn et al., 2004; Robertson et al., 2003; Tremblay et al., 2000; Vincent et al., 2003). No morphological node or notochord was evident in any mutant embryos examined. This observation is supported by the absence of Shh expression in cKO mutants (data not shown). In wild type E7.0 embryos, Foxa2 expression is restricted to the anterior visceral endoderm (AVE, Fig. 3C and arrow in C′) and DE (arrow in Fig. 3C and C″). In Yy1 mutants, Foxa2 is correctly localized to the AVE (arrow in Fig. 3G′) but is aberrantly expressed throughout distally located streak derived cells (arrow in Fig. 3G″). As development progresses Foxa2 is normally expressed in both the anterior DE and the notochord at E7.75 (Fig. 3D). In E7.5 mutants, Foxa2 is inappropriately expressed in a large swath of distally located streak derived cells at E7.5 (Fig. 3H). Taken together these data suggest that axial midline structures are not present but that the DE is specified and abundant in Yy1 cKO mutants.
Abnormal endoderm specification in YY1 cKO Embryos
Once the streak has extended distally in WT embryos, DE gene expression can be observed [reviewed in (Lewis and Tam, 2006)] as streak derived DE cells first intercalate into the visceral endoderm (Kwon et al., 2008). To assess DE specification, we examined markers including Cer1, Hex and Foxa2. In Yy1 mutants, Cer1 is expressed in the AVE (arrowhead Fig. 3L), but is completely absent in streak derivatives (compare Figs. 3I and L). Hex is expressed in the mutant AVE (arrowhead Fig. 3M), but unlike Cer1, Hex is also expressed in streak derivatives that remain in the middle germ layer instead of migrating/intercalating into the outermost layer (compare Figs. 3J′ and M′). Combined with the Foxa2 in situ results discussed above (Fig. 3G″ and H), this data suggests that loss of YY1 in the epiblast leads to endoderm specification without the ability to intercalate into the outer layer.
To examine the lack of DE intercalation, we first utilized the R26R allele to lineage trace Sox2-Cre positive epiblast cells. As expected, all three germ layers of WT Sox2-Cre+/−; R26R+/− E7.5 embryos showed X-gal activity indicating appropriate movement of epiblast derived DE into the outer layer (arrow in Fig. 3K). In contrast, X-gal positive cells were not present in the outer layer of mutant embryos, supporting the hypothesis that epiblast-derived cells do not properly integrate into the outer layer in mutant embryos (arrow in Fig. 3N). To assess the identity of cells surrounding the mutant epiblast, HNF4α expression was examined. In WT embryos, HNF4α positive visceral endoderm (VE) surrounds only the extraembryonic tissues of the conceptus [Sup. Fig. 2A–E and (Duncan et al., 1994)]. However, in mutant embryos, the thickened HNF4α positive VE completely surrounds the conceptus (Sup. Fig. 2F–J) indicating an absence of the DE-mediated dispersal of VE. Taken together, these data show that in the absence of YY1, the DE is specified but does not intercalate into the surrounding HNF4α expressing VE.
Anterior/posterior patterning in Yy1 mutant embryos
The AVE is a subset of VE initially located at the distal tip of the egg cylinder at E5.5 that migrates anteriorly prior to the onset of gastrulation (Beddington and Robertson, 1999; Rivera-Perez et al., 2003). Anterior specification is dependent on both AVE migration and the subsequent maintenance of AVE specific gene expression, events coordinated by reciprocal interactions between the AVE and epiblast [reviewed in (Lu et al., 2001)]. As discussed above, AVE expression of both Cer1 and Hex is appropriately localized in E7.0 Yy1 cKO mutants, indicating that the AVE has migrated anteriorly. However, expression of both genes is significantly reduced, again implicating YY1 in embryonic (epiblast) to extraembryonic (AVE) signaling (compare Fig. 3I–J with L–M).
