Background: A tetrameric branched peptide (BP) containing a TFLK motif was designed to induce MUC3 expression and inhibit bacteria adherence.
Results: TFLK-containing 10-mer BP induced MUC3 expression and dramatically inhibited bacteria adherence to the HT-29-Gal cells.
Conclusion: This peptide acts through transcriptional mechanisms.
Significance: This peptide has potential therapeutic value against gastrointestinal infection.
Keywords: Bacterial Adhesion, Mucins, Peptides, Transcription Factors, Transcriptomics, Branched Peptide, MUC3, Bacteria Adherence, Mucin
Abstract
We investigated whether a synthetic tetrameric branched peptide based on the conserved TFLK motif from mammary-associated serum amyloid A3 (M-SAA3) is more efficient than the monomeric peptide at up-regulating MUC3 expression and examined the possible mechanism(s) and biological significance of this process. We used standard solid-phase methods to synthesize a tetrameric branched peptide (sequence GWLTFLKAAG) containing a trilysine core, termed the TFLK-containing 10-mer BP. The aberrant expression of transcription factors was analyzed using a transcription factor protein/DNA array. MUC3 and relevant transcription factors were detected using real-time PCR and/or Western blots. The luciferase assay, EMSA, and ChIP assays were used to analyze the activity of the human MUC3 promoter. The bacterial adherence assay was used to evaluate the in vitro inhibition of enteropathogenic Escherichia coli or enterohemorrhage E. coli serotype O157:H7 (EHEC O157:H7) adherence to HT-29-Gal cells after treatment with the TFLK-containing 10-mer BP. In HT-29-Gal cells, the TFLK-containing 10-mer BP induced higher levels of MUC3 expression than the M-SAA3-derived N-terminal 10-mer monomeric peptide, and MUC3 expression was activated through transcriptional mechanisms, including the induction of multiple transcription factors and further binding with their cis-elements between nucleotides −242 and −62 within MUC3 promoter. Interestingly, the TFLK-containing 10-mer BP dramatically inhibited enteropathogenic E. coli and EHEC O157:H7 adherence to the HT-29-Gal cells compared with the controls. This finding suggests a potential therapeutic use for this peptide to prevent gastrointestinal infection.
Introduction
Mucosal surfaces are protected against xenobiotics and pathogenic microorganisms by a variety of innate and adaptive mechanisms. For example, mucins prevent the noxious interaction of the epithelial cells with microbial pathogens and toxic chemicals by providing a physicochemical barrier through specific mucin-microorganism interactions and cell-signaling regulation (1, 2). By limiting the adherence or invasion of microorganisms to the epithelial cell surface, mucin effectively hampers the ability of the bacteria and viruses to colonize and invade the cells, blocks the spread of the pathogens along the mucosal surfaces, and limits the amount of microbial-produced toxins that reaches the mucosal cells (3). Intestinal mucins provide protection against the adherence of enteric pathogens by steric hindrance or through specific bacterial or viral binding domains. The attachment of microbes to intestinal mucins depends on the composition and quantity of mucins expressed and secreted and on the motility and fluid flow of the intestinal tract (4). If the rate of pathogenic microbial adherence and colonization exceeds the rate of removal, disease is a likely result. These events contribute to the development of localized intestinal necrosis that promotes further growth and the translocation of pathogenic microbes that can lead to shock, sepsis, and sometimes death. Human MUC3 and rodent Muc3 are membrane-bound mucins that are moderately expressed in the colon but are found abundantly in both goblet cells and enterocytes in the small intestine (5). MUC3 binds numerous enteropathogenic bacteria and viruses and prevents their attachment to the intestinal cell surfaces (4). Recombinant Muc3 and recombinant human MUC17 mucins stimulate cell migration in human colon cell lines and protect cells from apoptosis (6, 7). Although known inducers of MUC3 expression are limited, a previous in vitro study demonstrated that co-incubation of the probiotic bacteria Lactobacillus spp. with human intestinal epithelial cells induced MUC3 expression and inhibited enteropathogenic Escherichia coli (EPEC)3 adherence (8, 9).
Numerous studies have shown that compared with formula-fed infants, infants fed colostrum are ∼7–10 times less likely to suffer gastrointestinal diseases (10, 11). Colostrum is a complex source of nutrients, immune factors, and bioactive substances that facilitates the successful transition of the mammalian neonate to extrauterine life (12, 13). However, despite the considerable number of studies investigating the stimulatory and inhibitory effects of colostrum, the numerous colostrum components and the underlying mechanisms that benefit the neonate are unknown.
In vitro studies demonstrated that the human mammary-associated serum amyloid A3 (M-SAA3)-derived N-terminal peptide increased MUC3 transcriptional expression in human intestinal epithelial cells and decreased EPEC adherence to these cells (14–16). M-SAA3 is secreted at highly elevated levels in colostrum. Each of these colostrum-derived M-SAA3 proteins contains a conserved TFLK motif positioned within the first eight residues of the N terminus regardless of the species. This TFLK motif has not been reported for any serum-derived serum amyloid A isoform in mammals and appears to be in part responsible for the bioactivity that results in the up-regulation of MUC3 transcriptional expression by human intestinal cells. The discovery of elevated M-SAA3 levels in the colostrum and the reduction of gastrointestinal diseases in infants fed colostrum implies a possible role for M-SAA3 in promoting the health of the neonate.
Recently, multimeric peptides have been described in the literature as an obvious solution to increase peptide size and affinity while maintaining the original amino acid sequence (17–19), and this approach is a key strategy to increase recognition surfaces and/or stability. Increasing the peptide size also increases the difficulty of identifying peptides with high affinity and selectivity; however, multimeric peptides comprising simple branched structures and a short arm may offer an optimal compromise between molecular size and the ease of preparation, yield, and purity of the final product. This strategy of combinatorial chemistry prompted us to explore whether the multimeric branched peptide based on the conserved TFLK motif from M-SAA3 induces increased expression of MUC3 in intestinal epithelial cells and exerted a more significant effect.
In this study we demonstrate that compared with the M-SAA3-derived N-terminal 10-mer monomeric peptide containing the TFLK motif (residues 1–10, sequence GWLTFLKAAG), the TFLK-containing 10-mer BP stimulates higher levels of human MUC3 expression in intestinal epithelial cells and dramatically inhibits the adherence of EPEC and EHEC O157:H7. Furthermore, we explored the mechanism(s) of transcriptional regulation of human MUC3 mucin by the TFLK-containing 10-mer BP in HT-29-Gal cells.
EXPERIMENTAL PROCEDURES
Peptide Synthesis
In this study the 10-mer branched peptide based on the conserved TFLK motif from M-SAA3 (referred as TFLK-containing 10-mer BP) was synthesized using the solid phase method at the Chinese Peptide Company (Hangzhou, China). The introduction of three symmetrically protected lysines into the peptide produces a trilysine core that is required for producing tetramerically branched peptides. This method utilizes standard solid-phase methods and an Applied Biosystems (Foster City, CA) Model 430A synthesizer. The syntheses were performed on a 0.25-mmol scale on a Wang resin using the Fmoc (N-(9-fluoenyl) methoxycarbonyl) synthetic scheme. The peptides were purified using analytical and preparative HPLC columns packed with C18-bonded silica gel and were characterized using amino acid compositional analysis and mass spectrometry. The M-SAA3-derived N-terminal 10-mer monomeric peptide was synthesized simultaneously and used as a control.
Cell Line and Culture
To examine MUC3 expression, the human intestinal epithelial cell line HT-29 (ATCC) was cultured in glucose-free McCoy's 5a medium (Invitrogen). The culture medium was supplemented with 5 mm galactose, 10% heat-inactivated qualified FBS, 100 units/ml penicillin G, 100 mg/ml streptomycin sulfate, and 0.25 mg/ml amphotericin B (Invitrogen). The glucose-free and galactose-substituted cell culture medium induces the differentiation of the HT-29 cells, referred to as HT-29-Gal cells (a mature and differentiated form of the HT-29 cell line) (20, 21). The synthetic branched peptide was added to the HT-29-Gal cells in culture for 0.5, 1, or 3 h. The cultures were incubated at 37 °C in a humidified atmosphere with 5% CO2. The cells were passaged after the release of the adherent cells using trypsin-EDTA (Invitrogen).
