Background: PfEMP1 proteins cause Plasmodium falciparum-infected erythrocytes to bind human tissues during malaria.
Results: The IT4VAR13 ectodomain is rigid, elongated, and monomeric, presenting a binding site for its ligand, ICAM-1.
Conclusion: The IT4VAR13 ectodomain is unlike that of VAR2CSA, a PfEMP1 that adopts a compact structure with multiple domains contributing to ligand binding.
Significance: PfEMP1 proteins have evolved diverse architectures to facilitate ligand recognition.
Keywords: Adhesion, Biophysics, Malaria, Parasitology, X-ray Scattering, ICAM-1, PfEMP1 Protein Architecture, Cytoadhesion
Abstract
The adhesion of Plasmodium falciparum-infected erythrocytes to human tissues or endothelium is central to the pathology caused by the parasite during malaria. It contributes to the avoidance of parasite clearance by the spleen and to the specific pathologies of cerebral and placental malaria. The PfEMP1 family of adhesive proteins is responsible for this sequestration by mediating interactions with diverse human ligands. In addition, as the primary targets of acquired, protective immunity, the PfEMP1s are potential vaccine candidates. PfEMP1s contain large extracellular ectodomains made from CIDR (cysteine-rich interdomain regions) and DBL (Duffy-binding-like) domains and show extensive variation in sequence, size, and domain organization. Here we use biophysical methods to characterize the entire ∼300-kDa ectodomain from IT4VAR13, a protein that interacts with the host receptor, intercellular adhesion molecule-1 (ICAM-1). We show through small angle x-ray scattering that IT4VAR13 is rigid, elongated, and monomeric. We also show that it interacts with ICAM-1 through the DBLβ domain alone, forming a 1:1 complex. These studies provide a first low resolution structural view of a PfEMP1 ectodomain in complex with its ligand. They show that it combines a modular domain arrangement consisting of individual ligand binding domains, with a defined higher order architecture that exposes the ICAM-1 binding surface to allow adhesion.
Introduction
Malaria, caused by the parasite Plasmodium falciparum, remains one of the deadliest diseases affecting humanity. In 2010, there were an estimated 216 million episodes and 655,000 fatalities (1). The most severe symptoms occur during the erythrocytic phase of the parasite life-cycle and are associated with sequestration of parasitized erythrocytes within the microvasculature (2). This protects the parasite from detection and destruction by the spleen and causes pathology due to accumulation of infected erythrocytes in tissues, resulting in inflammation and occlusion of blood flow. Cerebral malaria is a major complication in the development of severe disease and is linked to erythrocyte accumulation within cerebral vessels and characterized by neurological symptoms such as impaired consciousness and seizures (2, 3).
Cytoadhesion is mediated by parasite-encoded PfEMP1 proteins that are exposed on the surfaces of infected erythrocytes (4). These multidomain proteins are encoded by ∼60 highly divergent var genes (5). In most cases the parasite only expresses one PfEMP1 at a time (6). As well as allowing the parasite to evade immune responses, switching of var gene expression can alter its adhesion phenotype (4). The PfEMP1 proteins are major targets for acquired, protective immunity that prevents severe disease and are, therefore, targets for vaccine development (7).
Numerous human receptors for P. falciparum have been identified (8) with CD36 and intercellular adhesion molecule-1 (ICAM-1),2 most commonly found to interact with infected erythrocytes (9, 10). ICAM-1 is a transmembrane glycoprotein with five extracellular immunoglobulin-like domains (D1–D5) and a short cytoplasmic tail. It is expressed at basal levels on endothelial cells, is greatly up-regulated during malaria-induced inflammation (2), and is important for efficient adhesion in vitro (11). Studies differ in their conclusions on the importance of ICAM-1 binding for the development of severe or cerebral disease. One recent work shows a correlation between ICAM-1 binding and cerebral malaria (12), and another showed increased, although not statistically significant, ICAM-1 binding in isolates from patients with clinical malaria compared with asymptomatic malaria (13). Infected erythrocytes also co-localize with ICAM-1 in patients who died of cerebral malaria (2), and vessels with higher ICAM-1 levels have higher levels of sequestration (14). However, although ICAM-1 may contribute to cerebral accumulation, it is not required for binding to endothelial cells derived from human brain tissue (15).
PfEMP1s have large modular ectodomains containing different numbers and combinations of Duffy binding-like (DBL) domains and cysteine-rich interdomain regions (CIDR). DBL and CIDR domains have been classified into different types (α-ζ) based on sequence identity (16). The DBLβ domains have been shown to contribute to ICAM-1 binding (17, 18). However, it is uncertain whether single domains from PfEMP1 proteins fully mimic the ligand binding phenotypes of intact ectodomains. Indeed VAR2CSA, a PfEMP1 involved in pregnancy-associated malaria, binds its ligand, chondroitin sulfate proteoglycan with 100,000-fold greater affinity than any of its individual DBL domains (19, 20).
The multimeric state of PfEMP1s and the stoichiometry of engagement with their receptors are also unclear. Two DBL-containing proteins involved in invasion, Plasmodium vivax Duffy-binding protein (21) and P. falciparum erythrocyte binding antigen 175 (EBA-175) (22), exist as dimers in their crystal structures, and the putative interfaces used to interact with binding partners include contributions from both monomers. It has also been suggested that dimerization of DBL domains is necessary for ligand binding in PfEMP1 proteins (21).
There are currently no structures available for any PfEMP1 or constituent domain bound to its ligand, yet the molecular mechanisms of PfEMP1 recognition of host receptors are of paramount importance to understand the role of cytoadherence in severe malaria and the mechanisms of antigenic variation. They may also guide the development of vaccines through the selection of appropriate antigens. Here we present data that demonstrate that the PfEMP1-ICAM-1 interaction is mediated fully by a single DBLβ domain binding to the ICAM-1 N terminus and that these form a 1:1 complex. Small angle x-ray scattering provides a striking visual confirmation of this interaction, showing that the domains within the PfEMP1 ectodomain form a rigid, elongated architecture that undergoes minimal structural changes as ICAM-1 docks onto the DBLβ domain. Therefore, this PfEMP1 ectodomain is a modular receptor, with ICAM-1 binding mediated by a single DBL domain, and yet has higher order organization.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The ectodomain of IT4VAR13 (UNIPROT ID A3R6S0, residues 1–2691) was cloned into baculovirus transfer vector pAcGP67-A (BD Biosciences), with a C-terminal V5 epitope and hexahistidine tag. The vector was co-transfected with linearized BakPak6 baculovirus DNA (BD Biosciences) into Sf9 insect cells to generate recombinant virus particles. Histidine-tagged proteins secreted into the supernatant of infected High-Five insect cells were purified using Co2+-chelate agarose. Eluted products were dialyzed into phosphate-buffered saline.
The DBLβ domains from IT4VAR13 (residues 811–1201), IT4VAR16 (835–1228), IT4VAR27 (919–1323), IT4VAR31 (810–1212), and IT4VAR41 (836–1228) were cloned into a modified pET15b vector, and the hexahistidine-tagged proteins were expressed in Escherichia coli Origami B cells (Novagen) at 25 °C. Cells were pelleted and lysed, and proteins were purified using nickel-nitrilotriacetic acid-Sepharose (Qiagen). The hexahistidine tags were removed by incubation overnight at 4 °C with 1 mg tobacco etch virus (TEV) protease for every 10 mg of protein before passing through a nickel-nitrilotriacetic acid column to remove TEV, tag, and uncleaved material. The domains were further purified on a Superdex 200 16/60 size-exclusion chromatography column (GE Healthcare) in 20 mm Tris, pH 8.0, 150 mm NaCl.
ICAM-1D1D5 (UNIPROT ID P05362, 1–485) and ICAM-1D1D2 (1–212) fused to human IgG1 Fc were transiently expressed in COS-7 cells and purified by Protein A-affinity chromatography. The Fc tag was cleaved from ICAM-1D1D5-Fc using endoproteinase GluC. ICAM-1D1D2 (28–212) was transiently expressed in HEK293T cells and purified using Ni2+-affinity chromatography.
