Background: HP0892 of H. pylori shares high structural similarity with other toxin-antitoxin (TA) system toxins.
Results: RNase activity and cell toxicity of HP0892 is inhibited by HP0893 via strong protein-protein interaction.
Conclusion: HP0892-HP0893 pair is a TA system with a protein-protein interaction mode similar to other TA pairs.
Significance: It is highly probable that HP0892-HP0893 TA pair is involved in H. pylori virulence.
Keywords: Helicobacter pylori, Nuclear Magnetic Resonance, Protein Structure, Protein-Protein Interactions, Ribonuclease, HP0892, HP0893, YafQ, Toxin-Antitoxin System, Virulence
Abstract
Bacterial chromosomal toxin-antitoxin (TA) systems have been proposed not only to play an important role in the stress response, but also to be associated with antibiotic resistance. Here, we identified the chromosomal HP0892-HP0893 TA proteins in the gastric pathogen, Helicobacter pylori, and structurally characterized their protein-protein interaction. Previously, HP0892 protein was suggested to be a putative TA toxin based on its structural similarity to other RelE family TA toxins. In this study, we demonstrated that HP0892 binds to HP0893 strongly with a stoichiometry of 1:1, and HP0892-HP0893 interaction occurs mainly between the N-terminal secondary structure elements of HP0892 and the C-terminal region of HP0893. HP0892 cleaved mRNA in vitro, preferentially at the 5′ end of A or G, and the RNase activity of HP0892 was inhibited by HP0893. In addition, heterologous expression of HP0892 in Escherichia coli cells led to cell growth arrest, and the cell toxicity of HP0892 was neutralized by co-expression with HP0893. From these results and a structural comparison with other TA toxins, it is concluded that HP0892 is a toxin with intrinsic RNase activity and HP0893 is an antitoxin against HP0892 from a TA system of H. pylori. It has been known that hp0893 gene and another TA antitoxin gene, hp0895, of H. pylori, are both genomic open reading frames that correspond to genes that are potentially expressed in response to interactions with the human gastric mucosa. Therefore, it is highly probable that TA systems of H. pylori are involved in virulence of H. pylori.
Introduction
Programmed cell death (PCD)2 refers to any form of cell death mediated by an intracellular death program (1). PCD systems such as apoptosis (2) or autophagy (3) in eukaryotic multicellular organisms play significant roles in a variety of biological processes including development, cell homeostasis, and oncogenesis (4). Traditionally, PCD has been considered associated with only eukaryotic multicellular organisms; however, recently, PCDs have also been observed in bacteria (1). For most eubacteria and archaea, the most common mechanism involved in bacteria PCD is the toxin-antitoxin (TA) system (5).
TA systems are usually composed of a stable toxin that arrests cell growth and a labile antitoxin that counteracts the toxin. The former always exists as a stable protein, whereas the latter is either a protein (type II TA system) or an RNA species (type I or III TA system) (6). In the type I TA system, the antitoxins are small antisense RNAs forming duplexes with the toxin mRNAs, which repress translation of the toxin genes (7). In the type II system, the antitoxins are proteins that neutralize the toxins by a direct protein-protein interaction with the toxins (8). In type III TA systems, small RNA antitoxin combines with and neutralizes protein toxins (9, 10). In most cases, toxin and antitoxin genes are in an operon where the antitoxin gene often locates upstream of the toxin gene (6), usually overlapping or being separated by a small intergenic region, so that TA genes are co-transcribed. In type II TA systems, TA operons are usually negatively autoregulated at the transcription level by antitoxins and TA complexes, which bind to the TA locus promoters (8).
Several TA systems have been discovered in bacterial plasmids and on chromosomes. Plasmid-borne TA systems are involved in plasmid stabilization through toxin-mediated post-segregation killing of plasmid-free progeny cells (11): Cells that do not inherit a copy of a plasmid upon division are killed because the unstable antitoxin is degraded faster than the more stable toxin. On the contrary, the biological roles of chromosomal TA systems have been the subject of widespread discussion and speculation. Recently, nine possible functions including genomic junk, stabilization of genomic parasites, selfish alleles, gene regulation, growth control, and production of persisters were suggested (12), but a flexible response of a bacterial cell to stress conditions such as starvation is regarded to be the major function of chromosomal TA systems. Moreover, TA systems can contribute to the formation of persistent cells during exposure to antibiotics, and recently, persistence was suggested as a common function of TA systems (13, 14).
The Escherichia coli K12 chromosome possesses at least eight relatively well characterized TA systems: MazF-MazE, RelE-RelB, ChpBK-ChpBI, YafQ-DinJ, YoeB-YefM, HipA-HipB, YafO-YafN, and MqsR-MqsA (6). Among the toxins of these modules, the RelE, YafQ, and YoeB toxins are classified within the same family (RelE family) due to low but significant sequence similarities among them (8). These toxins function by inhibiting translation through mRNA cleavage (8). Among those toxins, RelE is a ribosome-dependent endoribonuclease that is only active when associating with a ribosome (15–17), whereas the others have intrinsic ribonuclease (RNase) activity (18, 19). However, YoeB and YafQ also associate with ribosome and are regarded to function as ribosome-dependent mRNA interferases, too (6, 19, 20).
Helicobacter pylori is a Gram-negative neutralophile that multiplies in an environmental pH from 6.0 to 8.0 with optimal growth close to pH 7.0 (21, 22). However, it has a unique ability to survive in an acidic environment of human stomach and to colonize the gastric mucosa (21). To live in the extremely acidic environment of the stomach, it has developed acid resistance mechanisms (21). For example, urease of H. pylori, which has optimum activity at neutral pH, produces NH3 from gastric juice urea, and neutralizes acidity around the bacterium (21). Because of these acid resistance mechanisms, H. pylori is thought to possess an intracellular pH near neutrality even in the acidic environment of stomach (23, 24). It can cause diverse gastric diseases such as peptic ulcers, chronic gastritis, mucosa-associated lymphoid tissue lymphoma, and gastric cancer (25–27). H. pylori is one of the most common chronic bacterial infections of humans, infecting more than half of the world population.
In our previous study (28), we found that HP0894 (a YafQ-homologous toxin) and HP0895 (an antitoxin for HP0894) proteins of H. pylori 26695 strain form a TA system. In another study of ours (29), we reported the solution structure of the conserved hypothetical protein HP0892 from this strain and found that its structure is very similar to those of HP0894 and other RelE family TA toxins. Those findings about HP0892 prompted us to speculate that HP0892 might also act as a TA toxin with intrinsic RNase activity like HP0894. In this study, we identified the biological function of HP0892 as a TA toxin and characterized some structural aspects of its toxin-antitoxin binding.
EXPERIMENTAL PROCEDURES
Molecular Cloning
Subcloning was carried out as described previously for HP0892 (29). The same procedure was used to prepare the cells harboring the recombinant plasmids for mutated HP0892. All resulting HP0892 constructs contained eight non-native residues (LEHHHHHH) at the C terminus (referred to as HP0892-His). Those residues facilitated subsequent protein purification. Additionally, the same procedure with minor change was used for HP0892 without any purification tag or non-native residues (in vector pET-21a; Novagen, Madison, WI) and for glutathione S-transferase (GST) fusion HP0893 (referred to as GST-HP0893) (in vector pGEX-4T-1; GE Healthcare) and HP0893 with 20 non-native residues including a His tag at the N terminus (referred to as His-HP0893) (in vector pET-28a; Novagen).
