Background: Nucleocytoplasmic trafficking of class IIa histone deacetylases is crucial for various biological processes.
Results: cAMP induces dephosphorylation at an SP motif conserved in HDAC4, HDAC5, and HDAC9 but not in HDAC7.
Conclusion: Dephosphorylation at the SP motif governs cAMP sensitivity and nuclear localization of class IIa histone deacetylases.
Significance: Cellular signaling pathways may act upon cAMP to promote dephosphorylation and nuclear localization of class IIa histone deacetylases.
Keywords: Cyclic AMP (cAMP), Histone Deacetylase, Nuclear Translocation, Phosphorylation, Protein Kinase A (PKA)
Abstract
Histone deacetylase 4 (HDAC4) and its paralogs, HDAC5, -7, and -9 (all members of class IIa), possess multiple phosphorylation sites crucial for 14-3-3 binding and subsequent nuclear export. cAMP signaling stimulates nuclear import of HDAC4 and HDAC5, but the underlying mechanisms remain to be elucidated. Here we show that cAMP potentiates nuclear localization of HDAC9. Mutation of an SP motif conserved in HDAC4, -5, and -9 prevents cAMP-stimulated nuclear localization. Unexpectedly, this treatment inhibits phosphorylation at the SP motif, indicating an inverse relationship between the phosphorylation event and nuclear import. Consistent with this, leptomycin B-induced nuclear import and adrenocorticotropic hormone (ACTH) treatment result in the dephosphorylation at the motif. Moreover, the modification synergizes with phosphorylation at a nearby site, and similar kinetics was observed for both phosphorylation events during myoblast and adipocyte differentiation. These results thus unravel a previously unrecognized mechanism whereby cAMP promotes dephosphorylation and differentially regulates multisite phosphorylation and the nuclear localization of class IIa HDACs.
Introduction
Lysine acetylation has recently emerged as a major post-translational modification important for chromatin regulation and many other cellular processes (1, 2) (reviewed in Ref. 3). In mammals, there are 18 known deacetylases responsible for removing the acetyl group from acetyllysine (4). Among them are histone deacetylases 4, 5, 7, and 9 (HDAC4, -5, -7, and -9),4 which are highly homologous and form a subgroup within class II. Cell-based studies have established that this group of enzymes plays important roles in skeletal muscle differentiation (5) and plasticity (6, 7), cardiac hypertrophy (8), angiogenesis (9–11), T-cell selection (12), and neurodegeneration (13). Not surprisingly, the knock-out mice display striking phenotypes; Hdac4 inactivation causes premature ossification of developing bones (14), Hdac4−/−;Hdac5−/− compound mice exhibit abnormal neurogenic muscle atrophy (15), Hdac5−/−;Hdac9−/− double knockouts show lethal ventricular septal defects and thin-walled myocardium (16), and Hdac7 deletion leads to embryonic lethality due to a failure in endothelial cell-cell adhesion and consequent rupture of blood vessels (17). Moreover, Hdac5 in mice controls behavioral adaptation to chronic emotional stimuli (18), murine Hdac7 has recently been identified as a novel oncogene (19), reduced human HDAC7 activity restores CFTR function in cystic fibrosis (20), and HDAC4 deletion in patients is linked to the brachydactyly mental retardation syndrome with characteristic bone malformation (21).
Phosphorylation at multiple phosphorylatable 14-3-3 binding sites and subsequent nuclear export of the deacetylases are important for regulating diverse physiological and pathological effects (reviewed in Refs. 22 and 23). A number of kinases are known to be responsible, including Ca2+/calmodulin-dependent protein kinases (CaMKs) (5, 24–27) and protein kinase D (PKD) (28–31). Stimuli that activate these kinases (e.g. an increase in intracellular [Ca2+] (32, 33) and VEGF treatment (11, 34)) induce class IIa HDAC phosphorylation and nuclear export, leading to derepression of MEF2-dependent transcription. In contrast, dephosphorylation of these serine residues by phosphatases, such as protein phosphatase 2A (PP2A) and myosin phosphatase (PP1β/MYPT1), promotes nuclear localization of class IIa HDACs and repression of MEF2-dependent transcription (10, 35–37). An important issue is how various cellular stimuli act through these kinases and phosphatases to link class IIa HDAC regulation to cellular signaling networks.
Upstream from such signaling networks, hormones act upon cells through G protein-coupled receptor-induced activation of adenylyl cyclase, which generates cAMP. This second messenger exerts many biological effects, in part through the activation of cAMP-dependent protein kinase (also called protein kinase A (PKA)) (38). cAMP signaling is tightly controlled temporally and spatially by phosphodiesterases that break down cAMP and protein kinase A anchoring proteins that serve as scaffolds to localize cAMP/PKA signaling nodes to discrete subcellular compartments (39, 40). PKA affects the function of many cell types, including skeletal muscle and neurons, in which one major target is the transcription factor CREB (41–43). Elevated cAMP levels also promote nuclear localization of HDAC5 in hippocampal neurons (44), suggesting that HDAC5 is another target. Thus, it would be interesting to elucidate the underlying mechanisms.
As shown for HDAC5, cAMP also promotes nuclear localization of HDAC4 in C2C12 myoblasts and MLB14 chondrocytes (45, 46). Two possible mechanisms have been suggested: (i) cAMP activates PKA, phosphorylates MEF2D, and then stimulates MEF2D binding and nuclear localization of HDAC4; and (ii) cAMP indirectly activates PP2A, which in turn dephosphorylates HDAC4 and inhibits its 14-3-3 binding and cytoplasmic localization (45, 46). We and others have recently demonstrated that LKB1 activates SIK2 and SIK3 to promote phosphorylation at 14-3-3 binding sites of class IIa HDACs and stimulate their cytoplasmic localization (47, 48, 89). As PKA phosphorylates LKB1 and SIKs (49–51), a third possible mechanism is that cAMP acts through the LKB1-SIK kinase cascade to regulate nucleocytoplasmic trafficking of HDAC4. We have investigated this intriguing possibility and report here that cAMP antagonizes phosphorylation of an SP motif conserved in HDAC4, -5, and -9 (but not HDAC7) to differentially regulate nuclear localization of these four deacetylases.
MATERIALS AND METHODS
Preparation of Plasmid Constructs
Plasmids constructs for GFP- and FLAG-tagged wild-type HDAC4, single point mutant S246A, and triple mutant (TM; S246A/S467A/S632A) have been described previously (52). An expression plasmid for HA-tagged mouse HDAC5 (53) was used to create GFP-tagged HDAC5 using a pEGFP-C2 (BD Biosciences) derivative. An expression plasmid for FLAG-tagged mouse HDAC7 (25) was utilized for construction of the expression plasmid for GFP-HDAC7. The GFP-HDAC9 expression construct was derived from an expression plasmid for FLAG-tagged human HDAC9 (54–56), by insertion of the coding sequence into a pEGFP-C2 derivative. cDNAs for SIK2, SIK3, and PKA were purchased from Open Biosystems and subcloned into pcDNA3.1 derivatives. Expression plasmids for point mutants of HDAC4 (S266A, S265A, S266D, and P267A), HDAC5 (S279A and S279D), HDAC7 (N197S and K196S/N197S), and SIK2 (S587A, T484A, and T484A/S587A) were generated by PCR-mediated site-directed mutagenesis using the Pfu polymerase system according to the manufacturer's instructions (Fermentas). All mutants were verified by automatic sequencing.
Cell Culture, Differentiation, and Transfection
HEK293, HeLa, and C2C12 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Sigma), and penicillin G and streptomycin (100 units/ml and 100 μg/ml, respectively; Invitrogen). For C2C12 differentiation, the medium was switched to DMEM containing horse serum (2%; Sigma), 100 units/ml penicillin G, and 100 μg/ml streptomycin and changed every 2 days until multinuclear myotubes were formed. Y1 adrenocorticotropic tumor cells (57) were grown in Ham's F12K medium supplemented with 2 mm glutamine, 15% horse serum, 2.5% FBS, 100 units/ml penicillin G, and 100 μg/ml streptomycin.
For cell treatment, the chemicals 8-Br-cAMP (1 mm), forskolin (10 μm), H-89 (10 μm), okadaic acid (100 nm), staurosporine (1 μm), KN-62 (10 μm), Bis I (10 μm), PD 98059 (10 μm), and kenpaullone (5 μm) were purchased from Calbiochem and used at the concentrations indicated. Leptomycin B (Sigma) was used at a final concentration of 10 ng/ml, whereas ACTH (Bachem) was used at 1 μm.
3T3-L1 preadipocytes (58) were grown in DMEM containing 10% fetal calf serum, 100 units/ml penicillin G, and 100 μg/ml streptomycin. 5 or 10% CO2 incubators were used. For differentiation, these cells were allowed to reach full confluence and maintained for an additional 2 days for subsequent differentiation by replacement of the growth medium with DMEM containing 10% FBS, 1 μm insulin, 0.25 μm dexamethasone, 0.5 mm isobutylmethylxanthine (Sigma), 100 units/ml penicillin G, and 100 μg/ml streptomycin. This was counted as differentiation day 0; on days 2, 4, and 6, the medium was switched to the same differentiation medium except that dexamethasone and isobutylmethylxanthine were omitted. Typically, foci of adipocytes containing lipid droplets within the cytoplasm appeared on day 4 and >90% cells became differentiated on day 8. The lipid droplets could be directly assessed under a light microscope or be stained with Oil Red O (Sigma).
For transfection, 40,000 HEK293 or 60,000 C2C12 cells were plated in one well of a 12-well cell culture plate. For HEK293, transfection was performed with 3 μl of Superfect (Qiagen) and 1.5 μg of total DNA. For C2C12, transfection was performed with 3 μl of Lipofectamine 2000 (Invitrogen) and 1.5 μg of DNA. Green fluorescence or immunofluorescence microscopy was then performed 24–48 h post-transfection. For Western blot and immunoprecipitation, 100,000 C2C12 cells were plated in 6-cm plates, and transfection was performed with 15 μl of Lipofectamine 2000 (Invitrogen) and 5 μg of plasmid DNA according to the manufacturer's instructions.
Immunofluorescence Microscopy
C2C12 and HeLa cells were seeded on glass coverslips. Once cells had reached ∼60% confluence, they were treated with 1 mm 8-Br-cAMP for 30 min. Coverslips were then washed twice with phosphate-buffered saline (PBS), fixed with 2% paraformaldehyde for 20 min, and washed three times with PBS. Cells were then permeabilized with 0.2% Triton X-100 for 10 min, followed by washes with 100 mm glycine, three times for 5 min each. After blocking with immunofluorescence buffer (PBS with 0.2% Triton X-100 and 0.05% Tween 20) containing 2% BSA for 45 min, cells were incubated overnight at 4 °C with the anti-HDAC4 antibody (1:500). After the primary antibody incubation, cells were rinsed with immunofluorescence buffer (3 times for 5 min each) and then incubated in the AlexaFluor 488 goat anti-rabbit IgG (H + L) secondary antibody (Invitrogen, Molecular Probes) (1:1000) at room temperature for 45 min, followed by washes with immunofluorescence buffer, three times for 5 min each. Cells were then counterstained with DAPI for 5 min at room temperature and then washed twice with double-distilled H2O (5 min each) and mounted on slides for microscopy.
Immunoprecipitation and Western Blotting Analysis
The day after transfection, C2C12 cells were washed twice with cold PBS and lysed in 0.25 ml of buffer K (20 mm sodium phosphate, pH 7.0, 150 mm KCl, 30 mm sodium pyrophosphate, 0.1% Nonidet P-40, 5 mm EDTA, 10 mm NaF, 0.1 mm Na3VO4, 25 mm β-glycerophosphate, and protease inhibitors). For affinity purification of FLAG-tagged proteins, 200 μl of extracts was added to 10 μl of M2 agarose beads (Sigma) and rotated at 4 °C for at least 2 h. After four washes with buffer K, bound proteins were eluted with 2 μl of FLAG peptide (Sigma) in 25 μl of buffer K.