Real-time PCR Analysis
The HT-29 cells were grown to 85% confluency in culture flasks containing either glucose-containing culture medium or the glucose-free galactose-containing medium. The synthetic branched peptides were added to the culture medium and incubated for 1 h at 37 °C in a humidified atmosphere with 5% CO2. The total RNA was isolated from the cells using TRIzol (TaKaRa Biotechnology Co. Ltd., Dalian, China). The RNA was subjected to real-time PCR or was stored at 70 °C. The mRNA was reversed-transcribed into cDNA using the PrimeScript RT reagent kit (TaKaRa Biotechnology Co. Ltd., Dalian, China). To determine the -fold changes in the expression of each gene, real-time PCR was performed using the first strand cDNA, the forward and reverse primers (supplemental Table 1), and the SYBR premix Ex TaqTM GreenII (Takara, Japan). The alteration of the MUC3 mRNA was quantified using real time PCR (CFX 96, Bio-Rad). The reaction and signal detection were measured using the Bio-Rad CFX manager software (Bio-Rad). The expression levels were calculated as the relative expression ratio compared with the expression levels of the control β-actin and GAPDH genes. The real-time PCRs were performed independently in triplicate.
Western Blotting
The cell pellets or nuclear proteins were extracted using NE-PER nuclear and cytoplasmic extraction reagents (Thermo Scientific Pierce Protein Research Products), lysed in SDS sample buffer, and subjected to SDS/PAGE (7.5 or 8% gels). Next, the proteins were transferred to PVDF membranes (Millipore, Bedford, MA). The membranes were blocked for 1 h in 5% nonfat dry milk in TBST (TBS containing 0.05% Tween 20) and hybridized at 4 °C overnight in TBST containing the following primary antibodies: anti-MUC3 (1:6000, Abcam, Cambridge, UK), anti-Sp1 (1:1500), anti-CDX2 (1:1500), anti-CREB1 (1:1000), or anti-USF1 (1:1500) (the anti-Sp1, anti-CDX2, anti-CREB1, and anti-USF1 antibodies were from Santa Cruz Biotechnology Inc). Lamin B1 and GAPDH were detected using the anti-lamin B1 (1:5000) and anti-GAPDH antibodies (1:5000) (Santa Cruz Biotechnology Inc.) and were used as the internal controls. The membranes were washed 3 times in TBST for 5 min each, and the secondary detection was performed using 1:50,000 dilutions of HRP-conjugated goat anti-rabbit or goat anti-mouse antibodies. The membranes were washed 3 times for 10 min each. The blots were processed using the SuperWest Pico chemiluminescent reagent (Pierce) and analyzed using ChemiDoxTM XRS+ with Image LabTM software.
Transfections and Luciferase Assays
Fragments of the MUC3 5′-flanking sequence were amplified using PCR and cloned into the luciferase reporter vector, pGL3-Basic (Promega, Madison, WI). Briefly, primers with containing XhoI and HindIII adapters were used to amplify the MUC3 promoter sequence from intestinal tissue DNA. The primer pairs used to produce each promoter fragment are listed in supplemental Table 2. The products were ligated into the pGL3-Basic vector that was digested with XhoI and HindIII. The 5′ serial deletions of the 1499-bp MUC3 promoter region were generated using the Erase-ABase system (Promega) in accordance with the manufacturer's recommendations. The plasmids for the transient transfections were purified using the EndoFree Plasmid Maxi kit (Qiagen, Valencia, CA). The day before the transfection, the cells were plated on 24-well plates at a density of 5 × 104 cells per well. The MUC3 promoter-luciferase constructs were transfected into the cells using LipofectamineTM 2000 (Invitrogen). To normalize for the transfection efficiency, the cells were simultaneously cotransfected with a pRL-TK vector expressing the Renilla luciferase enzyme (pRL, Promega). The cells were harvested after 24 h, and the luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) using a single sample luminometer. The cells were also transfected with the pRL-TK vector, and the MUC3 activity is presented as the percentage of the pGL3-control activity. For the synthetic branched peptide stimulation, the cells were exposed to the peptide at a concentration of 50 μg/ml for 1 h, 12 h after transfection, and the stimulation was repeated three times. The differences between the untreated cells and the synthetic branched peptide-treated cells were determined using Student's t test.
Transcription Factor (TF) Protein/DNA Array
The protein/DNA array (Combo Array, Panomics/Affymetrix) was performed at the Shanghai KANCHENG Biochip Company (Shanghai, China) in accordance with the protocols recommended for the protein/DNA array system. The nuclear protein extracts were semi-quantitatively assayed by measuring the DNA binding activity of 345 TFs. The nuclear protein extracts were prepared from the HT-29-Gal cells, and the HT-29-Gal cells were treated with the TFLK-containing 10-mer BP as previously described (22). Briefly, 5 μg of nuclear protein pooled from four HT-29-Gal cell extracts or from four TFLK-containing 10-mer BP-treated HT-29-Gal cell extracts were used for binding to a mixture of biotin-labeled TF-specific DNA probes in solution. The unbound DNA probes were washed away, and the TF-bound probes were denatured and hybridized to the array membrane. After the addition of streptavidin-HRP, signals were generated using enhanced chemiluminescence (ECL) and exposure to Hyperfilm ECL (Amersham Biosciences). Various exposure times were used to generate signals over a large dynamic range. The signal intensities were quantified using a Bio-Rad calibrated GS-800 scanner and the Quantity One software. Only the non-saturated signals were used for further analysis. The TF binding activity was considered significant when at least a 2-fold signal difference was observed between the HT-29-Gal cells and the HT-29-Gal cells treated with the TFLK-containing 10-mer BP.
Electrophoretic Mobility-shift Assay (EMSA)
The nuclear protein extract was prepared from the HT-29-Gal cell line. The following double-stranded DNA probes were used: wild-type Sp1 consensus oligonucleotide (5′-ATTCGATCGGGGCGGGGCGAGC-3′), wild-type CREB1 consensus oligonucleotide (5′-AGAGATTGCCTGACGTCAGAGAGCTAG-3′), wild-type USF-1 consensus oligonucleotide (5′-CACCCGGTCACGTGGCCTACACC-3′) (positive control), mutated Sp1 oligonucleotide (5′-ATTCGATCGGTTCGGGGCGAGC-3′), mutated CREB1 oligonucleotide (5′-AGAGATTGCCTGTGGTCAGAGAGCTAG-3′), and the mutated USF-1 oligonucleotide (5′-CACCCGGTCAATTGGCCTACACC-3′). The probes were purchased from Santa Cruz Biotechnology Inc. An isotopic EMSA was performed as described previously (23) using 5 μg of the nuclear cell extracts, 50 ng of the probe, and 1 μg of salmon testes DNA (Sigma) in a 5 × binding reaction buffer (100 mm Hepes, pH 7.9, 250 mm KCl, 2.5 mm DTT, 0.25 mm EDTA, 5 mm MgCl2, and 25% glycerol). After 20 min, the samples were separated on a 10% polyacrylamide gel (39:1 polyacrylamide:bisacrylamide) at a constant temperature of 4 °C in 1 × Tris borate-EDTA buffer (45 mA for 4.5 h). Similarly, for the supershift assay, 8 ng/μl specific antibody (anti-Sp1, anti-CREB1, or anti-USF-1) or the control antibody (anti-ER) was incubated for 20 min in the HT-29-Gal cell nuclear protein extracts before the addition of the probes. The samples were size-separated by electrophoresis on a 10% polyacrylamide minigel (39:1 polyacrylamide:bisacrylamide) at room temperature in 0.5 × Tris borate-EDTA buffer.
Chromatin Immunoprecipitation Assay (ChIP)
ChIP assays were performed using a ChIP assay kit according to the manufacturer's instructions (Upstate Biotechnology, Lake Placid, NY). Soluble chromatin was prepared from HT-29-Gal cells treated with or without TFLK-containing 10-mer BP. Chromatin was immunoprecipitated with antibodies against CDX2, Sp1, or CREB1. The final DNA extracts were amplified by PCR using primer pairs that included a CDX2, Sp1, or CREB1 consensus sequence in the human MUC3 promoter (supplemental Fig. 1). The primer sequences and the amplified PCR products that included CDX2 (ChIP1 and ChIP2), CREB1 (ChIP3), and or Sp1 (ChIP4) consensus sequence in the MUC3 promoter were presented in supplemental Table 3.