Preparation of Protein Samples
Overexpression was carried out as described previously (29) for HP0892-His. For the production of 15N- or 15N,13C-labeled proteins, M9 medium containing 15NH4Cl and/or [13C]glucose as stable isotope sources were used. The protein samples were purified using a Ni2+-affinity column following standard protocol. The same procedures were used to prepare His-HP0893 and mutated HP0892-His. For His-HP0893, because it is a putative DNA-binding protein, further purification using polyethyleneimine was carried out. Polyethyleneimine was added to the previously purified His-HP0893 solution at a final concentration of 0.08%, which was determined by a titration test. After ultracentrifugation, the supernatant was collected, dialyzed against a 50 mm borate buffer (pH 11), and loaded onto a disposable PD-10 column (GE Healthcare) packed with Q-Sepharose fast flow resin (GE Healthcare). The column was washed with 100 ml of the same buffer, and then the protein was eluted with a 50 mm borate buffer (pH 11) containing 1 m NaCl.
Construction of HP0892/HP0893 Co-expression Systems
The full-length hp0892 and hp0893 genes were amplified from H. pylori genomic DNA by PCR as different DNA fragments and were subsequently subcloned under the T7 promoter in pET-21a (ampicillin-resistant) and pET-29a (kanamycin-resistant) vectors (Novagen), respectively. These two vectors were co-transformed into E. coli BL21 (DE3) (HP0892/HP0893 co-expression system I). In this case, the HP0893 construct contained eight non-native residues including a His tag at the C terminus (referred to as HP0893-His). The HP0892 construct contained no additional residues. Using the same procedure, another co-expression system composed of HP0892 (in vector pET-21a) and HP0893 (in pET-28a) was constructed (HP0892/HP0893 co-expression system II). In this case, the HP0893 construct contained 20 non-native residues including a His tag at the N terminus (referred to as His-HP0893). The HP0892 construct contained no additional residues.
Pulldown Assay for HP0892/HP0893 Binding
E. coli BL21 (DE3) cells harboring GST-HP0893 expression plasmid (vector pGEX-4T-1) and HP0892-His expression plasmid (vector pET-21a) were cultured and expressed with 0.5 mm isopropyl-β-d-thiogalactopyranoside, separately. The cells were harvested by centrifugation and resuspended with cell lysis buffer (20 mm sodium phosphate buffer (pH 7.3)), separately. The two cell cultures were mixed and disrupted by sonication. The lysate was centrifuged at 10,000 rpm, 4 °C for 1 h, and the supernatant was loaded onto a glutathione-Sepharose column. The column was washed with the same cell lysis buffer until the UV peak stabilized. The adsorbed protein was eluted with a glutathione buffer (3.08 g of reduced glutathione in 1 liter of 50 mm Tris-HCl buffer (pH 8.0)). The eluted solution was analyzed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The fractions containing the target proteins were pooled, and buffer exchange was carried out with 20 mm Tris-HCl buffer (pH 7.9) containing 0.5 m NaCl. The solution was applied to a Ni2+-affinity column. The column was washed with the 20 mm Tris-HCl buffer (pH 7.9) containing 60 mm imidazole and 0.5 m NaCl. The adsorbed proteins were eluted with the 20 mm Tris-HCl (pH 7.9) buffer containing 0.5 m NaCl and a linear imidazole gradient (0∼500 mm imidazole) and analyzed by SDS-PAGE.
Co-expression and Co-purification of HP0892/HP0893
HP0892 and HP0893 were co-expressed and co-purified using the HP0892/HP0893 co-expression system I described above. Bacteria were grown at 37 °C in LB broth containing antibiotics (ampicillin 50 μg/ml and kanamycin 40 μg/ml). Induction, cell harvest, cell lysis, and purification with a Ni2+-affinity column were done following standard protocols. The fractions eluted from the Ni2+-affinity column (using stepwise imidazole gradient) were analyzed by SDS-PAGE.
Toxicity of HP0892 in E. coli
E. coli BL21 (DE3) cells harboring the HP0892 expression plasmids (HP0892/pET21a) and the HP0892/HP0893 co-expression plasmids (HP0892/HP0893 co-expression system II) were grown at 37 °C in 100 ml of LB broth containing antibiotics (50 μg/ml ampicillin for pET21a and HP0892/pET21a, and 50 μg/ml ampicillin and 40 μg/ml kanamycin for HP0892/HP0893 co-expression system II). At an A600 of approximately 0.3, 1 mm isopropyl-β-d-thiogalactopyranoside was added. Subsequently, samples for viable counts and SDS-PAGE analyses were taken at several time points. Viable counts were made by plating dilutions of the cultures onto LB plates containing the appropriate antibiotics. In addition, the cells harvested from 125 μl of the samples taken at each time point were analyzed by SDS-PAGE.
In Vitro RNA Cleavage by HP0892
A DNA fragment containing the T7 promoter and the hp0893 gene was obtained by PCR amplification of H. pylori genomic DNA. The hp0893 mRNA was synthesized in vitro from this DNA fragment using the T7-MEGAshortscript kit (Ambion, Austin, TX). Mixtures of hp0893 mRNA (∼310 bases, 300 ng), HP0892, HP0892 mutants, and/or HP0893 were prepared in several ratios in 20 mm Tris-HCl (pH 7.4) buffer containing 150 mm NaCl (total volume, 5 μl). The reaction was done at 37 °C for 60 min and was stopped by adding 5 μl of the sequencing loading buffer (Ambion). The samples were run on a 1.5% agarose gel containing ethidium bromide with Tris borate/EDTA buffer.
Primer Extension
The hp0893 mRNA was used as a primer template. Two different DNA primers were synthesized and 5′-labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs). Primers (0.8 pmol) were annealed to each template (1 pmol) in a 10-μl mixture samples by incubation at 90 °C for 5 min followed by slow cooling to room temperature. The reaction mixture was digested by adding 1 μl of various amounts of HP0892 at 37 °C for 1 h. Subsequent primer extension reactions were performed by adding 4 μl of 5× buffer (250 mm Tris-HCl (pH 8.3), 375 mm KCl, and 15 mm MgCl2), 5 mm DTT, 0.5 mm dNTP, and 20 units of SuperScript III reverse transcriptase (Invitrogen) for a final reaction volume of 20 μl. The cDNA was synthesized at 55 °C for 30 min and then purified by phenol/chloroform extraction and ethanol precipitation. The products were then subjected to 8% denaturing PAGE and analyzed by autoradiography.
NMR Spectroscopy and Titrations
For HP0893Ctp titration against HP0892, a 30-residue peptide corresponding to the C-terminal region (Glu-66 to Leu-95) of HP0893 was commercially synthesized (Anygen, Kwangju, Korea) and purified (96% purity). The two-dimensional 1H,15N TROSY spectra of 15N-labeled HP0892 alone (final concentration 0.1 mm) and with HP0893Ctp added incrementally (0.05, 0.1, and 0.2 mm) to the sample were obtained on a Bruker Avance 600 spectrometer. The averaged chemical shift changes were calculated by (30).