After the addition of 3× SDS sample buffer, soluble cell extracts and affinity-purified proteins were boiled for 5 min, separated by SDS-PAGE, and transferred to nitrocellulose membranes. Membranes were blocked in PBS plus 0.15% Tween 20 (PBS-T) with 20% horse serum for 1 h at room temperature and then incubated overnight at 4 °C with anti-FLAG (Sigma), -HDAC4, -phospho-Ser-246, -phospho-Ser-266, and -phospho-Thr-707 antibodies (see below). For detection of endogenous proteins in cell extracts, membranes were blocked in PBS-T with 5% milk for 1 h at room temperature and then incubated overnight at 4 °C with anti-HDAC4, -phospho-Ser-246, -phospho-Ser-266, -phospho-Thr-707, and -α-tubulin (Sigma) antibodies. Membranes were then washed in PBS-T for six times (8 min each), and then incubated in the appropriate secondary antibody conjugated to HRP for 1 h at room temperature. After another set of six 8-min washes in PBS-T, membranes were incubated in PBS (two times for 5 min each) and then visualized on film after 5 min of incubation in Supersignal enhanced chemiluminescent solution (Pierce).
Antibody Generation and Affinity Purification
The anti-HDAC4 antibody was affinity-purified from polyclonal rabbit antisera generated against an N-terminal fragment of HDAC4 (52). The antibody was highly selective for HDAC4 and did not cross-react with HDAC5, -7, or -9.5 The anti-phospho-Ser-266 (HDAC4) antibody was prepared by immunization of two rabbits with the phosphopeptide ERRSpSPLLRRKDC, where pS is phospho-Ser and the C-terminal Cys was added for conjugation to mcKLH and affinity purification gel. The phosphopeptide was conjugated to Imject maleimide-activated mcKLH (Pierce, Thermo Scientific) for injection into rabbits, which were housed and handled by the McGill Animal Facility according to approved animal use Protocols. The antiserum was used for affinity purification on SulfoLink gel (Pierce) linked to the phosphopeptide through its free Cys. The specificity of the affinity-purified antibody toward phospho-Ser-266 versus the non-phosphorylated peptide was confirmed by dot blotting. The anti-phospho-Ser-246 polyclonal antibody was prepared by immunization of two rabbits with the phosphopeptide LRKTApSEPNLKC, and the affinity-purified one was specific to HDAC4 phosphorylated at Ser-246.5 The anti-phospho-Thr-707 polyclonal rabbit antibody was prepared by immunization of one rabbit with the phosphopeptide CIRGRKApTLEE, and the affinity-purified one was specific to HDAC4 phosphorylated at Thr-707 (data not shown).
In Vitro Kinase Assay
Sf9 cells were infected with baculoviruses expressing FLAG-tagged HDAC4, and soluble extracts were prepared for affinity purification on anti-FLAG M2-agarose as described (59). In vitro kinase assays were performed in a total volume of 40 μl comprising 20 μl of IVK buffer (50 mm Tris, pH 7.5, 5 mm MgOAc, 5 mm MnCl2, 0.1 mm EGTA, 1 mm DTT, 10 mm NaF, 0.1 mm Na3VO4, and protease inhibitors) containing up to 4 μg of FLAG-HDAC4, the catalytic subunit of bovine heart PKA (Calbiochem (catalog no. 539576, 1 mg/ml, 20,000 units/μg) or Sigma), and 100 pmol of ATP. Reactions were carried out at 30 °C for 1 h and were stopped upon the addition of 20 μl of ice-cold 3× SDS sample buffer. Samples were boiled for 5 min and separated by SDS-PAGE, which was then either followed by staining with Colloidal Coomassie Blue or immunoblotting with anti-phospho-Ser-266 or anti-FLAG antibody. For immunoblotting, samples separated by SDS-PAGE were transferred to nitrocellulose membranes, which were blocked in PBS-T containing 20% horse serum for 1 h at room temperature. Membranes were incubated with anti-phospho-Ser-266 (1:5000) antibody overnight at 4 °C.
Statistical Analysis
Data are presented as means ± S.E. One-way analyses of variance were performed with a Bonferroni post hoc test. Unpaired two-tailed Student's t tests were performed with Prism (GraphPad). p < 0.05 was considered statistically significant.
RESULTS
Differential Regulation of Class IIa HDACs by cAMP Signaling
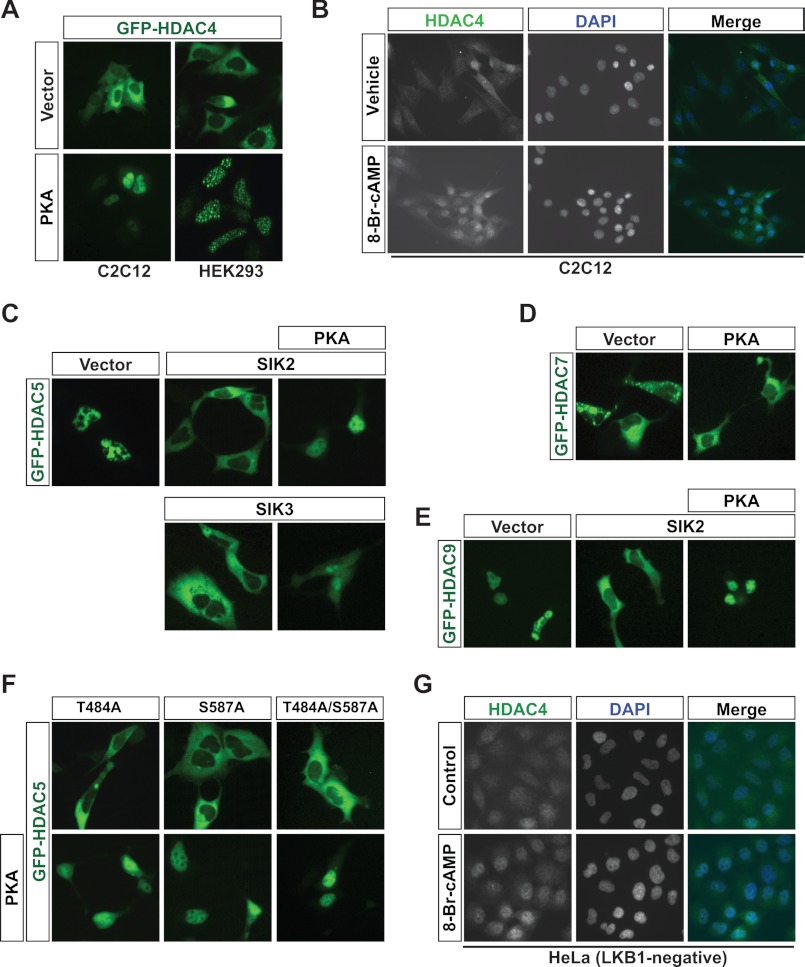
While investigating how PKA acts through the LKB1-SIK kinase cascade, we noticed that PKA promoted nuclear localization of GFP-HDAC4 (Fig. 1A). In addition, when C2C12 myoblasts were exposed to 8-Br-cAMP prior to immunofluorescence microscopy using an affinity-purified anti-HDAC4 antibody, HDAC4 localization shifted from a pancellular distribution to a predominantly nuclear pattern (Fig. 1B). This is reminiscent of what was reported previously (45). Unlike HDAC4, HDAC5 exhibited a basal nuclear localization pattern (Fig. 1C, left). Therefore, to investigate whether cAMP/PKA signaling has a similar effect on HDAC5, we first induced a cytoplasmic localization pattern for HDAC5. This was achieved by co-expression of GFP-HDAC5 along with SIK2, SIK3 (Fig. 1C), or CaMKIV (supplemental Fig. S1). As was the case for HDAC4, PKA prevented the cytoplasmic localization of GFP-HDAC5, whether SIK2/3 (Fig. 1C) or CaMKIV (supplemental Fig. S1) was co-expressed.
FIGURE 1.
PKA differentially stimulates the nuclear localization of class IIa HDACs. A, C2C12 (left) or HEK293 cells (right) were transiently transfected with a GFP-HDAC4 expression plasmid, together with either pcDNA3.1 or the expression plasmid encoding PKA. GFP-HDAC4 localization was visualized via live green fluorescence microscopy. B, immunofluorescence microscopy of endogenous HDAC4 subcellular localization in C2C12 cells by use of the anti-HDAC4 antibody after 30 min of treatment with 1 mm 8-Br-cAMP. C, as in A except that HEK293 cells were transiently transfected with a GFP-HDAC5 expression plasmid together with pcDNA3.1 or an expression plasmid encoding SIK2 (top) or SIK3 (bottom) in the presence or absence of the expression plasmid for PKA. D, as in A except that HEK293 cells were transiently transfected with the GFP-HDAC7 expression plasmid together with pcDNA3.1 or a PKA expression plasmid. E, same as in C (top) except that a GFP-HDAC9 expression plasmid was transfected. 300 ng of each plasmid was used. F, HEK293 cells were transfected with a GFP-HDAC5 expression plasmid and the indicated SIK2 mutant expression plasmid (T484A, S587A, or T484A/S587A) with (top) or without (bottom) the PKA expression plasmid. G, as in B except that LKB1-negative HeLa cells were used for immunofluorescence microscopy.
Unlike HDAC4 and HDAC5, it was unclear how HDAC7 and HDAC9 are subject to regulation by cAMP/PKA signaling. Thus, we analyzed the subcellular localization of GFP-HDAC7 and -HDAC9. Similar to GFP-HDAC4, GFP-HDAC7 was mainly cytoplasmic in HEK293 cells (Fig. 1D, left). Unexpectedly, the cytoplasmic localization pattern of GFP-HDAC7 was not significantly altered by coexpression of PKA (Fig. 1D, right), suggesting that HDAC7 displays resistance or reduced sensitivity to PKA signaling. We then analyzed the effect on the subcellular localization of GFP-HDAC9. Similar to HDAC5, HDAC9 exhibited a basal nuclear localization pattern (Fig. 1E, left). Thus, we first had to induce a cytoplasmic localization pattern for HDAC9 by coexpression along with SIK2 (Fig. 1E, middle). As was the case for HDAC5, PKA prevented the cytoplasmic localization of GFP-HDAC9 induced by SIK2 expression (Fig. 1E, right). Taken together, these results indicate that cAMP/PKA promotes nuclear import of HDAC4, -5, and -9 but has a lesser impact on HDAC7.
Class IIa HDAC Regulation by PKA Is Independent of SIK Inhibition
PKA has been shown to phosphorylate SIK2 on Ser-587, and mutation of this residue to alanine (S587A) renders SIK2 resistant to PKA-mediated inhibition (51). Thus, we tested whether PKA could still have its effect on HDAC5 localization when the S587A mutant was used to export HDAC5 to the cytoplasm. PKA was still able to prevent nuclear export of HDAC5 caused by SIK2 S587A (Fig. 1F). We found the same thing using another SIK2 mutant (T484A) in which a second putative PKA consensus phosphorylation motif was disrupted or when a double mutant bearing both these mutations (T484A/S587A) was used to relocate HDAC5 to the cytoplasm (Fig. 1F). To complement the transient transfection experiments, we monitored the localization of endogenous HDAC4 in response to 8-Br-cAMP in cells with defective SIK signaling. For this, we used HeLa cells, in which SIKs are inactive (60) due to the lack of expression of their upstream activator, the tumor suppressor kinase LKB1 (61). In these cells, HDAC4 was more nuclear in response to 8-Br-cAMP treatment (Fig. 1G). Although PKA does inhibit SIKs (51), these results indicate that SIK inhibition is not the underlying cause for the effect of cAMP/PKA and further suggest that HDAC4, -5, and -9 are the direct targets. This is also consistent with the finding that expression of PKA alone promoted nuclear import of GFP-HDAC4 (Fig. 1A).