Bacterial Strains, Growth Conditions, and Bacterial Adherence Assay
For the in vitro inhibition of EPEC or EHEC O157:H7 (purchased from the Chinese Center for Food Safety Risk Assessment, Beijing, China) adherence to the HT-29 intestinal cells and the HT-29-Gal cells, the cells were grown to near confluence in glucose-free, galactose-containing McCoy's 5a culture medium in 12-well tissue culture plates. The cells were washed, and the antibiotic-free culture medium containing the M-SAA3-derived N-terminal 10-mer monomeric peptide (50 or 200 μg/ml), the TFLK-containing 10-mer BP (50 or 200 μg/ml), or no peptide was added 1 h before incubation with the bacteria. After the 1-h incubation with or without stimulation with the peptides, the EPEC or EHEC O157:H7 bacteria (106 colony-forming units in 0.1 ml of PBS, pH 7.4) were added to each well, and the cells were incubated for an additional 3 h at 37 °C. The cells were washed with chilled PBS 4 times for 5 min each to remove the non-adherent EPEC or EHEC O157:H7 bacteria and to inhibit further growth of the adhering bacteria. The cells with adherent bacteria were released from the polystyrene wells using trypsin-EDTA. The bacteria were quantified by determining the colony-forming units after the plating of serial dilutions of the bacteria on MacConkey agar (BD Biosciences) and incubation overnight at 37 °C. The number of viable EPEC or EHEC O157:H7 bacteria that remained on the TFLK-containing 10-mer BP-treated HT-29-Gal cells was compared with the number of bacteria adhered to the HT-29-Gal cells not exposed to peptides or exposed to the M-SAA3-derived N-terminal 10-mer monomeric peptide and HT-29 cells. The individual experiments were performed in triplicate, and each experiment was repeated at least twice.
RESULTS
Schematic Representation of TFLK-containing 10-Mer BP and M-SAA3-derived N-terminal 10-mer Monomeric Peptide
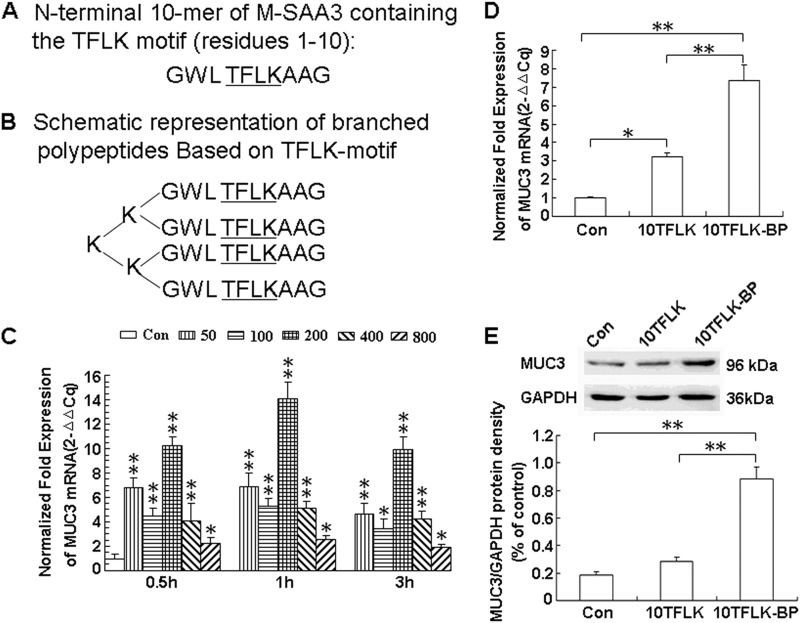
The M-SAA3-derived N-terminal 10-mer monomeric peptide (residues 1–10, sequence GWLTFLKAAG) is shown in Fig. 1A, and a schematic representation of the tetramerically branched polypeptide based on the conserved TFLK motif from M-SAA3 (TFLK-containing 10-mer BP) is shown in Fig. 1B. The four amino acids (the TFLK motif) are underlined and are conserved within the first eight N-terminal amino acid residues of M-SAA3.
FIGURE 1.
Shown are schematic representations of the M-SAA3-derived N-terminal 10-mer monomeric peptide (A) and the branched polypeptides based on the TFLK motif (B) and their induction of MUC3 expression in HT-29-Gal cells. TFLK-containing 10-mer BP increased MUC3 mRNA expression levels in HT-29-Gal cells dose-dependently compared with non-treated HT-29-Gal cells (C). TFLK-containing 10-mer BP (10TFLK-BP) (200 μg/ml) induced MUC3 mRNA expression levels in HT-29-Gal cells at significantly higher levels than in untreated HT-29-Gal cells (p < 0.01) and in HT-29-Gal cells pretreated with the M-SAA3-derived N-terminal 10-mer monomeric peptide (10TFLK) (D) (p < 0.01). Western blot experiments demonstrated similar results for MUC3 protein levels, the anti-MUC3 antibody used in the study recognized the epitope that was located at the C-terminal domain of human MUC3 and just after the proteolytic cleavage site (LRNGSIVV) and can only recognize the cleaved C-terminal fragment of human MUC3, so the molecular weight of MUC3 protein by Western blot was in 96 kDa (E) (p < 0.01). The M-SAA3-derived N-terminal 10-mer monomeric peptide (10TFLK) did not increase MUC3 protein levels compared with untreated HT-29-Gal cells (p > 0.05) (E); real-time PCR experiments, however, demonstrated a significant difference between these samples (p < 0.05) (D). * represents a significant difference (p < 0.05); ** represents a significant difference (p < 0.01).
TFLK-containing 10-Mer-BP Enhances MUC3 Expression in HT-29-Gal Cells
To determine the ideal concentration of the TFLK-containing 10-mer BP for the activation of MUC3 expression in the HT-29-Gal cells, we analyzed the MUC3 mRNA levels by real-time PCR in the HT-29-Gal cells stimulated with different concentrations of the TFLK-containing 10-mer BP (50, 100, 200, 400, and 800 μg/ml) and compared it to the untreated HT-29-Gal cells at three time points (0.5, 1, and 3 h). We found that 200 μg/ml TFLK-containing 10-mer BP produced the most efficient stimulation of MUC3 mRNA expression at the 1-h point (p < 0.01) (Fig. 1C).
As shown in Fig. 1D, the MUC3 mRNA levels in the HT-29-Gal cells after stimulation with the TFLK-containing 10-mer BP for 1 h (200 μg/ml) was significantly higher than the untreated HT-29-Gal cells (p < 0.01) and the HT-29-Gal cells stimulated with the M-SAA3-derived N-terminal 10-mer monomeric peptide (200 μg/ml) (p < 0.01). The MUC3 protein level in HT-29-Gal cells after stimulation with the TFLK-containing 10-mer BP was significantly higher than in the untreated HT-29-Gal cells and the HT-29-Gal cells after stimulation with the M-SAA3-derived N-terminal 10-mer monomeric peptide (p < 0.01) (Fig. 1E). There was no significant difference in the MUC3 protein levels between the untreated HT-29-Gal cells and the HT-29-Gal cells stimulated with the M-SAA3-derived N-terminal 10-mer monomeric peptide (p > 0.05) (Fig. 1E).
