Background: LpxC is an essential enzyme in the biosynthesis of lipid A of the Gram-negative bacterial outer membrane.
Results: Mutations that confer resistance to LpxC inhibitors in ThrS slow the cellular growth and in FabZ unexpectedly reduce the cellular LpxC activity.
Conclusion: Alteration of fatty acid and protein biosyntheses compensates for impaired lipid A biosynthesis.
Significance: Our results reveal an important role of cellular homeostasis in antibiotic resistance.
Keywords: Antibiotic Resistance, Antibiotics, Lipids, Lipid Synthesis, Lipopolysaccharide (LPS)
Abstract
LpxC, the deacetylase that catalyzes the second and committed step of lipid A biosynthesis in Escherichia coli, is an essential enzyme in virtually all Gram-negative bacteria and is one of the most promising antibiotic targets for treatment of multidrug-resistant Gram-negative infections. Despite the rapid development of LpxC-targeting antibiotics, the potential mechanisms of bacterial resistance to LpxC inhibitors remain poorly understood. Here, we report the isolation and biochemical characterization of spontaneously arising E. coli mutants that are over 200-fold more resistant to LpxC inhibitors than the wild-type strain. These mutants have two chromosomal point mutations that account for resistance additively and independently; one is in fabZ, a dehydratase in fatty acid biosynthesis; the other is in thrS, the Thr-tRNA ligase. For both enzymes, the isolated mutations result in reduced enzymatic activities in vitro. Unexpectedly, we observed a decreased level of LpxC in bacterial cells harboring fabZ mutations in the absence of LpxC inhibitors, suggesting that the biosyntheses of fatty acids and lipid A are tightly regulated to maintain a balance between phospholipids and lipid A. Additionally, we show that the mutation in thrS slows protein production and cellular growth, indicating that reduced protein biosynthesis can confer a suppressive effect on inhibition of membrane biosynthesis. Altogether, our studies reveal a previously unrecognized mechanism of antibiotic resistance by rebalancing cellular homeostasis.
Introduction
An alarming number of nosocomial pathogens have acquired resistance to a broad range of existing antibiotics, resulting in increased mortality and morbidity (1–6). To combat these intractable pathogens, antibiotics targeting previously unexploited pathways must be developed and their potential resistance mechanisms understood (7).
Gram-negative bacteria contain a unique saccharolipid called lipid A in the outer leaflet of the outer membrane. Lipid A (endotoxin) is the membrane anchor of lipopolysaccharide (LPS) that protects Gram-negative bacteria against external agents such as antibiotics and detergents and is responsible for the toxic effects associated with Gram-negative sepsis. Because lipid A is essential for bacterial growth, and because its biosynthesis has never been targeted by current antibiotics, disrupting lipid A biosynthesis is an attractive strategy for the development of novel antibiotics to treat multidrug-resistant Gram-negative pathogens.
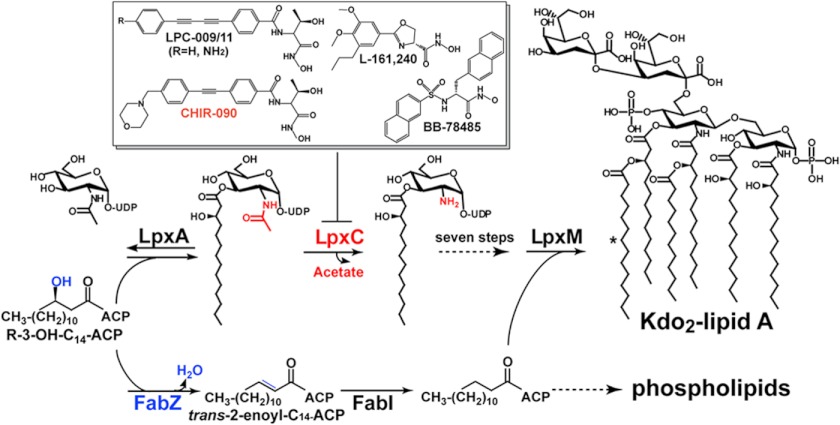
Biosynthesis of lipid A in Escherichia coli requires nine enzymes (8). Because the first reaction of biosynthesis, catalyzed by LpxA, is reversible and thermodynamically unfavorable (9), the committed step of the pathway is the second reaction catalyzed by the UDP-3-O-(hydroxymyristoyl)-N-acetylglucosamine deacetylase (LpxC)3 (Fig. 1) (10). LpxC does not share homology with any known mammalian protein, making it an excellent target for the design of novel antibiotics. Over the past 2 decades, there has been rapid progress in lead optimization for LpxC-targeting inhibitors (11–19). These lead compounds display excellent physiochemical properties and impressive bactericidal activity in vitro and in mouse models with little reported toxicity (11, 15–19).
FIGURE 1.
LpxC (labeled in red) catalyzes the committed step of lipid A biosynthesis in E. coli that leads to the formation of Kdo2-lipid A. The addition of the final myristoyl chain by LpxM is indicated by an asterisk. Inhibitors of LpxC include CHIR-090, L-161,240, BB-78485, LPC-009 (R = H), and LPC-011 (R = NH2). FabZ (labeled in blue) is a dehydratase in fatty acid biosynthesis and shares a substrate with LpxA. Point mutations that decrease FabZ activity have been proposed to increase the flux of (R)-3-hydroxymyristoyl-ACP toward lipid A biosynthesis.
In anticipation for the advancement of LpxC inhibitors into human clinical trials, we undertook studies to probe potential bacterial resistance mechanisms to these compounds. Although spontaneously resistant E. coli and Pseudomonas aeruginosa mutants have been previously reported (12, 20), these mutants only displayed moderate resistance, with an average 4–32-fold increase in minimum inhibitory concentrations (MIC) relative to wild type, and their biochemical consequences remain largely uncharacterized.
In this study, we report a two-step isolation of spontaneously resistant E. coli mutants that have >200-fold resistance to LpxC inhibitors. Detailed biochemical characterization of resistant mutants reveals an unexpected regulatory network balancing the biosynthesis of phospholipids and lipid A and a suppressive effect of impaired protein biosynthesis on inhibition of membrane synthesis.
EXPERIMENTAL PROCEDURES
Bacteria were grown in LB liquid or agar medium at 37 °C unless otherwise indicated. DNA primers were purchased from IDT Inc. (Coralville, IA), and sequences are annotated in Table 1. DNA sequencing was done at Eton Bioscience, Inc. (Research Triangle Park, NC) unless otherwise noted.
TABLE 1.
Sequence of primers used in this study
| Primer designation | Sequence (5′ to 3′) |
|---|---|
| 1 | CAAGCGTCTGAAATCGCTTGAGCG |
| 2 | GACAACGTGAGATTTCAGTACGGTACCC |
| 3 | AACCTTTCAGACGCACCGTGATG |
| 4 | ACGGAATTTAATTTCCTTAACCTGGATAACTTTTTGC |
| 5 | GTCTGGAGATCATTCGTCACTCCTGT |
| 6 | ATTTCTCAAGTCTGCTTTAACACGAATGCC |
| 7 | GGCGCAGCATATGTTGACTACTAACACTCATACTCTGCAGATTGAAGAG |
| 8 | GCAGAAGCTTTCAGGCCTCCCGGCTAC |
| 9 | GCAGAAGCTTGGCCTCCCGGCTACGAG |
| 10 | GTCACACTTTGCTATGCCAT |
| 11 | AATTCTGTTTTATCAGACCGCTT |
| 12 | TAATACGACTCACTATAGGG |
| 13 | CTAGTTATTGCTCAGCGGTG |
| 14 | GGCGCAGTCTAGATTAACTTTAAGAAGGAGATATACATATGCCTGTTATAACTCTTCCTGATGGCA |
| 15 | GCAGAAGCTTTTATTCCTCCAATTGTTTAAGACTGCGGCT |
| 16 | GGCGCAGTCTAGATTAACTTTAAGAAGGAGATATACATATGCCTGTTATAACTCTTCCTGATGGCA |
| 17 | GCAGAAGCTTTTCCTCCAATTGTTTAAGACTGCGGC |
| 18 | TAATACGACTCACTATAGGCTGATATAGCTCAGTTGGTAGAGCGCACCCTTGGTAAGGGTGAGGTCGGCAGTTCGAATCTGCCTATCAGC |
| 19 | TTTTGAATTCTAATACGACTCACTATAGGCTGATATAGCTCAGTTGGTAGAGCG |
| 20 | GGTGCTGATAGGCAGATTCGAACTGCCG |
| 21 | TGGTGCTGATAGGCAGATTCGAACTGCCG |
| 22 | GGCGCAGCATATGGTGAAGATTTATCTGGTCGGTGGTGC |
| 23 | GGCGCAGGGATCCTCATTCAGGCTTTGGGCAACGTT |
| 24 | GGCGCAGAAGCTTACTGCAAATAAGGATATAAAATGCCTGTTATAACTCTTCCT |
| 25 | GGCGCAGTCTAGATTATTCCTCCAATTGTTTAAGACTGCGGCT |
| 26 | GGCGCAGTCTAGATTAACTTTAAGAAGGAGATATACATTTGACTACTAACACTCATACTCTGCAGATTGAAGAG |
| 27 | GCAGAAGCTTTCAGGCCTCCCGGCTAC |
| 28 | GCGCGTCTAGAAAGGAGATATACATATGATCAAACAAAGGACACTTAAACGTATC |
| 29 | TTATGCCAGTACAGCTGAAGGC |
| 30 | GCCTTCAGCTGTACTGGCATAAAAGGAGATATACATGTGATTGATAAATCCGCCTTTGTG |
| 31 | GCGCGAAGCTTTTAACGAATCAGACCGCGCG |
| 32 | GGCGCAGTCTAGATTAACTTTAAGAAGGAGATATACATATGATCAAACAAAGGACACTTAAACGTATCGTTCAG |
| 33 | GCAGAAGCTTTTATGCCAGTACAGCTGAAGGCG |
Isolation of CHIR-090-resistant Mutants
109 cells of W3110 (Coli Genetic Stock Center, CGSC, Yale University) diluted from overnight cultures were plated on LB agar containing 1 μg/ml CHIR-090. After 24 h of growth, visible colonies were restreaked on the same medium and then purified three times on LB agar. Two colonies were isolated and designated “CRM1” and “CRM5.” 108 cells of CRM1 and CRM5, diluted from overnight cultures, were plated on LB agar containing 10 μg/ml CHIR-090. After 24 h of growth, visible colonies were isolated, restreaked, and purified using the same process described above. Two colonies, one descended from CRM1 and the other from CRM5, were designated “CRM1B” and “CRM5B,” respectively. Among all the mutants isolated in the second step, colonies of CRM1B and CRM5B grew most robustly and were visible after ∼24 h, whereas other mutant colonies were visible only after >36 h of growth.