For backbone resonance assignments of HP0892 bound with HP0893Ctp, the three-dimensional TROSY-type HNCA (trHNCA) of 13C,15N-labeled HP0892 (0.2 mm) mixed with HP0893Ctp (0.3 mm) was obtained on a Varian VNMRS 900 spectrometer. All NMR samples were dissolved in 20 mm sodium phosphate (pH 6.0) buffer without any additional salt. The NMR samples contained 10% D2O for the lock signal. All spectra were processed and analyzed using NMRPipe/NMRDraw (31) and NMRView (32), respectively.
Gel Filtration Chromatography
A mixture of HP0892 with HP0893 was prepared by co-lysis of cells harboring HP0892-His and His-HP0893, respectively, and subsequently, Ni2+-affinity column purification followed. The mixture and molecular mass markers were fractionated on a Superdex 75, 10/300 GL column (GE Healthcare) using an AKTA FPLC system (GE Healthcare). The mobile phase consisted of 50 mm sodium phosphate (pH 7.4) buffer containing 100 mm NaCl. The void volume was measured with blue dextran 2000 (2000 kDa). The proteins were eluted at flow rates of 0.5 ml/min and were monitored using UV light at 280 nm.
RESULTS
HP0892 and HP0893 Proteins Form a Stable Complex
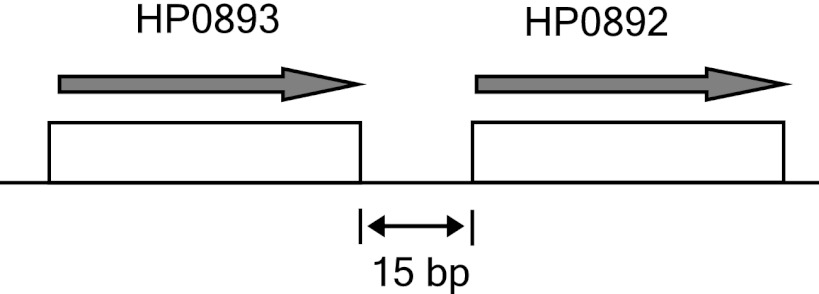
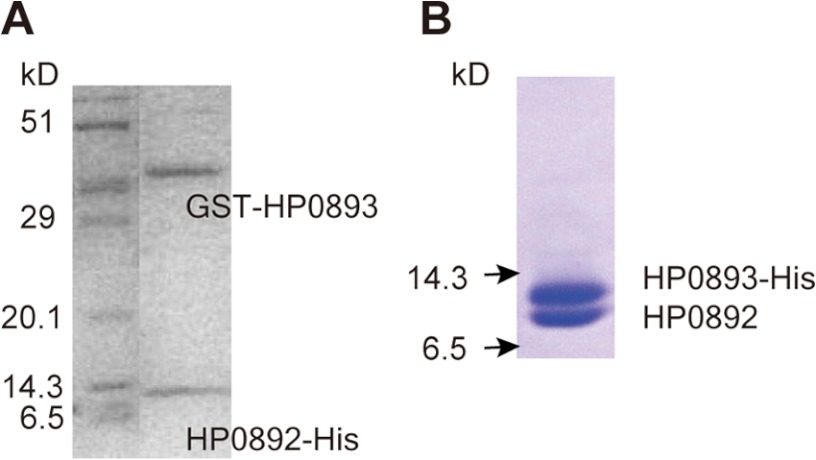
Generally, the antitoxin gene is usually located directly upstream of the toxin gene in TA systems. On the H. pylori chromosome, the hp0893 gene is located directly upstream of the hp0892 gene (Fig. 1). Although HP0893 has been known to be a hypothetical protein, we expected that if HP0892 is a TA toxin, then HP0893 would bind to HP0892, as a putative TA antitoxin of H. pylori. To identify the potential interaction between HP0892 and HP0893, samples of protein mixtures including HP0893 fused with GST (GST-HP0893) and HP0892 fused with His tag at the C terminus (HP0892-His) were purified by the pulldown assay method (see “Experimental Procedures”) using a glutathione-Sepharose and subsequently, Ni2+-affinity column. As shown in Fig. 2A, the SDS-PAGE analysis revealed the presence of both proteins, HP0892-His and GST-HP0893, in the same eluted fractions from the latter Ni2+-affinity column. For a control, we also monitored the interaction between HP0892-His and GST alone by the same pulldown assay method, but no interaction was observed (data not shown). To confirm the interaction between HP0892 and HP0893, both proteins were co-expressed from the HP0892/HP0893 co-expression system I (see “Experimental Procedures”) and then co-purified. For detection and purification, HP0893 was expressed as a C-terminal His tag fusion (HP0893-His) and HP0892 was expressed without a purification tag. SDS-PAGE analysis under a range of imidazole treatments revealed the presence of both expressed proteins (HP0893-His and HP0892) in the same eluted fractions from the Ni2+-affinity column (Fig. 2B). These results indicate that there is a stable and strong interaction between the HP0892 and HP0893 proteins.
FIGURE 1.
Structure of the region of the H. pylori chromosome encoding hp0892 and hp0893.
FIGURE 2.
Protein-protein binding between HP0892 and HP0893. A, SDS-PAGE of the elution fraction from the latter Ni2+-affinity column in the pulldown assay (see “Experimental Procedures”) using a GST and a Ni2+-affinity column for a solution harboring GST-HP0893 and HP0892-His. B, SDS-PAGE of elution fraction from a Ni2+-affinity column loaded with a solution harboring HP0892 and His-HP0893 co-expressed in cells.
Inhibitory Effect of HP0892 on E. coli Cell Growth Is Neutralized by HP0893
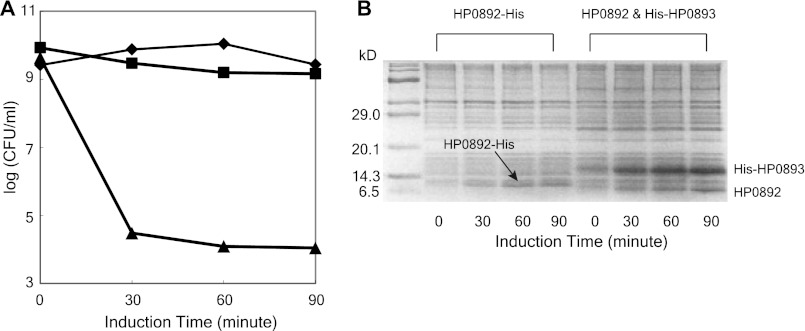
The potential of HP0892 as a toxin and of HP0893 as an antitoxin was examined by measuring their effects on the viability of E. coli cells grown in the presence of isopropyl-β-d-thiogalactopyranoside. As shown in Fig. 3, the number of viable E. coli cells decreased significantly with expression of HP0892, but co-expression of HP0893 neutralized the detrimental effect of HP0892 on E. coli cell growth.
FIGURE 3.