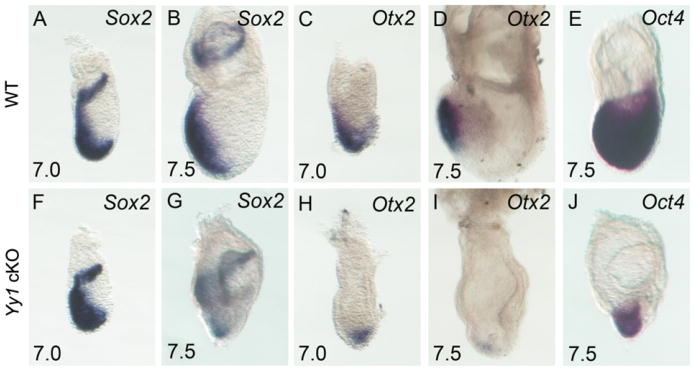
From E7.0–7.5 the expression domains of Sox2, Oct4 and Otx2 normally overlap in the WT prospective anterior ectoderm, specifying presumptive neural progenitors [Fig. 4A–E (Scholer et al., 1990; Wood and Episkopou, 1999)]. Surprisingly, at E7.0 and E7.5 appropriate Sox2 and Oct4 expression is observed in mutant embryos (Fig. 4). However, the Otx2 expression domain is reduced at E7.0 and fails to become anteriorly restricted (compare Fig. 4C and H). At E7.5 Otx2 is confined to a small population of distal epiblast cells in mutant embryos (compare Fig. 4D and I), results consistent with data demonstrating that YY1 activates the Otx2 locus (Takasaki et al., 2007). However, the normal Oct4 and Sox2 expression indicate that neural specification is initiated in the absence of embryonic YY1.
Fig. 4.
Neural progenitor specification is initiated in Yy1 cKO embryos. WISH of Sox2 (A, B, F, G), Otx2 (C, D, H, I) and Oct4 (E, J) in WT (A–E) and mutant embryos (F–J) at E7.0 (A, C, F, H) and E7.5 (B, D, E, G, I, J). Anterior Sox2 and Oct4 expressing cells demonstrate specification of neural progenitors, despite the drastic reduction in Otx2 expression.
YY1 binds directly to the Lefty2 locus in vivo
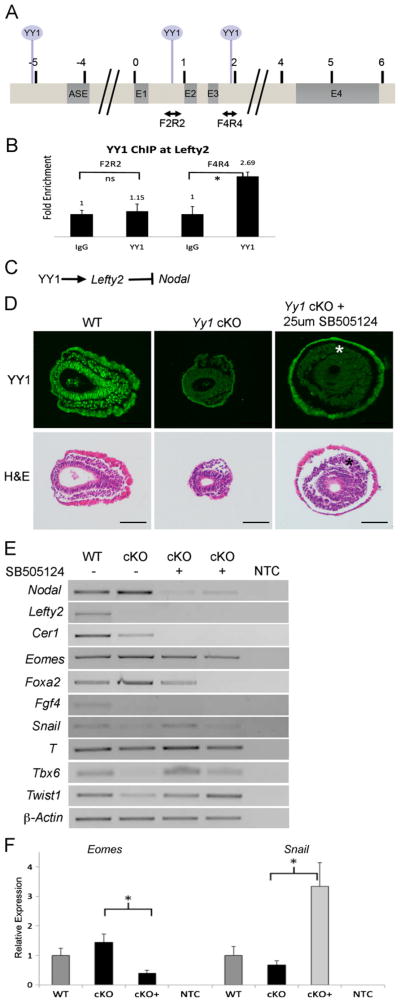
As many of the phenotypic features of the Yy1 cKO embryo could be attributed to increased Nodal signaling, we next chose to examine if YY1 interacted with either the Nodal or Lefty2 locus in E7.5 in vivo by chromatin immunoprecipitation (ChIP). Several predicted YY1 binding sites (Fig. 5A and S3) at the Lefty2 locus were examined and specific binding of YY1 to sequences within intron 3 (Fig. 5B) was found in E7.5 chromatin. Importantly, we detected no/weak interaction at intron 2 of Lefty2 (F2R2, Fig. 5A– B), indicating specificity of YY1 binding. Despite a similar experimental approach, specific interactions at the Nodal locus were not detected in E7.5 chromatin (data not shown). Taken together, our in situ and ChIP results suggest that in the gastrulating epiblast YY1 may bind to and activate Lefty2 transcription, which in turn represses Nodal [Fig. 5C, (Meno et al., 1999)].
Fig. 5.
YY1 regulates Nodal indirectly through Lefty2. (A) Map of the Lefty2 locus. Double-headed arrows indicate amplicons used for YY1 ChIP and circles above mark locations of predicted YY1 binding sites. (B) YY1 ChIP-qPCR shows that YY1 binds between exons 3 and 4 (F4R4) in vivo, but not between exons 1 and 2. (C) Schematic illustration of possible regulation of Nodal through activation of Lefty2. (D) Transverse sections of untreated wildtype and mutant embryos, as well as mutant embryos cultured with the TGFβ inhibitor SB505124 (top row is YY1 immunofluorescence and bottom row is H and E of the same sections). Treatment with SB505124 results in increased mesodermal-like streak derivatives in mutant embryos (asterisks). (E) RT-PCR indicates that mesodermal markers Tbx6, Twist1 and Snail are increased in the mutants treated with the inhibitor while Lefty2 and Fgf4 expression are unaffected as predicted. RT-PCR also confirms reduction of Nodal, Cer1, Eomes and Foxa2 by treatment with SB505124. F. RT-qPCR confirmation of reduced Eomes and enhanced Snail expression in mutant embryos treated with SB505124. Asterisks represent P values less than 0.05 by student's T-Test. Error bars represent SEM. Scale bars represent 75 μm.