An SP Motif of HDAC4 Is Essential for Regulation by PKA
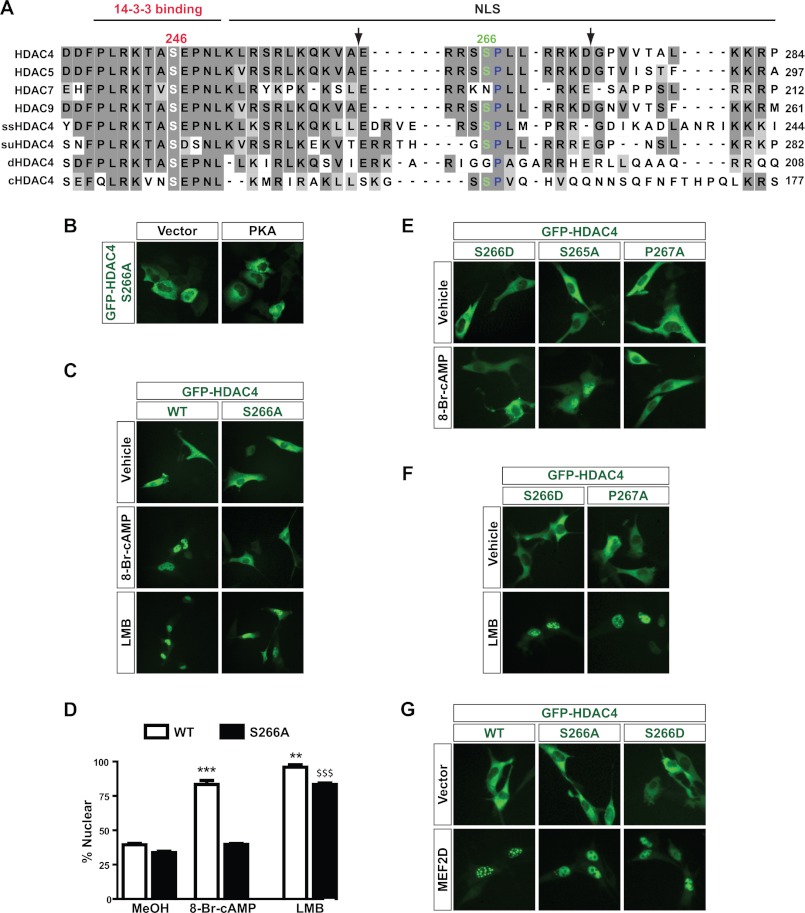
Compared with HDAC4, -5, and -9, HDAC7 was less sensitive to PKA (Fig. 1D), so our focus shifted to examine differences in the sequences of these HDACs that may ultimately be responsible for the differential sensitivity to cAMP/PKA signaling. For this, we performed a sequence alignment comparison. HDAC4 possesses two putative PKA sites, Ser-266 (RRXS) and Thr-707 (RKXT), conforming to the consensus sequence R(R/K)X(S/T). Strikingly, Ser-266 is conserved in HDAC5 (Ser-279) and HDAC9 (Ser-243) but is absent from HDAC7 (Fig. 2A). By contrast, the Thr-707 site is located within the deacetylase domain and is thus highly conserved among the four HDACs. In addition, the Ser-266 site is evolutionarily conserved from zebrafish to humans (supplemental Fig. S2) and is located within the nuclear localization signal and is adjacent to the N-terminal 14-3-3 binding site (Ser-246; Fig. 2A). Thus, Ser-266 appeared to be a strong target site from cAMP/PKA signaling. To test this, we engineered a GFP-S266A expression plasmid, in which Ser-266 of HDAC4 was mutated to alanine. This mutation had no effect on basal localization of HDAC4, but it prevented nuclear localization induced by PKA overexpression in HEK293 cells (Fig. 2B). HDAC4 S266A was also resistant to 8-Br-cAMP treatment in C2C12 cells (Fig. 2, C and D). These results suggest that Ser-266 may represent a novel PKA phosphorylation site, so we also created an S266D mutant, in which the negatively charged aspartate residue loosely mimics the negative charge of a phosphate group. In that case, we expected the S266D mutant to be constitutively nuclear, but we detected no change in basal HDAC4 localization (Fig. 2E). In fact, the S266D mutant behaved exactly as the S266A mutant did, because it was also resistant to cAMP-mediated nuclear localization (Fig. 2E). By contrast, substitution of the second putative PKA site, Thr-707, with alanine or glutamate had minimal effects (data not shown).
FIGURE 2.
Ser-266 and Pro-267 of HDAC4 form a novel regulatory motif. A, sequence comparison of the nuclear localization signal (NLS) and the adjacent 14-3-3 binding site of human HDAC4 with the corresponding region of human HDAC5, HDAC7, HDAC9, and HDAC4 orthologs from sea squirt (ss), sea urchin (su), Drosophila melanogaster (d), and Caenorhabditis elegans (c). Identical residues are highlighted in dark gray boxes, whereas similar residues are highlighted in light gray boxes. Ser-246 and Ser-266 of HDAC4 as well as the corresponding residues in its paralogs and orthologs are presented in white and green type, respectively, whereas Pro-267 is presented in blue type. The peptide used for generating the anti-phospho-Ser-266 antibody is indicated by two flanking arrows. B, HEK293 cells were transiently transfected with an expression plasmid encoding GFP-S266A together with pcDNA empty vector or the PKA expression plasmid. C, C2C12 cells were transfected with GFP-HDAC4 or -S266A expression plasmids and then treated with 1 mm 8-Br-cAMP for 30 min or 10 ng/ml LMB or vehicle (70% methanol) for 1 h before HDAC4 localization was analyzed through live green fluorescence microscopy. D, quantification of C. At least 100 cells were counted for each condition for three independent experiments. ***, p < 0.001 versus S266A for cAMP; **, p < 0.01 versus S266A for LMB; $$$, p < 0.001 versus S266A for vehicle and S266A for cAMP. E, as in C except that expression plasmids for GFP-S266D, -S265A, and -P267A were transfected prior to 8-Br-cAMP treatment. F, as in C except that expression plasmids for GFP-S266D and -P267A were transfected prior to LMB treatment. G, as in C except that GFP-HDAC4, -S266A, or -S266D was cotransfected with empty vector (top) or a MEF2D expression plasmid (bottom).
We also investigated the importance of the sequence surrounding Ser-266. Mutation of Ser-265 to alanine (S265A) had a very minor effect on localization because this mutant largely accumulated in the nucleus following 8-Br-cAMP treatment (Fig. 2E). In contrast, mutant P267A was unresponsive to 8-Br-cAMP treatment and remained in the cytoplasm like S266A (Fig. 2E). Consistent with these results, Ser-266 and Pro-267 are highly conserved during evolution (supplemental Fig. S2). Thus, Ser-266 and Pro-267 form an evolutionarily conserved SP motif that is crucial for cAMP-mediated nuclear localization of HDAC4.
We next tested whether mutants S266A and P267A were defective in general nuclear import or whether the effect was specific to cAMP/PKA-mediated nuclear localization. For this, we applied two stimuli known to induce nuclear localization of class IIa HDACs. First, we treated C2C12 cells with leptomycin B (LMB), an inhibitor of the nuclear export receptor CRM1 (62). Because class IIa HDACs constantly shuttle between the nucleus and cytoplasm, inhibition of nuclear export by LMB results in nuclear accumulation of these HDACs (52, 63). As expected, LMB treatment induced nuclear localization of HDAC4 (Fig. 2, C and D). The nuclear localization of mutant S266A was slightly reduced compared with wild-type HDAC4, but it was still predominantly nuclear in response to LMB (Fig. 2, C and D). The S266D and P267A mutants also accumulated in the nucleus after LMB treatment (Fig. 2F). Another stimulus known to cause nuclear accumulation of HDAC4 is overexpression of MEF2C (25, 64–67). This was also the case for MEF2D (Fig. 2G, left panels). More importantly, the S266A and S266D mutants also moved to the nucleus upon overexpression of MEF2D (Fig. 2G, middle and right panels). These results demonstrate that Ser-266 and Pro-267 are important for cAMP/PKA-mediated nuclear accumulation but not essential for general nuclear import of HDAC4.
Ser-279 of HDAC5 and Ser-243 of HDAC9 Are Crucial for cAMP Sensitivity
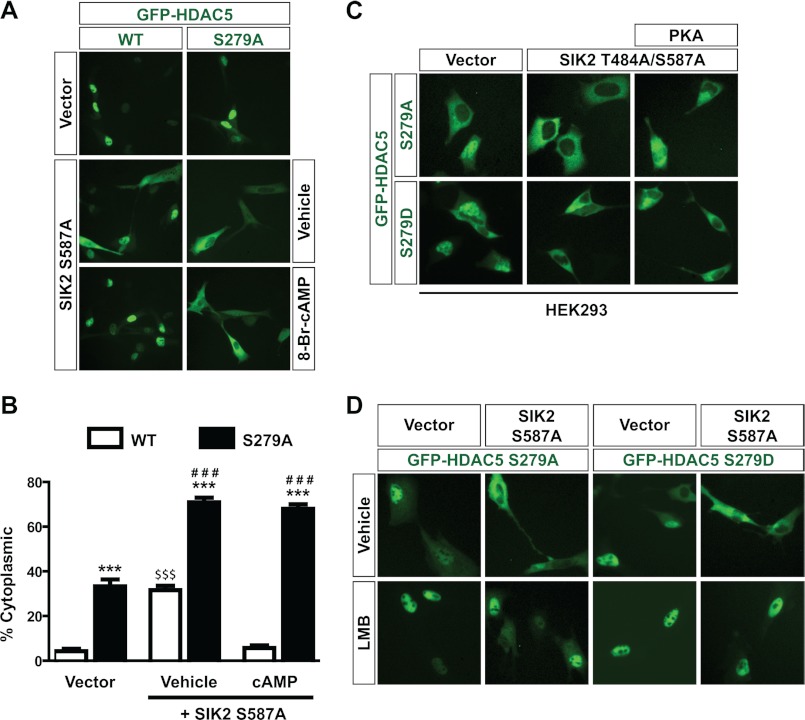
Because the cAMP/PKA-responsive SP motif of HDAC4 is conserved in HDAC5 (Fig. 2A), we sought to determine whether HDAC5 shuttling is regulated in a similar fashion. As in HEK293 cells (Fig. 1C), HDAC5 was predominantly nuclear in C2C12 cells (Fig. 3A). These cells were used because, unlike HEK293 cells, they are responsive to cAMP administration. Thus, we expressed the constitutively active SIK2 mutant S587A to promote localization of HDAC5 to the cytoplasm to determine the effect of cAMP on nuclear import of HDAC5. Similar to what was observed in HEK293 cells upon PKA overexpression (Fig. 1F), S587A expression stimulated cytoplasmic localization of GFP-HDAC5, and cAMP administration blocked this effect (Fig. 3, A and B). Next, we repeated the same experiment with GFP-S279A, in which Ser-279, equivalent to Ser-266 of HDAC4, was mutated to alanine. Strikingly, S279A was significantly more cytoplasmic than wild-type HDAC5 (Fig. 3, A and B). This was in contrast to what we observed with HDAC4 (Fig. 2, B–D), but this can be explained by the fact that HDAC4 was already fully cytoplasmic in the basal state, and thus the cytoplasmic localization could not be increased. In addition, S279A was also more cytoplasmic than wild-type HDAC5 in response to coexpression of the SIK2 mutant S587A (Fig. 3, A and B). These results indicate an interesting synergy between SIK2 S587A coexpression and Ser-279 mutation in promoting cytoplasmic localization of HDAC5.
FIGURE 3.
Ser-279 regulates subcellular localization of HDAC5. A, C2C12 cells were transiently transfected with the expression plasmid for GFP-HDAC5 or -S279A along with an empty vector or the expression plasmid for the constitutively active SIK2 S587A mutant. Cells were then treated with vehicle (H2O) or 1 mm 8-Br-cAMP for 30 min, followed by live green fluorescence microscopy. B, quantification of A. At least 100 cells were counted for each condition for three independent experiments. ***, p < 0.001 versus WT under the same conditions; ###, p < 0.001 versus S279A under vehicle conditions with empty vector transfected; $$$, p < 0.001 versus WT in two other conditions. C, HEK293 cells were transiently transfected with the expression plasmid for GFP-HDAC5 S279A or S279D together with empty vector or the expression plasmid for the SIK2 mutant T484A/S587A, with or without the PKA expression plasmid. D, as in A except that GFP-S279A or -S279D was transfected with an empty vector (left) or the expression plasmid for the SIK2 mutant S587A (right), prior to treatment with vehicle (70% methanol, top) or 10 ng/ml LMB (bottom) for 1 h.