MIC Assays
MICs were determined according to the National Committee for Clinical Laboratory Standards protocol (21), which was adapted to 96-well plates. Briefly, 106 bacterial cells in LB media containing various concentrations of the designated compound were incubated at 37 °C for 22 h. After incubation, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide solution (Sigma) was added (0.2 mg/ml final concentration) and incubated at 37 °C for an additional 12–16 h. The MIC was determined as the lowest concentration of compound that prevented color change (yellow to purple). To enhance inhibitor solubility, 10% DMSO was added to the growth media when assaying L-161,240, BB-78485, LPC-009, and LPC-011. MIC assays for ciprofloxacin (Mediatech, Manassas, VA) and nalidixic acid (Sigma) were repeated 10 times, and values were averaged.
Disc Diffusion Assays
Assays were done as described previously (22); the exact amounts of each compound are specified in Fig. 2.
FIGURE 2.
Disc diffusions assays. Spot 1 is 100% DMSO (2 μl); spot 2 is CHIR-090 (10 μg); spot 3 is L-161,240 (40 μg), and spot 4 is BB-78485 (40 μg). Compared with W3110 (far left), all mutants are fully resistant to LpxC inhibitors L-161,240 and BB-78485.
Whole Genome Sequencing
Genomic DNA was extracted from W3110 and CRM mutants using the Easy-DNA kit (Invitrogen) and was sequenced by Ambry Genetics (Aliso Viejo, CA). Resulting FASTQ files were analyzed using Maq-0.7.1 and aligned to E. coli K-12 W3110 (AC_000091.1). Additional point mutations present in CRM strains, but not present in the parental strain W3110, with quality scores >100 are shown in Table 2. These point mutations were independently verified by PCR amplification from genomic DNA and sequencing using primers 1–6.
TABLE 2.
Point mutations and MIC of mutant strains
wtfabZ is wild-type fabZ; wtthrS is wild-type thrS, and muthrS is mutant thrS. ND means not determined due to solubility limit of compound.
| Strain | Chromosomal mutations | Protein | CHIR-090 | LPC-009 | LPC-011 |
|---|---|---|---|---|---|
| μg/ml | μg/ml | μg/ml | |||
| W3110 | None | None | 0.2 | 0.04 | 0.02 |
| CRM1 | fabZ: 50T→A | FabZ: L17Q | 12.3 | 3.13 | 0.78 |
| CRM1B | fabZ: 50T→A | FabZ: L17Q | 56.3 | ND | ND |
| thrS: 1549T→G | ThrS: S517A | ||||
| CRM5 | fabZ: 212C→T | FabZ: A71V | 12.3 | 6.25 | 1.56 |
| CRM5B | fabZ: 212C→T | FabZ: A71V | 56.3 | ND | ND |
| thrS: 1549T→G | ThrS: S517A | ||||
| CRM1B wtfabZ | thrS: 1549T→G | ThrS: S517A | 1.25 | 0.625 | 0.313 |
| metN::kan | |||||
| CRM5B wtfabZ | thrS: 1549T→G | ThrS: S517A | 1.25 | 0.625 | 0.313 |
| metN::kan | |||||
| CRM5B | ΔmetN | None | 0.2 | 0.04 | 0.02 |
| wtfabZ, wtthrS | ydiU::kan | ||||
| W3110 wtthrS | None | None | 0.2 | 0.04 | 0.02 |
| W3110 muthrS | thrS: 1549T→G | ThrS: S517A | 1.25 | 0.625 | 0.313 |
Constructions of CRM wtfabZ and CRM wtfabZ, wtthrS Mutants
To create CRM strains containing wild-type fabZ, P1vir lysate was generated from the Keio mutant JW0195 (E. coli Genetic Stock center, Yale University) containing a kanamycin cassette 20 kb downstream of fabZ (23) and was used to transfect CRM1B and CRM5B. Colonies were plated and purified three times on LB agar containing 50 μg/ml kanamycin and 5 mm sodium citrate following established protocols (24). Genomic DNA was isolated from colonies, and the region around fabZ was amplified and sequenced using primers 1 and 2. Colonies harboring wild-type fabZ were designated “CRM1B wtfabZ” and “CRM5B wtfabZ” with respect to their parental strains. To create CRM5B wtfabZ, wtthrS, the kanamycin cassette in CRM5B, was excised using pCP20 as described previously (25). P1vir lysate was generated from the Keio mutant JW1696 (E. coli Genetic Stock center, Yale University) containing a kanamycin cassette 10 kb upstream of thrS (23). Colonies were selected and purified as described above. The area around thrS was amplified using primers 3 and 4 and sequenced using primers 3–6. A colony that harbored wild-type thrS was designated “CRM5B wtfabZ, wtthrS” (Table 1).
Construction of pBAD33.1 (fabZ), pBAD33.1 (lpxC), pWSK29 (fabZ), and pBAD33 (lpxCA)
Wild-type fabZ was amplified using genomic DNA from W3110 and primers 7 and 8. The PCR fragment was purified using QIAQuick gel extraction kit (Qiagen, Valencia, CA). A pBAD33.1 plasmid (26) was prepared using the QIAprep miniprep kit (Qiagen, Valencia, CA). Both the vector and PCR fragment were digested using restriction enzymes NdeI and HindIII (New England Biolabs, Ipswich, MA). The vector was treated with calf intestinal alkaline phosphatase (New England Biolabs). After PCR purification, the vector and DNA fragment were ligated using T4 DNA ligase (Invitrogen), transformed into XL1-Blue Competent cells (Stratagene, Santa Clara, CA), and grown on LB agar containing 25 μg/ml chloramphenicol (Sigma). Correct constructs were verified using primers 10 and 11 for DNA fragment amplification and sequencing. Confirmed constructs were transformed into chemically competent W3110 as described previously (24). Plasmid pBAD33.1 (lpxC) was constructed with the same method using primers 32 and 33 to amplify fabZ and using XbaI and HindIII restriction enzymes for cloning.
Plasmid pWSK29 (fabZ) was constructed similarly. Briefly, fabZ was amplified with primers 26 and 27 using W3110 genomic DNA as template, and the PCR fragment was purified and digested with XbaI and HindIII. The resulting DNA fragment was ligated to similarly digested pWSK29 vector to yield pWSK29 (fabZ) plasmid.
To construct pBAD33 (lpxCA) plasmid, lpxC and lpxA genes were amplified individually by PCR with primers 28 and 29 for lpxC, and primers 30 and 31 for lpxA. Both primer 28 and 30 contain ribosome-binding sites upstream of the start codons to facilitate gene transcription. Then a single DNA fragment including both lpxC and lpxA genes was amplified using the above two DNA fragments as templates with primers 28 and 31. The PCR products were purified and digested with XbaI and HindIII and ligated to similarly digested pBAD33 vector to yield pBAD33 (lpxCA).
Liquid Chromatography-Mass Spectrometry (LC-MS)
The method of normal phase LC-MS was described previously (27). Reverse phase LC-MS was performed using a Shimadzu LC system (consisting of a solvent degasser, two LC-10A pumps, and an SCL-10A system controller) coupled to a QSTAR XL quadrupole time-of-flight tandem mass spectrometer. LC was operated at a flow rate of 200 μl/min with a linear gradient as follows: 100% of mobile phase A was held isocratically for 2 min and then linearly increased to 100% mobile phase B over 14 min and held at 100% B for 4 min. Solvent A consisted of methanol/acetonitrile/aqueous 1 mm ammonium acetate (60:20:20, v/v/v). Solvent B consisted of 100% ethanol containing 1 mm ammonium acetate. A Zorbax SB-C8 reversed-phase column (5 μm, 2.1 × 50 mm) was obtained from Agilent (Palo Alto, CA). The post-column splitter diverted ∼10% of the LC flow to the ESI source of the mass spectrometer.