Effects of expressing HP0892 and HP0893 on E. coli cell growth. A, viable counts of E. coli cells expressing the empty vector pET21a (♦), the vector HP0892-His/pET21a (▴), and HP0892/pET21a and His-HP0893/pET29a (■) at the indicated induction times. B, SDS-PAGE analyses of HP0892 and HP0893 expressions at the indicated induction times.
In Vitro RNase Activity of HP0892 Is Inhibited by HP0893
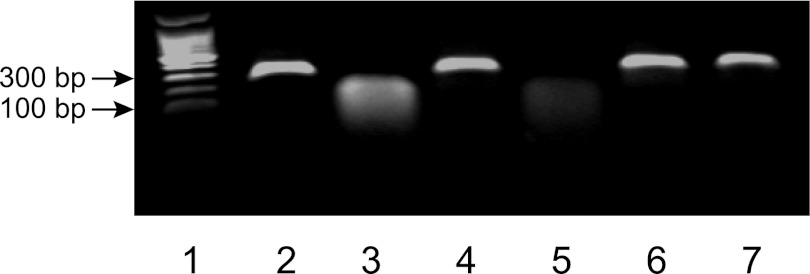
To determine whether HP0892 has intrinsic RNase activity, we tested the in vitro RNase activity of HP0892 and the inhibitory effect of HP0893. As mentioned previously in the Introduction, H. pylori is very likely to possess an intracellular pH near neutrality in even acidic environment of stomach, so this test and the following primer extension test were conducted at a neutral pH condition (pH 7.4). As shown in Fig. 4, in vitro synthesized mRNA was digested into smaller fragments following incubation with HP0892, whereas addition of HP0893 inhibited mRNA decay by HP0892. HP0893 alone did not digest mRNA. In addition, the RNase activity of a single mutant HP0892 (H86A) was markedly reduced, indicating that RNA cleavage by HP0892 was not due to contamination of RNases during purification and that the His-86 residue is a key catalytic residue of HP0892.
FIGURE 4.
Cleavage of hp0893 mRNA by HP0892 and the inhibitory effect of HP0893. In vitro transcribed mRNA was incubated at 37 °C for 60 min with various combinations of HP0892, HP0892 mutant (H86A), and/or HP0893. Lanes: 1, DNA ladder; 2, mRNA (300 ng, control); 3, mRNA and 4 μm HP0892; 4, mRNA and 4 μm HP0892 mutant (H86A); 5 and 6, mRNA, 4 μm HP0892 and 2 or 4 μm HP0893, respectively; 7, mRNA and 4 μm HP0893.
Primer Extension
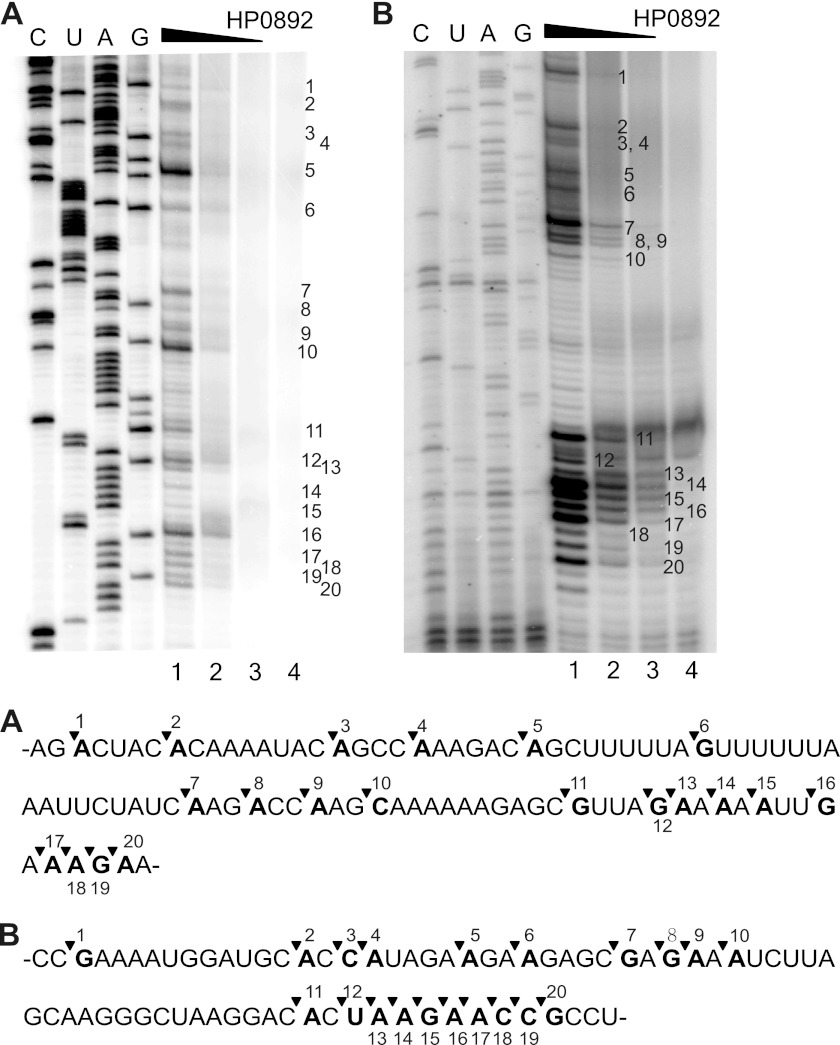
To identify the sequence of HP0892 cleavage site on mRNA, HP0892-mediated cleavage of hp0893 mRNA was analyzed by primer extension experiments. As shown in Fig. 5, cleavage occurred preferentially at the 5′ end of adenine (A) or guanine (G) residues, although not all of A or G, and AG-rich sequence regions were relatively more sensitive.
FIGURE 5.
Primer extension analyses of mRNA cleavage by HP0892. hp0893 mRNA with different DNA primers was used to produce A and B. Lanes 1–4, mRNA. Lane 5, 2.5, 1.25, or 0 μm, respectively, HP0892. DNA sequence ladders are shown at the left with upper lane labels for the sense strand. Major cleavage sites are indicated by sequential numbers along the right side of the images. The RNA recognition sequences for A and B, shown below, were derived from the RNA complement of the DNA sequencing ladder in the above images. The numbers and inverse triangles on the sequences correspond to the sequentially numbered major cleavage sites in the above images.
Binding Sites of HP0892 and HP0893
Because HP0893 exists as an aggregated form with a high molecular mass and is easily precipitated in solution at NMR scale concentrations, we were not able to get well dispersed NMR spectra of 15N-labeled HP0893 and could not investigate the binding aspect between HP0892 and HP0893 with the NMR titration method using full HP0893 protein sample. In the H. pylori HP0894-HP0895 TA complex, the 30-residue C-terminal region of the HP0895 antitoxin is responsible for binding with the HP0894 toxin (28), which is a structural and sequential homologue close to HP0892. In addition, in E. coli YoeB-YefM and RelE-RelB TA complexes, approximately a 30-residue C-terminal region of YefM or RelB antitoxin, also mainly takes part in the binding with YoeB or RelE toxin (18, 33), respectively, which are also structural homologues of HP0892. Therefore, we hypothesized that the C-terminal region of HP0893 is also responsible for binding with HP0892 and investigated its binding with the C-terminal region of HP0893 instead of the full-length HP0893.