We then analyzed the effect of cAMP signaling. After 8-Br-cAMP treatment, wild-type HDAC5 returned to the nucleus (even in the presence of coexpressed S587A), whereas mutant S279A remained fully cytoplasmic (Fig. 3, A and B). Thus, like Ser-266 in HDAC4, Ser-279 of HDAC5 is necessary for cAMP-mediated nuclear accumulation. Similar results were obtained in HEK293 cells in the presence of overexpressed PKA; this kinase was ineffective in preventing SIK2 T484A/S587A-induced export of the HDAC5 mutant S279A or S279D (Fig. 3C). These results contrast sharply with those presented in Fig. 1F, where PKA prevented SIK2 T484A/S587A-mediated nuclear export of wild-type HDAC5. Moreover, the basal localization of mutant S279D, much like S279A, was less nuclear than wild-type HDAC5 (supplemental Fig. S3, A and B). In agreement with this, in the context of its triple mutant, the HDAC4 mutation S266D made it more cytoplasmic (supplemental Fig. S3C). Moreover, as with HDAC4, substitution of Ser-279 with Ala or Asp had a minimal effect on general nuclear import, as seen using LMB (Fig. 3D).
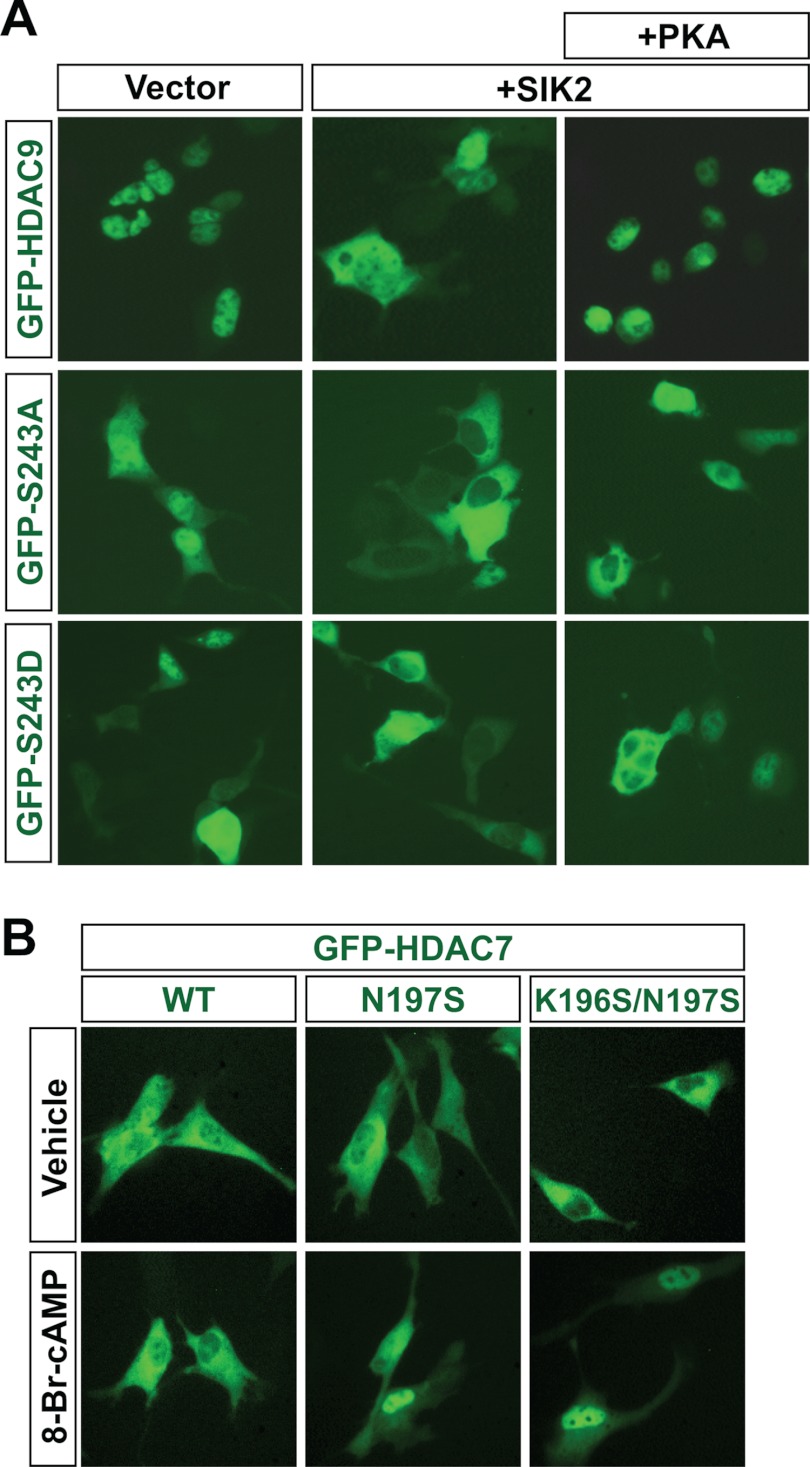
Because the cAMP/PKA-responsive SP motif of HDAC4 is also conserved in HDAC9 (Fig. 2A), we sought to determine whether its shuttling is similarly regulated. As in HDAC5 (Fig. 1C), HDAC9 was predominantly nuclear in HEK293 cells (Figs. 1E and 4A). As with the HDAC5 mutants, the S243A and S243D mutants of HDAC9 were more cytoplasmic than the wild type (Fig. 4A, left). In addition, both mutants were also more cytoplasmic than the wild-type in response to co-expression of wild-type SIK2 (Fig. 4A, middle), indicating a synergy between SIK2 coexpression and Ser-243 mutation in promoting cytoplasmic localization of HDAC9. In addition, PKA promoted nuclear localization of wild-type HDAC9 but had minimal effects on the two mutants (Fig. 4A, right). Therefore, the sensitivity of HDAC4, -5, and -9 to cAMP/PKA signaling depends on a similar mechanism.
FIGURE 4.
Analysis of Ser-243 in HDAC9 and restoration of an equivalent site in HDAC7. A, HEK293 cells were transiently transfected with the expression plasmid for GFP-HDAC9, -S243A, or -S243D along with an empty vector or the expression plasmid for wild-type SIK2, with or without the PKA expression plasmid. The transfection was performed as in Fig. 1E except that the amounts for the GFP-HDAC9 (wild type and mutants), SIK2, and PKA expression plasmids were 500, 45, and 200 ng, respectively. Representative images were taken 2 days after transfection. B, C2C12 cells were transiently transfected with the expression plasmid for GFP-HDAC7, -N197S, or -K196S/N197S, prior to treatment with 1 mm 8-Br-cAMP for 30 min.
Transferring the SP Motif to HDAC7 Confers Partial cAMP Sensitivity
As discussed earlier (Fig. 2A), HDAC7 is the only class IIa member that lacks the PKA consensus site corresponding to Ser-266 of HDAC4. To further test the importance of this motif in conferring cAMP/PKA sensitivity to class IIa HDACs, we engineered HDAC7 mutants to possess the same regulatory sequence as HDAC4. We generated an HDAC7 mutant, N197S, which contains a serine residue equivalent to Ser-266 of HDAC4 and HDAC5 Ser-279, and a second mutant, K196S/N197S, where the RRSSP motif in HDAC4/5/9 is fully restored. Whereas wild-type HDAC7 remained cytoplasmic in response to 8-Br-cAMP treatment, both HDAC7 N197S and HDAC7 K196S/N197S displayed a partial localization shift to the nucleus after 8-Br-cAMP treatment (Fig. 4B). These results confirm the importance of the SP motif for cAMP/PKA sensitivity of class IIa HDACs but also suggest that other factors may be involved.
The cAMP Effect on HDAC4 Localization Is Mediated by PKA
PKA is one of the major proteins activated by increased cAMP concentration, and both cAMP and PKA stimulated nuclear localization of HDAC4 (Fig. 1, A and B). However, it formally remains a possibility that the effect of cAMP signaling on HDAC4 subcellular localization is mediated through non-PKA targets. To test this, we employed the PKA inhibitor H-89 to see if the effect of 8-Br-cAMP on HDAC4 was altered. Pretreatment of C2C12 cells with H-89 abolished the change in HDAC4 localization upon 8-Br-cAMP treatment (supplemental Fig. S4). Thus, PKA is necessary for cAMP-induced nuclear localization of HDAC4. Next, we checked the involvement of PP2A, a target of PKA, in this process. Unlike the results with H-89, pretreating cells with okadaic acid (a PP2A inhibitor) had no effect on the change in HDAC4 localization induced by 8-Br-cAMP (supplemental Fig. S4), suggesting that either PP2A is not involved or acts together with other phosphatases. Moreover, mutant S246A (which is slightly less cytoplasmic than wild-type HDAC4) still became more nuclear in response to 8-Br-cAMP treatment (supplemental Fig. S4). These results indicate that the effect of cAMP on HDAC4 localization is not due to dephosphorylation at the 14-3-3 binding sites that induce nuclear export. Instead, as suggested by the results in Figs. 2, 3, and 4, cAMP/PKA signals specifically to the novel regulatory SP motif.
Ser-266 Dephosphorylation in Response to cAMP Signaling
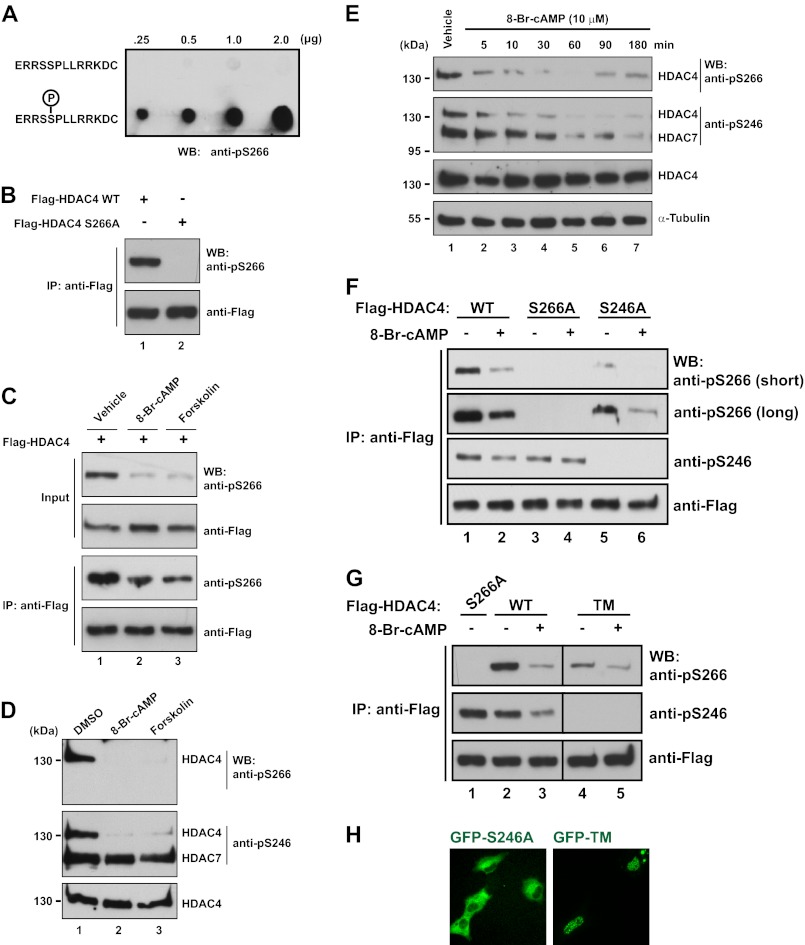
Due to the dramatic effects of HDAC4 Ser-266 mutation (and HDAC5 S279A and HDAC9 Ser-243 mutation) on the responsiveness to cAMP signaling (Figs. 2–4), we hypothesized that this represents a novel phosphorylation site. To investigate this possibility, we generated a phospho-specific antibody that detected HDAC4 only when Ser-266 is phosphorylated (Fig. 5A). This antibody recognized a single band of the correct size in whole cell extracts from cells that had been transfected with FLAG-HDAC4 but not FLAG-S266A (Fig. 5B), demonstrating its specificity in vivo. In similar experiments, the affinity-purified anti-phospho-Thr-707 antibody did not detect any signals (data not shown), further indicating that Thr-707 is not phosphorylated. The epitope region for the anti-phospho-Ser-266 antibody is identical among HDAC4, -5, and -9 (Fig. 2A), so this antibody should recognize the latter two as well. However, under similar conditions, only weak signals or no signals were detected with HDAC5 and -9 (supplemental Fig. S5), suggesting that, compared with HDAC4, they are less phosphorylated at the equivalent sites. This is consistent with the fact that they tend to be more nuclear than HDAC4. Unexpectedly, activation of PKA by treatment with 8-Br-cAMP or forskolin did not lead to an increase but rather caused a decrease in Ser-266 phosphorylation (Fig. 5C and supplemental Fig. S5C). 8-Br-cAMP or forskolin treatment also decreased Ser-266 phosphorylation of endogenous HDAC4 (Fig. 5D, top).