FabZ Assay
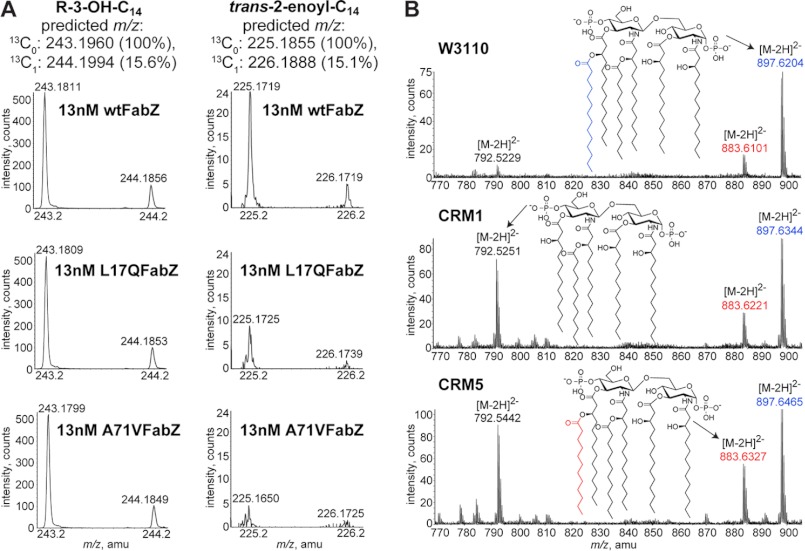
Genomic DNAs extracted from W3110, CRM1, and CRM5 were used as templates to amplify wild-type fabZ, fabZ T50A, and fabZ C212T, respectively, using primers 7 and 9. Plasmids containing C-terminally His-tagged wild-type fabZ, fabZ T50A, or fabZ C212T were constructed in the pET21b(+) vector (EMD Biosciences, San Diego). Correct constructs were confirmed using primers 12–13 by sequencing and were transformed into C41(DE3) cells (28, 29). C-terminally His-tagged FabZ was expressed and purified to >90% homogeneity as determined from SDS-PAGE analysis as described previously (30). (RS)-3-Hydroxymyristoyl-ACP was prepared as described previously (31). Each 40-μl reaction mixture included 100 mm sodium phosphate, pH 7, 1 mm 2-β-mercaptoethanol, and 50 μm (RS)-3-hydroxymyristoyl-ACP. Reactions were initiated with the addition of 12.75–25.5 nm of the wild-type enzyme or 12.75–100 nm of mutant enzymes, and time points were taken over the course of 20 min at 37 °C. Reactions were quenched by addition of 1× volume of 1.25 m NaOH and placed on ice. After 10 min, an additional 1× volume of 1.25 m acetic acid was added. After 10 min, a 2.2× final volume of methanol and a 2.2× final volume of chloroform were added to the mixture for conversion into a two-phase Bligh/Dyer at room temperature (32). The mixture was vortexed vigorously, centrifuged at 13,000 rpm for 5 min, and the lower phase collected. The sample was then dried under nitrogen and resuspended in 2:1 ratio of chloroform/methanol mixture. Samples were injected on the C8 reverse phase column and run on LC-MS as described above. Both (R)-3-hydroxymyristate and trans-2-enoyl-myristate eluted at 3–3.5 min on the column (Fig. 3A). Peaks were quantified using Analyst QS software (Applied Biosystems, Carlsbad, CA), and product (P) conversion was determined as [P]/([S] + [P]), where S is substrate. All quantifications were subtracted from the no enzyme control. Product conversion was linear with respect to both time and enzyme concentrations used.
FIGURE 3.
LC-MS of FabZ mutants. A, FabZ enzymatic activity assay based on reverse phase LC-MS. Mass spectra (after blank buffer subtraction) averaged from those acquired during 3–3.5 min show reduced product conversion (decreased counts of trans-2-enoyl-myristate) in the samples with L17Q FabZ (L17QFabZ) or A71V FabZ (A71VFabZ) compared with the wild-type enzyme (wtFabZ) at 10 min. B, mass spectra (after background subtraction) of lipid A species eluted during 36–38 min on normal phase LC-MS show increased amounts of penta-acylated lipid A as well as lipid A with a lauroyl instead of a myristoyl chain in FabZ mutant strains. Lipid A species are detected by ESI/MS in the negative ion mode as the doubly charged [M − 2H]2− ions.
LpxC Activity Assay
Preparation of cell-free extracts and LpxC activity assays were performed as described previously (31). To prepare membrane-free lysates, the cell-free extracts were further centrifuged at 45,000 rpm (∼140,000 × g) in a Beckman type 70.1 Ti rotor for 1 h at 4 °C. Aliquots were stored at −80 °C.
Western Blots
The LpxC antibody was used as described previously (31). Preparation of cell extracts and Western blots was performed as described previously (31). 0.2 μg of purified LpxC was used as a positive control. W3110 cells harboring pBAD33.1 constructs (vector control or harboring wild-type fabZ) were grown to an absorbance at 600 nm (A600) ∼0.2 in LB media containing 25 μg/ml chloramphenicol; 0.2% l-arabinose (Sigma) was added to the LB media in the indicated samples only (Fig. 4C). Cells were harvested ∼50 min later, centrifuged down, and resuspended in Laemmli sample buffer (Bio-Rad) normalized to A600 = 1 per 100 μl of buffer. Samples were then heated to >95 °C for 10 min and loaded onto 15% Tris-HCl precast gels (Bio-Rad). Overexpression of FabZ was confirmed by the appearance of a band of the expected molecular mass (∼17,000 daltons) that only appeared in cells harboring wild-type fabZ on the plasmid and grown in the presence of 0.2% l-arabinose as visualized by SDS-PAGE.
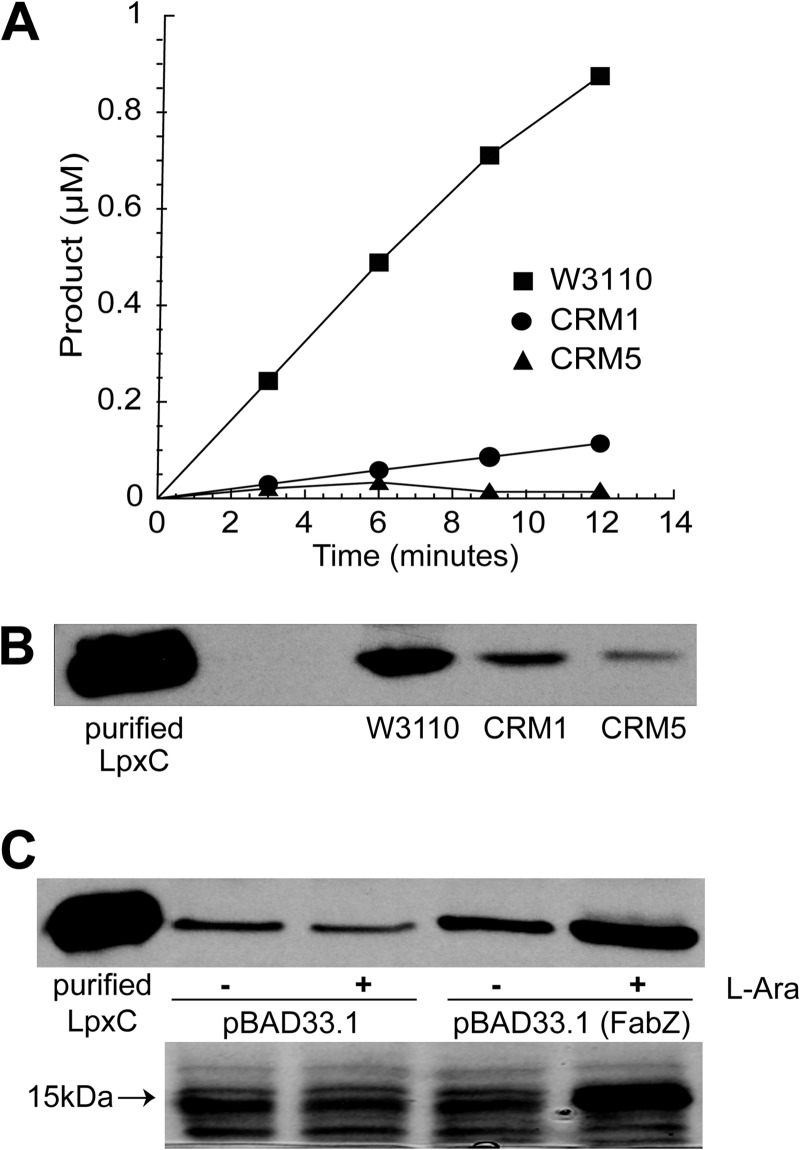
FIGURE 4.
LpxC activity and levels in FabZ mutants. A, enzymatic LpxC activity assays show significantly less product (UDP-3-O-(R-3-hydroxymyristoyl)-glucosamine) conversion in CRM1 and CRM5 relative to W3110. Each spot was connected for better visualization. B, Western blot using an LpxC antibody shows CRM1 and CRM5 have less LpxC protein amount relative to W3110. C, Western blot detect increased LpxC protein amount when FabZ is overexpressed in the presence of 0.2% l-arabinose (l-Ara) on a pBAD33.1 plasmid in W3110 (top). SDS-PAGE analysis of the protein gel shows overexpression of FabZ (∼17,000 daltons) only in the presence of 0.2% l-arabinose (bottom).
Extraction and Visualization of Lipid Profile
Phospholipids and lipid A were extracted as described previously (26). Samples were analyzed using normal phase LC-MS as described above.
Construction of pWSK29 (thrS)
Primers 14 and 15 were used to amplify thrS from W3110 genomic DNA and cloned into a pWSK29 vector (24) using restriction sites XbaI and HindIII. Cells were grown on LB agar containing 100 μg/ml ampicillin. Constructs were verified using primers 12 and 13 and transformed into chemically competent W3110, CRM1B wtfabZ, and CRM5B wtfabZ cells following the previously described method (24). MICs were determined in LB media containing 100 μg/ml ampicillin.
Preparation of ThrS Constructs
Genomic DNAs extracted from W3110 and CRM5B were used as templates to amplify wild-type thrS and thrS T1549G, respectively, using primers 16 and 17 and restriction sites XbaI and HindIII. Cloning of these pET21b vector-based constructs was done as described above for the FabZ assay. Expression and purification of ThrS proteins were done as described previously (33). Enzymes were purified to over 90% homogeneity as determined by SDS-PAGE analysis.