Circular Dichroism
IT4VAR13 and IT4VAR13DBLβ at 0.4 mg ml−1 were dialyzed into 50 mm phosphate buffer, pH 7.2. Spectra were recorded using a Aviv Model 410 spectrometer (Aviv Biomedical) at 25 °C. Measurements were taken in a 0.1-cm path length cell at 0.5-nm intervals between 180 and 290 nm with a 1-s averaging time for each data point. Three consecutive recordings were made, averaged, and corrected for absorption by buffer alone. Secondary structure estimation was performed using the CDSSTR method from DichroWeb (23).
Thermal Shift
IT4VAR13DBLβ was mixed at 0.5 mg ml−1 with SYPRO orange dye (Invitrogen) (1:250) in 20 mm Tris, pH 8.0, 150 mm NaCl. Buffer-alone controls were run in the same plate. The samples were heated in an iCycler iQ real-time PCR detection system (Bio-Rad) from 35 to 50 °C in 0.5 °C increments. The fluorescence intensity was measured with a charge-coupled device using an excitation and emission wavelength of 490 and 525 nm, respectively.
Surface Plasmon Resonance (SPR)
SPR experiments were carried out using a BIAcore T100 instrument (GE Healthcare). All experiments were performed in 10 mm HEPES, pH 7.2, 150 mm NaCl, 50 μm EDTA, and 0.05% Tween 20 at 20 °C. Protein A was immobilized on a CM5 chip (GE Healthcare) by amine coupling. ICAM-1-Fc was captured onto the Protein A surface. Concentration series of full-length IT4VAR13 or DBLβ domains were flowed over the ICAM-1-Fc-bound surface at 30 ml/min for 240 s followed by buffer for 300 s. After each run, the biosensor chip was regenerated using 20 mm glycine, pH 1.5, which breaks the Protein A-Fc interaction. The specific binding response to ICAM-1 was obtained by subtracting the response given by analytes to an uncoupled Protein A surface. No binding was observed to Fc alone. The kinetic sensorgrams were fitted to a global 1:1 interaction model to allow calculation of ka, kd, and KD using BIAevaluation software 2.0.3 (GE Healthcare).
Analytical Size-exclusion Chromatography (SEC)
SEC was performed using a Superdex 200 10/300 column (GE Healthcare) equilibrated with 200 mm ammonium acetate, pH 7.5, at a flow rate of 0.5 ml/min with absorbance at 280 nm used for detection. Each sample had at a total protein concentration of 1 μm, and 200 μl was injected for each run.
Analytical Ultracentrifugation
Sedimentation velocity experiments were conducted using a Beckman Optima XL-1 analytical ultracentrifuge (Beckman Coulter) at 20 °C. All samples were prepared in 50 mm Tris, pH 8.0, 150 mm NaCl. Reference and sample were loaded into a double-sector centerpiece and mounted in a Beckman An-60 Ti rotor. Sample concentrations were selected to give an absorbance of 1.0 at a wavelength of 280 nm. This required 3.2 μm for IT4VAR13, 5.5 μm for IT4VAR13DBLβ, and 10 μm for ICAM-1D1D5. Components were mixed in 1:1 molar ratios to form complexes, again giving total absorbance of 1.0 at 280 nm wavelength. Centrifugation rates were chosen based on the predicted size of the species, and absorbance at 280 nm was measured across the sample cell every 2 min for 70–200 scans during centrifugation. Multiple scans were fitted to a continuous size distribution using SEDFIT (24). The solvent density and viscosity and the partial specific volumes for the different samples were calculated using SEDNTERP (25). The solvent viscosity was 0.010312 poise, and solvent density was 1.00585 g ml−1. The partial specific volumes based on the amino acid sequence for the proteins were 0.7231 cm3 g−1 (IT4VAR13), 0.7222 cm3 g−1 (IT4VAR13DBLβ), and 0.7358 cm3 g−1 (ICAM-1D1D5).
Small-angle X-ray Scattering (SAXS)
SAXS data were collected at the ID14-3 beamline at the European Synchrotron Radiation Facility using a wavelength (l) of 0.931 Å. The sample-to-detector distance was 2.43 m, resulting in scattering vectors, q, ranging from 0.04 to 0.61 Å−1. The scattering vector is defined as 4p sinq/l, where 2q is the scattering angle. Scattering was detected using a Pilatus image reader.
Samples were prepared at 5.0, 2.5, 1.25, 0.63, and 0.31 mg ml−1 in 20 mm Tris, pH 8.0, 150 mm NaCl. Complexes were mixed in a 1:1 molar ratio for 30 min to give a total combined protein concentration of 5 mg ml−1 before producing the dilution series. All experiments were performed at 20 °C. Ten consecutive frames each with an exposure time of 10 s were recorded for proteins and buffer. Each frame was carefully inspected for protein radiation damage before averaging unaffected images.
SAXS data were normalized to the intensity of the incident beam and averaged, and background was subtracted using PRIMUS (26, 27). They were investigated for aggregation using Guinier plots (28). The scattering curves were extrapolated to zero concentration, and composite curves were generated by scaling and merging low concentration data with high concentration data to counter effects due to protein-protein concentration.
The distance distribution function (P(r)) was derived using indirect Fourier transform (29), from which the radius of gyration (Rg) and the maximum particle dimension (Dmax) were estimated. The Porod volume of the hydrated particle was calculated as described (30). Ab initio shape reconstructions were calculated using DAMMIF (31) and averaged with DAMAVER (32). DAMMIF was run in slow mode using 50 spherical harmonics for the processing of IT4VAR13.
Docking into envelopes was performed using atomic resolution structures of ICAM-1 immunoglobulin-like domains 1–2 (PDB ID 1IAM) and domains 3–5 (PDB ID 1P53). Three-dimensional models of IT4VAR13 domains were generated using I-TASSER (33). DBL domains were homology modeled using EBA-175 (PDB ID 1ZRL) and DBL3x (PDB ID 3BQK) as templates. Partial CIDR domains were homology-modeled using PDB ID 3C64 as a template. These atomic models were docked into the envelopes using SITUS and SCULPTOR, a program designed for modeling atomic resolution structures and low resolution biophysical data (34). The fit of the models to the envelopes was calculated using SUPCOMB (35). To validate the final models, HydroPro (36) was used to calculate theoretical hydrodynamic parameters to compare with the experimentally derived ones. As the atomic resolution structure of only one of the binding partners is known, rigid body modeling was not used.