Nodal reduction rescues mesoderm induction in Yy1 cKO embryos
Regulation of Nodal signaling is critical for many aspects of early embryonic development (Brennan et al., 2001; Conlon et al., 1994; Meno et al., 1997). To assess the role of Nodal overexpression in Yy1 mutant embryos, we suppressed Nodal activity by pharmacological inhibition with SB505124, an inhibitor of TGFβ type 1 receptor signaling (DaCosta Byfield et al., 2004). We found that 25 μM SB505124 was sufficient to block Nodal signaling in WT embryos as evidenced by a reduction in Nodal transcripts [due to the absence of Nodal autoregulation (Brennan et al., 2001; Norris et al., 2002)], but still allows for embryo survival in culture (Fig. S4). E6.5 litters were dissected and cultured for 24 h with or without SB505124. Yy1 cKO embryos cultured with SB505124 showed an obvious increase in YY1 negative (epiblast derived) mesenchymal cells surrounding the epiblast (Fig. 5D, asterisks) suggesting an increase in the movement of cells through the streak. To further evaluate the identity of these cells, we assessed expression of transcripts that are altered in the absence of YY1.
Further indicating enhanced mesoderm specification, analysis of SB505124 treated cKO embryos demonstrated a slight increase in T expression as well as a dramatic increase in Tbx6, Twist1 and Snail transcripts (Fig. 5E–F), which are all severely reduced or absent in untreated mutant embryos. Consistent with findings that Foxa2 and Eomes expression levels correlate with Nodal activity (Arnold et al., 2008; Vincent et al., 2003), treatment of Yy1 cKO embryos with SB505124 resulted in reduced expression of both genes (Fig. 5E–F). Importantly, reduction of Nodal signaling had no effect on the loss of Lefty2 in cKO embryos, supporting a direct role for YY1 in the regulation of Lefty2 that is independent of Nodal activity. Likewise, SB505124 treatment of cKO embryos had no effect on the loss of Fgf4 in cKO embryos, indicating that both Nodal dependant and Nodal independent events contribute to the gastrulation defects occurring in the absence of YY1 in the epiblast.
Discussion
Here we present the results of an epiblast-specific deletion of Yy1, a multifunctional gene that has a plethora of functions ascribed to it in many cell types in vitro. YY1 is expressed throughout the epiblast and extraembryonic regions of the embryo prior to and during gastrulation. Sox2-Cre mediated deletion of Yy1 leads to a loss of YY1 protein throughout the epiblast. Mutant embryos display noticeable phenotypes by E7.0, indicating that YY1 is critical for gastrulation. Two main observations can be made from the analysis of E7.0–7.5 mutant embryos. The first is that although PS formation is initiated, it does not extend anteriorly and the streak derivatives fail to repress E-Cadherin resulting in an accumulation of streak derivatives. Secondly, Nodal fails to undergo proper spatiotemporal regulation and remains expressed throughout much of the epiblast at E7.5. We suggest that while many aspects of the cKO phenotype can be attributed to over expression of Nodal (due to the absence of Lefty2 activation by YY1), other Nodal-independent transcriptional and morphogenetic abnormalities (loss of Fgf4 and PS defects) also occur when YY1 is not present in the epiblast.
Critical requirement of YY1 in EMT
The most obvious morphological feature of Yy1 cKO embryos is the accumulation of PS cells that fail to migrate properly. The retention of E-Cadherin by these streak derivatives causes reduced mobility which in turn presumably abrogates (1) the migration of mesoderm; (2) the proximal displacement of EXE; and (3) DE intercalation into the overlying VE.