Preparation of RNA Substrate
Primers 18–20 were used to create the DNA template for in vitro transcription of tRNA(GGU) lacking the 3′-terminal adenosine for synthesis of radiolabeled substrate. Primers 18, 19, and 21 were used to create fully formed tRNA(GGU) for synthesis of cold carrier substrate. The amplified DNA fragments were run on 2% agarose gel and purified as described above. RNA transcription reactions and isopropyl alcohol purifications were done using MEGAscript® kit high yield transcription kit (Applied Biosystems, Carlsbad, CA). The resulting RNA constructs were >90% homogeneous as visualized by 8% denatured polyacrylamide gel containing 7 m urea. For preparation of radiolabeled substrate, E. coli tRNA nucleotidyltransferase enzyme was cloned from W3110 genomic DNA using primers 22 and 23 and restriction sites NdeI and BamHI in a pET16b vector (EMD Biosciences, San Diego). The construct was expressed and purified as described previously (34). Radiolabeling of 3′-adenosine was done as described previously (34) with modifications. Briefly, a 50-μl reaction mixture containing 2 μm tRNA(GGU) lacking 3′-adenosine, 50 μm sodium pyrophosphate, 3 μm ATP, 250 μCi of [α-32P]ATP (9.25 MBq) (PerkinElmer Life Sciences), 10 mm magnesium chloride, and 50 mm glycine at pH 9.0 was reacted with 0.2 μm of tRNA nucleotidyltransferase. The reaction mixture was incubated at 37 °C for 7 min, at which time the reaction mixture underwent phenol/chloroform extraction, and the aqueous layer was put through two consecutive Micro Bio-Spin P-6 columns (Bio-Rad). 1-μl aliquots, taken before addition of enzyme, after 7 min of reaction, and after column purification, were spotted on a TLC PEI-cellulose-F plate (Merck) and run in a solvent system consisting of saturated ammonium sulfate to assess conversion and purity of the final tRNA(GGU) substrate. All tRNA substrates were stored in RNase-free water at −20 °C.
ThrS Assay
tRNA substrates were refolded before use in the assay by heating to 95 °C for 3 min and then slowly cooling down to 37 °C over the course of 30 min. Each reaction mixture contained 50 mm Tris at pH 8, 20 mm potassium chloride, 10 mm magnesium chloride, 5 mm DTT, 100 μm ATP, 50 μm l-threonine, 10 μm cold carrier tRNA(GGU), and 10,000 cpm/reaction of [α-32P]tRNA(GGU) at 37 °C. Reactions were started with the addition of 5–10 nm wild-type ThrS or 5–100 nm mutant ThrS. Time points were taken over the course of 10 min, and reactions were quenched in 4× volume of S1 nuclease (10 units total diluted in 1× reaction buffer) (Fermentas, Glen Burnie, MD) at room temperature for 10 min. 1 μl of each sample was spotted on a PEI-cellulose-F plate and was run in a solvent system consisting of 5:10:85 volume ratios of glacial acetic acid, 1 m ammonium chloride/water, respectively. Product formation was visualized by using a PhosphorImager screen (Fig. 5B), and product conversion was calculated using ImageQuant TL (Amersham Biosciences). Product conversion was linear with respect to time and enzyme concentrations used.
FIGURE 5.
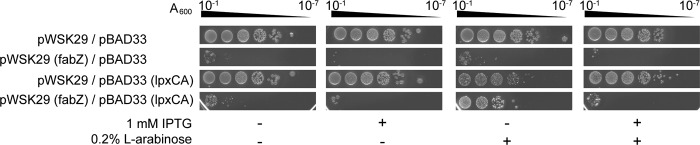
MacConkey plate tests. W3110 host strains carrying both pWSK29 and pBAD33 vector controls or the indicated protein-expressing constructs were grown to A600 >1.0 and diluted in LB media. Then 3 μl of diluted samples were spotted on MacConkey plates. The A600 labels indicate the calculated absorbance readings based on the folds of dilution. Protein expression was induced by including 1 mm IPTG, 0.2% l-arabinose, or both in the agar plates as indicated. Each plate was scanned after extended incubation (∼40 h) at 37 °C for better visualization.
Construction of W3110 muthrS
Primers 24 and 25 were used to amplify thrS T1549G using genomic DNA extracted from CRM5B and ligated into a pMAK705 plasmid (17) using restriction enzymes HindIII and XbaI. The verified construct was transformed into W3110 cells. Cells were grown on LB agar containing 25 μg/ml chloramphenicol at 42 °C. In these conditions, the pMAK705 plasmid replicon cannot replicate (17), and surviving cells must have incorporated the plasmid into the chromosome through homologous recombination with the chromosomal copy of wild-type thrS. Purified colonies were then grown at 30 °C in same medium. At this permissive temperature, the pMAK705 plasmid replicon can replicate, and surviving cells must have excised the plasmid, leaving either the original chromosomal wild-type copy of thrS or the original plasmid-borne copy of thrS T1549G remaining on the chromosome. Single cells were then purified three times at 42 °C in the absence of any antibiotic to dilute out the plasmid. Complete loss of the plasmid was confirmed by cells' inability to grow on LB agar containing 25 μg/ml chloramphenicol at 30 °C. PCR analysis and DNA sequencing were employed to confirm the genotypes using primers 3–6. A construct with wild-type thrS remaining on the chromosome was designated “W3110 wtthrS,” and a construct with thrS T1549G on the chromosome was designated “W3110 muthrS” (Table 2).
Protein Labeling Experiments
Strains W3110, W3110 wtthrS, and W3110 muthrS were grown in M9 media supplemented with 0.2% glucose, 1 mm magnesium chloride, 100 μm calcium chloride, 3 mm iron chloride, 10 μg/ml thiamine until the A600 reached 0.2. Then 5 μCi of 14C-labeled l-amino acids (1.85 Mbq) (PerkinElmer Life Sciences) was added to the growth culture. 1-ml samples were collected at 0.5, 2, 4, and 8 min by quenching with 200 μg/ml chloramphenicol and placed on ice. Samples were centrifuged to pellet the cells; the supernatant was decanted, and the remaining cell pellet was resuspended with Laemmli buffer using the ratio of 109 cells/100 μl. Samples were heated to >95 °C for 10 min, and 10 μl of each sample was loaded on 12% Tris-HCl gels (Bio-Rad) and run in 1× Tris-Glycine Buffer (Bio-Rad) at 200 V for 40 min. Incorporation of 14C-labeled l-amino acids into proteins was visualized by a PhosphorImager, and counts were quantified using ImageQuant TL. Counts for each time point (for W3110 wtthrS and W3110 muthrS) were normalized to the counts detected in W3110 to measure a relative ratio of protein biosynthesis.
Growth Curves
Bacterial cultures were diluted 1:100 from overnight cultures and grown until log phase (A600 of ∼0.5). Cells were then diluted 10-fold into fresh LB media, and A600 was measured over the course of ∼12 generations. To ensure continuous log phase growth, once bacterial cultures reached an A600 of ∼0.5, cells were continuously diluted 10-fold into fresh media. Doubling time for each strain was calculated using Kaleidagraph (Synergy Software, Reading, PA) by exponentially fitting the plot of log A600 versus minutes using Equation 1,
where y is log(A600), x is time (minutes), m1 is A600 of initial cells, and m2 is doubling time (minutes).
MacConkey Agar Plate Tests
Wild-type W3110 was transformed with both pWSK29 and pBAD33 containing either the vector plasmids or their derivative genes, as specified in Fig. 5. Each strain was grown in LB medium to A600 >1.0 at 37 °C. Bacterial cultures were first diluted to A600 = 1, and then a series of 10-fold dilutions were prepared, and 3 μl were spotted on MacConkey agar plates, supplemented with 100 μg/ml ampicillin, 25 μg/ml chloramphenicol, and with or without 1 mm IPTG or 0.2% arabinose or both. The spots were air-dried, and the plates were incubated at 37 °C for ∼40 h for better colony visibility in the scanned images.
RESULTS
Isolation of Spontaneously Resistant Mutants to CHIR-090
CHIR-090 is one of the most thoroughly characterized LpxC-targeting antibiotics to date (13, 14, 20, 22, 35–37). It is a slow, tight binding inhibitor for E. coli LpxC, and it displays potent antibiotic activity for a wide range of Gram-negative human pathogens. It also elicits a very low rate of spontaneous resistance mutations in E. coli (<10−9). Accordingly, efforts to isolate and characterize spontaneously arising mutants with high levels of CHIR-090 resistance in one step have not been successful.
To overcome this limitation and isolate spontaneously CHIR-090-resistant mutants with a clean background, we employed a two-step approach, starting from the genetically well characterized E. coli strain W3110. First, medium level resistant mutants were generated by exposing wild-type W3110 cells in the presence of a low concentration of CHIR-090 (1 μg/ml, 5× MIC). Surviving CHIR-090-resistant mutants (CRMs) were isolated at a frequency of ∼10−9. These isolates were used as starting cells for a second round of selection in the presence of a high concentration of CHIR-090 (10 μg/ml, 50× MIC). Surviving mutants in the second step were isolated at a frequency of ∼10−7. Among these isolates, the two fastest growing resistant mutants (CRM1B and CRM5B) together with their parental strains (CRM1 and CRM5) were chosen for further characterization.