FIGURE 5.
Ser-266 is a novel phosphorylation site regulated by cAMP signaling. A, different amounts of regular peptide (top) or phosphopeptide (bottom) were spotted on a nitrocellulose membrane, air-dried, blocked for 2 h in PBS-T containing 20% horse serum, and then incubated overnight at 4 °C with the anti-phospho-Ser-266 antibody (1:5000 dilution). B, HEK293 cells were transfected with an expression plasmid for FLAG-HDAC4 or -S266A. 48 h later, cells were harvested in buffer K, and FLAG-tagged proteins were immunoprecipitated (IP) with M2-agarose beads and eluted with FLAG peptide. Eluted proteins were separated by SDS-PAGE, and immunoblotting (WB) was performed with anti-phospho-Ser-266 and anti-FLAG antibodies as indicated. C, C2C12 cells were transfected with the FLAG-HDAC4 expression plasmid and treated with DMSO (lane 1), 1 mm 8-Br-cAMP (lane 2), or 10 μm forskolin (lane 3) for 30 min. Cells were harvested for immunoprecipitation and immunoblotting as in B. D, C2C12 cells were treated with DMSO (lane 1), 1 mm 8-Br-cAMP (lane 2), or 10 μm forskolin (lane 3) for 30 min. Soluble extracts were subjected to SDS-PAGE, and immunoblotting was performed with anti-phospho-Ser-266, anti-phospho-Ser-246, and anti-HDAC4 primary antibodies as specified. The positions of bands representing HDAC4 and HDAC7 detected by the anti-phospho-Ser-246 antibody are indicated to the immediate right of the blot. Protein size in kDa is indicated to the left of the blots. E, C2C12 cells were treated with vehicle (H2O) for 30 min or 1 mm 8-Br-cAMP for the indicated number of minutes. Immunoblotting of endogenous proteins was performed as in D. F, C2C12 cells were transfected with an expression plasmid for FLAG-HDAC4, -S266A, or -S246A and treated with 1 mm 8-Br-cAMP (lanes 2, 4, and 6) or vehicle (H2O; lanes 3, 5, and 7). Cells were harvested in buffer K, and FLAG-tagged proteins were immunoprecipitated on M2-agarose beads and eluted with FLAG peptide. Eluted proteins were separated by SDS-PAGE, and immunoblotting was performed with anti-phospho-Ser-266, anti-phospho-Ser-246, and anti-FLAG antibodies. For the immunoblotting with the anti-phospho-Ser-266, images with short and long exposure are shown. G, as in F except that plasmids for FLAG-HDAC4, -S266A, and TM expression were transfected as indicated. H, C2C12 cells were transfected with expression plasmids for GFP-HDAC4 S246A or TM. Subcellular localization was monitored by live green fluorescence microscopy.
We then examined the phosphorylation status of the nearby 14-3-3 binding site, Ser-246 (Fig. 2A). Endogenous Ser-246 phosphorylation was also decreased after 30 min of 8-Br-cAMP or forskolin treatment (Fig. 5D, middle). However, phosphorylation of the equivalent residue of HDAC7 (Ser-155) was unaltered by these treatments (Fig. 5D, middle). To better gauge the timing of phosphorylation changes, we performed time course experiments in which C2C12 cells were exposed to 8-Br-cAMP for a period of 3 h. Endogenous HDAC4 Ser-266 phosphorylation was significantly lower by 10 min (Fig. 5E). This effect was somewhat transient because the level returned slightly by 90 min. In contrast, Ser-246 phosphorylation exhibited a slightly slower rate of recovery, with phosphorylation still depressed at 180 min (Fig. 5E). Interestingly, HDAC7 Ser-155 phosphorylation (as measured with the anti-phospho-Ser-246 antibody) did eventually decline but at a significantly slower rate than Ser-246 phosphorylation (Fig. 5E, with t½ = 20–30 min versus 2–5 min), indicating that although cAMP does promote dephosphorylation of HDAC7 Ser-155, this deacetylase is more resistant than the other class IIa members.
Related to the subcellular localization results shown in supplemental Fig. S4, H-89 treatment partially abrogated the cAMP-induced decline in Ser-266 phosphorylation, whereas okadaic acid treatment had no effect (supplemental Fig. S6). This suggests that cAMP at least partially acts through PKA and that PP2A may not be the sole phosphatase involved.
Phosphorylation Synergy between Ser-246 and Ser-266 of HDAC4
We next tested whether the effect of cAMP on Ser-266 phosphorylation is altered by mutation of Ser-246. Strikingly, we noticed a major decrease in basal Ser-266 phosphorylation using an HDAC4 construct with an S246A mutation (Fig. 5F, compare lanes 1 and 5). Although the basal Ser-266 phosphorylation level was much lower than for wild-type HDAC4, there was still a further decrease in Ser-266 phosphorylation after 8-Br-cAMP treatment (Fig. 5F, lanes 1 and 2 versus lanes 5 and 6), suggesting that Ser-246 phosphorylation influences basal Ser-266 phosphorylation but not its response to cAMP. A similar effect was observed with the TM mutant (Fig. 5G). This mutant contains S467A and S632A in addition to S246A (52). These changes were not simply due to subcellular localization changes because HDAC4 S246A was still largely cytoplasmic in C2C12 cells, whereas HDAC4 TM was predominantly nuclear (Fig. 5H) (52). Conversely, when Ser-266 was mutated, the change in basal Ser-246 phosphorylation was modest (Fig. 5F, compare lanes 1 and 3), or there was a modest increase in Ser-246 phosphorylation (Fig. 5G, lanes 1 and 2). However, the response of Ser-246 phosphorylation to cAMP treatment was slightly muted (Fig. 5F, lanes 1 and 2 versus lanes 3 and 4). These results indicate that there is a cross-talk between Ser-246 and Ser-266.
Dephosphorylation at Ser-266 but Not Ser-246 during Nuclear Import of HDAC4
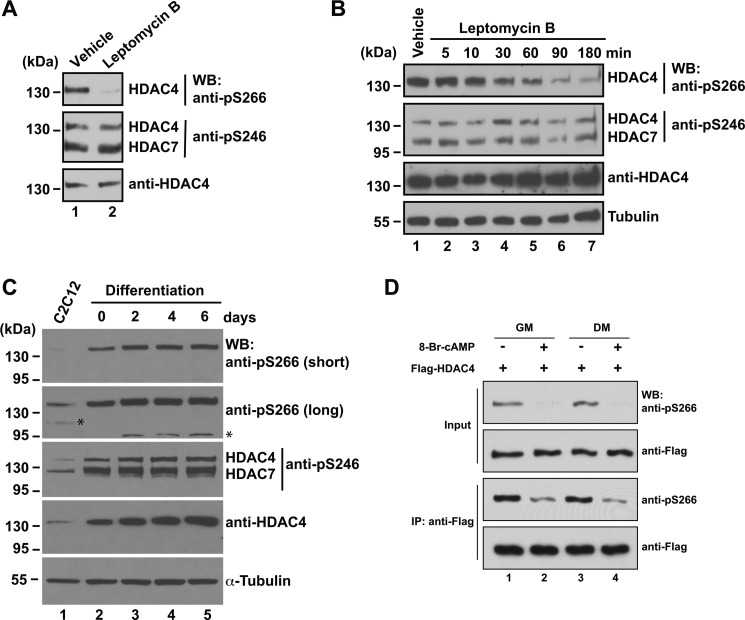
To further elucidate the molecular mechanisms underlying Ser-266 phosphorylation, we treated cells with LMB to investigate whether the phosphorylation was affected by altered HDAC4 localization. As expected, LMB treatment in C2C12 cells resulted in nuclear accumulation of HDAC4 (Fig. 2C). This was associated with a dramatic decrease in Ser-266 phosphorylation, whereas Ser-246 phosphorylation remained unaltered (Fig. 6A). Time course experiments showed that Ser-266 phosphorylation continued to decrease to 3 h of LMB treatment, whereas Ser-246 phosphorylation did not change during this period (Fig. 6B). These findings suggest that during nuclear import of HDAC4, dephosphorylation occurs at Ser-266 but not Ser-246.
FIGURE 6.
Dynamic 266 phosphorylation during nuclear import and myogenesis. A, C2C12 cells were treated with vehicle (70% methanol) or 10 ng/ml leptomycin B for 1 h. Soluble extracts were separated by SDS-PAGE, and immunoblotting (WB) was performed with anti-phospho-Ser-266, anti-phospho-Ser-246, and anti-HDAC4 antibodies as indicated. The positions of bands representing HDAC4 and HDAC7 detected by the anti-phospho-Ser-246 antibody are indicated to the immediate right of the blot. Protein size in kDa is indicated to the left of the blots. B, time course experiment as in Fig. 5E except that the vehicle (70% methanol) treatment was for 1 h, and 10 ng/ml leptomycin B was used instead of 8-Br-cAMP. C, proliferating C2C12 myoblasts (lane 1) were grown to 100% confluence and induced to differentiate for up to 6 days. Cells were collected at the indicated days (lanes 2–5) for extract preparation and immunoblotting with the specified antibodies. For the immunoblot with the anti-phospho-Ser-266 antibody, short and long exposure times are shown (top two panels), with the asterisks denoting nonspecific bands on the long exposure image. D, as in Fig. 5C except that cells were cultured in growth medium (GM), containing 10% FBS, or in differentiation medium (DM), containing 2% horse serum, for 5 h prior to 8-Br-cAMP treatment. IP, immunoprecipitation.
Similar Ser-246 and Ser-266 Phosphorylation Dynamics during Cell Differentiation
cAMP plays an active role during myogenesis (68, 69), so we next analyzed Ser-266 phosphorylation during differentiation of C2C12 myoblasts. As these cells became confluent and were induced to differentiate, HDAC4 expression increased (Fig. 6C, bottom two panels). During the course, phosphorylation of HDAC4 at Ser-246 and HDAC7 at the equivalent site (Ser-155) increased (middle panel). Similarly, phosphorylation of HDAC4 at Ser-266 also increased at the early phase of myogenesis but slightly peaked on day 2 and then decreased slightly (top two panels). 8-Br-cAMP treatment induced dephosphorylation in C2C12 cells even when they were grown in differentiation medium (Fig. 6D and supplemental Fig. S5C), suggesting that cAMP signaling to Ser-266 phosphorylation remains intact during differentiation. Although it is generally considered that class IIa HDACs are subject to nuclear export during myogenesis (23, 70), it has also been reported that HDAC4 translocates to the nucleus during the course (27). Of relevance, the increased HDAC4 protein level (Fig. 6C, bottom two panels) during the differentiation is also in stark contrast to the general assumption that class IIa HDAC functions are repressed during myogenesis. Nonetheless, our findings suggest that HDAC4 phosphorylation at Ser-266 follows a dynamic pattern that is more complicated than anticipated.
To investigate the phosphorylation dynamics in another differentiation program, we employed 3T3-L1 preadipocytes, which can be induced to differentiate into adipocytes (58). Phosphorylation at both Ser-246 and Ser-266 decreased upon treatment with the initial differentiation mix containing insulin, isobutylmethylxanthine, and dexamethasone, but the level recovered as the differentiation progressed and the medium was switched to the one containing insulin only (data not shown). Furthermore, in all cell and mouse tissue extracts that we have surveyed, phosphorylation at Ser-246 and Ser-266 displayed similar patterns (data not shown). Therefore, in myoblast, adipocyte, and perhaps other differentiation programs, phosphorylation at Ser-246 and Ser-266 follows similar dynamic patterns.