Functionally redundant pathways are known to regulate EMT, and while some mechanisms are cell type specific, most utilize a conserved pathway involving repression of E-Cadherin by a member of the Slug family (Barrallo-Gimeno and Nieto, 2005). Many cofactors have been shown to be required for Slug's regulation of E-Cadherin. These include AJUBA, SUZ12 and the SIN3A/HDAC1/HDAC2 histonemodifying complex (Calder et al., 2008; Herranz et al., 2008; Peinado et al., 2004). In mouse the Slug homolog Snail represses E-Cadherin in cells undergoing EMT during gastrulation, allowing PS formation to proceed (Sefton et al., 1998). Similar to what is found in Yy1 cKO embryos, Snail null mouse embryos do not complete gastrulation and show aberrant E-Cadherin expression in streak derivatives (Carver et al., 2001), suggesting that one role of YY1 is to activate Snail in the PS. Indeed, YY1 has been shown to bind the Snail enhancer and activate the locus in Hela cells (Palmer et al., 2009), supporting the hypothesis that YY1 may be an activator of the Snail locus in vivo as well. Furthermore similar to several mouse mutants in which EMT is disrupted, the PS forms in Yy1 cKO embryos but fails to extend appropriately (Arnold et al., 2008; Carver et al., 2001; Ciruna and Rossant, 2001; Sun et al., 1999). Additionally, both Fgf4 and Fgf8 are known to be required for Snail expression (Ciruna and Rossant, 2001), and transcripts from both of these genes are absent in Yy1 mutants. These divergent mechanisms regulating Snail and EMT are not mutually exclusive, making it difficult to assess if loss of Snail is a direct or indirect consequence of cells lacking YY1.
YY1 mediates Nodal signaling
One exciting molecular phenotype apparent in Yy1 cKO mutant embryos is the mis-regulation of Nodal signaling. Appropriate spatiotemporal Nodal expression is critical for many aspects of normal development. Deletion of Yy1 leads to an expansion of Nodal throughout the epiblast at E7.5 with concomitant loss of two antagonists of the nodal pathway, Cer1 and Lefty2. It is important to note that loss of Lefty2 alone results in accumulation of PS cells, lack of axial midline structures and increased Foxa2 expression (Meno et al., 1999) – features that we also document in Yy1 cKO mutants (Figs. 1–3). Furthermore, combined loss of Cer1 and Lefty1, has been shown to result in an accumulation of cells in the streak and an increase in endodermal gene expression (Perea-Gomez et al., 2002; Yamamoto et al., 2004). The similarities of gastrulation phenotypes between Yy1 cKO and Nodal antagonist mutant embryos suggest that mis-regulation of Nodal may be one of the primary molecular defects underlying the gastrulation phenotype we observe. Consistent with this possibility, our ChIP results indicate that YY1 binds directly to the Lefty2 locus in E7.5 embryos (Fig. 5A–B), suggesting that YY1 normally activates Lefty2 and indirectly regulates Nodal signals (Fig. 5C). Additionally, we find that the suppression of Nodal in the context of a YY1 null epiblast results in enhanced mesoderm formation and increased expression of mesodermal markers including Tbx6, Twist and Snail (Fig. 5E and F). Furthermore, we find that the expression of Foxa2 and Eomes in cKO embryos correlate with Nodal levels, suggesting that overexpression of these genes in the cKO embryo is a result of Nodal overexpression. These results support the hypothesis that over-expression of Nodal signaling is an important component of the cKO phenotype and suggest that one role of YY1 may be repression of Nodal through activation of Lefty2.
Several reports have shown that differential post-translational modifications of YY1 can alter binding specificity (Hiromura et al., 2003; Takasaki et al., 2007; Yao et al., 2001; Zheng et al., 2010). Therefore we cannot rule out the possibility that YY1 may also act as a direct repressor of Nodal. It may be informative to determine precisely which post-translationally modified YY1 proteins are present in the epiblast, distinct germ layers and organizing centers during development and differentiation.
Despite the finding that anterior DE markers are specified, we observe a lack of intercalation of YY1 negative cells into the VE layer in cKO embryos (Fig. 3 and S2). A similar phenotype is found in other mutations that down-regulate FGF signaling in the streak (Ciruna et al., 1997; Sun et al., 1999), suggesting that in YY1 mutants this phenotype may be due to the observed loss of FGF4/8 (Sun et al., 1999). The similarities between Fgf8 knockout embryos and Yy1 epiblast mutants suggest that loss of Fgf8 or Fgf4 may also be a major cause of the observed Yy1 cKO phenotype (in addition to elevated Nodal signaling). In silico analyses indicate an absence of YY1 binding sites at the Fgf8 locus (data not shown). Therefore we do not believe that YY1 is directly responsible for the loss of Fgf8 expression, but that there may be a molecular intermediate between YY1 and regulation of FGF signaling
Reduction of Nodal in the cKO embryos had a noticeable effect on many genes implicated in primitive streak formation and mesoderm induction but did not change the level of Fgf4, which is absent in all cKO embryos examined. This result raises the interesting possibility that Fgf4 is directly downstream of YY1. Loss of Fgf4 results in a perimplantation phenotype that is similar to the YY1 null embryos (Donohoe et al., 1999; Tanaka et al., 1998), suggesting that loss of Fgf4 may in part explain the YY1 null phenotype as well.