RESULTS
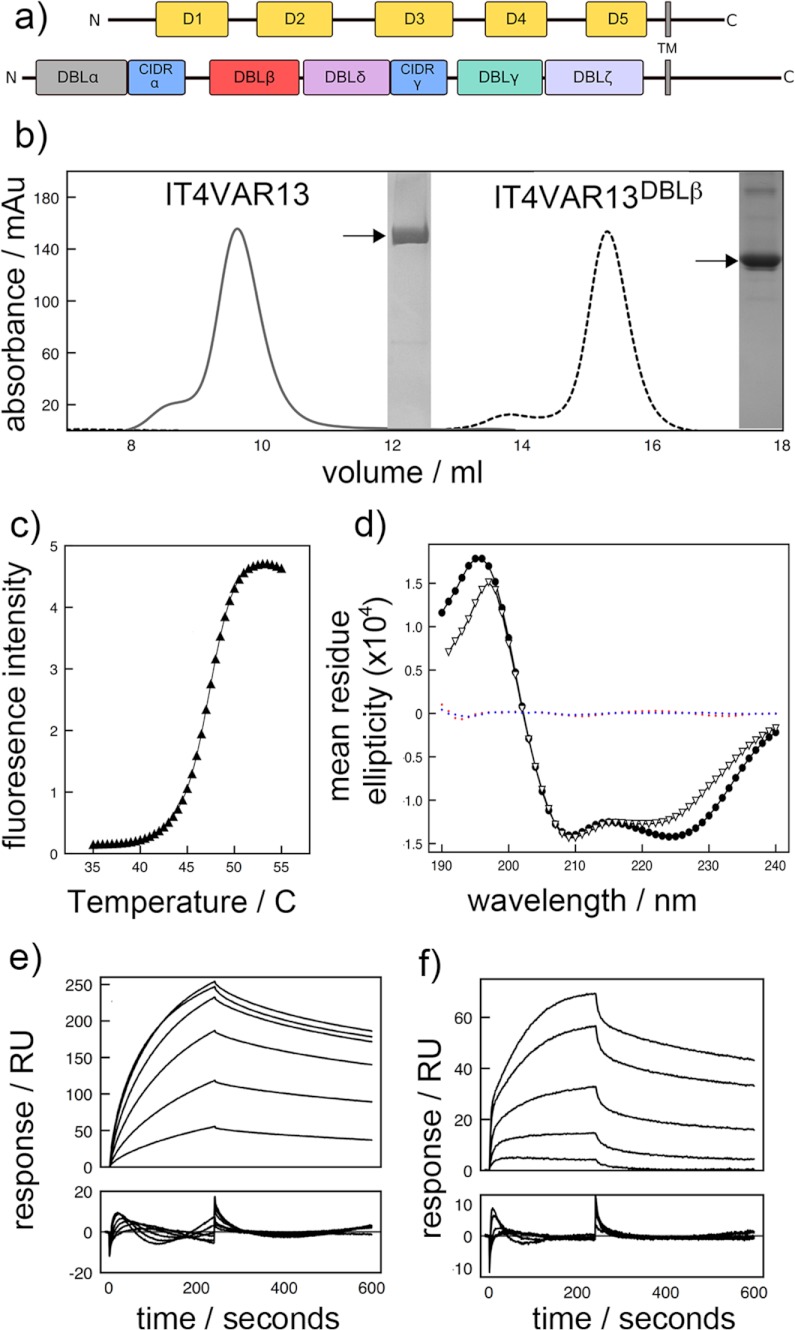
A baculovirus system was used to express an entire PfEMP1 ectodomain, containing five DBL and two CIDR domains, from the var13 gene of the IT4 isolate of P. falciparum (IT4VAR13). The DBLβ domain (IT4VAR13DBLβ) has been reported to bind ICAM-1 (16) and was expressed in E. coli. Both were purified to homogeneity and are stable and correctly folded as revealed by circular dichroism (Fig. 1).
FIGURE 1.
The DBLβ domain of IT4VAR13 binds ICAM-1D1D2 with nanomolar affinity. a, shown is a schematic of IT4VAR13 and ICAM-1. IT4VAR13 contains seven domains: five DBL domains (DBLα (gray), -β (red), -δ (magenta), -γ (teal), and -ζ (light blue)) and two CIDR domains (CIDRα and -γ (both blue)). ICAM-1 has five immunoglobulin-like domains (D1–D5 (yellow)). Both proteins have a single transmembrane (TM) domain. IT4VAR13 domain boundaries were set according to Rask et al. (16), and ICAM-1 domain boundaries were according to PFAM. b, shown is a size-exclusion chromatogram of IT4VAR13 and IT4VAR13DBLβ with purity shown by SDS-PAGE. IT4VAR13 is predominantly monomeric but with a shoulder indicating the presence of higher order aggregates that elute near the void volume. IT4VAR13DBLβ is similarly predominantly monomeric, with a low percentage of dimer. mAu, milliabsorbance units. c, shown is a representative fluorescence melting curve for IT4VAR13DBLβ in 20 mm Tris, pH 8.0, 150 mm NaCl. The temperature was altered in 0.5 °C increments. The y axis shows fluorescence (arbitrary units). IT4VAR13DBLβ has a melting temperature (Tm) of 47.5 °C. d, secondary structure analysis by CD is shown. Spectra were recorded between 190 and 240 nm for IT4VAR13 (●) and IT4VAR13DBLβ (▿). For each protein, three measurements were averaged, normalized for buffer absorption, and deconvoluted using an experimental model. Fitting residuals for IT4VAR13 (blue) and IT4VAR13DBLβ (red) are shown. IT4VAR13 is composed of 46% α-helix and 18% β-strand, and IT4VAR13DBLβ is composed of 57% α-helix and 6% β-strand. Shown are SPR sensorgrams (upper panels) with fitting residuals (lower panels) for the binding of IT4VAR13 to ICAM-1D1D5-Fc at concentrations of 5, 10, 20, 30, 40, and 50 nm (e) and IT4VAR13DBLβ to ICAM-1D1D5-Fc (10, 20, 30, 40, and 50 nm) (f). Binding was conducted with a 4-min association phase and 6-min dissociation phase at a constant flow rate of 30 ml min−1. In each case the lower panel shows residuals from binding.
A Single DBLβ Domain Fully Mediates the Binding of the IT4VAR13 Ectodomain to ICAM-1
PfEMP1s are considered as modular proteins with discrete ligand binding domains. However, recent studies show that the full-length VAR2CSA ectodomain binds chondroitin sulfate proteoglycan with a 100,000-fold higher affinity and greater specificity than constituent domains, suggesting a binding site containing more than just a single domain (19, 20). This raised the question of whether other PfEMP1 proteins also use combinations of domains to form ligand binding surfaces, a question that influences selection of vaccine components. To test this, we used SPR to obtain kinetic parameters for the binding of IT4VAR13 and IT4VAR13DBLβ to ICAM-1.
Domains 1 to 5 of ICAM-1 (ICAM-1D1D5) were expressed with a C-terminal Fc tag and captured onto the surface of a Protein A chip. This mimicked the orientation of presentation on the endothelial cell surface, exposing the complete ICAM-1 ectodomain for binding. It also allowed regeneration between experiments with low pH used to break the Protein A-Fc interaction before capture of an equivalent quantity of fresh ICAM-1D1D5-Fc.
Sensograms revealed clear binding of both IT4VAR13 and IT4VAR13DBLβ to ICAM-1D1D5 (Fig. 1, e and f), although VAR2CSADBL4e, a domain not predicted to bind ICAM-1 (19), showed no response (data not shown). Binding data were fitted globally to a one-site kinetic model with low residuals (Fig. 1, e and f, Table 1). IT4VAR13 bound ICAM-1D1D5 with low nanomolar affinity (KD = 2.8 nm) and with rapid association (ka = 2.7 × 105 m−1 s−1) and slow dissociation (kd = 7.5 × 10−4 s−1) kinetics (Fig. 1e). This is a tighter affinity than that of ICAM-1 for either rhinovirus (37) or its native ligand, lymphocyte function-association antigen-1 (38), but is similar to that of VAR2CSA for placental chondroitin sulfate proteoglycan (19, 20).
TABLE 1.
Kinetic parameters derived from SPR
Kinetic parameters for the binding of IT4VAR13 and IT4VAR13DBLβ to ICAM-1D1D5-Fc were derived from fitting to a one-site interaction model. The S.E. of each experiment is shown as reported by the BIAevaluation software.
| MW | ICAM-1 capture levels | ka | kd | KD | |
|---|---|---|---|---|---|
| RU | m−1 s−1 × 105 | s−1 × 10−4 | m × 10−9 | ||
| IT4VAR13-ICAM-1D1D5-Fc | 313.9 | 119.9 ± 2.8 | 2.7 ± 0.004 | 7.5 ± 0.01 | 2.8 |
| IT4VAR13DBLβ-ICAM-1D1D5-Fc | 45.3 | 102.8 + 0.5 | 3.5 ± 0.013 | 9.0 ± 0.03 | 2.6 |
The binding of IT4VAR13DBLβ to ICAM-1D1D5-Fc was investigated using the same chip surface. The binding curves showed a partially biphasic interaction with a major component that fitted with an affinity (KD = 2.6 nm) and kinetic parameters (ka = 3.5 × 105 m−1 s−1, kd = 1.0 × 10−4 s−1), almost identical to the parameters observed for the entire IT4VAR13 ectodomain (Fig. 1f). We attribute the minor binding component, with extremely fast on and off rates, to nonspecific adhesion of IT4VAR13DBLβ to the ICAM-1D1D5-Fc-coated surface. Indeed, although IT4VAR13 is monomeric in solution, IT4VAR13DBLβ shows some propensity to dimerize and aggregate with removal of the DBLβ domain from its ectodomain context presumably exposing normally buried surfaces and causing some “stickiness” (see Fig. 4). Nevertheless, the similarity in binding parameters of the principal interactions of IT4VAR13DBLβ and IT4VAR13 for ICAM-1 is consistent with a modular architecture for IT4VAR13 in which a single DBLβ domain contributes the ICAM-1 binding site.