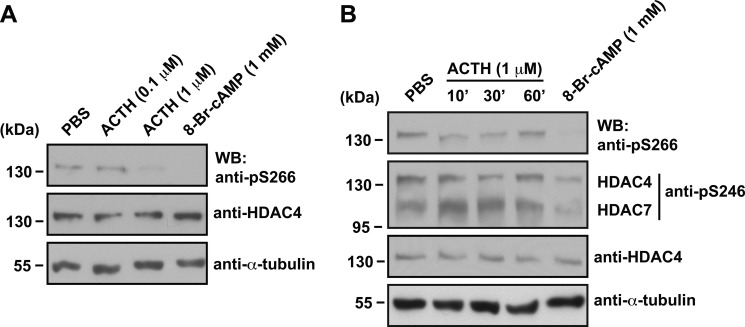
ACTH Regulates Endogenous HDAC4 Phosphorylation at Ser-266
Next we investigated whether Ser-266 phosphorylation status is affected by treatment with a hormone that induces an increase in cAMP concentration. ACTH acts on adrenal cells by increasing the intracellular cAMP concentration (57, 71). We treated Y1 adrenocortical tumor cells with different amounts of ACTH to examine whether endogenous HDAC4 phosphorylation at Ser-266 is influenced. Only the higher dose (1 μm ACTH) induced a drop in Ser-266 phosphorylation (Fig. 7A). This dose was then used to perform a time course experiment. ACTH treatment resulted in a decrease in Ser-266 phosphorylation at 10 and 30 min, but the effect wore off by 60 min (Fig. 7B). The effect was less potent than that observed with 8-Br-cAMP (Fig. 7B), which probably reflects the enhanced resistance toward phosphodiesterase-mediated degradation possessed by 8-Br-cAMP compared with native cAMP (72). Nevertheless, these findings point to Ser-266 phosphorylation as a physiologically relevant target of diverse cAMP/PKA signaling modules.
FIGURE 7.
ACTH inhibits endogenous Ser-266 phosphorylation level in Y1 cells. A, Y1 cells were treated with vehicle (PBS) or different concentrations (0.1 or 1 μm) of adrenocorticotropic hormone (ACTH), or 1 mm 8-Br-cAMP for 30 min. Whole cell extracts were subject to SDS-PAGE, and immunoblotting (WB) was performed using anti-phospho-Ser-266, anti-HDAC4, or anti-α-tubulin primary antibodies. Protein size in kDa is indicated to the left of the blots. B, Y1 cells were treated with vehicle (PBS) for 60 min, with 1 μm ACTH for different times (10, 30, or 60 min), or with 1 mm 8-Br-cAMP for 30 min. Immunoblotting was performed as in A except that the anti-phospho-Ser-246 antibody was also used, with the position of bands corresponding to HDAC4 or HDAC7 detected by this antibody indicated to the right of the blot.
DISCUSSION
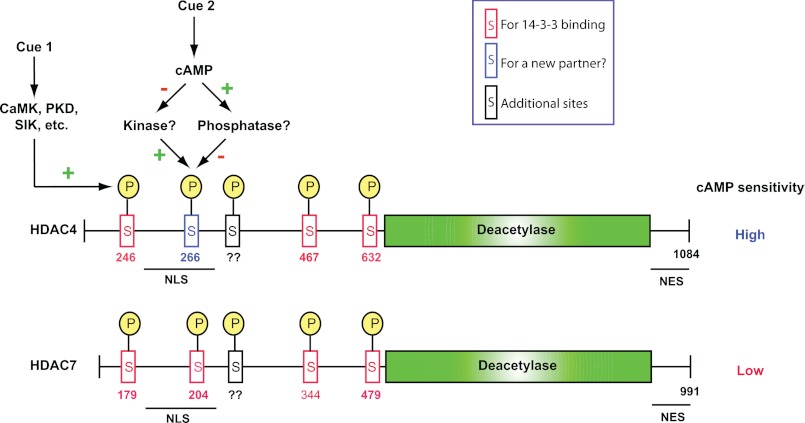
Herein we have demonstrated that cAMP/PKA acts on class IIa HDACs through a novel phosphorylation site, Ser-266, -279, and -243 in HDAC4, -5, and -9, respectively (Fig. 2A). Substitution of the site with alanine rendered these three deacetylases unresponsive or less responsive to cAMP treatment or PKA expression (Figs. 2, 3, and 4A). Interestingly, replacement with aspartate had a similar effect (Figs. 2, 3, and 4A and supplemental Fig. S3), suggesting that the hydroxy group of the serine residues is important for nuclear import. Alternatively, the negative charge per se may not be sufficient for mediating the impact of phosphorylation. Although this was somewhat unexpected, there is precedent for the inability of aspartate to mimic phosphorylation. For example, S133D mutation has no effect on CREB transcriptional activity (42). In addition to Ser-266 mutants, the HDAC4 mutant P267A was cAMP-resistant (Fig. 2, E and F). This proline residue is invariant among paralogs and orthologs of HDAC4 (Fig. 2A and supplemental Fig. S2), so Ser-266 and Pro-267 form a conserved SP motif that is important for HDAC4 regulation. This motif is present in HDAC5 and HDAC9 but not in HDAC7 (Fig. 2A and supplemental Fig. S2), thereby conferring different cAMP sensitivity among the four class IIa members (Fig. 8). On the other hand, LMB-induced nuclear import was not impaired in the mutants (Figs. 2F and 3D). Related to this, the S266A mutation has no effect on nuclear accumulation of HDAC4 in response to treatment of a nuclear export inhibitor (36). Thus, although it is not so important for nuclear import in general, dephosphorylation at the SP motif is crucial for cAMP/PKA-induced nuclear accumulation of HDAC4, -5, and -9 and perhaps also the orthologs that contain such a motif (Fig. 2A and supplemental Fig. S2). Notably, our results are consistent with a recent study (73) demonstrating that cAMP promotes dephosphorylation of HDAC5 at Ser-279, a site that is equivalent to Ser-266 of HDAC4 (Fig. 2A).
FIGURE 8.
Model depicting differential regulation of HDAC4 and 7 by cAMP signaling. Cue 1 activates class IIa HDAC kinases and promotes phosphorylation at the 14-3-3 binding sites (red boxes labeled with the red letter S; for simplicity, the arrow is only pointed to phospho-Ser-246 but not phospho-Ser-467 or -632 of HDAC4) and subsequent cytoplasmic localization of the HDACs. cAMP signaling (cue 2) promotes dephosphorylation and nuclear localization. Whereas HDAC4 contains three 14-3-3 binding sites, HDAC7 possesses an extra site at the C-terminal end of the nuclear localization signal. Ser-266 of HDAC4 (blue box with the blue letter S) confers hypersensitivity to cAMP signaling, but HDAC7 lacks an equivalent residue and is more resistant to dephosphorylation at its 14-3-3 sites. There may be a cross-talk between phosphorylation at Ser-246 and that at Ser-266 of HDAC4. Through two distinct pathways, cues 1 and 2 may synergize or antagonize with each other. Although PP2A is a potential candidate mediating the cAMP effect, additional studies are needed to establish this. HDAC5 and HDAC9 are more similar to HDAC4 than to HDAC7. See “Discussion” for details about potential kinases and phosphatases. NLS, nuclear localization signal; NES, nuclear export signal.
Similar to HDAC4, -5, and -9, dephosphorylation of NFAT transcription factors at conserved SP motifs promotes nuclear import (74, 75). Although not fully understood, dephosphorylation is assumed to expose the nuclear localization signals of NFATs. In both cases, phosphorylation blocks nuclear import. The difference is that alanine substitution of the serine residues on the deacetylases stimulates cytoplasmic localization (Figs. 2, 3, and 4A and supplemental Fig. S3), whereas such mutations have an opposite effect on NFATs. Such similarity and difference in signal-dependent nucleocytoplasmic trafficking are intriguing. It is also tempting to speculate that these two examples may serve as prototypes for additional cases to be found when numerous phosphorylation sites to be identified in other proteins by proteomic studies are fully characterized.
Guided by the conserved RRXS motif (Fig. 2A and supplemental Fig. S2), we initially set out to characterize it as a PKA phosphorylation site and thus expected the phosphorylation to increase in response to PKA expression or cAMP treatment. Surprisingly, Ser-266 phosphorylation decreased upon cAMP signaling (Fig. 5) or PKA expression (data not shown). Moreover, PKA did not appear to phosphorylate HDAC4 in vitro (supplemental Fig. S7). Although these results are unexpected, they are consistent with the effect of the mutation P267A (Fig. 2, E and F). Many kinases phosphorylate serine/threonine residues immediately preceding proline, but this residue at the +1 position is strongly disfavored among PKA substrates and acts to “veto” PKA-mediated phosphorylation when artificially placed in this position of PKA substrate peptides (76, 77). Moreover, the SP motif is more conserved than the arginine residues at −1 and −2 positions required for PKA phosphorylation (Fig. 2A and supplemental Fig. S2). Ser-266 conforms to the consensus phosphorylation sites for several other kinases, including CaMKs, PKC, ERK1/2, and GSK-3, but we did not observe any decrease in phosphorylation after treatment with inhibitors of these kinases (supplemental Fig. S8). Moreover, expression of CaMKIV in HEK293 cells promoted export of HDAC5 but had minimal effects on Ser-279 phosphorylation (data not shown). Mirk/dyrk1B was shown to phosphorylate Ser-243 of an alternatively spliced isoform corresponding to the N-terminal half of HDAC9 (78). However, expression of this and related kinases did not alter the subcellular localization of class IIa HDACs in our assay conditions (data not shown). Expression of SIK2 in HEK293 cells only slightly increased Ser-279 phosphorylation of HDAC5 (data not shown). Together, these results suggest the involvement of a new kinase (Fig. 8). Indeed, Cdk5 phosphorylates Ser-279 of HDAC5 (73). Whether there are additional kinases acting at the SP motif is an interesting question worthy of further investigation (Fig. 8).
As for phosphatases, PP2A forms stable complexes with class IIa HDACs and inhibits their phosphorylation at 14-3-3 binding sites (10, 35, 36, 79). Okadaic acid, an inhibitor of PP2A, did not have any effect on Ser-266 phosphorylation in response to cAMP treatment (supplemental Fig. S6), suggesting that PP2A may not be the sole phosphatase. Another candidate is myosin phosphatase (PP1β/MYPT1), which has been shown to dephosphorylate HDAC7 (37). Whether cAMP/PKA decreases Ser-266 phosphorylation through inhibition of a Ser-266 kinase(s) and/or through activation of a phosphatase(s) (Fig. 8) remains an interesting issue that will shed new light on unknown links between class IIa HDACs and cellular signaling networks.
Ser-266 of HDAC4 has recently been identified as a phosphorylation site in a phosphoproteomic study (80). Ser-279 of HDAC5 was just reported to be a target of cAMP signaling and PKA phosphorylation (81) and was independently identified as a phosphorylation site by mass spectrometry (79). Together, these findings provide further support for the importance of phosphorylation at Ser-266 of HDAC4 and Ser-279 of HDAC5. Unexpectedly, our results indicate that cAMP signaling does not stimulate but rather inhibits the phosphorylation (Fig. 8), thus unraveling a novel mechanism through which the HDAC regulation is linked to signaling networks downstream from hormones such as ACTH (Fig. 7) and parathyroid hormone-related protein (46). Our results also suggest that the phosphorylation event does not act alone but cross-talks with other sites (e.g. with Ser-246) (Fig. 5, F–H). Although not investigated here, it is reasonable to assume a cross-talk with the other two 14-3-3 binding sites in HDAC4, -5, or -9 (Fig. 8). Related to this, mutation of HDAC5 Ser-279 (Fig. 3A) or HDAC9 Ser-243 (Fig. 4A) synergized with expression of SIK2 in promoting nuclear export.