Characterization of the Human MUC3 Promoter
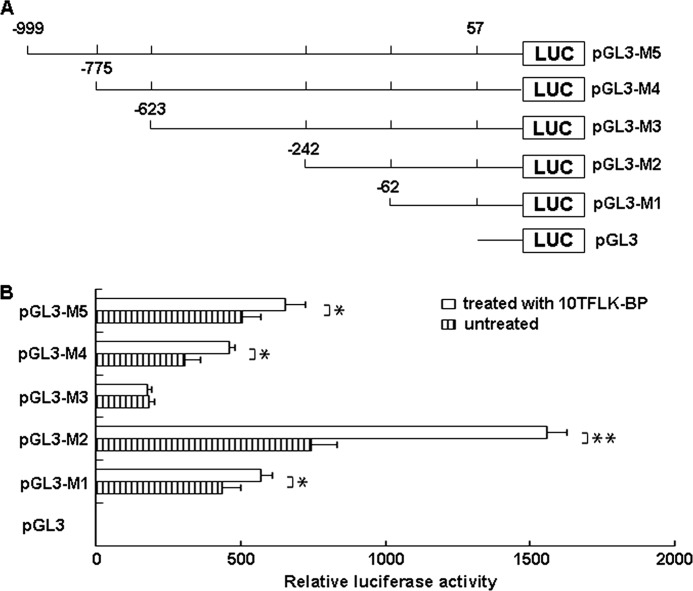
As shown in Fig. 2, the human MUC3 promoter region produced a high luciferase activity in the HT-29-Gal cells in the presence and absence of the TFLK-containing 10-mer BP. The data indicated that this region of the human MUC3 promoter (from nucleotides −999 to +57) contains the regulatory elements responsible for the expression of human MUC3. To locate the specific elements involved in MUC3 expression, we generated the following series of truncations of the promoter region: pGL3-M4 (−775 to +57), pGL3-M3 (−623 to +57), pGL2-M2 (−242 to +57), and pGL3-M1 (−62 to +57) (Fig. 2A). Truncation of the upstream region to within 242 bp of the putative transcription start sites (TSS) produced the strongest effect on the luciferase activity; however, further truncation of the upstream region to within 62 bp of the TSS resulted in a significant loss of activity, indicating that multiple elements between nucleotides −242 and −62 contribute to the activity of the promoter (Fig. 2B). The truncation of the upstream region to within 999, 775, and 623 bp of the TSS produced a modest effect on the promoter activity compared with truncation of the upstream region to within 242 bp of the TSS. Therefore, multiple elements between nucleotides −999 and −242 exerted an inhibitory effect on the activity of the promoter (Fig. 2B).
FIGURE 2.
Transcriptional regulation of human MUC3 by the TFLK-containing 10-mer BP. A, shown is a schematic representation of the human MUC3 promoter constructs. The five deletion luciferase constructs that were generated to identify the sites of transcriptional regulation within the MUC3 promoter responded to the TFLK-containing 10-mer BP (10TFLK-BP). B, HT-29-Gal cells were transfected with the different luciferase constructs and treated or not treated with the TFLK-containing 10-mer BP. The cells were harvested after 48 h, and relative luciferase activity was determined using the Dual-Luciferase Reporter Assay System on a single sample luminometer. The data represent the mean ± S.E. of three independent experiments. * represents a significant difference (p < 0.05); ** represents a significant difference (p < 0.01).
TFLK-containing 10-mer BP Transcriptionally Activated the MUC3 Promoter in HT-29-Gal Cells
The TFLK-containing 10-mer BP had a significant effect on increasing the luciferase activity in HT-29-Gal cells for the promoter truncations pGL3-M1 (p < 0.05), pGL3-M2 (p < 0.01), pGL3-M4 (p < 0.05), and pGL3-M5 (p < 0.05) (Fig. 2B). The data indicated that multiple elements between −999 and −775, −775 and −623, −242 and −62, and −62 and +57 contributed to the activity of the human MUC3 promoter when stimulated with TFLK-containing 10-mer BP in the HT-29-Gal cells.
Transcriptomic Responses to the TFLK-containing 10-Mer BP
The Protein/DNA Combo array analyzing the transcriptomic changes in the HT-29-Gal cells in response to the TFLK-containing 10-mer BP indicated the transcription factors (81 in total) that were up-regulated 2.0-fold in the TFLK-containing 10-mer BP-treated HT-29-Gal cells versus the untreated HT-29-Gal cells are shown in supplemental Table 4. The transcription factors (12 in total) that were down-regulated 2.0-fold in the TFLK-containing 10-mer BP-treated HT-29-Gal cells versus the untreated HT-29-Gal cells are shown in supplemental Table 5. The 11 transcription factors, AP-1, GAS/ISRE, Pbx1, AP-2, GATA, CREB1, USF-1, RIPE3a1, Pax3, Sp1 and PRDII-BF1, were up-regulated 10-fold.
Analysis of MUC3 Promoter Indicating That Sp1, CREB1, and CDX2 Might Be Responsible for the Up-regulation of MUC3
The genomic region flanking the MUC3 promoter (from −999 to +57) containing the consensus sequences for possible TF-binding sites predicted using TFSEARCH (Version 1.3) are depicted in supplemental Fig. 1. These elements, including CREB1, Nkx-2, CDX2, SRY, δE, HSF2, C/EBP, GATA, Sox5, HFH-2, AML-1a, CDXA, c-Re1, IK-2/3, HNF-3b, Oct-1, MZF-1, Tst-1, N-Myc, USF, and Sp1, are present in the promoter region (from −999 to +57) of the human MUC3 gene.
We focused on the Sp1, CREB1, USF1, and CDX2 elements for because the Sp1, CREB1, and USF1 cis-elements are located between −242 and −64 of the TSS of MUC3 and were significantly induced after stimulation with the TFLK-containing 10-mer BP and because CDX2 is critical for the development of the intestinal epithelium (24–27).
TFLK-containing 10-mer BP Activates CDX2, CREB1, Sp1, and USF1 in HT-29-Gal Cells Accompanied by the Up-regulation of MUC3
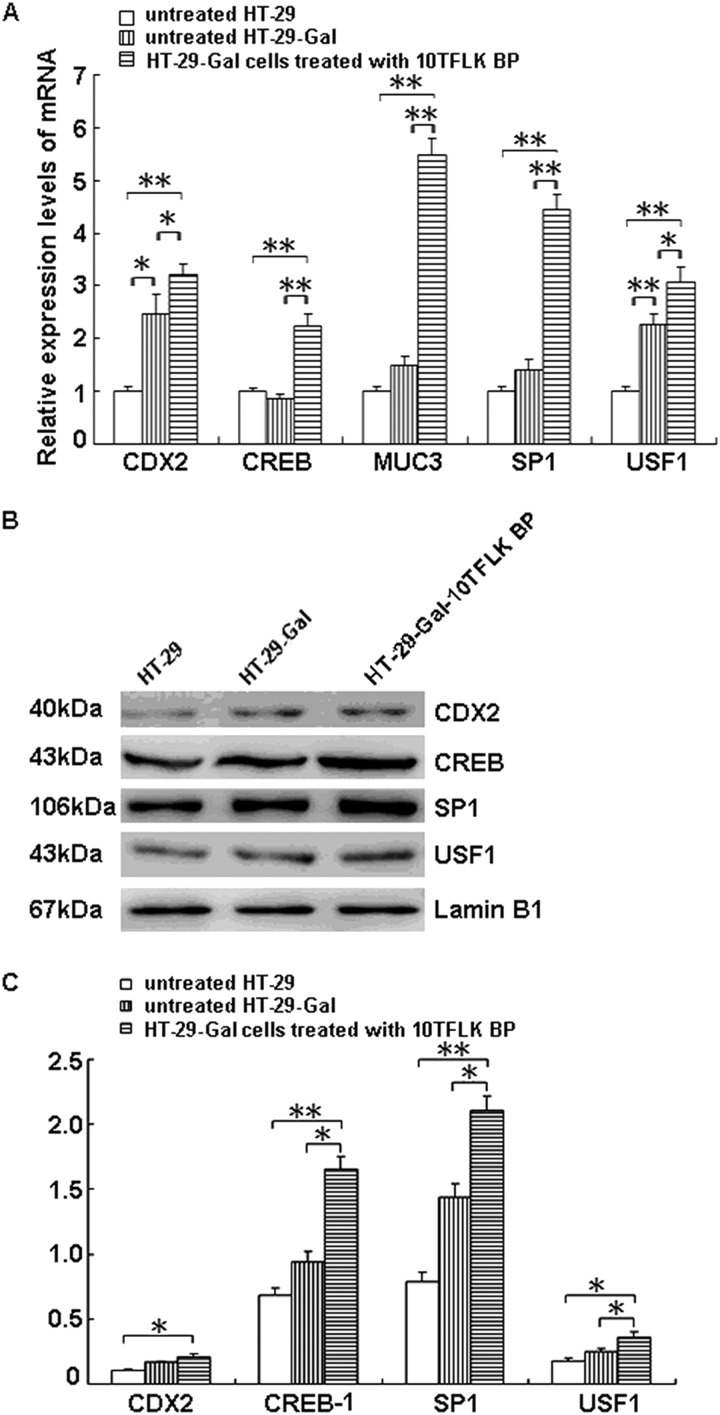
The mRNA levels of CDX2, CREB1, Sp1, and USF1 quantified by real-time PCR analysis were significantly increased (3.2-, 2.3-, 4.4-, and 3.0-fold, respectively) in the HT-29-Gal cells after stimulation with the TFLK-containing 10-mer BP compared with the untreated HT-29-Gal cells (p < 0.05) in concordance with the increased MUC3 mRNA level (5.5-fold) (p < 0.05) (Fig. 3A). The Western blots detected significant increases in the CDX2, CREB1, Sp1, and USF1 nuclear protein levels in the HT-29-Gal cells after stimulation with the TFLK-containing 10-mer BP compared with the untreated HT-29 cells (p < 0.05 or p < 0.01) and the untreated HT-29-Gal cells (p < 0.05) (Fig. 3, B and C).