FIGURE 4.
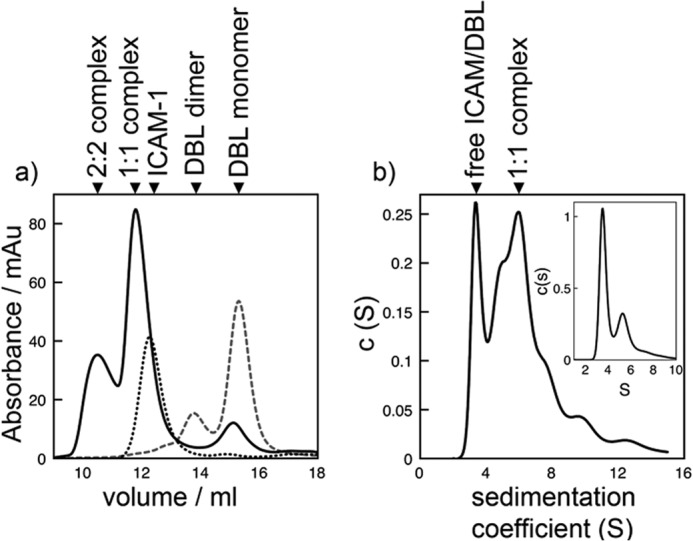
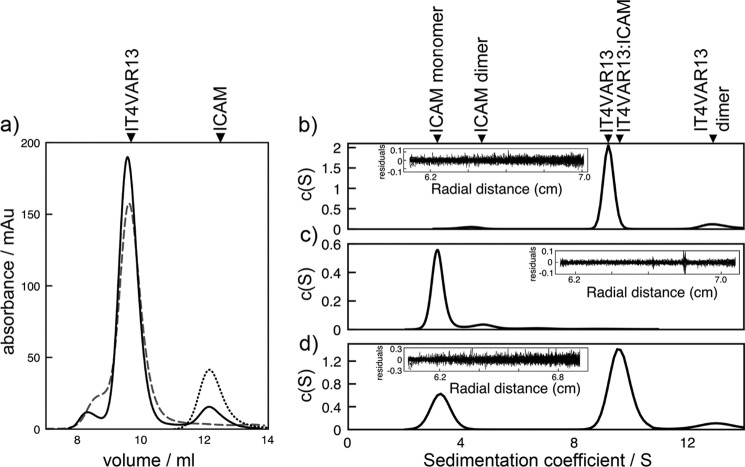
Stoichiometry of the interaction between IT4VAR13DBLβ and ICAM-1. Stoichiometry was determined by analytical SEC (a) and ultracentrifugation (b). mAu, milliabsorbance units. a, shown is a size-exclusion chromatogram of IT4VAR13DBLβ (gray, wide dashes), ICAM-1D1D5 (black, short dashes) and IT4VAR13DBLβ·ICAM-1D1D5 (black, continuous line). b, shown is continuous sedimentation coefficient distribution that best describes the AUC data for IT4VAR13DBLβ incubated with ICAM-1D1D5 for 30 min before centrifugation with the continuous sedimentation coefficient distribution for IT4VAR13DBLβ inset for comparison. The distribution for ICAM-1D1D5 is in Fig. 3c.
Comparison of ICAM-1 Binding by DBLβ Domains from the IT4 Strain
As well as IT4VAR13, six other PfEMP1 proteins from the P. falciparum IT4 isolate have been shown to bind ICAM-1 (IT4VAR1, -14, -16, -27, -31, -41) (17). The seven DBLβ domains from these PfEMP1 proteins share 46% sequence identity (Fig. 2a). The DBLβ domains of four of these were expressed in E. coli and purified to homogeneity (IT4VAR16, -27, -31, and -41), and ICAM-1 binding was examined using SPR (Fig. 2, b–e). The data were fitted to a one-site binding model, and the kinetic parameters determined are given in Table 2. In all cases comparison of the maximum binding levels with the amount of ICAM-1D1D5 coupled to the chip surface suggests formation of 1:1 complexes. ITVAR31DBLβ bound ICAM-1D1D5-Fc with the lowest affinity (144 nm). This is a more than 50-fold lower affinity than the strongest ICAM-1 binder (IT4VAR13DBLβ, 2.8 nm) and is consistent with the observation that parasite lines expressing ITVAR31 bind ICAM-1 weakly (42, 43).
FIGURE 2.
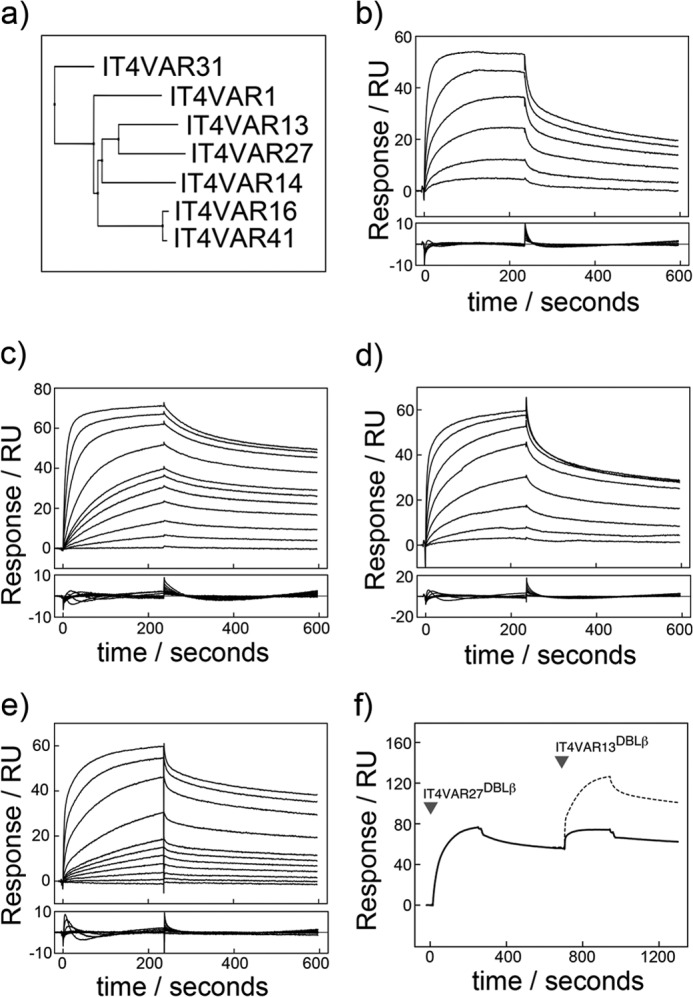
Characterization by SPR of the interactions of ICAM-1 with multiple DBLβ domains from the P. falciparum IT4 isolate. a, shown is a phylogenetic tree of seven PfEMP1 DBLβ domains known to bind ICAM-1 from the P. falciparum IT4 isolate. Shown are sensorgrams (upper panel) with resulting residuals when fit to a one-site kinetic model (lower panel). Shown are SPR sensorgrams (upper panels) with fitting residuals (lower panels) for the binding of IT4VAR16DBLβ (50, 100, 250, 500, 1000, and 2000 nm) (b), IT4VAR27DBLβ (1, 5, 10, 20, 30, 40, 50, 100, 250, 500, and 1000 nm) (c), IT4VAR31DBLβ (0.05, 0.1, 0.25, 1, 2, 5, and 10 μm) (d), and IT4VAR41DBLβ (1, 5, 10, 20, 30, 40, 50, 100, 250, 500, and 1000 nm) (e) to ICAM-1D1D2-Fc with an association phase of 4 min and a dissociation phase of 6 min at a flow rate of 30 μl min−1. f, DBLβ domains from IT4VAR13 and IT4VAR27 recognize ICAM-1 with overlapping binding sites. Expected binding levels assuming IT4VAR13DBLβ bound to ICAM-1 in a mode independent and unaffected by the binding of IT4VAR27DBLβ are shown with a dashed line. Actual binding levels shown by a solid line.