In stark contrast, during nuclear import, dephosphorylation occurred at Ser-266 but not Ser-246 of HDAC4 (Fig. 6, A and B). This suggests that during nuclear import, Ser-266 phosphorylation has a role independent of Ser-246 phosphorylation. Related to this, HDAC5 is more nuclear than HDAC4 (Fig. 1) and is also much less phosphorylated at Ser-279 than at Ser-266 of HDAC4 (supplemental Fig. S5). It is likely that HDAC5 is phosphorylated to a higher level under different cellular and/or signaling contexts. Thus, phosphorylation at the SP motif marks the status of cytoplasmic localization.
Additional phosphorylation sites have been identified in class IIa HDACs, including Ser-298 of HDAC4 (36), Ser-253 of the HDAC9 splice variant (78, 82), and multiple residues of HDAC4 and HDAC5 reported by phosphoproteomic studies (see the Human Protein Reference Database) (79, 80, 83, 84), suggesting that complex multisite phosphorylation programs may respond to cAMP and other upstream signaling cues (Fig. 8). These programs may be further linked to ubiquitination (85, 86) and sumoylation (87, 88). The conserved SP motif identified herein (Fig. 2A and supplemental Fig. S2) is likely to play a significant role in fine-tuning the modification programs for regulating important biological functions of class IIa HDACs in diverse physiological and pathological processes.
Supplementary Material
This project was supported by operating grants from the Canadian Institutes of Health Research and the Canadian Cancer Society (to X. J. Y.).

This article contains supplemental Figs. S1–S8.
- HDAC
- histone deacetylase
- CaMK
- Ca2+/calmodulin-dependent protein kinase
- PP2A and PP1β
- protein phosphatase 2A and 1β, respectively
- CREB
- cAMP-response element-binding protein
- SIK
- salt-inducible kinase
- TM
- Hdac4 triple mutant (S246A/S467A/S632A)
- 8-Br-cAMP
- 8-bromo-cyclic AMP
- LMB
- leptomycin B
- NFAT
- nuclear factor of activated T cells.
REFERENCES
- 1. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 2. Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., Li H., Xie L., Zhao W., Yao Y., Ning Z. B., Zeng R., Xiong Y., Guan K. L., Zhao S., Zhao G. P. (2010) Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327, 1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang X. J., Seto E. (2008) Lysine acetylation. Codified crosstalk with other posttranslational modifications. Mol. Cell 31, 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Khochbin S., Verdel A., Lemercier C., Seigneurin-Berny D. (2001) Functional significance of histone deacetylase diversity. Curr. Opin. Genet. Dev. 11, 162–166 [DOI] [PubMed] [Google Scholar]
- 5. McKinsey T. A., Zhang C. L., Lu J., Olson E. N. (2000) Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408, 106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim M. S., Fielitz J., McAnally J., Shelton J. M., Lemon D. D., McKinsey T. A., Richardson J. A., Bassel-Duby R., Olson E. N. (2008) Protein kinase D1 stimulates MEF2 activity in skeletal muscle and enhances muscle performance. Mol. Cell Biol. 28, 3600–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu Y., Contreras M., Shen T., Randall W. R., Schneider M. F. (2009) α-Adrenergic signalling activates protein kinase D and causes nuclear efflux of the transcriptional repressor HDAC5 in cultured adult mouse soleus skeletal muscle fibres. J. Physiol. 587, 1101–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang C. L., McKinsey T. A., Chang S., Antos C. L., Hill J. A., Olson E. N. (2002) Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110, 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ha C. H., Jhun B. S., Kao H. Y., Jin Z. G. (2008) VEGF stimulates HDAC7 phosphorylation and cytoplasmic accumulation modulating matrix metalloproteinase expression and angiogenesis. Arterioscler. Thromb. Vasc. Biol. 28, 1782–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martin M., Potente M., Janssens V., Vertommen D., Twizere J. C., Rider M. H., Goris J., Dimmeler S., Kettmann R., Dequiedt F. (2008) Protein phosphatase 2A controls the activity of histone deacetylase 7 during T cell apoptosis and angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 105, 4727–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang S., Li X., Parra M., Verdin E., Bassel-Duby R., Olson E. N. (2008) Control of endothelial cell proliferation and migration by VEGF signaling to histone deacetylase 7. Proc. Natl. Acad. Sci. U.S.A. 105, 7738–7743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Parra M., Verdin E. (2010) Regulatory signal transduction pathways for class IIa histone deacetylases. Curr. Opin. Pharmacol. 10, 454–460 [DOI] [PubMed] [Google Scholar]
- 13. Majdzadeh N., Morrison B. E., D'Mello S. R. (2008) Class IIA HDACs in the regulation of neurodegeneration. Front Biosci. 13, 1072–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vega R. B., Matsuda K., Oh J., Barbosa A. C., Yang X., Meadows E., McAnally J., Pomajzl C., Shelton J. M., Richardson J. A., Karsenty G., Olson E. N. (2004) Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 119, 555–566 [DOI] [PubMed] [Google Scholar]
- 15. Moresi V., Williams A. H., Meadows E., Flynn J. M., Potthoff M. J., McAnally J., Shelton J. M., Backs J., Klein W. H., Richardson J. A., Bassel-Duby R., Olson E. N. (2010) Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell 143, 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang S., McKinsey T. A., Zhang C. L., Richardson J. A., Hill J. A., Olson E. N. (2004) Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell Biol. 24, 8467–8476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chang S., Young B. D., Li S., Qi X., Richardson J. A., Olson E. N. (2006) Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell 126, 321–334 [DOI] [PubMed] [Google Scholar]
- 18. Renthal W., Maze I., Krishnan V., Covington H. E., 3rd, Xiao G., Kumar A., Russo S. J., Graham A., Tsankova N., Kippin T. E., Kerstetter K. A., Neve R. L., Haggarty S. J., McKinsey T. A., Bassel-Duby R., Olson E. N., Nestler E. J. (2007) Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron 56, 517–529 [DOI] [PubMed] [Google Scholar]
- 19. Rad R., Rad L., Wang W., Cadinanos J., Vassiliou G., Rice S., Campos L. S., Yusa K., Banerjee R., Li M. A., de la Rosa J., Strong A., Lu D., Ellis P., Conte N., Yang F. T., Liu P., Bradley A. (2010) PiggyBac transposon mutagenesis. A tool for cancer gene discovery in mice. Science 330, 1104–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hutt D. M., Herman D., Rodrigues A. P., Noel S., Pilewski J. M., Matteson J., Hoch B., Kellner W., Kelly J. W., Schmidt A., Thomas P. J., Matsumura Y., Skach W. R., Gentzsch M., Riordan J. R., Sorscher E. J., Okiyoneda T., Yates J. R., 3rd, Lukacs G. L., Frizzell R. A., Manning G., Gottesfeld J. M., Balch W. E. (2010) Reduced histone deacetylase 7 activity restores function to misfolded CFTR in cystic fibrosis. Nat. Chem. Biol. 6, 25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Williams S. R., Aldred M. A., Der Kaloustian V. M., Halal F., Gowans G., McLeod D. R., Zondag S., Toriello H. V., Magenis R. E., Elsea S. H. (2010) Haploinsufficiency of HDAC4 causes brachydactyly mental retardation syndrome, with brachydactyly type E, developmental delays, and behavioral problems. Am. J. Hum. Genet. 87, 219–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Verdin E., Dequiedt F., Kasler H. G. (2003) Class II histone deacetylases. Versatile regulators. Trends Genet. 19, 286–293 [DOI] [PubMed] [Google Scholar]
- 23. Yang X. J., Seto E. (2008) The Rpd3/Hda1 family of lysine deacetylases. From bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9, 206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Backs J., Song K., Bezprozvannaya S., Chang S., Olson E. N. (2006) CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J. Clin. Invest. 116, 1853–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kao H. Y., Verdel A., Tsai C. C., Simon C., Juguilon H., Khochbin S. (2001) Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 276, 47496–47507 [DOI] [PubMed] [Google Scholar]
- 26. Little G. H., Bai Y., Williams T., Poizat C. (2007) Nuclear calcium/calmodulin-dependent protein kinase IIδ preferentially transmits signals to histone deacetylase 4 in cardiac cells. J. Biol. Chem. 282, 7219–7231 [DOI] [PubMed] [Google Scholar]
- 27. Zhao X., Ito A., Kane C. D., Liao T. S., Bolger T. A., Lemrow S. M., Means A. R., Yao T. P. (2001) The modular nature of histone deacetylase HDAC4 confers phosphorylation-dependent intracellular trafficking. J. Biol. Chem. 276, 35042–35048 [DOI] [PubMed] [Google Scholar]
- 28. Dequiedt F., Van Lint J., Lecomte E., Van Duppen V., Seufferlein T., Vandenheede J. R., Wattiez R., Kettmann R. (2005) Phosphorylation of histone deacetylase 7 by protein kinase D mediates T cell receptor-induced Nur77 expression and apoptosis. J. Exp. Med. 201, 793–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Harrison B. C., Kim M. S., van Rooij E., Plato C. F., Papst P. J., Vega R. B., McAnally J. A., Richardson J. A., Bassel-Duby R., Olson E. N., McKinsey T. A. (2006) Regulation of cardiac stress signaling by protein kinase d1. Mol. Cell Biol. 26, 3875–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vega R. B., Harrison B. C., Meadows E., Roberts C. R., Papst P. J., Olson E. N., McKinsey T. A. (2004) Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol. Cell Biol. 24, 8374–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carnegie G. K., Soughayer J., Smith F. D., Pedroja B. S., Zhang F., Diviani D., Bristow M. R., Kunkel M. T., Newton A. C., Langeberg L. K., Scott J. D. (2008) AKAP-Lbc mobilizes a cardiac hypertrophy signaling pathway. Mol. Cell 32, 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chawla S., Vanhoutte P., Arnold F. J., Huang C. L., Bading H. (2003) Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J. Neurochem. 85, 151–159 [DOI] [PubMed] [Google Scholar]
- 33. Wu X., Zhang T., Bossuyt J., Li X., McKinsey T. A., Dedman J. R., Olson E. N., Chen J., Brown J. H., Bers D. M. (2006) Local InsP3-dependent perinuclear Ca2+ signaling in cardiac myocyte excitation-transcription coupling. J. Clin. Invest. 116, 675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ha C. H., Wang W., Jhun B. S., Wong C., Hausser A., Pfizenmaier K., McKinsey T. A., Olson E. N., Jin Z. G. (2008) Protein kinase D-dependent phosphorylation and nuclear export of histone deacetylase 5 mediates vascular endothelial growth factor-induced gene expression and angiogenesis. J. Biol. Chem. 283, 14590–14599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Illi B., Dello Russo C., Colussi C., Rosati J., Pallaoro M., Spallotta F., Rotili D., Valente S., Ragone G., Martelli F., Biglioli P., Steinkuhler C., Gallinari P., Mai A., Capogrossi M. C., Gaetano C. (2008) Nitric oxide modulates chromatin folding in human endothelial cells via protein phosphatase 2A activation and class II histone deacetylases nuclear shuttling. Circ. Res. 102, 51–58 [DOI] [PubMed] [Google Scholar]
- 36. Paroni G., Cernotta N., Dello Russo C., Gallinari P., Pallaoro M., Foti C., Talamo F., Orsatti L., Steinkühler C., Brancolini C. (2008) PP2A regulates HDAC4 nuclear import. Mol. Biol. Cell 19, 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Parra M., Mahmoudi T., Verdin E. (2007) Myosin phosphatase dephosphorylates HDAC7, controls its nucleocytoplasmic shuttling, and inhibits apoptosis in thymocytes. Genes Dev. 21, 638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Francis S. H., Corbin J. D. (1994) Structure and function of cyclic nucleotide-dependent protein kinases. Annu. Rev. Physiol. 56, 237–272 [DOI] [PubMed] [Google Scholar]