Mutants Are Resistant to a Variety of LpxC Inhibitors
We first evaluated the four isolated mutants for their ability to survive in the presence of well characterized LpxC inhibitors, including CHIR-090, the diacetylene-based compounds LPC-009 (15) and LPC-011 (16), and the earlier generation of LpxC inhibitors L-161,240 (11) and BB-78485 (Fig. 1) (12). The CRM1 and CRM5 mutants, isolated after exposure to 1 μg/ml CHIR-090, show ∼50-fold higher MIC to LpxC inhibitors CHIR-090, LPC-009, and LPC-011, whereas the two mutants CRM1B and CRM5B, isolated after exposure to 10 μg/ml CHIR-090, show a remarkable >200-fold higher MIC against CHIR-090 relative to the parental strain W3110 (Table 2). Likewise, our disc diffusion assays show that all of these mutants are highly resistant to the earlier generation of LpxC inhibitors L-161,240 and BB-78585 (Fig. 2). However, because of the high level of resistance and limitations in compound solubility of LPC-009, LPC-011, L-161,240, and BB-78485, the exact MICs cannot be determined for these compounds. It is important to note that these LpxC inhibitors are based on a variety of chemical scaffolds. The observation that our isolated mutants are universally resistant to these compounds, regardless of their differing chemical structures, suggests that the mutants' resistance mechanisms are unlikely to be caused by specific mutations in LpxC that reduce the potency of LpxC inhibitors but rather caused by more general resistance mechanisms that counter the suppression of lipid A biosynthesis.
Mutations of fabZ and thrS Confer CHIR-090 Resistance Additively and Independently
To pinpoint the specific mutations that confer resistance, we obtained whole genome sequencing data for the four isolated mutant strains and compared them with the parental strain W3110. Our results show that CRM1 and CRM5 each has a single point mutation, T50A and C212T, in fabZ, respectively, whereas both CRM1B and CRM5B, compared with their parental strains, have an additional point mutation, T1569G, in thrS (Table 2).
To determine whether point mutations in fabZ and thrS cause CHIR-090 resistance in a synergistic manner, we replaced the mutated fabZs with wild-type fabZ on the chromosome of the double mutants CRM1B and CRM5B. The resulting strains, CRM1B wtfabZ and CRM5B wtfabZ, which contain the single point mutation in thrS, show 6-fold higher MIC to CHIR-090 (1.25 μg/ml) compared with the wild-type strain W3110 (0.2 μg/ml). Further loss of the remaining point mutation in thrS resulted in a strain (CRM5 wtfabZ, wtthrS) with wild-type MIC (Table 2). These results show that the point mutations in fabZ and in thrS confer ∼50- and ∼4–8-fold resistance, respectively, and that fabZ and thrS mutations account for >200-fold resistance in an additive and independent manner.
Point Mutations in fabZ Decrease Enzymatic Activity
FabZ is an intermediate enzyme of fatty acid biosynthesis catalyzing the dehydration of (R)-3-hydroxymyristoyl-ACP (38) in fatty acid elongation and shares a common substrate with LpxA, the enzyme that precedes LpxC in lipid A biosynthesis. The end products of fatty acid biosynthesis are used in phospholipid biosynthesis. Because (R)-3-hydroxymyristoyl-ACP is used in the biosynthesis of both fatty acids and lipid A, reactions catalyzed by FabZ and LpxA are considered the branch point between phospholipid and lipid A biosynthesis (Fig. 1).
In our isolated CRM strains, mutation of T50A or C212T in fabZ results in altered FabZ protein with either Leu-17 replaced by Gln or Ala-71 replaced by Val, respectively. In the crystal structure of P. aeruginosa FabZ (Protein Data Bank code 1U1Z), which shares 54% sequence identity with E. coli FabZ (39), both of these corresponding residues form the lining of the proposed substrate-binding site, and they likely contribute to the binding of fatty acid. To examine the effects of the isolated E. coli FabZ point mutations on enzymatic activity, we purified wild-type and mutant FabZs and developed a liquid chromatography-mass spectrometry (LC-MS)-based assay to determine their specific activities. In this assay, hydroxymyristoyl-ACP was incubated with purified wild-type FabZ or its mutants. Reaction samples were quenched with base to remove the ACP portion. The liberated fatty acids were extracted using two-phase Bligh-Dyer, injected on a reverse phase column, and detected by mass spectrometry in the negative ion mode. The ion intensities of the substrate hydroxymyristate (observed as the [M − H]− ion at m/z 243.18) and product trans-2-enoyl-myristate (observed as the [M − H]− ion at m/z 225.17) in each sample reaction were quantified to calculate product conversion. Under our assay conditions, both mutants had reduced specific activities (Fig. 3A); the L17Q mutant is ∼2.5-fold less active compared with wild-type and the A71V is ∼8-fold less active (Table 3). Importantly, CHIR-090 has no inhibitory effect on wild-type or mutant FabZ enzymes at concentrations up to 200 μm, suggesting that neither FabZ nor its mutants are secondary targets of LpxC inhibitors and that FabZ mutations indirectly compensate for impaired lipid A biosynthesis.
TABLE 3.
Specific activities of FabZ proteins
| FabZ protein | Specific activity |
|---|---|
| μmol/min/mg | |
| Wild type | 1.87 ± 0.346 |
| L17Q | 0.74 ± 0.063 |
| A71V | 0.23 ± 0.015 |
We next assessed whether reduced FabZ activities result in detectable changes in vivo. Analysis of the global lipid composition using LC-MS showed that the FabZ mutants CRM1 and CRM5 did not cause a profound change in the profiles of free fatty acids or major phospholipids (phosphatidylethanolamine, phosphatidylglycerol, and cardiolipin), except for a slight increase of monounsaturated (14:1, 16:1 and 18:1) fatty acyl chain levels (∼15%) relative to the wild-type cells (data not shown). This observation suggests that in mutant cells reduced FabZ activity is compensated by its isozyme, FabA, in fatty acid biosynthesis, which shares broad overlapping chain length specificities with FabZ (30). FabA additionally catalyzes the isomerization of E-2-decenoyl-ACP to Z-3-decenoyl-ACP (40), leading to increased production of unsaturated fatty acids and phospholipids.
In contrast to the modest change of fatty acid and phospholipid profiles, the FabZ mutants have a more pronounced effect on the lipid A profile. Although the canonical hexa-acylated lipid A remains as the predominant lipid species, there is an enrichment of alternative lipid structures (referred to as “minor” lipids due to their small populations in the wild-type E. coli cells). For example, LC-MS analysis of the lipid A species extracted from CRM1 and CRM5 showed enhanced levels of penta-acylated lipid A (observed as the [M − 2H]2− ion at m/z 792.5) as well as hexa-acylated lipid A decorated with lauroyl (observed as the [M − 2H]2− ion at m/z 883.6) instead of a myristoyl chain relative to the wild-type lipid A (observed as the [M − 2H]2− ion at m/z 897.6) (Fig. 3B). Because myristoyl-ACP is used by LpxM in the last step of lipid A biosynthesis to generate the hexa-acylated lipid A structure (Fig. 1) (41, 42), the observation of an altered lipid A profile is consistent with reduced cellular concentrations of myristoyl-ACP due to decreased FabZ activity toward long chain β-hydroxyacyl-ACPs and accumulation of lauroyl-ACP in CRM1 and CRM5 cells due to the compensating FabA activity that is most efficient for generating intermediate chain length (8–12 carbons) β-hydroxyacyl-ACPs (30).
FabZ and LpxC Activities Are Co-regulated
As a central regulator of lipid A biosynthesis, the specific activity of LpxC has been reported to increase by 5–10-fold in the presence of LpxC inhibitors to counter reduced lipid A biosynthesis. Such an up-regulation of LpxC activity is due to an increase in the total amount of LpxC protein in cell extracts (31). To examine if the FabZ mutants similarly have elevated LpxC activities that might confer the observed resistance to LpxC inhibitors, we measured the LpxC specific activities in membrane-free lysates of CRM1 and CRM5 cells grown in the absence of LpxC inhibitors. Much to our surprise, we instead observed a significant drop in LpxC specific activities (Fig. 4A) in the CRM mutants. Western blotting using an LpxC-specific antibody reveals a corresponding drop in LpxC protein concentrations (Fig. 4B), suggesting that the reduced enzymatic activity is due to a decrease of the cellular protein concentration. Thus, impairment of FabZ activity causes a reduction of the cellular LpxC level and activity. To evaluate if there is a direct correlation between the FabZ and LpxC activities, we overexpressed FabZ on a plasmid in wild-type cells and again measured the level of LpxC protein. In this case, an increase of LpxC protein concentration was detected in extracts of cells overexpressing FabZ (Fig. 4C). Taken together, these experimental observations strongly suggest that the cellular activities of FabZ and LpxC are co-regulated in vivo.
After demonstrating a direct correlation of the cellular activities between LpxC and FabZ, we went on to probe if such a correlation is functionally important in vivo using a MacConkey plate assay (Fig. 5). The MacConkey agar contains high concentrations of bile salt, which is tolerated by wild-type E. coli strains due to their bile-resistant outer membrane barriers (43). When fatty acid biosynthesis was artificially increased through leaky overexpression of FabZ on a plasmid (strain W3110/pWSK29 (fabZ)/pBAD33), cells lost the ability to grow on MacConkey plates; this situation was exacerbated by further induction of FabZ with IPTG; however, when LpxA and LpxC were likewise overexpressed together on a plasmid in these cells (W3110/pWSK29 (fabZ)/pBAD33 (lpxCA)) via arabinose induction, this lethal phenotype was rescued, presumably because the additional overexpression of enzymes in lipid A biosynthesis compensated for the increased fatty acid biosynthesis as a result of FabZ overexpression. When the FabZ level was further elevated with addition of 1 mm IPTG, the LpxA-LpxC-dependent rescue became less effective, presumably due to further disturbance of the balance between the two biosynthetic pathways. Taken together, these observations highlight the functional importance of co-regulation between fatty acid and lipid A biosynthesis. Interestingly, overexpression of pBAD33 (lpxCA) alone has limited impact on cell survival and bacterial fitness under high bile salt conditions, indicating the existence of a regulatory mechanism of lipid A biosynthesis that is likely involving rapid degradation of LpxC by the FtsH protease in E. coli (44).