TABLE 2.
Kinetic parameters derived from SPR
Kinetic parameters were derived from a global fit to a one-site model. The S.E. of each experiment is shown as reported by the BIAevaluation software. Theoretical interaction stoichiometry was determined using n = (RUmax × MWICAM-1-Fc)/(RUICAM-1-Fc × MWDBLβ), where MWICAM-1-Fc = 78.2 kDa and assuming all ICAM-1 on the surface is fully accessible and functional.
| DBLβ MW | ICAM-1-Fc capture levels | ka | kd | KD | n | |
|---|---|---|---|---|---|---|
| RU | m−1 s−1 × 104 | s−1 × 10−4 | m × 10−9 | |||
| IT4VAR13DBLβ-ICAM-1D1D5-Fc | 45.3 | 102.8 + 0.5 | 3.5 ± 0.013 | 9.0 ± 0.03 | 2.6 | 1.16 |
| IT4VAR16DBLβ-ICAM-1D1D5-Fc | 44.9 | 100.5 ± 0.8 | 4.1 ± 0.04 | 20.8 ± 0.09 | 51.1 | 0.93 |
| IT4VAR27DBLβ-ICAM-1D1D5-Fc | 45.8 | 94.1 ± 0.6 | 13.0 ± 0.07 | 84.2 ± 0.04 | 6.5 | 1.30 |
| IT4VAR31DBLβ-ICAM-1D1D5-Fc | 46.0 | 102.5 ± 0.3 | 1.0 ± 0.01 | 14.5 ± 0.12 | 144 | 0.99 |
| IT4VAR41DBLβ-ICAM-1D1D5-Fc | 44.8 | 95.5 ± 0.6 | 6.0 ± 0.01 | 83.1 ± 0.02 | 14.0 | 1.09 |
The ability of different DBL domains to bind simultaneously to ICAM-1D1D5 was also studied using SPR. IT4VAR27DBLβ was initially bound to ICAM-1D1D5 followed by an injection of IT4VAR13DBLβ (Fig. 2f). Had the two domains been capable of binding both simultaneously and independently, the expected response would have been considerably higher than that observed. Instead, the increase in material bound to the surface upon injection of IT4VAR13DBLβ was comparable to the decrease caused by the dissociation of IT4VAR27DBLβ, making it likely that IT4VAR13DBLβ binds to an overlapping binding site on ICAM-1.
IT4VAR13 Is Monomeric and Binds to ICAM-1 with a 1:1 Stoichiometry
The oligomeric states of PfEMP1s are uncertain. However, crystal structures suggest that DBL domain-containing proteins involved in erythrocyte invasion function as dimers (21, 22), and PfEMP1s have been predicted to do the same (21). Here we have used both analytical SEC and sedimentation velocity analytical ultracentrifugation (AUC) to assess the oligomeric states of IT4VAR13 and IT4VAR13DBLβ both alone and in complex with ICAM-1.
IT4VAR13 is predominantly monomeric in solution with SEC revealing a single major peak (Fig. 3a). This was confirmed by AUC, which showed 92% of the protein to be monomeric, with a sedimentation coefficient of 9.2 S, corresponding to a mass of around 285 kDa (Fig. 3b), whereas 8% was found in a peak consistent with a dimer. A frictional coefficient, f/f0, of 1.7 suggests that the ectodomain is elongated but unlikely to exist in a fully extended conformation. Isolated IT4VAR13DBLβ shows a greater propensity to form dimers both by SEC and AUC (Fig. 4), but despite the removal of neighboring domains, some 75–80% of the protein in still in the form of a monomer, with a sedimentation coefficient of 3.2 S consistent with a mass of around 50 kDa. IT4VAR13DBLβ is, therefore, no different from the PfEMP1 DBL domains structurally characterized to date, which all exist as monomers within their crystals (44–47).
FIGURE 3.
Stoichiometry of the interaction between IT4VAR13 and ICAM-1. Stoichiometry was determined by analytical SEC (a) and ultracentrifugation (b-d). mAu, milliabsorbance units. a, shown is a size-exclusion chromatogram of IT4VAR13 (gray, wide dashes), ICAM-1D1D5 (black, short dashes), and IT4VAR13·ICAM-1D1D5 (black, continuous line) from a calibrated Superdex S200 column. Shown are continuous sedimentation coefficient distributions that best describe the AUC data, with fitting residuals inset for IT4VAR13 (b) ICAM-1D1D5 (c), and IT4VAR13 (d) incubated with ICAM-1D1D5 for 30 min before centrifugation.
ICAM-1D1D5 prepared by removal of the Fc tag was also predominantly monomeric, as expected (48). SEC generated a single peak (Fig. 3a). This was confirmed by AUC (Fig. 3c, Table 3), which revealed more than 80% of the sample to form a primary species with a sedimentation coefficient of 3.2 S, corresponding with a mass of around 60 kDa, consistent with a glycosylated ICAM-1D1D5 monomer (Fig. 3c). Smaller peaks corresponding to masses of around 120 (10%) and 200 kDa (<5%) are most likely due to oligomers. ICAM-1D1D5 has an f/f0 of 1.7 consistent with electron micrographs that show that ICAM-1 assumes a bent rod-like shape 18.7 nm in length (49).
TABLE 3.
Summary of AUC sedimentation velocity results
Theoretical molecular weights of monomeric and 1:1 species are given in parentheses.
| Frictional coefficient (f/f0) | Sedimentation coefficient (S) | MWapp | Predicted species | |
|---|---|---|---|---|
| IT4VAR13 (314 kDa) | 1.66 | 4.2 | 89.1 | Truncation |
| 9.2 | 286.9 | IT4VAR13 monomer | ||
| 13.1 | 488.7 | IT4VAR13 dimer | ||
| ICAM-1D1D5 (50 kDa) | 1.67 | 3.2 | 63.4 | Monomer |
| 4.8 | 118.0 | Dimer | ||
| 6.8 | 197.0 | Tetramer | ||
| IT4VAR13:ICAM-1D1D5 (364 kDa) | 1.74 | 3.3 | 71.0 | ICAM-1D1D5 monomer |
| 9.6 | 333.1 | 1:1 IT4VAR13:ICAM-1D1D5 | ||
| 13.0 | 479.4 | IT4VAR13 dimer | ||
| IT4VAR13DBLβ (45 kDa) | 1.37 | 3.1 | 51.9 | Monomer |
| 4.8 | 114.0 | Dimer | ||
| IT4VAR13DBLβ:ICAM-1D1D5 (95 kDa) | 1.53 | 3.4 | 51.0 | Unbound monomeric IT4VAR13DBLβ/ ICAM-1D1D5 |
| 5.9 | 120.0 | 1:1 IT4VAR13DBLβ-ICAM-1D1D5 |
To investigate whether binding to ICAM-1 induces multimerization, IT4VAR13 was mixed with a small excess of ICAM-1D1D5 (in a ratio of 1:1.1) and analyzed by SEC. The ICAM-1 peak (12.3 ml) was depleted in size, suggesting most of it entered the complex, and the major peak was shifted slightly, from 9.6 ml (for IT4VAR13) to 9.5 ml (for IT4VAR13·ICAM-1D1D5). AUC confirmed the formation of a complex, indicating the presence of a predominant species with a sedimentation coefficient of 9.6 S and apparent mass of around 335 kDa. This is consistent with a single ICAM-1D1D5 bound to one IT4VAR13 ectodomain.
We confirmed this 1:1 stoichiometry by analyzing the IT4VAR13DBLβ·ICAM-1D1D5 complex. In this case, a greater variety of species was observed, consistent with the observation that the DBLβ domain alone multimerizes more readily than intact ectodomain. By both SEC and AUC, the predominant species observed in a mixture of IT4VAR13DBLβ and ICAM-1D1D5 is a 1:1 complex. In SEC, ICAM-1D1D5 was mixed with a small excess (1:1.1) of IT4VAR13DBLβ, leading to the formation of a predominant 1:1 complex, with a smaller amount of 2:2 complex. In AUC, two main peaks were observed, with sedimentation coefficients of 3.4 S (∼50 kDa and consistent with free IT4VAR13DBLβ and ICAM-1D1D5) and 5.9 S (∼120 kDa, consistent with a complex of one IT4VAR13DBLβ and one ICAM-1D1D5).