- 39. Baillie G. S., Scott J. D., Houslay M. D. (2005) Compartmentalisation of phosphodiesterases and protein kinase A. Opposites attract. FEBS Lett. 579, 3264–3270 [DOI] [PubMed] [Google Scholar]
- 40. Wong W., Scott J. D. (2004) AKAP signalling complexes. Focal points in space and time. Nat. Rev. Mol. Cell Biol. 5, 959–970 [DOI] [PubMed] [Google Scholar]
- 41. Daniel P. B., Walker W. H., Habener J. F. (1998) Cyclic AMP signaling and gene regulation. Annu. Rev. Nutr. 18, 353–383 [DOI] [PubMed] [Google Scholar]
- 42. Montminy M. (1997) Transcriptional regulation by cyclic AMP. Annu. Rev. Biochem. 66, 807–822 [DOI] [PubMed] [Google Scholar]
- 43. Shaywitz A. J., Greenberg M. E. (1999) CREB. A stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem. 68, 821–861 [DOI] [PubMed] [Google Scholar]
- 44. Belfield J. L., Whittaker C., Cader M. Z., Chawla S. (2006) Differential effects of Ca2+ and cAMP on transcription mediated by MEF2D and cAMP-response element-binding protein in hippocampal neurons. J. Biol. Chem. 281, 27724–27732 [DOI] [PubMed] [Google Scholar]
- 45. Du M., Perry R. L., Nowacki N. B., Gordon J. W., Salma J., Zhao J., Aziz A., Chan J., Siu K. W., McDermott J. C. (2008) Protein kinase A represses skeletal myogenesis by targeting myocyte enhancer factor 2D. Mol. Cell Biol. 28, 2952–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kozhemyakina E., Cohen T., Yao T. P., Lassar A. B. (2009) Parathyroid hormone-related peptide represses chondrocyte hypertrophy through a protein phosphatase 2A/histone deacetylase 4/MEF2 pathway. Mol. Cell Biol. 29, 5751–5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mihaylova M. M., Vasquez D. S., Ravnskjaer K., Denechaud P. D., Yu R. T., Alvarez J. G., Downes M., Evans R. M., Montminy M., Shaw R. J. (2011) Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 145, 607–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang B., Moya N., Niessen S., Hoover H., Mihaylova M. M., Shaw R. J., Yates J. R., 3rd, Fischer W. H., Thomas J. B., Montminy M. (2011) A hormone-dependent module regulating energy balance. Cell 145, 596–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Collins S. P., Reoma J. L., Gamm D. M., Uhler M. D. (2000) LKB1, a novel serine/threonine protein kinase and potential tumour suppressor, is phosphorylated by cAMP-dependent protein kinase (PKA) and prenylated in vivo. Biochem. J. 345, 673–680 [PMC free article] [PubMed] [Google Scholar]
- 50. Sapkota G. P., Kieloch A., Lizcano J. M., Lain S., Arthur J. S., Williams M. R., Morrice N., Deak M., Alessi D. R. (2001) Phosphorylation of the protein kinase mutated in Peutz-Jeghers cancer syndrome, LKB1/STK11, at Ser431 by p90RSK and cAMP-dependent protein kinase, but not its farnesylation at Cys433, is essential for LKB1 to suppress cell growth. J. Biol. Chem. 276, 19469–19482 [DOI] [PubMed] [Google Scholar]
- 51. Screaton R. A., Conkright M. D., Katoh Y., Best J. L., Canettieri G., Jeffries S., Guzman E., Niessen S., Yates J. R., 3rd, Takemori H., Okamoto M., Montminy M. (2004) The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell 119, 61–74 [DOI] [PubMed] [Google Scholar]
- 52. Wang A. H., Kruhlak M. J., Wu J., Bertos N. R., Vezmar M., Posner B. I., Bazett-Jones D. P., Yang X. J. (2000) Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol. Cell Biol. 20, 6904–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Verdel A., Khochbin S. (1999) Identification of a new family of higher eukaryotic histone deacetylases. Coordinate expression of differentiation-dependent chromatin modifiers. J. Biol. Chem. 274, 2440–2445 [DOI] [PubMed] [Google Scholar]
- 54. Zhou X., Marks P. A., Rifkind R. A., Richon V. M. (2001) Cloning and characterization of a histone deacetylase, HDAC9. Proc. Natl. Acad. Sci. U.S.A. 98, 10572–10577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Petrie K., Guidez F., Howell L., Healy L., Waxman S., Greaves M., Zelent A. (2003) The histone deacetylase 9 gene encodes multiple protein isoforms. J. Biol. Chem. 278, 16059–16072 [DOI] [PubMed] [Google Scholar]
- 56. Yuan Z., Peng L., Radhakrishnan R., Seto E. (2010) Histone deacetylase 9 (HDAC9) regulates the functions of the ATDC (TRIM29) protein. J. Biol. Chem. 285, 39329–39338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Szyf M., Slack A. D. (2000) Mechanisms of epigenetic silencing of the c21 gene in Y1 adrenocortical tumor cells. Endocr. Res. 26, 921–930 [DOI] [PubMed] [Google Scholar]
- 58. Magun R., Boone D. L., Tsang B. K., Sorisky A. (1998) The effect of adipocyte differentiation on the capacity of 3T3-L1 cells to undergo apoptosis in response to growth factor deprivation. Int. J. Obes. Relat. Metab. Disord. 22, 567–571 [DOI] [PubMed] [Google Scholar]
- 59. Wang A. H., Bertos N. R., Vezmar M., Pelletier N., Crosato M., Heng H. H., Th'ng J., Han J., Yang X. J. (1999) HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol. Cell Biol. 19, 7816–7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Katoh Y., Takemori H., Lin X. Z., Tamura M., Muraoka M., Satoh T., Tsuchiya Y., Min L., Doi J., Miyauchi A., Witters L. A., Nakamura H., Okamoto M. (2006) Silencing the constitutive active transcription factor CREB by the LKB1-SIK signaling cascade. FEBS J. 273, 2730–2748 [DOI] [PubMed] [Google Scholar]
- 61. Tiainen M., Ylikorkala A., Mäkelä T. P. (1999) Growth suppression by Lkb1 is mediated by a G1 cell cycle arrest. Proc. Natl. Acad. Sci. U.S.A. 96, 9248–9251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kudo N., Matsumori N., Taoka H., Fujiwara D., Schreiner E. P., Wolff B., Yoshida M., Horinouchi S. (1999) Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. U.S.A. 96, 9112–9117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. McKinsey T. A., Zhang C. L., Olson E. N. (2000) Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc. Natl. Acad. Sci. U.S.A. 97, 14400–14405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Miska E. A., Karlsson C., Langley E., Nielsen S. J., Pines J., Kouzarides T. (1999) HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 18, 5099–5107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lemercier C., Verdel A., Galloo B., Curtet S., Brocard M. P., Khochbin S. (2000) mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J. Biol. Chem. 275, 15594–15599 [DOI] [PubMed] [Google Scholar]
- 66. Borghi S., Molinari S., Razzini G., Parise F., Battini R., Ferrari S. (2001) The nuclear localization domain of the MEF2 family of transcription factors shows member-specific features and mediates the nuclear import of histone deacetylase 4. J. Cell Sci. 114, 4477–4483 [DOI] [PubMed] [Google Scholar]
- 67. Wang A. H., Yang X. J. (2001) Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol. Cell Biol. 21, 5992–6005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Winter B., Braun T., Arnold H. H. (1993) cAMP-dependent protein kinase represses myogenic differentiation and the activity of the muscle-specific helix-loop-helix transcription factors Myf-5 and MyoD. J. Biol. Chem. 268, 9869–9878 [PubMed] [Google Scholar]
- 69. Choi R. C., Siow N. L., Zhu S. Q., Tsim K. W. (2000) The cAMP-dependent protein kinase mediates the expression of AChE in chick myotubes. Neuroreport 11, 801–806 [DOI] [PubMed] [Google Scholar]
- 70. Haberland M., Montgomery R. L., Olson E. N. (2009) The many roles of histone deacetylases in development and physiology. Implications for disease and therapy. Nat. Rev. Genet. 10, 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Garren L. D., Gill G. N., Masui H., Walton G. M. (1971) On the mechanism of action of ACTH. Recent Prog. Horm. Res. 27, 433–478 [DOI] [PubMed] [Google Scholar]
- 72. Wood H. N., Braun A. C. (1973) 8-Bromoadenosine 3′:5′-cyclic monophosphate as a promoter of cell division in excised tobacco pith parenchyma tissue. Proc. Natl. Acad. Sci. U.S.A. 70, 447–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Taniguchi M., Carreira M. B., Smith L. N., Zirlin B. C., Neve R. L., Cowan C. W. (2012) Histone deacetylase 5 limits cocaine reward through cAMP-induced nuclear import. Neuron 73, 108–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Crabtree G. R. (1999) Generic signals and specific outcomes: signaling through Ca2+, calcineurin, and NF-AT. Cell 96, 611–614 [DOI] [PubMed] [Google Scholar]
- 75. Gwack Y., Sharma S., Nardone J., Tanasa B., Iuga A., Srikanth S., Okamura H., Bolton D., Feske S., Hogan P. G., Rao A. (2006) A genome-wide Drosophila RNAi screen identifies DYRK-family kinases as regulators of NFAT. Nature 441, 646–650 [DOI] [PubMed] [Google Scholar]
- 76. Shabb J. B. (2001) Physiological substrates of cAMP-dependent protein kinase. Chem. Rev. 101, 2381–2411 [DOI] [PubMed] [Google Scholar]
- 77. Zhu G., Fujii K., Belkina N., Liu Y., James M., Herrero J., Shaw S. (2005) Exceptional disfavor for proline at the P + 1 position among AGC and CAMK kinases establishes reciprocal specificity between them and the proline-directed kinases. J. Biol. Chem. 280, 10743–10748 [DOI] [PubMed] [Google Scholar]
- 78. Deng X., Ewton D. Z., Mercer S. E., Friedman E. (2005) Mirk/dyrk1B decreases the nuclear accumulation of class II histone deacetylases during skeletal muscle differentiation. J. Biol. Chem. 280, 4894–4905 [DOI] [PubMed] [Google Scholar]
- 79. Greco T. M., Yu F., Guise A. J., Cristea I. M. (2011) Nuclear import of histone deacetylase 5 by requisite nuclear localization signal phosphorylation. Mol. Cell Proteomics 10, M110.004317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3. [DOI] [PubMed] [Google Scholar]
- 81. Ha C. H., Kim J. Y., Zhao J., Wang W., Jhun B. S., Wong C., Jin Z. G. (2010) PKA phosphorylates histone deacetylase 5 and prevents its nuclear export, leading to the inhibition of gene transcription and cardiomyocyte hypertrophy. Proc. Natl. Acad. Sci. U.S.A. 107, 15467–15472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Harrison B. C., Huynh K., Lundgaard G. L., Helmke S. M., Perryman M. B., McKinsey T. A. (2010) Protein kinase C-related kinase targets nuclear localization signals in a subset of class IIa histone deacetylases. FEBS Lett. 584, 1103–1110 [DOI] [PubMed] [Google Scholar]
- 83. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sui S., Wang J., Yang B., Song L., Zhang J., Chen M., Liu J., Lu Z., Cai Y., Chen S., Bi W., Zhu Y., He F., Qian X. (2008) Phosphoproteome analysis of the human Chang liver cells using SCX and a complementary mass spectrometric strategy. Proteomics 8, 2024–2034 [DOI] [PubMed] [Google Scholar]
- 85. Li X., Song S., Liu Y., Ko S. H., Kao H. Y. (2004) Phosphorylation of the histone deacetylase 7 modulates its stability and association with 14-3-3 proteins. J. Biol. Chem. 279, 34201–34208 [DOI] [PubMed] [Google Scholar]
- 86. Potthoff M. J., Wu H., Arnold M. A., Shelton J. M., Backs J., McAnally J., Richardson J. A., Bassel-Duby R., Olson E. N. (2007) Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J. Clin. Invest. 117, 2459–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tatham M. H., Jaffray E., Vaughan O. A., Desterro J. M., Botting C. H., Naismith J. H., Hay R. T. (2001) Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276, 35368–35674 [DOI] [PubMed] [Google Scholar]
- 88. Kirsh O., Seeler J. S., Pichler A., Gast A., Müller S., Miska E., Mathieu M., Harel-Bellan A., Kouzarides T., Melchior F., Dejean A. (2002) The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 21, 2682–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Walkinshaw D. R., Weist R., Kim G.-W., You L., Xiao L., Nie J., Li C. S., Zhao S., Yang X. J. (February 7, 2013) The tumor suppressor kinase LKB1 activates SIK2 and SIK3 to stimulate nuclear export of class IIa histone deacetylases. J. Biol. Chem. 10.1074/jbc.M113.456996 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.