Point Mutation in thrS Slows Down Protein Synthesis
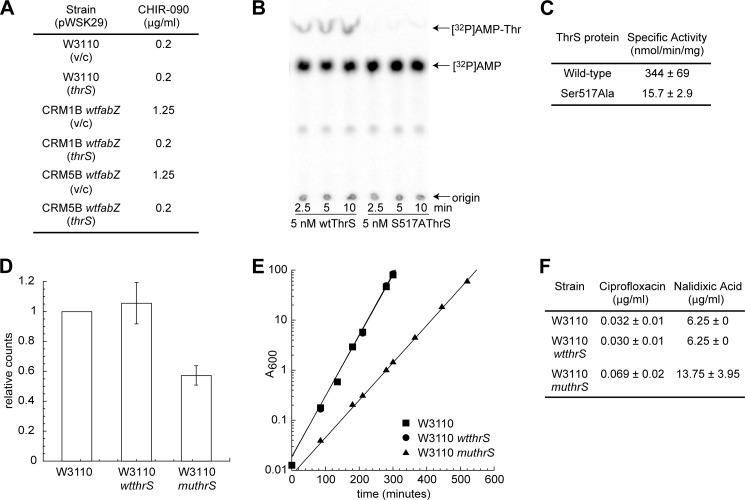
ThrS is the threonine-tRNA ligase (45), and our data show that this mutation results in ∼4–8-fold resistance to inhibition of lipid A biosynthesis by CHIR-090 (Table 2). Because ThrS plays no direct role in membrane biosynthesis, it is intriguing that a point mutation in this gene would confer resistance to LpxC inhibitors. To verify that this point mutation indeed confers resistance, we complemented the thrS mutants (CRM1B wtfabZ and CRM5B wtfabZ) with the wild-type thrS gene on a plasmid. The resulting mutants completely lost resistance to CHIR-090 (Fig. 6A), confirming that the point mutation in thrS is the bona fide resistance factor.
FIGURE 6.
Analyses of ThrS mutant. A, MICs of wild-type and CRM strains with either wild-type thrS encoded on a pWSK29 plasmid or vector control (v/c). B, PEI-cellulose plate shows S517A ThrS has significantly reduced Thr-tRNA charging activity compared with the wild-type enzyme (wtThrS). C, S517A ThrS has ∼20-fold less Thr-tRNA charging activity compared with the wild-type enzyme. D, incorporation of 14C-labeled l-amino acids into proteins in cell extracts was significantly lower for W3110 muthrS compared with wild-type strains W3110 and W3110 wtthrS. The counts for each strain are normalized to the counts detected for W3110. Values plotted are the average values taken at time points 0.5, 2, 4, and 8 min. Error bars represent the standard deviation. E, growth curves show W3110 muthrS grows significantly slower compared with the wild-type strains W3110 and W3110 wtthrS. Each plot was exponentially fitted to calculate doubling times as follows: 24.9 ± 0.39 min for W3110, 24.7 ± 0.57 min for W3110 wtthrS, and 40.3 ± 1.2 min for W3110 muthrS. F, MICs show W3110 muthrS is ∼2-fold resistant to the fluoroquinolones ciprofloxacin and nalidixic acid relative to wild-type strains W3110 and W3110 wtthrS.
The identified T1569G mutation in thrS causes a replacement of Ser-517 by Ala. In the crystal structure of E. coli ThrS (Protein Data Bank code 1QF6) (46), this residue is located in the ATP binding pocket, and its hydroxyl group lies within hydrogen bonding distance (2.8 Å) away from the N3 position of the adenine ring. Hence, the S517A mutation likely results in the loss of a hydrogen bond, which is expected to negatively impact the overall catalytic activity of ThrS. To measure the ThrS enzymatic activity in vitro, we purified wild-type and mutant ThrS enzymes to homogeneity, and 32P radiolabeled the α-phosphate on the 3′-terminal adenosine in tRNAThr to directly monitor the Thr-tRNA charging activity. In this assay, ATP, l-Thr, and [32P]tRNAThr were incubated with purified wild-type or mutant enzymes. Reaction samples were then digested with S1 nuclease to cleave off the radiolabeled 3′-terminal adenosine and release [32P]AMP or [32P]AMP-Thr. These two species were separated using thin layer chromatography (TLC). Under our assay conditions, S517A ThrS has an ∼20-fold decrease in Thr-tRNAThr charging activity compared with wild-type ThrS (Fig. 6, B and C).
Because ThrS binds threonine and CHIR-090 has a threonyl scaffold (Fig. 1), we examined if either the wild-type ThrS or its mutant could be a secondary target of CHIR-090. We used the same radiometric assay to probe for any inhibitory effects of ThrS in the presence of CHIR-090. Our results showed there was no change in specific activity when wild-type and mutant enzymes were incubated with up to 200 μm CHIR-090, suggesting that neither the wild-type nor the mutant ThrS is a secondary target of CHIR-090.
To probe the functional consequence of the S517A ThrS point mutation in vivo, we moved the mutant thrS gene into a clean W3110 genetic background without leaving any selective markers or scar sequences on the chromosome. Using this mutant (W3110 muthrS), any phenotype observed will be directly and solely caused by the point mutation in thrS. As a control, we also included a strain that went through the same construction process as W3110 muthrS; however, this cell (W3110 wtthrS) does not have any point mutations in thrS on the chromosome and should be genetically identical to W3110.
Because Thr-tRNAThr is an essential component of the protein synthetic machinery, we reasoned that a significant decrease in ThrS activity could lead to a reduction of the overall rate of protein synthesis. Indeed, when we pulse-labeled whole cells with 14C-labeled l-amino acids, we found that the total counts of 14C-labeled l-amino acids incorporated into proteins in W3110 muthrS is ∼2-fold less relative to wild-type strains W3110 and W3110 wtthrS when visualized on an SDS gel (Fig. 6D). Consistent with reduced protein synthesis, W3110 muthrS also grows ∼2-fold slower compared with W3110 and W3110 wtthrS (Fig. 6E). Interestingly, in addition to being resistant to inhibitors of lipid A biosynthesis (Table 2), this mutant has ∼2-fold resistance to ciprofloxacin and nalidixic acid (Fig. 6F), suggesting that reduced protein synthesis also confers slight resistance to inhibitors of DNA synthesis.
DISCUSSION
Through a two-step gradual exposure method, we have isolated spontaneously resistant mutants that display >200-fold resistance to CHIR-090 and other LpxC inhibitors with differing chemical scaffolds. This is the highest level of resistance observed against LpxC inhibitors to date. Interestingly, these spontaneously resistant mutants cannot be isolated in a single step, suggesting that they arise from cumulative mutations of multiple gene products, such as fabZ and thrS.
Co-regulation of Fatty Acid and Lipid Biosynthesis
The major contributor to the resistance of LpxC inhibitors arises from mutations in fabZ, accounting for as much as a 50-fold increase in MIC values. Although mutations in fabZ have been previously implicated in the resistance of a weaker LpxC inhibitor BB-78485, none of the isolated FabZ mutants were biochemically characterized (12).
Here, using an LC-MS-based assay, we show that L17Q and A71V mutations decrease FabZ enzymatic activity by ∼2.5- and ∼8-fold, respectively. In addition, we see an accumulation of partially acylated and lauroyl-acylated lipid A in FabZ mutant cells. Such an observation is consistent with decreased cellular levels of myristoyl-ACP, which is the downstream product of FabZ.
Most unexpectedly, we observed a significant decrease of LpxC specific activity and protein level in the FabZ mutant cells; conversely, when FabZ is overexpressed, an increased amount of LpxC is detected. As LpxC catalyzes the committed step of lipid A biosynthesis, these data suggest that altered fatty acid synthesis is matched by a proportional change in lipid A biosynthesis. Because fatty acid biosynthesis directly contributes to the generation of phospholipids, a balance in the biosyntheses of fatty acid/phospholipid and lipid A is critical for maintaining bacterial membrane integrity. Indeed, disruption of this balance by overexpressing FabZ (which increases fatty acid/phospholipid biosynthesis) renders bacterial cells unable to survive in high bile salt media. This lethal phenotype is rescued by overexpressing enzymes in lipid A biosynthesis. Likewise, the isolated FabZ mutant cells with impaired fatty acid biosynthesis also have reduced LpxC activity, resulting in reduced lipid A biosynthesis. Accordingly, these cells maintain full membrane integrity, and they display wild-type MIC values to polymyxin B, an antibiotic that specifically targets LPS, and remain resistant to vancomycin (MIC of 250 μg/ml), a glycopeptide that is impermeable through an intact enterobacterial outer membrane.
Because FabZ shares a common substrate with LpxA, it was previously speculated that decreased FabZ enzymatic activity would increase the concentrations of the LpxA substrate and thus buildup of the LpxC substrate. Accumulation of the LpxC substrate would enhance the overall efficiency of catalysis and directly compete with the LpxC inhibitor, countering the suppressive effect of lipid A biosynthesis (12). However, such a proposal implies unbalanced biosyntheses of lipid A and phospholipids, compromised bacterial outer membrane integrity, and significantly enhanced susceptibility to other antibiotics, none of which has been observed experimentally.