Eomes is normally expressed in the EXE prior to gastrulation and then in the PS as it elongates (Ciruna and Rossant, 1999; Russ et al., 2000). Studies in the mouse have demonstrated that Nodal signaling from the epiblast is required to maintain Eomes in the EXE (Brennan et al., 2001; Guzman-Ayala et al., 2004). The data we present supports this hypothesis, demonstrating that loss of YY1 in the epiblast leads to prolonged high levels of epiblast derived Nodal signaling, which subsequently produces high levels of Eomes in the adjacent EXE. Eomes, like Nodal, is required for DE specification in the epiblast (Arnold and Robertson, 2009) and although both factors are required for proper gastrulation, the extent to which they interact in the proximal epiblast and PS is unclear. Our study supports a role for YY1 in the negative regulation of Nodal and supports the idea that alterations in Eomes expression are downstream of changes in Nodal expression (Figs. 2, 3 and 5).
Also of note are findings that DRAP1, a transcriptional corepressor, prevents excess Nodal accumulation in the epiblast (Iratni et al., 2002). Through interactions with FOXH1, DRAP1 attenuates the positive Nodal auto-regulatory feedback loop, such that loss of Drap1 results in increased expression of Nodal. Similar to Yy1 cKO embryos, Drap1 mutants demonstrate an accumulation of cells adjacent to the streak, an abundance of Nodal in the epiblast, dramatic reduction of T, an increase in FoxA2 and a loss of Lefty2. Furthermore, reduction of Nodal in Drap1 null embryos restores T expression, similar to what we present with YY1 cKO mutants (Fig. 5). Drap1 is expressed specifically in the epiblast during gastrulation. We observe no alteration in Drap1 mRNA levels in Yy1 cKO embryos (data not shown) raising the possibility that YY1 may physically interact with DRAP1 in the epiblast.
In conclusion we show that YY1 is required in vivo for proper morphogenetic movements during gastrulation as well as to maintain appropriate spatiotemporal Nodal expression, possibly through activation of Lefty2.
Supplementary Material
Acknowledgments
We thank Dr. Dominique Alfandari for generosity with antibodies, and Drs. Jaime Rivera and James Li for WISH probes. We thank Jake Hiller and Mara Geurrero for help with help optimizing antibody protocols. This work was supported in part by NIH R01DK087753 to KDT and a Massachusetts Life Sciences Grant to JM.
Footnotes
Appendix A. Supporting information: Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2012.05.031.
Contributor Information
Kimberly D. Tremblay, Email: kdtrembl@vasci.umass.edu.
Jesse Mager, Email: jmager@vasci.umass.edu.
References
- Affar el B, Gay F, Shi Y, Liu H, Huarte M, Wu S, Collins T, Li E. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol Cell Biol. 2006;26:3565–3581. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SJ, Hofmann UK, Bikoff EK, Robertson EJ. Pivotal roles for eomesodermin during axis formation, epithelium-to-mesenchyme transition and endoderm specification in the mouse. Development. 2008;135:501–511. doi: 10.1242/dev.014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML. Transcription factor YY1 functions as a PcG protein in vivo. Embo J. 2003;22:1347–1358. doi: 10.1093/emboj/cdg124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- Beddington RS, Rashbass P, Wilson V. Brachyury–a gene affecting mouse gastrulation and early organogenesis. Dev Suppl. 1992:157–165. [PubMed] [Google Scholar]
- Beddington RS, Robertson EJ. Axis development and early asymmetry in mammals. Cell. 1999;96:195–209. doi: 10.1016/s0092-8674(00)80560-7. [DOI] [PubMed] [Google Scholar]
- Bedford FK, Ashworth A, Enver T, Wiedemann LM. HEX: a novel homeobox gene expressed during haematopoiesis and conserved between mouse and human. Nucleic Acids Res. 1993;21:1245–1249. doi: 10.1093/nar/21.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RS, Robertson EJ. Nodal signalling in the epiblast patterns the early mouse embryo. Nature. 2001;411:965–969. doi: 10.1038/35082103. [DOI] [PubMed] [Google Scholar]
- Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- Calder MD, Madan P, Watson AJ. Bovine oocytes and early embryos express Staufen and ELAVL RNA-binding proteins. Zygote. 2008;16:161–168. doi: 10.1017/S096719940700456X. [DOI] [PubMed] [Google Scholar]
- Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21:8184–8188. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- Ciruna BG, Rossant J. Expression of the T-box gene Eomesodermin during early mouse development. Mech Dev. 1999;81:199–203. doi: 10.1016/s0925-4773(98)00243-3. [DOI] [PubMed] [Google Scholar]
- Ciruna BG, Schwartz L, Harpal K, Yamaguchi TP, Rossant J. Chimeric analysis of fibroblast growth factor receptor-1 (Fgfr1) function: a role for FGFR1 in morphogenetic movement through the primitive streak. Development. 1997;124:2829–2841. doi: 10.1242/dev.124.14.2829. [DOI] [PubMed] [Google Scholar]
- Conlon FL, Lyons KM, Takaesu N, Barth KS, Kispert A, Herrmann B, Robertson EJ. A primary requirement for nodal in the formation and maintenance of the primitive streak in the mouse. Development. 1994;120:1919–1928. doi: 10.1242/dev.120.7.1919. [DOI] [PubMed] [Google Scholar]
- Crossley PH, Martin GR. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- DaCosta Byfield S, Major C, Laping NJ, Roberts AB. SB-505124 is a selective inhibitor of transforming growth factor-beta type I receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2004;65:744–752. doi: 10.1124/mol.65.3.744. [DOI] [PubMed] [Google Scholar]
- Dong XP, Pfister H. Overlapping YY1- and aberrant SP1-binding sites proximal to the early promoter of human papillomavirus type 16. J Gen Virol. 1999;80:2097–2101. doi: 10.1099/0022-1317-80-8-2097. Pt 8. [DOI] [PubMed] [Google Scholar]
- Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SA, Manova K, Chen WS, Hoodless P, Weinstein DC, Bachvarova RF, Darnell JE., Jr Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc Natl Acad Sci USA. 1994;91:7598–7602. doi: 10.1073/pnas.91.16.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn NR, Vincent SD, Oxburgh L, Robertson EJ, Bikoff EK. Combinatorial activities of Smad2 and Smad3 regulate mesoderm formation and patterning in the mouse embryo. Development. 2004;131:1717–1728. doi: 10.1242/dev.01072. [DOI] [PubMed] [Google Scholar]
- Fritsch C, Brown JL, Kassis JA, Muller J. The DNA-binding polycomb group protein pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development. 1999;126:3905–3913. doi: 10.1242/dev.126.17.3905. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Dehart DB, Sulik KK, Hogan BL. Distinct requirements for extra-embryonic and embryonic bone morphogenetic protein 4 in the formation of the node and primitive streak and coordination of left-right asymmetry in the mouse. Development. 2002;129:4685–4696. doi: 10.1242/dev.129.20.4685. [DOI] [PubMed] [Google Scholar]
- Galvin KM, Shi Y. Multiple mechanisms of transcriptional repression by YY1. Mol Cell Biol. 1997;17:3723–3732. doi: 10.1128/mcb.17.7.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia E, Marcos-Gutierrez C, del Mar Lorente M, Moreno JC, Vidal M. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. Embo J. 1999;18:3404–3418. doi: 10.1093/emboj/18.12.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girton JR, Jeon SH. Novel embryonic and adult homeotic phenotypes are produced by pleiohomeotic mutations in Drosophila. Dev Biol. 1994;161:393–407. doi: 10.1006/dbio.1994.1040. [DOI] [PubMed] [Google Scholar]
- Griffith GJ, Trask MC, Hiller J, Walentuk M, Pawlak JB, Tremblay KD, Mager J. Yin-yang1 is required in the Mammalian oocyte for follicle expansion. Biol Reprod. 2011;84:654–663. doi: 10.1095/biolreprod.110.087213. [DOI] [PubMed] [Google Scholar]
- Guzman-Ayala M, Ben-Haim N, Beck S, Constam DB. Nodal protein processing and fibroblast growth factor 4 synergize to maintain a trophoblast stem cell microenvironment. Proc Natl Acad Sci USA. 2004;101:15656–15660. doi: 10.1073/pnas.0405429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Lewis P, Pevny L, McMahon AP. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech Dev. 2002;119(Suppl 1):S97–S101. doi: 10.1016/s0925-4773(03)00099-6. [DOI] [PubMed] [Google Scholar]
- He Y, Casaccia-Bonnefil P. The Yin and Yang of YY1 in the nervous system. J Neurochem. 2008;106:1493–1502. doi: 10.1111/j.1471-4159.2008.05486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Kim JY, Dupree J, Tewari A, Melendez-Vasquez C, Svaren J, Casaccia P. Yy1 as a molecular link between neuregulin and transcriptional modulation of peripheral myelination. Nat Neurosci. 2010;13:1472–1480. doi: 10.1038/nn.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, Dave N, Escriva M, Hernandez-Munoz I, Di Croce L, Helin K, Garcia de Herreros A, Peiro S. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiromura M, Choi CH, Sabourin NA, Jones H, Bachvarov D, Usheva A. YY1 is regulated by O-linked N-acetylglucosaminylation (O-glcNAcylation) J Biol Chem. 2003;278:14046–14052. doi: 10.1074/jbc.M300789200. [DOI] [PubMed] [Google Scholar]