FIGURE 3.
Expression of multiple transcription factors after stimulation with the TFLK-containing 10-mer BP. The mRNA levels of CDX2, CREB1, SP1, USF1, and MUC3 in HT-29-Gal cells treated with the TFLK-containing 10-mer BP (10TFLK BP) (200 μg/ml), untreated HT-29-Gal cells, and untreated HT-29 cells were determined using real-time PCR (A). The protein levels of CDX2, CREB1, SP1, and USF1 in HT-29-Gal cells treated with the TFLK-containing 10-mer BP (10TFLK BP) (200 μg/ml), untreated HT-29-Gal cells, and untreated HT-29 cells were determined using Western blotting (B). The protein levels of each transcription factor relative to the nuclear protein internal control, lamin B1, in three independent experiments, are shown in panel C. * represents a significant difference (p < 0.05); ** represents a significant difference (p < 0.01).
Transcription Factors Sp1, CREB1, and CDX2 Binding to the MUC3 Promoter Increased in HT-29-Gal Cells Treated with TFLK-containing 10-mer BP
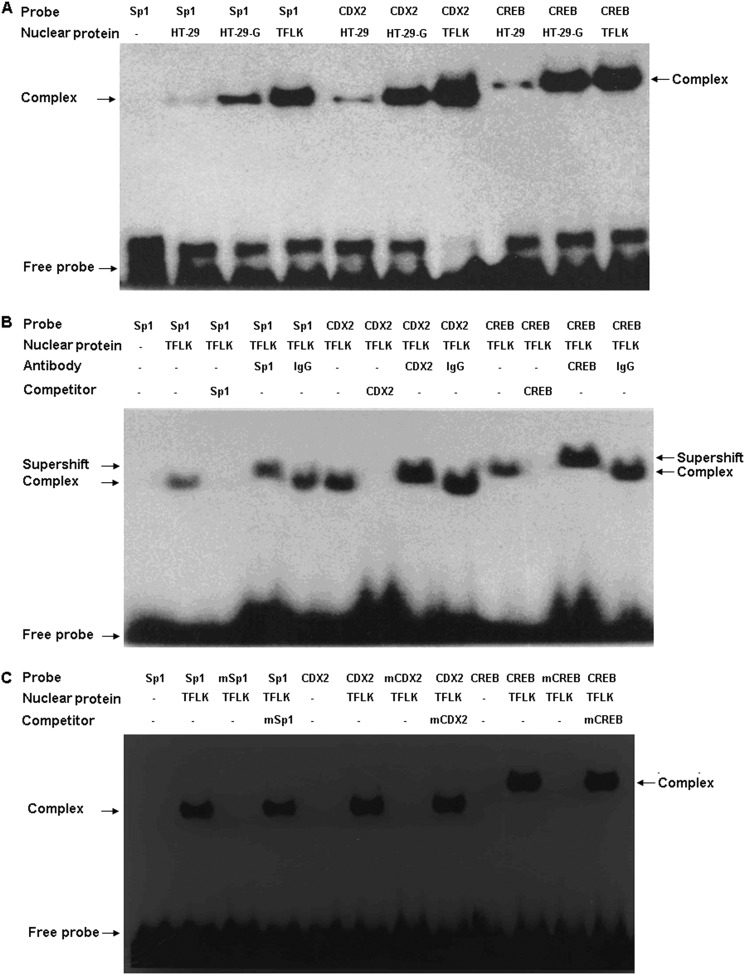
The Sp1, CREB1, and CDX2 transcription factors were induced in the HT-29-Gal cells after stimulation with the TFLK-containing 10-mer BP (USF1 was not included in this experiment because of the lack of a suitable antibody). It is unknown whether the transcription factors play a role in the up-regulation of the human MUC3 gene in HT-29-Gal cells stimulated with the TFLK-containing 10-mer BP. To investigate whether there were changes in the binding of these transcription activators to the human MUC3 promoter, we performed EMSA using the nuclear protein extracts from the HT-29-Gal cells with or without treatment with the TFLK-containing 10-mer BP and from HT-29 cells. The binding of Sp1, CDX2, and CREB1 to the human MUC3 promoter increased significantly, 18.5-, 19.9- and 17.4-fold, respectively, in the HT-29-Gal cells treated with the TFLK-containing 10-mer BP compared with the untreated HT-29 cells, and 6.4-, 5.5- and 1.7-fold, respectively, compared with the untreated HT-29-Gal cells (Fig. 4A). The binding of the transcription factors to the human MUC3 promoter was specific, as confirmed by the mutation and supershift EMSA experiments (Fig. 4, B and C).
FIGURE 4.
TFLK-containing 10-mer BP-treated HT-29-Gal cells demonstrate the increased binding activities of Sp1, CREB1, and CDX2 to the MUC3 promoter. Shown are representative EMSAs of the Sp1, CREB1, and CDX2 response elements in the human MUC3 promoter using the nuclear extract from HT-29 cells (HT-29), HT-29-Gal cells (HT-29-G), and HT-29-Gal cells treated with 200 μg/ml TFLK-containing 10-mer BP (TFLK), the complexes formed with Sp1, CREB1, or CDX2 probes, and the nuclear extract from HT-29-Gal cells treated with 200 μg/ml TFLK-containing 10-mer BP were more than that formed with Sp1, CREB1, or CDX2 probes and the nuclear extracts from non-treated HT-29 cells and untreated HT-29-Gal cells (A). The supershift EMSA was performed to confirm that the induced Sp1, CREB1, or CDX2 complexes were specific and contained Sp1, CREB1, or CDX2, and the antibodies against Sp1, CREB1, or CDX2 led to the supershift (marked as an arrow) compared with the relative complexes (B and C). Competitive inhibition using the mutated Sp1, CREB1, or CDX2 probes led to the failure of complexes forming. The wild types of Sp1, CREB1, or CDX2 probes, antibodies against Sp1, CREB1, or CDX2 (and nonimmune serum, IgG), and mutants (Sp1 mutation (mSp1); CREB1 mutation (mCREB); CDX2 mutation (mCDX2) were used in the supershift EMSAs are shown.
ChIP Assays Indicated Sp1, CREB1, and CDX2 Binding to the MUC3 Promoter and Sp1 and CREB1 Binding to the MUC3 Promoter Increased in HT-29-Gal Cells Treated with TFLK-containing 10-mer BP
To determine if CDX2, Sp1, and CREB1 can bind directly to the potential sites in the human MUC3 promoter and their bindings increased due to TFLK-containing 10-mer BP treatment in HT-29-Gal cells, we performed ChIP. As shown in Fig. 5A, two CDX2, one CREB1, and one Sp1 potential binding site were located in the proximal promoter of the human MUC3 gene. The binding of these transcription factors in their consensus sites within the MUC3 promoter was exactly present when compared with the negative controls. The binding of CDX2 to its potential sites was not increased in HT-29-Gal cells after treatment with TFLK-containing 10-mer BP; however, the binding of CREB1 and Sp1 to their potential sites was increased in HT-29-Gal cells after treatment with TFLK-containing 10-mer BP compared with untreated HT-29-Gal cells (Fig. 5B).
FIGURE 5.