Instead, our results point to a previously unrecognized regulatory mechanism between fatty acid biosynthesis and lipid A biosynthesis that maintains a proper ratio between phospholipids and lipid A, the two essential components of Gram-negative bacterial membranes. How bacterial cells achieve this balance is unclear at this point. As a regulatory enzyme in lipid A biosynthesis, LpxC has been previously shown to be degraded by the membrane-bound protease FtsH (47). Disruption of the essential ftsH gene, which causes accumulation of LpxC activity, requires a gain-of-function mutation of fabZ in the sfhC21 allele that causes a L85P substitution and up-regulation of the FabZ dehydratase activity (44), suggesting that maintaining balance between fatty acid and lipid A biosynthesis is required for cell survival. The loss of FtsH function also leads to the activation of a Cpx two-component regulatory system (48), which consists of a kinase sensor CpxA and a response regulator CpxR. The Cpx system is known as a cell envelope stress response system that regulates diverse physiological functions (49). Interestingly, based on chromosomal sequence analysis, fabZ is expressed from hlpA operon promoter, which contains the sequence recognized by the phosphorylated CpxR activator, suggesting that FabZ expression is under the regulation of the Cpx stress response system. To further understand the regulation between LpxC and FabZ, we measured fabZ expression at the transcriptional level when LpxC was overexpressed from plasmid pBAD33 (lpxC) compared with vector control using RT-quantitative PCR to monitor the fabZ mRNA amount. The housekeeping gene gapA was used as the internal control. We noted that fabZ mRNA levels increased by ∼2.5-fold in response to LpxC overexpression after several generations (2.5–5.0 h) of logarithmic growth (data not shown). Conceivably, the overexpression of LpxC results in increased membrane stress due to the resulting unbalanced lipid A production. This membrane stress is sensed by a stress response system, such as the Cpx system, resulting in the up-regulation of FabZ expression and fatty acid synthesis. These observations suggest a potential regulatory mechanism between FabZ and LpxC through a cellular membrane stress sensing response system, and they strongly support the regulatory connection between fatty acid and lipid A biosyntheses to maintain membrane homeostasis in Gram-negative bacteria.
Reduction of Protein Synthesis Suppresses Inhibition of Lipid A Biosynthesis
In addition to FabZ, our data also identified a point mutation in the tRNA synthetase ThrS, which confers resistance to inhibitors of lipid A biosynthesis. The ThrS S517A mutation causes an ∼20-fold decrease in the Thr-tRNAThr charging activity compared with wild-type enzyme, and in accordance with the important metabolic role of threonine in rapidly growing cells (50, 51), it slows down protein synthesis and cellular growth. In such a state of hypo-metabolic activity, bacterial cells are more tolerant to not only inhibition of lipid A biosynthesis but also inhibition of DNA biosynthesis (Fig. 6F), suggesting a general defense mechanism caused by reduced protein synthesis or metabolism that may not be limited to mutations of ThrS.
Indeed, we were able to isolate other mutants that also have >200-fold resistance to CHIR-090. For example, the CRM1E mutant strain that arose from the parental strain CRM1 has a point mutation in FabZ (L17Q) but no point mutations in ThrS. Instead, this mutant has a second point mutation in RpsA, the 30 S ribosomal protein S1 required for translation of most mRNAs (52), and this mutant similarly shows reduced cellular growth. Although the exact locations of other isolated second mutations remain to be pinpointed, these mutant cells also do not have point mutations in ThrS, and they all grow significantly slower than the wild-type cells (>2-fold doubling time). Altogether, these observations suggest that potential point mutations within multiple targets that slow down protein synthesis or metabolism are able to counter the effect of impaired lipid A biosynthesis. Further supporting this notion, inclusion of sub-lethal doses of erythromycin, an inhibitor of protein synthesis, results in a minor but consistent increase (∼1.5-fold) of the MIC value of CHIR-090.
Although defects in protein synthesis have previously been reported to suppress inhibition of DNA synthesis (53), the same has not been observed for membrane biosynthesis. Hence, our observations that defects in protein biosynthesis confer significant levels of resistance to inhibitors of membrane biosynthesis as well as DNA biosynthesis highlight the regulatory interactions among all three major cellular processes in E. coli as follows: membrane biosynthesis, protein biosynthesis, and DNA biosynthesis.
Role of Homeostasis in Antibiotic Resistance
Our studies document a previously unrecognized co-regulation of LpxC and FabZ activities and demonstrate the functional importance of balanced phospholipid and lipid A biosyntheses. We additionally show that alteration of protein biosynthesis suppresses the inhibitory effect of membrane biosynthesis. Both of these resistance mechanisms fall outside of the well established paradigm of bacterial resistance involving enhanced efflux of compound and modification of compound or target protein, and they highlight maintenance of cellular homeostasis as an additional mechanism for generating antibiotic resistance.
Acknowledgments
We thank all members of the Raetz laboratory for stimulating discussions. We are especially grateful to Drs. Xiaofei Liang and Eric Toone for the generous gifts of LpxC inhibitors used in this study, Ali Masoudi for help in the preparation of acyl-ACP, Dr. William T. Doerrler for providing the polyclonal LpxC antibody, and Drs. Michael D. Been and John Hsieh for help in ThrS assay design. The Mass Spectrometry Facility, Department of Biochemistry, Duke University Medical Center, was supported by National Institutes of Health Lipid Maps Large Scale Collaborative Grant GM-069338.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-51310 (to C. R. H. R.) and Grant AI-055588 (to P. Z.).
- LpxC
- UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase
- FabZ
- R-3-hydroxymyristoyl acyl carrier protein dehydratase
- ThrS
- threonine-tRNA ligase
- ACP
- acyl carrier protein
- MIC
- minimum inhibitory concentration
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- CRM
- CHIR-090-resistant mutant.
REFERENCES
- 1. Pop-Vicas A. E., D'Agata E. M. (2005) The rising influx of multidrug-resistant Gram-negative bacilli into a tertiary care hospital. Clin. Infect. Dis. 40, 1792–1798 [DOI] [PubMed] [Google Scholar]
- 2. O'Fallon E., Pop-Vicas A., D'Agata E. (2009) The emerging threat of multidrug-resistant Gram-negative organisms in long-term care facilities. J. Gerontol. A Biol. Sci. Med. Sci. 64, 138–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cosgrove S. E., Kaye K. S., Eliopoulous G. M., Carmeli Y. (2002) Health and economic outcomes of the emergence of third-generation cephalosporin resistance in Enterobacter species. Arch. Intern. Med. 162, 185–190 [DOI] [PubMed] [Google Scholar]
- 4. Harris A., Torres-Viera C., Venkataraman L., DeGirolami P., Samore M., Carmeli Y. (1999) Epidemiology and clinical outcomes of patients with multiresistant Pseudomonas aeruginosa. Clin. Infect. Dis. 28, 1128–1133 [DOI] [PubMed] [Google Scholar]
- 5. Evans H. L., Lefrak S. N., Lyman J., Smith R. L., Chong T. W., McElearney S. T., Schulman A. R., Hughes M. G., Raymond D. P., Pruett T. L., Sawyer R. G. (2007) Cost of Gram-negative resistance. Crit. Care Med. 35, 89–95 [DOI] [PubMed] [Google Scholar]
- 6. Slama T. G. (2008) Gram-negative antibiotic resistance. There is a price to pay. Crit. Care 12, S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Walsh C. (2000) Molecular mechanisms that confer antibacterial drug resistance. Nature 406, 775–781 [DOI] [PubMed] [Google Scholar]
- 8. Raetz C. R., Whitfield C. (2002) Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anderson M. S., Bull H. G., Galloway S. M., Kelly T. M., Mohan S., Radika K., Raetz C. R. (1993) UDP-N-acetylglucosamine acyltransferase of Escherichia coli. The first step of endotoxin biosynthesis is thermodynamically unfavorable. J. Biol. Chem. 268, 19858–19865 [PubMed] [Google Scholar]
- 10. Jackman J. E., Raetz C. R., Fierke C. A. (1999) UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase of Escherichia coli is a zinc metalloenzyme. Biochemistry 38, 1902–1911 [DOI] [PubMed] [Google Scholar]
- 11. Onishi H. R., Pelak B. A., Gerckens L. S., Silver L. L., Kahan F. M., Chen M. H., Patchett A. A., Galloway S. M., Hyland S. A., Anderson M. S., Raetz C. R. (1996) Antibacterial agents that inhibit lipid A biosynthesis. Science 274, 980–982 [DOI] [PubMed] [Google Scholar]