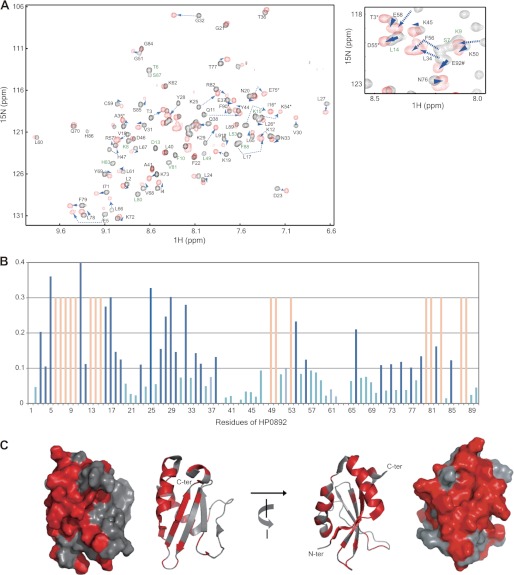
To confirm the binding of this C-terminal region of HP0893 with HP0892, we synthesized a 30-residue peptide, HP0893Ctp (Glu-66 to Leu-95), corresponding to the C-terminal region of HP0893, and investigated the interaction between this peptide and HP0892 with the NMR titration method. A series of two-dimensional 1H,15N TROSY spectra of 15N-labeled HP0892 were recorded with successive additions of unlabeled HP0893Ctp (0, 0.5, 1, and 2 molar equivalents). As shown in Fig. 6, obvious chemical shift changes in slow or intermediate exchange mode on the NMR time scale were observed for a lot of residues of HP0892, which implies that the dissociation constants (Kd) of HP0893Ctp (and consequently HP0893) with HP0892 are relatively small (∼10−8 m) (34) and their binding is strong. Chemical shift changes from the residues in slow exchange mode were completed with an equimolar amount of HP0893Ctp. These results and the relative intensities of the protein bands corresponding to HP0892 and HP0893 in the SDS-PAGE gel from the previous pulldown assay (Fig. 2B) indicate that the binding stoichiometry of HP0892 and HP0893 is 1:1. Considering the overall results, we can reasonably suggest that the C-terminal region of HP0893 is responsible for its binding with HP0892.
FIGURE 6.
Chemical shift perturbations of HP0892 residues by HP0893Ctp binding. A, two-dimensional 1H,15N TROSY spectra of 100 μm 15N-labeled HP0892 with HP0893Ctp added at 0 (black) and 1 (red) molar ratios of HP0893Ctp/HP0892 are superimposed. A central spectral region is enlarged aside. The residues whose resonances disappeared (intermediate exchange mode) in the spectrum at 1 molar ratio are labeled in green. B, averaged chemical shift changes (Δδave) of individual residues of HP0892 upon binding with HP0893Ctp (1:1 molar ratio). The changes of the residues in the obvious slow exchange mode (with Δδave more than 1.0 ppm) are colored in dark blue, those in the intermediate exchange mode in light red, and those with Δδave <1.0 ppm in light blue. C, ribbon and surface displays of HP0892 structure colored according to chemical shift perturbations. Residues whose chemical shift perturbations are in the obvious slow or intermediate exchange mode are colored in red. The program PyMOL (35) was used to visualize the structures.
A plot of the chemical shift changes against the residues of HP0892 is shown in Fig. 6B. For this plot, the backbone N and HN resonances of uncomplexed HP0892 were assigned from the data obtained in an earlier study (29). Regarding backbone resonance assignments for HP0893Ctp-bound HP0892, sequential Cα connections determined from three-dimensional trHNCA of 13C,15N-labeled HP0892 in complex with HP0893Ctp and comparison of Cα resonance values between uncomplexed and HP0893Ctp-complexed HP0892 were used. The N-terminal residues Thr-3 to Asn-19, Asp-23, Lys-25, Leu-27 to Val-30, Gly-32, Leu-34, Thr-35, and Gln-38, and the middle range residues Leu-49, Lys-50, Leu-53, Lys-54, and Phe-56, and the C-terminal residues Leu-66, Ile-71, Lys-73, Glu-75, Thr-77, Phe-79 to Leu-83, Ser-85, Ser-87, and Phe-88 showed obvious chemical shift changes in the slow or intermediate exchange mode. These three residue groups in the HP0892 sequence could be mapped structurally into a nearly consecutive region on a three-dimensional HP0892 NMR structure (Fig. 6C). It is reasonably suggested that those residue groups form the binding site for HP0893. Overall, the chemical shift changes of the residues in the N-terminal secondary structure elements (one β-strand and two α-helixes in Leu-2 to Gln-38) were more substantial than those of the other residues (Fig. 6B), which indicates that this N-terminal region of HP0892 is the main region responsible for its binding with HP0893.
Oligomerization State of HP0892-HP0893 Complex
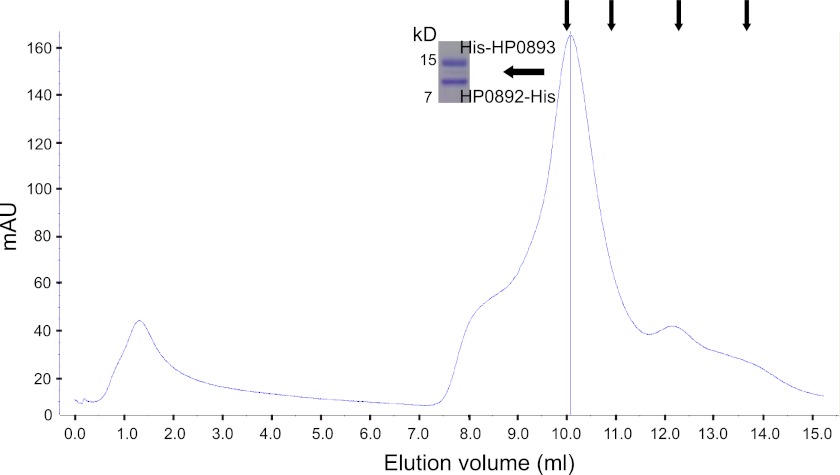
To evaluate the oligomerization state of the HP0892-HP0893 complex, gel filtration chromatography was performed on a mixture of the HP0892-His and His-HP0893 (nearly the same amount of HP0892-His and His-HP0893 was in the mixture) using 20 mm sodium phosphate buffer (pH 7.4) containing 100 mm NaCl as a mobile phase. As shown in Fig. 7, the HP0892-HP0893 (molar ratio 1:1) complex (HP0892-His, 11.48 kDa; His-HP0893, 13.52 kDa) eluted as a single peak with an apparent molecular mass of 73.69 kDa, which fit well with an oligomerization state of (HP0892-HP0893)3 heterohexamer (74.99 kDa).
FIGURE 7.