- Hyde-DeRuyscher RP, Jennings E, Shenk T. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 1995;23:4457–4465. doi: 10.1093/nar/23.21.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iratni R, Yan YT, Chen C, Ding J, Zhang Y, Price SM, Reinberg D, Shen MM. Inhibition of excess nodal signaling during mouse gastrulation by the transcriptional corepressor DRAP1. Science. 2002;298:1996–1999. doi: 10.1126/science.1073405. [DOI] [PubMed] [Google Scholar]
- Kwon GS, Viotti M, Hadjantonakis AK. The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extra-embryonic lineages. Dev Cell. 2008;15:509–520. doi: 10.1016/j.devcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Galvin KM, Shi Y. Evidence for physical interaction between the zinc-finger transcription factors YY1 and Sp1. Proc Natl Acad Sci USA. 1993;90:6145–6149. doi: 10.1073/pnas.90.13.6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SL, Tam PP. Definitive endoderm of the mouse embryo: formation, cell fates, and morphogenetic function. Dev Dyn. 2006;235:2315–2329. doi: 10.1002/dvdy.20846. [DOI] [PubMed] [Google Scholar]
- Liu H, Schmidt-Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H, Pelanda R, Reth M, Skok J, Rajewsky K. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007;21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CC, Brennan J, Robertson EJ. From fertilization to gastrulation: axis formation in the mouse embryo. Curr Opin Genet Dev. 2001;11:384–392. doi: 10.1016/s0959-437x(00)00208-2. [DOI] [PubMed] [Google Scholar]
- Meno C, Gritsman K, Ohishi S, Ohfuji Y, Heckscher E, Mochida K, Shimono A, Kondoh H, Talbot WS, Robertson EJ, Schier AF, Hamada H. Mouse Lefty2 and zebrafish antivin are feedback inhibitors of nodal signaling during vertebrate gastrulation. Mol Cell. 1999;4:287–298. doi: 10.1016/s1097-2765(00)80331-7. [DOI] [PubMed] [Google Scholar]
- Meno C, Ito Y, Saijoh Y, Matsuda Y, Tashiro K, Kuhara S, Hamada H. Two closely-related left-right asymmetrically expressed genes, lefty-1 and lefty-2: their distinct expression domains, chromosomal linkage and direct neuralizing activity in Xenopus embryos. Genes Cells. 1997;2:513–524. doi: 10.1046/j.1365-2443.1997.1400338.x. [DOI] [PubMed] [Google Scholar]
- Mohd-Sarip A, Venturini F, Chalkley GE, Verrijzer CP. Pleiohomeotic can link polycomb to DNA and mediate transcriptional repression. Mol Cell Biol. 2002;22:7473–7483. doi: 10.1128/MCB.22.21.7473-7483.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MJ, Woltering JM, In der Rieden PM, Durston AJ, Thiery JP. YY1 regulates the neural crest-associated slug gene in Xenopus laevis. J Biol Chem. 2004;279:46826–46834. doi: 10.1074/jbc.M406140200. [DOI] [PubMed] [Google Scholar]
- Norris DP, Brennan J, Bikoff EK, Robertson EJ. The Foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo. Development. 2002;129:3455–3468. doi: 10.1242/dev.129.14.3455. [DOI] [PubMed] [Google Scholar]
- Palmer MB, Majumder P, Cooper JC, Yoon H, Wade PA, Boss JM. Yin yang 1 regulates the expression of snail through a distal enhancer. Mol Cancer Res. 2009;7:221–229. doi: 10.1158/1541-7786.MCR-08-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Gomez A, Vella FD, Shawlot W, Oulad-Abdelghani M, Chazaud C, Meno C, Pfister V, Chen L, Robertson E, Hamada H, Behringer RR, Ang SL. Nodal antagonists in the anterior visceral endoderm prevent the formation of multiple primitive streaks. Dev Cell. 2002;3:745–756. doi: 10.1016/s1534-5807(02)00321-0. [DOI] [PubMed] [Google Scholar]
- Rezai-Zadeh N, Zhang X, Namour F, Fejer G, Wen YD, Yao YL, Gyory I, Wright K, Seto E. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev. 2003;17:1019–1029. doi: 10.1101/gad.1068003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Perez JA, Mager J, Magnuson T. Dynamic morphogenetic events characterize the mouse visceral endoderm. Dev Biol. 2003;261:470–487. doi: 10.1016/s0012-1606(03)00302-6. [DOI] [PubMed] [Google Scholar]