Therefore, IT4VAR13 and ICAM-1D1D5 are both predominantly monomeric in solution and combine to form a 1:1 complex. When removed from its ectodomain context, IT4VAR13DBLβ shows an increased and presumably artifactual propensity to multimerize but still forms a predominately 1:1 complex with ICAM-1D1D5. In addition, the stoichiometry of ∼1 in SPR experiments confirmed the formation of complexes containing equal quantities of IT4VAR13 and ICAM-1D1D5. Therefore, ITVAR13 is a monomeric ectodomain that interacts with a single ICAM-1.
Low Resolution Structures of IT4VAR13DBLβ Bound to ICAM-1 Fragments
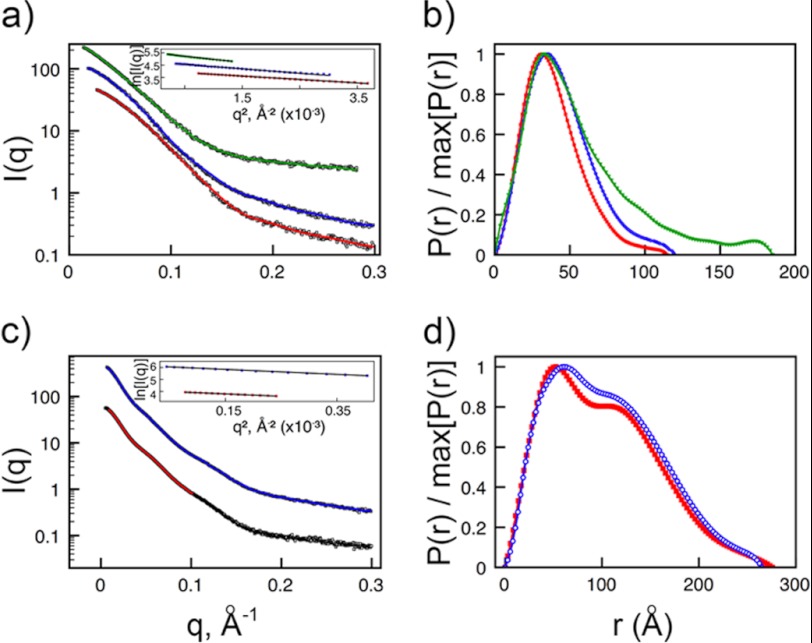
There are currently no structures of PfEMP1 proteins or domains bound to protein ligands. To understand better the architecture of the complex between PfEMP1 and ICAM-1, we performed SAXS with IT4VAR13DBLβ either alone or in complex with ICAM-1D1D2 or ICAM-1D1D5 (Fig. 5). The radius of gyration, determined from Guinier plots, is 3.4 nm for the IT4VAR13DBLβ·ICAM-1D1D2 complex and 4.2 nm for the IT4VAR13DBLβ·ICAM-1D1D5 complex compared with 3.0 nm for IT4VAR13DBLβ alone (Table 4). Concomitant increases in the Porod volume and apparent molecular weight were consistent with SEC and AUC data in suggesting that the interaction is predominantly 1:1. When compared with the distance distribution function for IT4VAR13DBLβ, both complexes have a slightly skewed profile with a tail extending toward a Dmax of 12 nm for the shorter complex and 18.5 nm for the longer complex, indicating that ICAM-1 protrudes from the DBLβ domain, resulting in an elongated particle (Fig. 5b).
FIGURE 5.
SAXS analysis of IT4VAR13·ICAM-1 complexes. a, shown is theoretical scattering calculated from ab initio reconstructions (continuous lines with IT4VAR13DBLβ in red, IT4VAR13DBLβ·ICAM-1D1D2 in blue, and IT4VAR13DBLβ-ICAM-1D1D5 in green) superimposed onto experimental scattering intensity curves (squares). Guinier plots are inset. b, shown are distance distribution function, P(r), plots for IT4VAR13DBLβ (squares, red); IT4VAR13DBLβ·ICAM-1D1D2 (circles, blue), and IT4VAR13DBLβ·ICAM-1D1D5 (triangles, green). c, theoretical scattering is calculated from ab initio reconstructions for full-length IT4VAR13 ectodomain (continuous lines with IT4VAR13 in red and IT4VAR13-ICAM-1D1D5 in blue) superimposed onto the experimental scattering intensity curves (squares). d, shown are P(r) plots for IT4VAR13 (squares, red) and IT4VAR13·ICAM-1D1D5 (circles, blue). The P(r) functions were calculated from the scattering intensity I(q) and normalized to unity at their maxima.
TABLE 4.
Summary of experimental SAXS values
The theoretical radius of gyration (Rg) was calculated using HydroPro (36) using atomic coordinates from docking into ab initio SAXS shape reconstructions. Experimental Rg values were derived from the Guinier plot using AutoRg (27). The maximum particle diameter (Dmax) was calculated using GNOM, and the Porod volume of the hydrated particle (Volume) was calculated as described (30). The apparent molecular weight was estimated from Porod volume/1.7. Theoretical molecular weights of monomeric and 1:1 species are given in parentheses. Twenty low resolution shape reconstructions were derived from the experimental data using ab initio modeling and the mean normalized spatial discrepancy (NSD), used to quantify the agreement between individual reconstructions (35). The χ2 for the fit of the best model to the experimental data is shown.
| Rg (theoretical) | Rg | Dmax | Volume | MWapp | Reconstructions NSD | χ2 | |
|---|---|---|---|---|---|---|---|
| nm | nm | nm | nm3 | ||||
| IT4VAR13DBLβ (45 kDa) | 2.6 | 2.97 ± 0.02 | 12.5 | 106.0 | 62.3 | 0.46 ± 0.03 | 2.7 |
| IT4VAR13DBLβ-ICAM-1D1D2 (70 kDa) | 3.1 | 3.41 ± 0.01 | 12.0 | 141.4 | 83.2 | 0.55 ± 0.02 | 4.4 |
| IT4VAR13DBLβ-ICAM-1D1D5 (95 kDa) | 4.0 | 4.21 ± 0.00 | 18.5 | 154.9 | 91.1 | 0.68 ± 0.04 | 4.6 |
| IT4VAR13 (314 kDa) | 8.1 | 8.1 ± 0.00 | 27.5 | 556.3 | 327.2 | 0.78 ± 0.06 | 1.3 |
| IT4VAR13- ICAM-1D1D5 (364 kDa) | 8.3 | 8.6 ± 0.00 | 26.5 | 649.4 | 382.0 | 1.06 ± 0.03 | 3.7 |
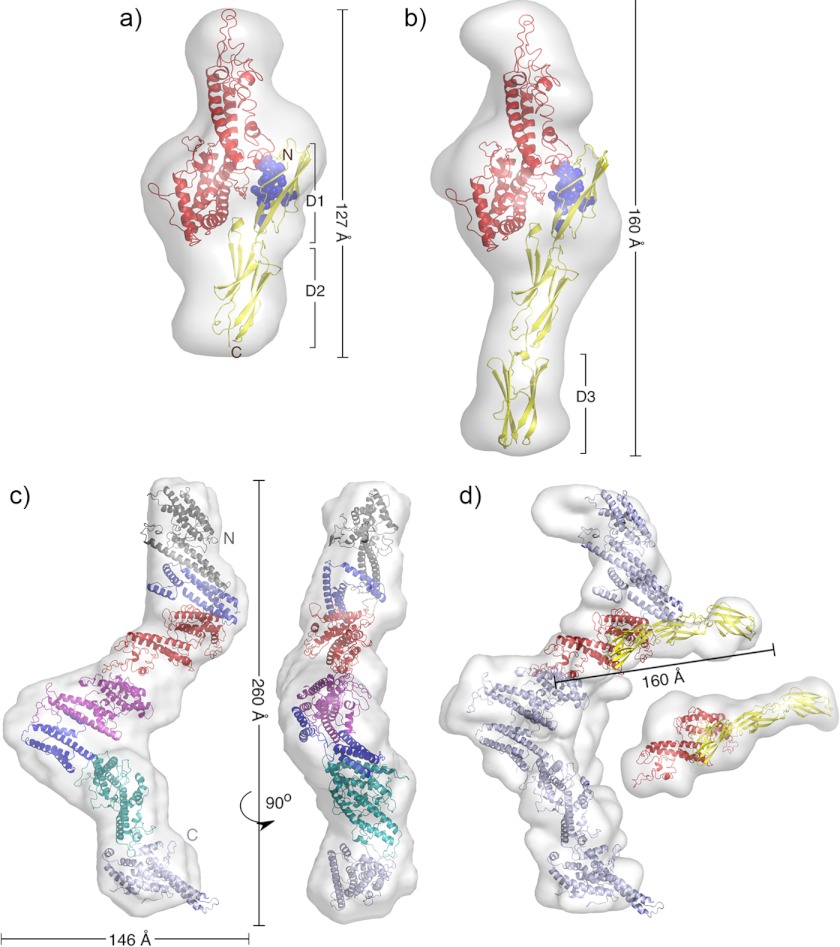
For each complex, 20 low resolution shape reconstructions were derived from the experimental data using ab initio modeling. As only one of the binding partners (ICAM-1) has a known high resolution structure, rigid body modeling was not employed, and the resultant envelopes were derived solely from scattering data. The models were averaged, and structures of IT4VAR13DBLβ (from a homology model based on EBA-175 (PDB ID 1ZRL) and DBL3x (PDB ID 3BQK) and ICAM-1D1D2 (PDB ID 1IAM)) were simultaneously docked into the shorter envelope with a 1:1 stoichiometry (Fig. 6a).