Gel filtration chromatogram of the HP0892-His/His-HP0893 mixture. Arrows above the chromatograms indicate the positions of the elution peaks of the molecular mass markers (conalbumin, 75 kDa; ovalbumin, 44 kDa; carbonic anhydrase, 29 kDa; RNase A, 13.7 kDa). SDS-PAGE of the elution fraction corresponding to the HP0892-HP0893 complex is shown beside the elution peak.
DISCUSSION
In our previous study, based on the structural and sequential similarities between H. pylori HP0892 and other RelE family toxin molecules such as E. coli YoeB, Pyrococcus horikoshii aRelE (the archaeal homologues of E. coli RelE), and H. pylori HP0894, we reported that there is a reasonable possibility that HP0892 may be a TA toxin. In TA systems, the antitoxin gene is usually located directly upstream of the toxin gene, and on the H. pylori chromosome, the hp0893 gene is located directly upstream of the hp0892 gene (Fig. 1) in an operon. Therefore, we expected that if HP0892 is a TA toxin, it would bind to HP0893. The 95-residue H. pylori-specific hypothetical protein HP0893 from H. pylori 26695 shares no detectable sequence similarity with other antitoxins. However, as revealed in the present study, HP0892 strongly binds to HP0893, with presumably a 1:1 binding stoichiometry. HP0892 has RNase activity on mRNA, and HP0893 inhibits the decay of mRNA by HP0892. Furthermore, His-86 of HP0892, which is homologous to key catalytic residues of His-87 of YafQ (19), His-83 of YoeB (18), Arg-81 of RelE (33), Arg-85 of aRelE (36), and His-84 of HP0894 (28), is also essential for its RNase activity. HP0892 expression has a toxic effect on E. coli cell growth, but the co-expression of HP0893 neutralizes the cell toxicity of HP0892. Especially, HP0892 shares detectable and relatively high sequence similarity with YafQ toxin among the three E. coli RelE family toxins (Fig. 8). These results, including sequential and structural analyses, suggest that HP0892 is a YafQ-homologous toxin with intrinsic RNase activity and HP0893 is the antitoxin against HP0892.
FIGURE 8.
Sequence homology of HP0892. A, multiple sequence alignments of H. pylori HP0892, H. pylori HP0894, and E. coli YafQ. B, average distance tree of H. pylori HP0892, H. pylori HP0894, E. coli YafQ, E. coli YoeB, and E. coli RelE. Sequence alignment was done with ClustalW (37) and colored according to Blosum scores. The figure was generated by JalView (38). The red rectangles in A indicate the residues forming putative catalytic triad of HP0892 and HP0894 (28) and the functionally important catalytic residues of YafQ revealed by a mutagenesis analysis (39).
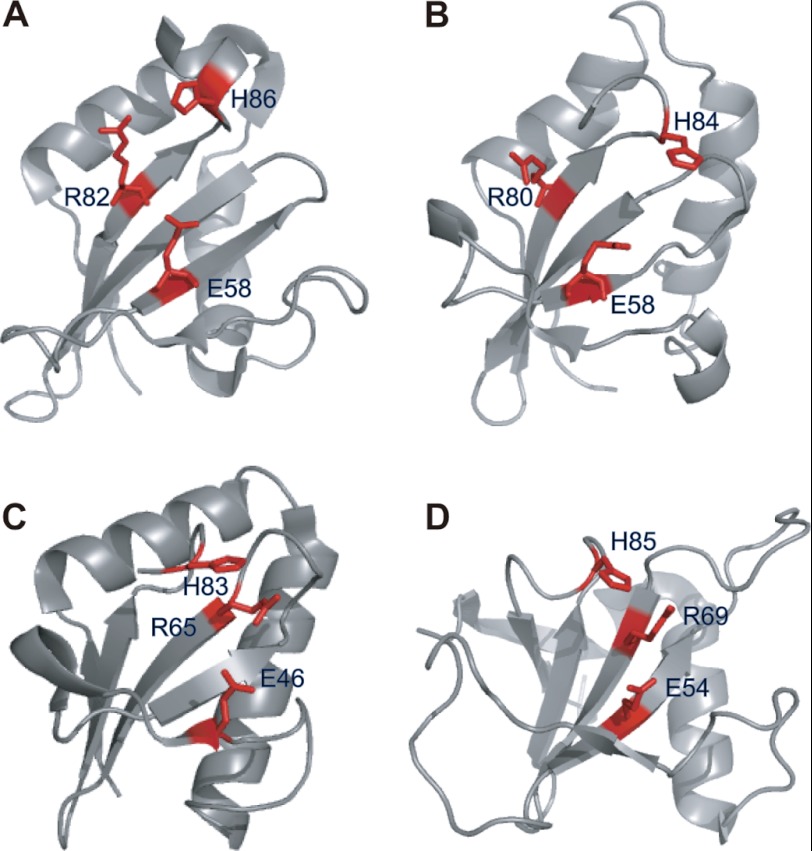
The comparison of HP0892 with its structural homologues harboring intrinsic RNase activity indicated that some of the key catalytic (or putative catalytic) residues involved in the RNase activity in RNase Sa, E. coli YoeB, and H. pylori HP0894 are conserved in HP0892 (Fig. 9). His-85 and Glu-54 of RNase Sa (42), His-83 and Glu-46 of YoeB (18), and His-84 and Glu-58 in HP0894 (28) are replaced with His-86 and Glu-58 in HP0892, respectively. However, Arg-69 of RNase Sa (42) and Arg-65 of YoeB (18), which form catalytic triads with the above two residues, respectively, are not conserved in HP0892 or HP0894 (28). Instead, Arg-82 of HP0892, like Arg-80 of HP0894 (28), seems to play the role corresponding to those of Arg-69 of RNase Sa or Arg-65 of YoeB.
FIGURE 9.
Comparison of structural and catalytic residues of HP0892 with those of its structural homologues. Ribbon displays of H. pylori HP0892 (PDB ID 2OTR) (29) (A); H. pylori HP0894 (PDB ID 1Z8M) (40) (B); E. coli YoeB (PDB ID 2A6R) (18) (C); and Streptomyces aureofaciens RNase Sa (PDB ID 1RGE) (41, 42) (D) are shown. The labeled functional or predicted key residues (see “Discussion”) are colored red. The structures were visualized by the program PyMOL (35).
By contrast, in the sequence homology analysis among HP0892, HP0894, and YafQ, the residues forming putative catalytic triad of HP0892 (Glu-58, Arg-82, and His-86) or HP0894 (Glu-58, Arg-80, and His-84) are relatively well conserved in YafQ (Asp-61, Arg-83, and His-87). However, whereas the Arg and His residues are absolutely conserved among them, the Glu residue, which seems to play the role of general base, of HP0892 or HP0894 is replaced with a slightly different residue, Asp-61 in YafQ. According to a mutagenesis study on E. coli YafQ toxin by Armalyate et al. (39), Asp-61 of YafQ plays a moderate role for its catalytic activity. On the contrary, Asp-67 and His-50 were shown to be critical for the catalytic activity of YafQ; thus, both or either of the two residues was proposed to play the role of general base instead of Asp-61. The above change of catalytic residues in YafQ compared with HP0892, HP0894, YoeB, and RNase Sa might be due to the lack of absolute conservation of Glu as the general base residue. In the study by Armalyate et al. (39), other catalytic residues of YafQ were also presented (Fig. 8). Overall, despite the small differences in local organization, the amino acid residues forming active site (catalytic residues and residues around them, some of which can take part in the recognition of specific RNA sequence (29)) of YafQ are well conserved in HP0892 and HP0894 (Fig. 8). Therefore, there is a high probability that HP0892 and YafQ share nearly the same functional mechanism. Moreover, HP0892, HP0894, and YafQ commonly display a preference for purine immediately downstream from the cleavage site when cleaving mRNAs in vitro (19, 28, 39), which can explain the result of the similar active site organizations among them. These similarities in the active site organization and the sequence specificity for RNA cleavage confirm that HP0892 is a type of YafQ-homologous toxin, similar to HP0894.