- Rivera-Perez JA, Magnuson T. Primitive streak formation in mice is preceded by localized activation of Brachyury and Wnt3. Dev Biol. 2005;288:363–371. doi: 10.1016/j.ydbio.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Robertson EJ, Norris DP, Brennan J, Bikoff EK. Control of early anterior-posterior patterning in the mouse embryo by TGF-beta signalling. Philos Trans R Soc Lond B Biol Sci. 2003;358:1351–1357. doi: 10.1098/rstb.2003.1332. discussion 1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner MH, Vigano MA, Ozato K, Timmons PM, Poirier F, Rigby PW, Staudt LM. A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature. 1990;345:686–692. doi: 10.1038/345686a0. [DOI] [PubMed] [Google Scholar]
- Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, Barton SC, Surani MA, Ryan K, Nehls MC, Wilson V, Evans MJ. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hogan BL. Enhancer analysis of the mouse HNF-3 beta gene: regulatory elements for node/notochord and floor plate are independent and consist of multiple sub-elements. Genes Cells. 1996;1:59–72. doi: 10.1046/j.1365-2443.1996.04004.x. [DOI] [PubMed] [Google Scholar]
- Schier AF. Nodal signaling in vertebrate development. Annu Rev Cell Dev Biol. 2003;19:589–621. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
- Schlisio S, Halperin T, Vidal M, Nevins JR. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. Embo J. 2002;21:5775–5786. doi: 10.1093/emboj/cdf577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholer HR, Dressler GR, Balling R, Rohdewohld H, Gruss P. Oct-4: a germline-specific transcription factor mapping to the mouse t-complex. EMBO J. 1990;9:2185–2195. doi: 10.1002/j.1460-2075.1990.tb07388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefton M, Sanchez S, Nieto MA. Conserved and divergent roles for members of the Snail family of transcription factors in the chick and mouse embryo. Development. 1998;125:3111–3121. doi: 10.1242/dev.125.16.3111. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta. 1997;1332:F49–66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- Shi Y, Seto E, Chang LS, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Sun X, Meyers EN, Lewandoski M, Martin GR. Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes Dev. 1999;13:1834–1846. doi: 10.1101/gad.13.14.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki N, Kurokawa D, Nakayama R, Nakayama J, Aizawa S. Acetylated YY1 regulates Otx2 expression in anterior neuroectoderm at two cis-sites 90 kb apart. EMBO J. 2007;26:1649–1659. doi: 10.1038/sj.emboj.7601619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Thomas P, Brickman JM, Popperl H, Krumlauf R, Beddington RS. Axis duplication and anterior identity in the mouse embryo. Cold Spring Harb Symp Quant Biol. 1997;62:115–125. [PubMed] [Google Scholar]
- Tremblay KD, Hoodless PA, Bikoff EK, Robertson EJ. Formation of the definitive endoderm in mouse is a Smad2-dependent process. Development. 2000;127:3079–3090. doi: 10.1242/dev.127.14.3079. [DOI] [PubMed] [Google Scholar]
- Vincent SD, Dunn NR, Hayashi S, Norris DP, Robertson EJ. Cell fate decisions within the mouse organizer are governed by graded Nodal signals. Genes Dev. 2003;17:1646–1662. doi: 10.1101/gad.1100503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, Herrmann BG. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- Wood HB, Episkopou V. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech Dev. 1999;86:197–201. doi: 10.1016/s0925-4773(99)00116-1. [DOI] [PubMed] [Google Scholar]
- Wu S, Hu YC, Liu H, Shi Y. Loss of YY1 impacts the heterochromatic state and meiotic double-strand breaks during mouse spermatogenesis. Mol Cell Biol. 2009;29:6245–6256. doi: 10.1128/MCB.00679-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovleva T, Kolesnikova L, Vukojevic V, Gileva I, Tan-No K, Austen M, Luscher B, Ekstrom TJ, Terenius L, Bakalkin G. YY1 binding to a subset of p53 DNA-target sites regulates p53-dependent transcription. Biochem Biophys Res Commun. 2004;318:615–624. doi: 10.1016/j.bbrc.2004.04.065. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Saijoh Y, Perea-Gomez A, Shawlot W, Behringer RR, Ang SL, Hamada H, Meno C. Nodal antagonists regulate formation of the anteroposterior axis of the mouse embryo. Nature. 2004;428:387–392. doi: 10.1038/nature02418. [DOI] [PubMed] [Google Scholar]
- Yao YL, Yang WM, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol. 2001;21:5979–5991. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Chu J, Zeng Y, Loh HH, Law PY. Yin Yang 1 phosphorylation contributes to the differential effects of mu-opioid receptor agonists on microRNA-190 expression. J Biol Chem. 2010;285:21994–22002. doi: 10.1074/jbc.M110.112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.