- 12. Clements J. M., Coignard F., Johnson I., Chandler S., Palan S., Waller A., Wijkmans J., Hunter M. G. (2002) Antibacterial activities and characterization of novel inhibitors of LpxC. Antimicrob. Agents Chemother. 46, 1793–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barb A. W., Zhou P. (2008) Mechanism and inhibition of LpxC. An essential zinc-dependent deacetylase of bacterial lipid A synthesis. Curr. Pharm. Biotechnol. 9, 9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bodewits K., Raetz C. R., Govan J. R., Campopiano D. J. (2010) Antimicrobial activity of CHIR-090, an inhibitor of lipopolysaccharide biosynthesis, against the Burkholderia cepacia complex. Antimicrob. Agents Chemother. 54, 3531–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee C. J., Liang X., Chen X., Zeng D., Joo S. H., Chung H. S., Barb A. W., Swanson S. M., Nicholas R. A., Li Y., Toone E. J., Raetz C. R., Zhou P. (2011) Species-specific and inhibitor-dependent conformations of LpxC. Implications for antibiotic design. Chem. Biol. 18, 38–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liang X., Lee C. J., Chen X., Chung H. S., Zeng D., Raetz C. R., Li Y., Zhou P., Toone E. J. (2011) Syntheses, structures, and antibiotic activities of LpxC inhibitors based on the diacetylene scaffold. Bioorg. Med. Chem. 19, 852–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brown M. F., Reilly U., Abramite J. A., Arcari J. T., Oliver R., Barham R. A., Che Y., Chen J. M., Collantes E. M., Chung S. W., Desbonnet C., Doty J., Doroski M., Engtrakul J. J., Harris T. M., Huband M., Knafels J. D., Leach K. L., Liu S., Marfat A., Marra A., McElroy E., Melnick M., Menard C. A., Montgomery J. I., Mullins L., Noe M. C., O'Donnell J., Penzien J., Plummer M. S., Price L. M., Shanmugasundaram V., Thoma C., Uccello D. P., Warmus J. S., Wishka D. G. (2012) Potent inhibitors of LpxC for the treatment of Gram-negative infections. J. Med. Chem. 55, 914–923 [DOI] [PubMed] [Google Scholar]
- 18. Montgomery J. I., Brown M. F., Reilly U., Price L. M., Abramite J. A., Arcari J., Barham R., Che Y., Chen J. M., Chung S. W., Collantes E. M., Desbonnet C., Doroski M., Doty J., Engtrakul J. J., Harris T. M., Huband M., Knafels J. D., Leach K. L., Liu S., Marfat A., McAllister L., McElroy E., Menard C. A., Mitton-Fry M., Mullins L., Noe M. C., O'Donnell J., Oliver R., Penzien J., Plummer M., Shanmugasundaram V., Thoma C., Tomaras A. P., Uccello D. P., Vaz A., Wishka D. G. (2012) Pyridone methylsulfone hydroxamate LpxC inhibitors for the treatment of serious Gram-negative infections. J. Med. Chem. 55, 1662–1670 [DOI] [PubMed] [Google Scholar]
- 19. Warmus J. S., Quinn C. L., Taylor C., Murphy S. T., Johnson T. A., Limberakis C., Ortwine D., Bronstein J., Pagano P., Knafels J. D., Lightle S., Mochalkin I., Brideau R., Podoll T. (2012) Structure based design of an in vivo active hydroxamic acid inhibitor of P. aeruginosa LpxC. Bioorg. Med. Chem. Lett. 22, 2536–2543 [DOI] [PubMed] [Google Scholar]
- 20. Caughlan R. E., Jones A. K., Delucia A. M., Woods A. L., Xie L., Ma B., Barnes S. W., Walker J. R., Sprague E. R., Yang X., Dean C. R. (2012) Mechanisms decreasing in vitro susceptibility to the LpxC inhibitor CHIR-090 in the Gram-negative pathogen Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 56, 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. National Committee for Clinical Laboratory Standards (1997) Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, 4th Ed., Approved Standard M7-A4, National Committee for Clinical Laboratory Standards, Wayne, PA [Google Scholar]
- 22. McClerren A. L., Endsley S., Bowman J. L., Andersen N. H., Guan Z., Rudolph J., Raetz C. R. (2005) A slow, tight-binding inhibitor of the zinc-dependent deacetylase LpxC of lipid A biosynthesis with antibiotic activity comparable with ciprofloxacin. Biochemistry 44, 16574–16583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang R. F., Kushner S. R. (1991) Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100, 195–199 [PubMed] [Google Scholar]
- 25. Lu Y. H., Guan Z., Zhao J., Raetz C. R. (2011) Three phosphatidylglycerol-phosphate phosphatases in the inner membrane of Escherichia coli. J. Biol. Chem. 286, 5506–5518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chung H. S., Raetz C. R. (2010) Interchangeable domains in the Kdo transferases of Escherichia coli and Haemophilus influenzae. Biochemistry 49, 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guan Z., Eichler J. (2011) Liquid chromatography/tandem mass spectrometry of dolichols and polyprenols, lipid sugar carriers across evolution. Biochim. Biophys. Acta 1811, 800–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miroux B., Walker J. E. (1996) Over-production of proteins in Escherichia coli. Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298 [DOI] [PubMed] [Google Scholar]
- 29. Inoue H., Nojima H., Okayama H. (1990) High efficiency transformation of Escherichia coli with plasmids. Gene 96, 23–28 [DOI] [PubMed] [Google Scholar]
- 30. Heath R. J., Rock C. O. (1996) Roles of the FabA and FabZ β-hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis. J. Biol. Chem. 271, 27795–27801 [DOI] [PubMed] [Google Scholar]
- 31. Sorensen P. G., Lutkenhaus J., Young K., Eveland S. S., Anderson M. S., Raetz C. R. (1996) Regulation of UDP-3-O-[R-3-hydroxymyristoyl]-N-acetylglucosamine deacetylase in Escherichia coli. The second enzymatic step of lipid a biosynthesis. J. Biol. Chem. 271, 25898–25905 [DOI] [PubMed] [Google Scholar]
- 32. Bligh E. G., Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 33. Bovee M. L., Pierce M. A., Francklyn C. S. (2003) Induced fit and kinetic mechanism of adenylation catalyzed by Escherichia coli threonyl-tRNA synthetase. Biochemistry 42, 15102–15113 [DOI] [PubMed] [Google Scholar]
- 34. Ledoux S., Uhlenbeck O. C. (2008) [3′-32P]-labeling tRNA with nucleotidyltransferase for assaying aminoacylation and peptide bond formation. Methods 44, 74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barb A. W., McClerren A. L., Snehelatha K., Reynolds C. M., Zhou P., Raetz C. R. (2007) Inhibition of lipid A biosynthesis as the primary mechanism of CHIR-090 antibiotic activity in Escherichia coli. Biochemistry 46, 3793–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barb A. W., Jiang L., Raetz C. R., Zhou P. (2007) Structure of the deacetylase LpxC bound to the antibiotic CHIR-090. Time-dependent inhibition and specificity in ligand binding. Proc. Natl. Acad. Sci. U.S.A. 104, 18433–18438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cole K. E., Gattis S. G., Angell H. D., Fierke C. A., Christianson D. W. (2011) Structure of the metal-dependent deacetylase LpxC from Yersinia enterocolitica complexed with the potent inhibitor CHIR-090. Biochemistry 50, 258–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mohan S., Kelly T. M., Eveland S. S., Raetz C. R., Anderson M. S. (1994) An Escherichia coli gene (FabZ) encoding (3R)-hydroxymyristoyl acyl carrier protein dehydrase. Relation to fabA and suppression of mutations in lipid A biosynthesis. J. Biol. Chem. 269, 32896–32903 [PubMed] [Google Scholar]
- 39. Kimber M. S., Martin F., Lu Y., Houston S., Vedadi M., Dharamsi A., Fiebig K. M., Schmid M., Rock C. O. (2004) The structure of (3R)-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Pseudomonas aeruginosa. J. Biol. Chem. 279, 52593–52602 [DOI] [PubMed] [Google Scholar]
- 40. Magnuson K., Jackowski S., Rock C. O., Cronan J. E., Jr. (1993) Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol. Rev. 57, 522–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vorachek-Warren M. K., Ramirez S., Cotter R. J., Raetz C. R. (2002) A triple mutant of Escherichia coli lacking secondary acyl chains on lipid A. J. Biol. Chem. 277, 14194–14205 [DOI] [PubMed] [Google Scholar]
- 42. Clementz T., Zhou Z., Raetz C. R. (1997) Function of the Escherichia coli msbB gene, a multicopy suppressor of htrB knockouts, in the acylation of lipid A. Acylation by MsbB follows laurate incorporation by HtrB. J. Biol. Chem. 272, 10353–10360 [DOI] [PubMed] [Google Scholar]
- 43. Nikaido H. (1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology (Neidhardt F. C., Curtiss R., 3rd, Ingraham J. L., Lin E. C. C., Low K. B., Magasanik B., Jr., Reznikoff W. S., Riley M., Schaechter M., Umbarger H. E., eds) 2nd Ed., pp. 29–47, American Society for Microbiology, Washington, D. C. [Google Scholar]
- 44. Ogura T., Inoue K., Tatsuta T., Suzaki T., Karata K., Young K., Su L. H., Fierke C. A., Jackman J. E., Raetz C. R., Coleman J., Tomoyasu T., Matsuzawa H. (1999) Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol. Microbiol. 31, 833–844 [DOI] [PubMed] [Google Scholar]
- 45. Johnson E. J., Cohen G. N., Saint-Girons I. (1977) Threonyl-transfer ribonucleic acid synthetase and the regulation of the threonine operon in Escherichia coli. J. Bacteriol. 129, 66–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sankaranarayanan R., Dock-Bregeon A. C., Romby P., Caillet J., Springer M., Rees B., Ehresmann C., Ehresmann B., Moras D. (1999) The structure of threonyl-tRNA synthetase-tRNA(Thr) complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell 97, 371–381 [DOI] [PubMed] [Google Scholar]
- 47. Langklotz S., Schäkermann M., Narberhaus F. (2011) Control of lipopolysaccharide biosynthesis by FtsH-mediated proteolysis of LpxC is conserved in enterobacteria but not in all gram-negative bacteria. J. Bacteriol. 193, 1090–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shimohata N., Chiba S., Saikawa N., Ito K., Akiyama Y. (2002) The Cpx stress response system of Escherichia coli senses plasma membrane proteins and controls HtpX, a membrane protease with a cytosolic active site. Genes Cells 7, 653–662 [DOI] [PubMed] [Google Scholar]
- 49. Dartigalongue C., Missiakas D., Raina S. (2001) Characterization of the Escherichia coli sigma E regulon. J. Biol. Chem. 276, 20866–20875 [DOI] [PubMed] [Google Scholar]
- 50. Almaas E., Kovács B., Vicsek T., Oltvai Z. N., Barabási A. L. (2004) Global organization of metabolic fluxes in the bacterium Escherichia coli. Nature 427, 839–843 [DOI] [PubMed] [Google Scholar]
- 51. Wang J., Alexander P., McKnight S. L. (2011) Metabolic specialization of mouse embryonic stem cells. Cold Spring Harbor Symp. Quant. Biol. 76, 183–193 [DOI] [PubMed] [Google Scholar]
- 52. Sørensen M. A., Fricke J., Pedersen S. (1998) Ribosomal protein S1 is required for translation of most, if not all, natural mRNAs in Escherichia coli in vivo. J. Mol. Biol. 280, 561–569 [DOI] [PubMed] [Google Scholar]
- 53. Bollenbach T., Quan S., Chait R., Kishony R. (2009) Nonoptimal microbial response to antibiotics underlies suppressive drug interactions. Cell 139, 707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]