Our results show that the direct interaction of the HP0892-HP0893 TA pair occurs mainly between the 30-residue C-terminal tail of HP0893 and the N-terminal secondary structure elements and an adjacent C-terminal region of HP0892. In addition, some residues in the core region of HP0892 also take part in the interaction. Similar interaction modes are seen in H. pylori HP0894-HP0895, E. coli RelE-RelB, and YoeB-YefM complexes (18, 28, 33). Overall, anchoring of the C-terminal region of the antitoxin to the N-terminal secondary structure of the toxin (33) is the common critical factor for strong binding between the toxin and the antitoxin in RelE family TA complexes such as E. coli RelE-RelB, YoeB-YefM, H. pylori HP0894-HP0895, and HP0892-HP0893 complexes.
However, there is a difference in the oligomeric state between HP0892-HP0893 and other TA complex. In other TA complexes such as RelE-RelB (43), YoeB-YefM (18), YafQ-DinJ (44), or MazF-MazE (45) TA pairs in E. coli, the antitoxin molecules exist in dimeric form. The dimeric form of the antitoxins is related to the DNA binding mode of those TA complexes (36, 45). By contrast, in the HP0892-HP0893 complex, it seems that HP0893 antitoxin exists in trimeric form, and the overall complex has a heterohexameric arrangement ((HP0892-HP0893)3). Therefore, the HP0892-HP0893 TA complex can bind to their promoter region on the chromosomal DNA with a different mode than other TA complexes in the process of the negative autoregulation of the transcription of their genes.
According to a study by Graham et al. (46), in common with hp0895 (the antitoxin gene in the HP0894/HP0895 TA system) (28), hp0893 is also one of the H. pylori genomic open reading frames that correspond to genes that are potentially expressed in response to interactions with the human gastric mucosa. In addition, according to gene comparison studies by Terry et al. (47), both hp0892 and hp0893 belong to H. pylori genes absent from a set of five-cag pathogenicity island-negative strains, and may represent a marker for the identification of virulent strains or novel virulence factors. Interestingly, until now, there have been no other TA pairs in H. pylori 26695 except HP0892-HP0893 and HP0894-HP0895 TA pairs, which have been predicted from bioinformatics studies or identified experimentally (48). These indicate that the TA systems in H. pylori, especially the HP0892-HP0893 pair, are related to the status of infections of H. pylori in the human gastric mucosa, probably through negative regulation of the toxin molecules by the antitoxins.
Because TA systems exist only in prokaryote and are directly involved in cell death, TA systems are promising antibiotic drug targets. Thus, the HP0892-HP0893 TA pair in this study as well as the HP0894-HP0895 TA pair may be an appropriate new target for antibacterial agents for H. pylori. The information in this study on the binding aspect of these two proteins may be helpful in the design and development of new antibiotic drugs.
Acknowledgments
We thank the National Center for Inter-University Research Facilities at Seoul National University and Korea Institute of Science and Technology for providing high field NMR equipment.
This work was supported by National Research Foundation of Korea Grants 20110001207 and 2012R1A2A1A01003569 funded by the Korean Government (MEST); Korea Healthcare Technology R&D Project, Ministry for Health, Welfare, and Family Affairs, Republic of Korea Grants A084420 and A092006; 2011 BK21 Project for Medicine, Dentistry, and Pharmacy; and Korea Institute of Science and Technology Grant 2V02900.
- PCD
- programmed cell death
- PDB
- Protein Data Bank
- TA
- toxin-antitoxin
- trHNCA
- TROSY-type HNCA
- TROSY
- transverse relaxation optimized spectroscopy.
REFERENCES
- 1. Engelberg-Kulka H., Amitai S., Kolodkin-Gal I., Hazan R. (2006) Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2, e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kerr J. F., Wyllie A. H., Currie A. R. (1972) Apoptosis: a basic biological phenomenon with wide-ranging implication in tissue kinetics. Br. J. Cancer 26, 239–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gozuacik D., Kimchi A. (2007) Autophagy and cell death. Curr. Top. Dev. Biol. 78, 217–245 [DOI] [PubMed] [Google Scholar]
- 4. Zhou F., Yang Y., Xing D. (2011) Bcl-2 and Bcl-xL play important roles in the crosstalk between autophagy and apoptosis. FEBS J. 278, 403–413 [DOI] [PubMed] [Google Scholar]
- 5. Hu M. X., Zhang X., Li E. L., Feng Y. J. (2010) Recent advancements in toxin and antitoxin systems involved in bacterial programmed cell death. Int. J. Microbiol. 2010, 781430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yamaguchi Y., Inouye M. (2011) Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol. 9, 779–790 [DOI] [PubMed] [Google Scholar]
- 7. Gerdes K., Wagner E. G. (2007) RNA antitoxins. Curr. Opin. Microbiol. 10, 117–124 [DOI] [PubMed] [Google Scholar]
- 8. Gerdes K., Christensen S. K., Løbner-Olesen A. (2005) Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3, 371–382 [DOI] [PubMed] [Google Scholar]
- 9. Fineran P. C., Blower T. R., Foulds I. J., Humphreys D. P., Lilley K. S., Salmond G. P. (2009) The phage abortive infection system, toxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. U.S.A. 106, 894–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blower T. R., Fineran P. C., Johnson M. J., Toth I. K., Humphreys D. P., Salmond G. P. (2009) Mutagenesis and functional characterization of the RNA and protein components of the toxIN abortive infection and toxin-antitoxin locus of Erwinia. J. Bacteriol. 191, 6029–6039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Engelberg-Kulka H., Glaser G. (1999) Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53, 43–70 [DOI] [PubMed] [Google Scholar]
- 12. Magnuson R. D. (2007) Hypothetical functions of toxin-antitoxin systems. J. Bacteriol. 189, 6089–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maisonneuve E., Shakespeare L. J., Jørgensen M. G., Gerdes K. (2011) Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. U.S.A. 108, 13206–13211 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14. Lewis K. (2008) Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 322, 107–131 [DOI] [PubMed] [Google Scholar]
- 15. Neubauer C., Gao Y. G., Andersen K. R., Dunham C. M., Kelley A. C., Hentschel J., Gerdes K., Ramakrishnan V., Brodersen D. E. (2009) The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell 139, 1084–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Christensen S. K., Gerdes K. (2003) RelE toxins from bacteria and archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol. Microbiol., 48, 1389–1400 [DOI] [PubMed] [Google Scholar]