FIGURE 6.
SAXS-derived architectures of IT4VAR13-ICAM-1 complexes. Models of the IT4VAR13DBLβ·ICAM-1D1D2 (a) and IT4VAR13DBLβ·ICAM-1D1D5 (b) complexes are based on ab initio SAXS envelopes. IT4VAR13DBLβ (red) was homology-modeled using EBA-175 (PDB ID 1ZRL) and DBL3x (PDB ID 3BQK) as templates. ICAM-1 domains 1 (D1) and 2 (D2) were extracted from PDB ID 1IAM, and domain 3 was from PDB ID 1P53 (all yellow). ICAM-1 residues identified as reducing infected erythrocyte adhesion under flow conditions are shown as blue spheres. c, a low resolution structure of IT4VAR13 was determined from SAXS data showing front and side views. All domains were modeled using homology to known structures. d, a low resolution structure of the IT4VAR13·ICAM-1D1D5 complex was determined from SAXS data. ICAM-1D1D5 exclusively contacts IT4VAR13DBLβ.
The envelope obtained for IT4VAR13DBLβ·ICAM-1D1D5 was longer in comparison (160 Å compared with 127 Å), consistent with additional domains extending away from DBLβ. The IT4VAR13DBLβ·ICAM-1D1D2 complex was modeled into the globular domain with the ICAM-1 D3 domain positioned below the D2 domain (Fig. 6b). However, domains D4 and D5 could not be positioned into the envelope, most likely due to flexibility of ICAM-1. Although there is substantial contact between the D1 and D2 domains, there is a flexible hinge between D2 and D3, and the ICAM-1D3D5 crystal structure shows a further kink of ∼160° between domains D3 and D4 (49, 50). Although D1 and D2 are rigidly held in place relative to the DBLβ domain by extensive D1-DBLβ contacts, there is enough conformational flexibility in the D2-D3 linker to allow multiple conformations of D3-D5 relative to the DBLβ-D1D2 complex, disordering these domains relative to D2. As the SAXS profile is a weighted average over all accessible structures (51), such flexibility broadens the density around D3 and causes D4 and D5 to be absent from the averaged envelope. A similar degree of disorder was seen in cryo-EM images of ICAM-1D1D5 bound to human rhinovirus (52) and in higher resolution reconstructions of the coxsackievirus A21-ICAM-1D1D5 complex (53) with D4 and D5 not observed due to disorder in either case.
In both cases docking positioned the convex surface of DBLβ in contact with the D1 domain of ICAM-1. DBL domains have previously been described as containing three subdomains (54). Both subdomain 2 and the proximal end of subdomain 3 are positioned close to ICAM-1 here, suggesting a large protein-protein interface and consistent with previous analysis from chimeric proteins and truncations (55–57). Residues equivalent to the mutations that affect the binding of IT4VAR16 to ICAM-1 (17) are located within this interface. Mutational studies that suggested that the interface lies on the “BED” side (that containing β-strands B, E, and D) of the ICAM-1 D1 domain (40, 41) are also consistent with this solution. The D2 domain is in close proximity to DBLβ and may also be capable of forming some interactions with subdomain 2. The observed interface is similar to that proposed by Bertonati and Tramontano (58) from in silico docking, albeit with the orientation that ICAM-1 approaches the DBL domain rotated ∼75°.
The Low Resolution Structure of IT4VAR13 and Its Complex with ICAM-1
To see how domains are organized within the PfEMP1 ectodomain and how they accommodate binding to ICAM-1, we collected SAXS data for both IT4VAR13 and the IT4VAR13·ICAM-1D1D5 complex (Fig. 5c). The Rg for the complex (8.6 nm) is larger than that observed for IT4VAR13 (8.1 nm), but both species have a similar Dmax, indicating that ICAM-1 does not bind in a head-to-head arrangement that increases the maximum particle diameter (Fig. 5d). A Kratky plot (data not shown) indicates that the ectodomain forms a rigid structure.
Low resolution shape reconstructions were derived from the experimental data using ab initio modeling. The IT4VAR13 envelope has dimensions of 260 × 146 × 50 Å (Fig. 6c) and adopts a zig-zag conformation that deviates considerably from a rod-like structure, as expected from an f/f0 of 1.7. This is very different from the compact architecture of VAR2CSA, the only other ectodomain characterized to date. However, neither IT4VAR13 nor VAR2CSA consists of a series of independent domains joined by flexible linkers. To provide more detailed structural insight, homology models of the individual domains were built using known crystal structures, allowing 75% of the IT4VAR13 ectodomain to be modeled. Models for these seven domains were docked into the envelope, having been restrained such that each domain follows in sequential order as dictated by the primary structure.
The SAXS-derived structure of the IT4VAR13·ICAM-1D1D5 complex, determined independently from that of IT4VAR13 and solely from scattering data, reveals an ectodomain architecture very similar to that in the absence of ligand, suggesting few if any conformational changes take place on ligand binding (Fig. 6d). The main difference is the additional mass immediately adjacent to DBLβ, which is consistent in size and position with the mass attributed to ICAM D1-D3 in reconstructions of data from the IT4VAR13DBLβ·ICAM-1D1D5 complex. Therefore, we observed contact only between ICAM-1 and DBLβ and see no significant architectural rearrangements of the rigid ectodomain and no multimerization upon ligand binding.
DISCUSSION
Sequestration of parasitized erythrocytes to the brain microvasculture through the interaction of PfEMP1s with host receptors including ICAM-1 is associated with cerebral malaria. Cytoadherence is a potential target of novel therapeutics to combat malaria, especially as it persists after conventional drugs have killed the parasite (59). To develop a greater understanding of the molecular underpinnings of pathogen-host receptor interactions, we used biophysical techniques to characterize the interaction between a PfEMP1 and human ICAM-1.
Here we show using SEC, AUC, and SAXS that IT4VAR13 is monomeric, and we present its low resolution structure. The protein has a rigid elongated shape that shows considerable deviation from a canonical rod with the DBL2δ and CIDR2γ domains protruding from the longest axis. The SAXS-derived structures of VAR2CSA (20, 60) show a compact organization that dispelled the notion that PfEMP1 domains are ordered as “beads on a string.” Here, we show that PfEMP1s can form alternative global shapes, dependent on the combination of domains present. VAR2CSA is unusual among PfEMP1 proteins, as it comprises six DBL domains and a single CIDRpam domain. IT4VAR13 has a more typical domain architecture, and the rigid, elongated arrangement may be more frequently observed.