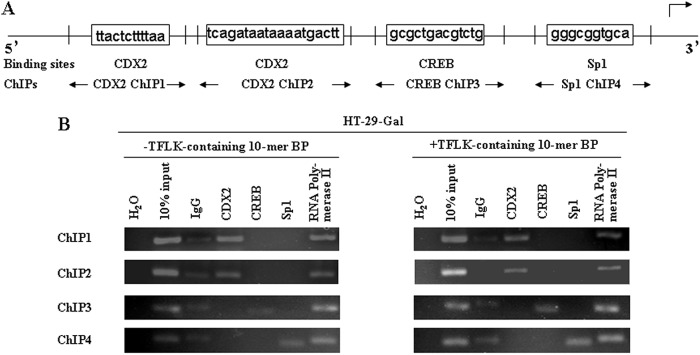
CDX2, Sp1, or CREB1 bind to the MUC3 promoter in response to TFLK-containing 10-mer BP treatment. Chromatin was isolated from HT-29-Gal cells that were either untreated or treated with 200 μg/ml TFLK-containing 10-mer BP. Chromatin was then subjected to immunoprecipitation using antibodies for IgG (negative control), CDX2, Sp1, CREB1, or RNA Polymerase II (positive control). Input represents 10% of the DNA used in the immunoprecipitation. Four loci were tested in the MUC3 promoter (two CDX2, one CREB1, and one Sp1 consensus sequences), ChIP1, ChIP2, ChIP3, and ChIP4. The binding sites for first CDX2 (−960 to −951), second CDX2 (−523 to −505), CREB1 (−148 to −136), and Sp1 (−92 to −84) in the MUC3 promoter region are presented (A). Final DNA extracts were amplified by PCR using primers that included CDX2 (ChIP1 and ChIP2), CREB1 (ChIP3), and or Sp1 (ChIP4) consensus sequence in the MUC3 promoter (B).
TFLK-containing 10-Mer BP Significantly Inhibited the Adherence of EPEC and O157 EHEC O157:H7 to HT-29-Gal Cells
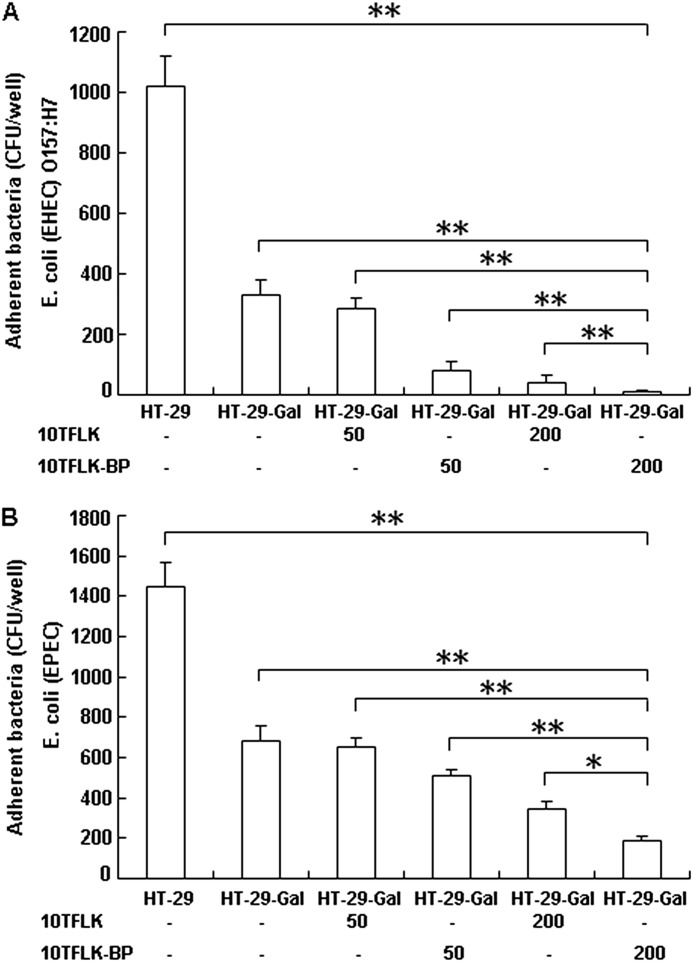
To evaluate the effect of the TFLK-containing 10-mer BP on the adherence of E. coli (EPEC) and E. coli (EHEC) O157:H7 to human intestinal epithelial cells, the TFLK-containing 10-mer BP (50 or 200 μg/ml) or the M-SAA3-derived N-terminal 10-mer monomeric peptide (50 or 200 μg/ml) were added to the culture medium of HT-29-Gal or HT-29 cells, and the effect of the peptides on the adherence of the bacteria was examined. As shown in Fig. 6A, pretreatment of the HT-29-Gal cells with 200 μg/ml TFLK-containing 10-mer BP produced the greatest reduction in adherence of the E. coli (EHEC) O157:H7 compared with the untreated HT-29-Gal cells, the HT-29-Gal cells pretreated with 50 μg/ml TFLK-containing 10-mer BP, and the HT-29-Gal cells pretreated with 50 or 200 μg/ml of the M-SAA3-derived N-terminal 10-mer monomeric peptide (p < 0.01). Similarly, pretreatment of the HT-29-Gal cells with 200 μg/ml TFLK-containing 10-mer BP produced the greatest reduction in the adherence of the E. coli (EPEC) compared with the untreated HT-29-Gal cells, the HT-29-Gal cells pretreated with 50 μg/ml TFLK-containing 10-mer BP or 50 μg/ml M-SAA3-derived N-terminal 10-mer monomeric peptide (p < 0.01), and the HT-29-Gal cells pretreated with 200 μg/ml M-SAA3-derived N-terminal 10-mer monomeric peptide (p < 0.05) (Fig. 6B). The results suggested that the TFLK-containing 10-mer BP of the N-terminal human serum amyloid A3 is more efficient than the M-SAA3-derived N-terminal 10-mer monomeric peptide at inducing MUC3 expression and leading to the significant inhibition of E. coli (EPEC) and E. coli (EHEC) O157:H7 adherence to intestinal epithelial cells.
FIGURE 6.
EPEC and EHEC O157:H7 adherence to HT-29-Gal cells after stimulation with the TFLK-containing 10-mer BP or the M-SAA3-derived N-terminal 10-mer monomeric peptide. HT-29-Gal cells or HT-29 cells received a single dose of the TFLK-containing 10-mer BP (10TFLK-BP) at 50 or 200 μg/ml or the M-SAA3-derived N-terminal 10-mer monomeric peptide (10TFLK) at 50 or 200 μg/ml. The cells were incubated at 37 °C for 1 h before the addition of 106 colony-forming units (CPU)/well of EPEC or EHEC O157:H7. After incubation for 3 h, the unbound bacteria were removed by 4 washes with chilled PBS (pH 7.4). The EHEC O157:H7 (A) or EPEC (B) adhering to the HT-29-Gal or HT-29 cells were quantified by determining the colony-forming units/well. The results are expressed as the mean ± S.E. of two independent experiments that were run in triplicate. * represents a significant difference (p < 0.05); ** represents a significant difference (p < 0.01).
Comparing the adherence of the E. coli (EPEC) and E. coli (EHEC) O157:H7 to the HT-29 and HT-29-Gal cells, it was interesting that the HT-29-Gal cells (a more mature and differentiated form of the HT-29 cell line) exhibited a significant reduction in the adherence of both the E. coli (EPEC) and E. coli (EHEC) O157:H7 strains. Therefore, the differentiation of the intestinal epithelial cells is associated with the adherence of E. coli (EPEC) or E. coli (EHEC) O157:H7.
DISCUSSION
Based on the study by Mack et al. (14), the M-SAA3 N-terminal 10-mer peptide (GWLTFLKAAG) containing the intact TFLK-motif increased MUC3 mRNA expression. M-SAA3 is a constituent of colostrum in humans and other mammalian species and plays a protective gastrointestinal role through the overexpression of intestinal mucin. It is likely that the presence of M-SAA3 in human colostrum is one of the reasons why colostrum-fed infants are ∼7–10 times less likely to suffer gastrointestinal diseases than formula-fed infants (10). A study by Domènech et al. (28) produced the recombinant goat milk serum amyloid A isoform 3 encompassing the TFLK region and showed that it was responsible for the up-regulation of mucins in the intestine; however, the protein did not promote macrophage phagocytosis. The experiments ruled out the possibility of determining the serum-derived forms of SAA that can opsonize Gram-negative bacteria facilitating their phagocytosis by circulating macrophages or intestinal epithelial cells (28). The exact mechanism of the intact TFLK motif in the M-SAA3-mediated induction of MUC3 expression and the inhibition of bacterial adherence remain unknown. Therefore, in our study we investigated a more efficient design for a polypeptide based on the conserved TFLK-motif from M-SAA3 that would induce increased MUC3 mucin expression in the intestinal epithelial cells and would be a more effective treatment to prevent enteric pathogen adherence. We also investigated the biological mechanism of the induction of human MUC3 expression by the TFLK-containing 10-mer BP.