- 17. Pedersen K., Zavialov A. V., Pavlov M. Y., Elf J., Gerdes K., Ehrenberg M. (2003) The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112, 131–140 [DOI] [PubMed] [Google Scholar]
- 18. Kamada K., Hanaoka F. (2005) Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol. Cell 19, 497–509 [DOI] [PubMed] [Google Scholar]
- 19. Prysak M. H., Mozdzierz C. J., Cook A. M., Zhu L., Zhang Y., Inouye M., Woychik N. A. (2009) Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol. Microbiol. 71, 1071–1087 [DOI] [PubMed] [Google Scholar]
- 20. Zhang Y., Inouye M. (2009) The inhibitory mechanism of protein synthesis by YoeB, an Escherichia coli toxin. J. Biol. Chem. 284, 6627–6638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sachs G., Weeks D. L., Melchers K., Scott D. R. (2003) The gastric biology of Helicobacter pylori. Annu. Rev. Physiol. 65, 349–369 [DOI] [PubMed] [Google Scholar]
- 22. Scott D. R., Weeks D., Hong C., Postius S., Melchers K., Sachs G. (1998) The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology 114, 58–70 [DOI] [PubMed] [Google Scholar]
- 23. Cover T. L., Berg D. E., Blaser M. J., Mobley H. L. T. (2001) in Principles of Bacterial Pathogenesis (Eduardo A., ed) pp. 509–558, Academic Press, Waltham, MA [Google Scholar]
- 24. Matin A., Zychlinsky E., Keyhan M., Sachs G. (1996) Capacity of Helicobacter pylori to generate ionic gradients at low pH is similar to that of bacteria which grow under strongly acidic conditions. Infect. Immun. 64, 1434–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rothenbacher D., Brenner H. (2003) Burden of Helicobacter pylori and H. pylori-related diseases in developed countries: recent developments and future implications. Microbes Infect. 5, 693–703 [DOI] [PubMed] [Google Scholar]
- 26. Wotherspoon A. C., Doglioni C., Diss T. C., Pan L., Moschini A., de Boni M., Isaacson P. G. (1993) Regression of primary low-grade B-cell gastric lymphoma of mucosa-associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet 342, 575–577 [DOI] [PubMed] [Google Scholar]
- 27. Peek R. M., Jr., Blaser M. J. (2002) Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer 2, 28–37 [DOI] [PubMed] [Google Scholar]
- 28. Han K. D., Matsuura A., Ahn H. C., Kwon A. R., Min Y. H., Park H. J., Won H. S., Park S. J., Kim D. Y., Lee B. J. (2011) Functional identification of toxin-antitoxin molecules from Helicobacter pylori 26695 and structural elucidation of the molecular interactions. J. Biol. Chem. 11, 4842–4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Han K. D., Park S. J., Jang S. B., Lee B. J. (2008) Solution structure of conserved hypothetical protein HP0892 from Helicobacter pylori. Proteins 70, 599–602 [DOI] [PubMed] [Google Scholar]
- 30. Schumann F. H., Riepl H., Maurer T., Gronwald W., Neidig K. P., Kalbitzer H. R. (2007) Combined chemical shift changes and amino acid specific chemical shift mapping of protein-protein interactions. J. Biomol. NMR 39, 275–289 [DOI] [PubMed] [Google Scholar]
- 31. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 32. Johnson B. A., Blevins R. A. (1994) NMR view: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 33. Li G. Y., Zhang Y., Inouye M., Ikura M. (2009) Inhibitory mechanism of Escherichia coli RelE-RelB toxin-antitoxin module involves a helix displacement near an mRNA interferase active site. J. Biol. Chem. 284, 14628–14636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pellecchia M., Sem D. S., Wüthrich K. (2002) NMR in drug discovery. Nat. Rev. Drug Discov. 1, 211–219 [DOI] [PubMed] [Google Scholar]
- 35. DeLano W. L. (2002) DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 36. Takagi H., Kakuta Y., Okada T., Yao M., Tanaka I., Kimura M. (2005) Crystal structure of archaeal toxin-antitoxin RelE-RelB complex with implications for toxin activity and antitoxin effects. Nat. Struct. Mol. Biol. 12, 327–331 [DOI] [PubMed] [Google Scholar]
- 37. Thompson J. D., Higgins D. G., Gibson T. J. (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Clamp M., Cuff J., Searle S. M., Barton G. J. (2004) The Jalview Java alignment editor. Bioinformatics 20, 426–427 [DOI] [PubMed] [Google Scholar]
- 39. Armalyte J., Jurenaite M., Beinoraviciūte G., Teiserskas J., Suziedeliene E. (2012) Characterization of Escherichia coli dinJ-yafQ toxin-antitoxin system using insights from mutagenesis data. J. Bacteriol. 194, 1523–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Han K. D., Park S. J., Jang S. B., Son W. S., Lee B. J. (2005) Solution structure of conserved hypothetical protein HP0894 from Helicobacter pylori. Proteins 61, 1114–1116 [DOI] [PubMed] [Google Scholar]
- 41. Sevcik J., Dauter Z., Lamzin V. S., Wilson K. S. (1996) Ribonuclease from Streptomyces aureofaciens at atomic resolution. Acta Crystallogr. D Biol. Crystallogr. 52, 327–344 [DOI] [PubMed] [Google Scholar]
- 42. Yakovlev G. I., Mitkevich V. A., Shaw K. L., Trevino S., Newsom S., Pace C. N., Makarov A. A. (2003) Contribution of active site residues to the activity and thermal stability of ribonuclease Sa. Protein Sci. 12, 2367–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li G. Y., Zhang Y., Inouye M., Ikura M. (2008) Structural mechanism of transcriptional autorepression of the Escherichia coli RelB/RelE antitoxin/toxin module. J. Mol. Biol. 380, 107–119 [DOI] [PubMed] [Google Scholar]
- 44. Motiejūnaite R., Armalyte J., Markuckas A., Suziedeliene E. (2007) Escherichia coli dinJ-yafQ genes act as a toxin-antitoxin module. FEMS Microbiol. Lett. 268, 112–119 [DOI] [PubMed] [Google Scholar]
- 45. Kamada K., Hanaoka F., Burley S. K. (2003) Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol. Cell 11, 875–884 [DOI] [PubMed] [Google Scholar]
- 46. Graham J. E., Peek R. M., Jr., Krishna U., Cover T. L. (2002) Global analysis of Helicobacter pylori gene expression in human gastric mucosa. Gastroenterology 123, 1637–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Terry C. E., McGinnis L. M., Madigan K. C., Cao P., Cover T. L., Liechti G. W., Peek R. M., Jr., Forsyth M. H. (2005) Genomic comparison of cag pathogenicity island (PAI)-positive and -negative Helicobacter pylori strains: identification of novel markers for cag PAI-positive strains. Infect. Immun. 73, 3794–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shao Y., Harrison E. M., Bi D., Tai C., He X., Ou H. Y., Rajakumar K., Deng Z. (2011) TADB: a web-based resource for type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 39, D606–611 [DOI] [PMC free article] [PubMed] [Google Scholar]