Combinatorial chemistry has had a profound effect on the approach of medical chemists to drug discovery, especially on the strategies for the synthesis and screening of synthetic peptides developed over the last two decades (29, 30). The linear, cyclic, multimeric, and branched structures have been used as scaffolds into which natural or non-natural amino acids have been inserted to increase the chemical diversity or to achieve a more rigid conformation and more stable structures (17). Several reports have demonstrated that increasing the copy number of a precursor peptide by multimerization is a simple and effective way to enhance immunogenicity, affinity, and stability. It is known that the multimerization of a bioactive monomeric peptide essentially leads to the multiplication of the interacting groups/sites of the starting unit but does not create new recognition surfaces. Instead, multimerization provides new molecules with an increased avidity deriving from the multiple identical and cooperative sites. The tetrameric branched tripeptide 4-23-5 binds to the vascular endothelial growth factor receptor-1 (VEGFR-1) and inhibits the formation of the structures similar to capillaries that are formed by primary human endothelial cells. The monomeric and dimeric variants were inactive, and the trimeric variants were partially active (31). Pini et al. (32) report that the tetrabranched peptide based on the nonnatural antimicrobial peptide KKIRVRLSA is a good candidate for the development of a new antibacterial drug. Based on the finding by Mack et al. (16) showing that the M-SAA3 N-terminal 10-mer peptide induces human MUC3 transcriptional expression and inhibits EPEC adherence, we generated a synthetic tetrameric structure of branched polylysine based on the conserved TFLK-motif from M-SAA3 (GWLTFLKAAG). This tetrameric peptide is more efficient at inducing human MUC3 expression in the HT-29-Gal cells than the M-SAA3-derived N-terminal 10-mer monomeric peptide at both transcriptional and protein levels. The results indicated that the synthetic branched peptide offers the numerous advantages over biotherapeutics because it is easier and cheaper to produce, is more stable, and is generally free of biological contaminants. Furthermore, the tetrameric peptide offers more opportunities for delivery.
Although the function of the M-SAA3 N-terminal 10-mer peptide in inducing MUC3 expression and inhibiting EPEC adherence is clear, the mechanism behind the biological phenomena is not understood. This polypeptide, as an exogenous stimulator, induces the intestinal epithelial cells to express MUC3 mucin; however, how it interacts with the cell surface (possibly through a receptor) and how it transmits activating signals into the cells are unknown. Remarkably, the results from the transcription factor protein/DNA array comparing the HT-29-Gal cells stimulated with the TFLK-containing 10-mer BP with the untreated HT-29-Gal cells indicated that at least 81 TFs were up-regulated 2.0-fold (supplemental Table 4), and only 12 TFs were down-regulated 2.0-fold (supplemental Table 5). Our study of the MUC3 promoter and previous studies of the MUC3 or Muc3 promoters (33, 34) showed that the TFLK-containing 10-mer BP produced the highest luciferase activity in HT-29-Gal cells when using the pGL3-M2 construct (Fig. 2); therefore, the multiple cis-elements in the −242 to −64 region of the TSS of human MUC3 contribute to the activity of the human MUC3 promoter. The N-myc, USF1, CREB1, MZF1, and Sp1 cis-elements were shown to bind to the −242 to −64 region of the human MUC3 TSS. N-myc was not induced by the TFLK-containing 10-mer BP (supplemental Table 4); however, the levels of USF1, CREB1, and Sp1 increased more than 10-fold after stimulation with the TFLK-containing 10-mer BP, and the MZF-1 levels increased only 2.65-fold. The ChIP results in Fig. 5 suggest that the enhanced activity of the pGL3-M2 construct results in part from enhanced binding of Sp1 and CREB1 to the indicated sites in the proximal promoter. Together the results from Figs. 2, 4, and 5 suggest a general role for CDX2 in MUC3 up-regulation and a specific role for Sp1 and CREB1 in MUC3 up-regulation after stimulation with TFLK-containing 10-mer BP. To our knowledge, this is the first study to report a mechanism for the biological function of the TFLK motif.
Notably, the induction of human MUC3 mucin by the TFLK-containing 10-mer BP led to the dramatic inhibition of EPEC or EHEC O157:H7 adherence in HT-29-Gal cells. This finding suggests that the TFLK-containing 10-mer BP has potential applications for molecular therapeutic approaches in the prevention of gastrointestinal infection. Furthermore, the TFLK-containing 10-mer BP was more efficient at inhibiting adherence of the bacteria than the M-SAA3-derived N-terminal 10-mer monomeric peptide. EPEC is an important cause of infant diarrhea in developing countries (35). This bacteria produces a characteristic intestinal histopathological lesion on enterocytes known as “attaching and effacing” (A/E), and these two steps are mediated by a type III secretory system. EHEC O157:H7 causes illnesses ranging from mild diarrhea to severe diseases such as hemorrhagic colitis and hemolytic uremic syndrome (36). EHEC O157:H7, which produces Shiga toxin (Stx), is the major EHEC O157:H7 serotype responsible for public health problems worldwide (37). We propose that human MUC3 production induced by the TFLK-containing 10-mer BP improves the intestinal defense mediated by epithelial cells and thereby protects the host against lethal infection.
Collectively, our results suggest that the TFLK-containing 10-mer BP based on the conserved TFLK-motif from M-SAA3 is a new specific compound design that has potential applications for clinical uses to prevent gastrointestinal infection through the multiple transcription factor-mediated up-regulation of human MUC3 mucin.
Supplementary Material
This work was supported by National Natural Science Foundation of China Grants 30800519 and 81170340.

This article contains supplemental Tables 1–5 and Fig. 1.
- EPEC
- enteropathogenic E. coli
- M-SAA3
- mammary-associated serum amyloid A3
- BP
- branched peptide
- TSS
- transcription start sites
- TF
- transcription factor.
REFERENCES
- 1. Kim Y. S., Ho S. B. (2010) Intestinal goblet cells and mucins in health and disease. Recent insights and progress. Curr. Gastroenterol. Rep. 12, 319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lindén S. K., Florin T. H., McGuckin M. A. (2008) Mucin dynamics in intestinal bacterial infection. PLoS One 3, e3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lieleg O., Lieleg C., Bloom J., Buck C. B., Ribbeck K. (2012) Mucin biopolymers as broad-spectrum antiviral agents. Biomacromolecules 13, 1724–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mack D. R., Michail S., Wei S., McDougall L., Hollingsworth M. A. (1999) Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am. J. Physiol. 276, G941–G950 [DOI] [PubMed] [Google Scholar]
- 5. Velcich A., Palumbo L., Jarry A., Laboisse C., Racevskis J., Augenlicht L. (1995) Patterns of expression of lineage-specific markers during the in vitro induced differentiation of HT29 colon carcinoma cells. Cell Growth Differ. 6, 749–757 [PubMed] [Google Scholar]
- 6. Ho S. B., Luu Y., Shekels L. L., Batra S. K., Kandarian B., Evans D. B., Zaworski P. G., Wolfe C. L., Heinrikson R. L. (2010) Activity of recombinant cysteine-rich domain proteins derived from the membrane-bound MUC17/Muc3 family mucins. Biochim. Biophys. Acta 1800, 629–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Luu Y., Junker W., Rachagani S., Das S., Batra S. K., Heinrikson R. L., Shekels L. L., Ho S. B. (2010) Human intestinal MUC17 mucin augments intestinal cell restitution and enhances healing of experimental colitis. Int. J. Biochem. Cell Biol. 42, 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caballero-Franco C., Keller K., De Simone C., Chadee K. (2007) The VSL#3 probiotic formula induces mucin gene expression and secretion in colonic epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 292, G315–G322 [DOI] [PubMed] [Google Scholar]
- 9. Mack D. R., Ahrne S., Hyde L., Wei S., Hollingsworth M. A. (2003) Extracellular MUC3 mucin secretion follows epithelial cells in vitro adherence of Lactobacillus strains to intestinal. Gut 52, 827–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Playford R. J., Macdonald C. E., Johnson W. S. (2000) Colostrum and milk-derived peptide growth factors for the treatment of gastrointestinal disorders. Am. J. Clin. Nutr. 72, 5–14 [DOI] [PubMed] [Google Scholar]
- 11. Bodammer P., Maletzki C., Waitz G., Emmrich J. (2011) Prophylactic application of bovine colostrum ameliorates murine colitis via induction of immunoregulatory cells. J. Nutr. 141, 1056–1061 [DOI] [PubMed] [Google Scholar]
- 12. Rawal P., Gupta V., Thapa B. R. (2008) Role of colostrum in gastrointestinal infections. Indian J. Pediatr. 75, 917–921 [DOI] [PubMed] [Google Scholar]
- 13. Møller H. K., Thymann T., Fink L. N., Frokiaer H., Kvistgaard A. S., Sangild P. T. (2011) Bovine colostrum is superior to enriched formulas in stimulating intestinal function and necrotising enterocolitis resistance in preterm pigs. Br. J. Nutr. 105, 44–53 [DOI] [PubMed] [Google Scholar]
- 14. Mack D. R., McDonald T. L., Larson M. A., Wei S., Weber A. (2003) The conserved TFLK motif of mammary-associated serum amyloid a3 is responsible for up-regulation of intestinal MUC3 mucin expression in vitro. Pediatr. Res. 53, 137–142 [DOI] [PubMed] [Google Scholar]
- 15. Gardiner G. E., O'Flaherty S., Casey P. G., Weber A., McDonald T. L., Cronin M., Hill C., Ross R. P., Gahan C. G., Shanahan F. (2009) Evaluation of colostrum-derived human mammary-associated serum amyloid A3 (M-SAA3) protein and peptide derivatives for the prevention of enteric infection. In vitro and in murine models of intestinal disease. FEMS Immunol. Med. Microbiol. 55, 404–413 [DOI] [PubMed] [Google Scholar]
- 16. Larson M. A., Wei S. H., Weber A., Mack D. R., McDonald T. L. (2003) Human serum amyloid A3 peptide enhances intestinal MUC3 expression and inhibits EPEC adherence. Biochem. Biophys. Res. Commun. 300, 531–540 [DOI] [PubMed] [Google Scholar]
- 17. Ruvo M., Sandomenico A., Tudisco L., De Falco S. (2011) Branched peptides for the modulation of protein-protein interactions. More arms are better than one? Curr. Med. Chem. 18, 2429–2437 [DOI] [PubMed] [Google Scholar]
- 18. Jin Z. H., Furukawa T., Waki A., Akaji K., Coll J. L., Saga T., Fujibayashi Y. (2010) Effect of multimerization of a linear Arg-Gly-Asp peptide on integrin binding affinity and specificity. Biol. Pharm. Bull. 33, 370–378 [DOI] [PubMed] [Google Scholar]
- 19. Falciani C., Lozzi L., Pini A., Corti F., Fabbrini M., Bernini A., Lelli B., Niccolai N., Bracci L. (2007) Molecular basis of branched peptides resistance to enzyme proteolysis. Chem Biol. Drug Des. 69, 216–221 [DOI] [PubMed] [Google Scholar]
- 20. Mack D.R., Hollingsworth M. A. (1994) Alteration in expression of MUC2 and MUC3 mRNA levels in HT29 colonic carcinoma cells. Biochem. Biophys. Res. Commun. 199, 1012–1018 [DOI] [PubMed] [Google Scholar]
- 21. Gout S., Marie C., Lainé M., Tavernier G., Block M. R., Jacquier-Sarlin M. (2004) Early enterocytic differentiation of HT-29 cells. Biochemical changes and strength increases of adherens junctions. Exp. Cell Res. 299, 498–510 [DOI] [PubMed] [Google Scholar]
- 22. Sakai H., Hirahara M., Chiba Y., Misawa M. (2011) Antigen challenge influences various transcription factors of rat bronchus. Protein/DNA array study. Int. Immunopharmacol. 11, 1133–1136 [DOI] [PubMed] [Google Scholar]
- 23. Chai J., He Y., Cai S. Y., Jiang Z., Wang H., Li Q., Chen L., Peng Z., He X., Wu X., Xiao T., Wang R., Boyer J. L., Chen W. (2012) Elevated hepatic multidrug resistance-associated protein 3/ATP-binding cassette subfamily C 3 expression in human obstructive cholestasis is mediated through tumor necrosis factor α and c-Jun NH2-terminal kinase/stress-activated protein kinase-signaling pathway. Hepatology 55, 1485–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Böhm S. K., Gum J. R., Jr., Erickson R. H., Hicks J. W., Kim Y. S. (1995) Human dipeptidyl peptidase IV gene promoter: tissue-specific regulation from a TATA-less GC-rich sequence characteristic of a housekeeping gene promoter. Biochem. J. 311, 835–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Traber P. G., Silberg D. G. (1996) Intestine-specific gene transcription. Annu. Rev. Physiol. 58, 275–297 [DOI] [PubMed] [Google Scholar]
- 26. Erickson R. H., Lai R. S., Lotterman C. D., Kim Y. S. (2000) Identification of upstream stimulatory factor as an activator of the human dipeptidyl peptidase IV gene in Caco-2 cells. Gene 258, 77–84 [DOI] [PubMed] [Google Scholar]
- 27. Gum J. R., Hicks J. W., Kim Y. S. (1997) Identification and characterization of the MUC2 (human intestinal mucin) gene 5′-flanking region. Promoter activity in cultured cells. Biochem. J. 325, 259–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Domènech A., Raynes J. G., Rodríguez E. G., Arís A., Bach A., Serrano A. (2012) Recombinant expression of goat milk serum amyloid A. Preliminary studies of the protein and derived peptides on macrophage phagocytosis. Protein Pept. Lett. 19, 299–307 [DOI] [PubMed] [Google Scholar]
- 29. Edwards P. J. (2009) Current parallel chemistry principles and practice. Application to the discovery of biologically active molecules. Curr. Opin. Drug Discov. Devel. 12, 899–914 [PubMed] [Google Scholar]
- 30. Shin D. S., Kim D. H., Chung W. J., Lee Y. S. (2005) Combinatorial solid phase peptide synthesis and bioassays. J. Biochem. Mol. Biol. 38, 517–525 [DOI] [PubMed] [Google Scholar]
- 31. Ponticelli S., Marasco D., Tarallo V., Albuquerque R. J., Mitola S., Takeda A., Stassen J. M., Presta M., Ambati J., Ruvo M., De Falco S. (2008) Modulation of angiogenesis by a tetrameric tripeptide that antagonizes vascular endothelial growth factor receptor 1. J. Biol. Chem. 283, 34250–34259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pini A., Falciani C., Mantengoli E., Bindi S., Brunetti J., Iozzi S., Rossolini G. M., Bracci L. (2010) A novel tetrabranched antimicrobial peptide that neutralizes bacterial lipopolysaccharide and prevents septic shock in vivo. FASEB J. 24, 1015–1022 [DOI] [PubMed] [Google Scholar]
- 33. Shekels L. L., Ho S. B. (2003) Characterization of the mouse Muc3 membrane bound intestinal mucin 5′-coding and promoter regions. Regulation by inflammatory cytokines. Biochim. Biophys. Acta 1627, 90–100 [DOI] [PubMed] [Google Scholar]
- 34. Gum J. R., Jr., Hicks J. W., Crawley S. C., Dahl C. M., Yang S. C., Roberton A. M., Kim Y. S. (2003) Initiation of transcription of the MUC3A human intestinal mucin from a TATA-less promoter and comparison with the MUC3B amino terminus. J. Biol. Chem. 278, 49600–49609 [DOI] [PubMed] [Google Scholar]
- 35. Guerrant R. L., Hughes J. M., Lima N. L., Crane J. (1990) Diarrhoea in developed and developing countries. Magnitude, special setting, and etiologies. Rev. Infect. Dis. 12, S41–S51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jarvis K.G., Girón J. A., Jerse A. E., McDaniel T. K., Donnenberg M. S., Kaper J. B. (1995) Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. U.S.A. 92, 7996–8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ho N. K., Ossa J. C., Silphaduang U., Johnson R., Johnson-Henry K. C., Sherman P. M. (2012) Enterohemorrhagic Escherichia coli O157:H7 Shiga toxins inhibit γ interferon-mediated cellular activation. Infect. Immun. 80, 2307–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.