If PfEMP1 proteins are generally modular arrays of ligand binding domains, why have a rigid architecture, with all of the resulting structural constraints on protein evolution? A possible reason is to reduce the surface area that is exposed to immune detection. A rigid architecture, formed by the close packing of domains, will hide surfaces that would otherwise be exposed in a flexible molecule. PfEMP1 proteins are under diversifying selection to evade immune detection and yet must maintain binding properties needed for cytoadhesion. Perhaps rigid ectodomains help reduce immune detection without imposing costly constraints on the evolution of immune diversity or the maintenance of ligand binding.
Previous studies identified DBLβ as an ICAM-1 binding domain. Here we use SPR to confirm this observation and show that isolated DBLβ domains bind to ICAM-1 with the same affinity as intact ectodomain, suggesting that it alone mediates ICAM-1 binding. Indeed, comparison of SAXS reconstructions of IT4VAR13 in the presence and absence of ICAM-1 show a single addition of mass due to ICAM-1 that lies immediately adjacent to the DBLβ domain. Although these SAXS-derived envelopes are low resolution, docking in the ICAM-1 structure, and a model of DBLβ gives an interface consistent with previous mutagenesis studies that predict that the convex surface of DBLβ interacts with the BED face of ICAM-1 (17, 39).
The elongated ectodomain structure positions the DBLβ domain ∼150 Å above the erythrocyte membrane surface. This, together with the clustering of PfEMP1s on knobs that protrude 110–160 nm from the red blood cell will position the ICAM-1 binding site for ready access to its ligand (61). The interaction of a domain close to the tip of the ectodomain with domains at the tip of ICAM-1, therefore, allows efficient recognition. The elongated nature of the ectodomain also makes it possible for other domains in the PfEMP1 to mediate interactions with other receptors, allowing binding synergy.
The tight nanomolar affinity (∼3 nm) and slow dissociation rate (Kd = 7.5 × 10−4 s−1) is consistent with the need for the interaction to be strong enough to pull the infected erythrocyte out of circulation to permit accumulation despite blood flow or to cause rolling adhesion under shear stress. This interaction is stronger and has a slower dissociation constant than interactions involved in selectin-mediated rolling adhesion of leukocytes from the bloodstream (61). Affinities in the nanomolar range (3–144 nm), characterized by slow dissociation kinetics, were also observed for other DBLβ domains from the P. falciparum IT4 isolate, suggesting a consistent mode of interaction with ICAM-1 within the family.
The stoichiometry of the engagement of DBL domains with their receptors has been under debate. Studies of DBL domains from proteins that mediate interactions involved in invasion have led to the suggestion that dimerization is necessary for ligand binding (21). For the interaction between PfEMP1 and ICAM-1, SPR, AUC, SEC, and SAXS all reveal a 1:1 stoichiometry, and multimerization is not required for, or driven by, binding.
These studies reveal many differences between IT4VAR13 and VAR2CSA, the only other intact PfEMP1 ectodomain with a low resolution structure determined to date. VAR2CSA adopts a compact structure with more than a single DBL domain required to form a high affinity, specific binding site for its carbohydrate ligand (19, 20). In contrast, IT4VAR13 is rigid but elongated, positioning a single ICAM-1 binding DBLβ domain close to its tip for efficient recognition of this protein ligand. This highlights some of the degree of diversity available to this adaptable adhesion protein, with changes in domain organization and use modulating the ability of these antigens to recognize their ligands as they perform their dual role of adhesion and immune evasion.
Acknowledgments
We acknowledge the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities, and we thank Adam Round and Andrew McCarthy for assistance in using beamline ID14-3. We thank Radu Aricescu for facilitating protein expression.
This work was supported by Wellcome Trust Grant 087692/Z/08/Z and the University of Copenhagen Program of Excellence.
- ICAM-1
- intercellular adhesion molecule-1
- DBL
- Duffy binding-like
- CIDR
- cysteine-rich interdomain region
- SPR
- surface plasmon resonance
- SEC
- size-exclusion chromatography
- SAXS
- small-angle x-ray scattering
- AUC
- analytical ultracentrifugation
- RU
- response units.
REFERENCES
- 1. WHO (2011) World Malaria Report 2011, Geneva, Switzerland [Google Scholar]
- 2. Turner G. D., Morrison H., Jones M., Davis T. M., Looareesuwan S., Buley I. D., Gatter K. C., Newbold C. I., Pukritayakamee S., Nagachinta B. (1994) An immunohistochemical study of the pathology of fatal malaria. Evidence for widespread endothelial activation and a potential role for intercellular adhesion molecule-1 in cerebral sequestration. Am. J. Pathol. 145, 1057–1069 [PMC free article] [PubMed] [Google Scholar]
- 3. Pongponratn E., Turner G. D., Day N. P., Phu N. H., Simpson J. A., Stepniewska K., Mai N. T., Viriyavejakul P., Looareesuwan S., Hien T. T., Ferguson D. J., White N. J. (2003) An ultrastructural study of the brain in fatal Plasmodium falciparum malaria. Am. J. Trop. Med. Hyg. 69, 345–359 [PubMed] [Google Scholar]
- 4. Smith J. D., Chitnis C. E., Craig A. G., Roberts D. J., Hudson-Taylor D. E., Peterson D. S., Pinches R., Newbold C. I., Miller L. H. (1995) Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82, 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Su X. Z., Heatwole V. M., Wertheimer S. P., Guinet F., Herrfeldt J. A., Peterson D. S., Ravetch J. A., Wellems T. E. (1995) The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 82, 89–100 [DOI] [PubMed] [Google Scholar]
- 6. Chen Q., Fernandez V., Sundström A., Schlichtherle M., Datta S., Hagblom P., Wahlgren M. (1998) Developmental selection of var gene expression in Plasmodium falciparum. Nature 394, 392–395 [DOI] [PubMed] [Google Scholar]
- 7. Duffy P. E., Craig A. G., Baruch D. I. (2001) Variant proteins on the surface of malaria-infected erythrocytes. Developing vaccines. Trends Parasitol. 17, 354–356 [DOI] [PubMed] [Google Scholar]
- 8. Rowe J. A., Claessens A., Corrigan R. A., Arman M. (2009) Adhesion of Plasmodium falciparum-infected erythrocytes to human cells. Molecular mechanisms and therapeutic implications. Expert Rev. Mol. Med. 11, e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gamain B., Gratepanche S., Miller L. H., Baruch D. I. (2002) Molecular basis for the dichotomy in Plasmodium falciparum adhesion to CD36 and chondroitin sulfate A. Proc. Natl. Acad. Sci. U.S.A. 99, 10020–10024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berendt A. R., Simmons D. L., Tansey J., Newbold C. I., Marsh K. (1989) Intercellular adhesion molecule-1 is an endothelial cell adhesion receptor for Plasmodium falciparum. Nature 341, 57–59 [DOI] [PubMed] [Google Scholar]
- 11. Gray C., McCormick C., Turner G., Craig A. (2003) ICAM-1 can play a major role in mediating P. falciparum adhesion to endothelium under flow. Mol. Biochem. Parasitol. 128, 187–193 [DOI] [PubMed] [Google Scholar]
- 12. Ochola L. B., Siddondo B. R., Ocholla H., Nkya S., Kimani E. N., Williams T. N., Makale J. O., Liljander A., Urban B. C., Bull P. C., Szestak T., Marsh K., Craig A. G. (2011) Specific receptor usage in Plasmodium falciparum cytoadherence is associated with disease outcome. PLoS One 6, e14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Newbold C., Warn P., Black G., Berendt A., Craig A., Snow B., Msobo M., Peshu N., Marsh K. (1997) Receptor-specific adhesion and clinical disease in. Plasmodium falciparum. Am. J. Trop. Med. Hyg. 57, 389–398 [DOI] [PubMed] [Google Scholar]
- 14. Silamut K., Phu N. H., Whitty C., Turner G. D., Louwrier K., Mai N. T., Simpson J. A., Hien T. T., White N. J. (1999) A quantitative analysis of the microvascular sequestration of malaria parasites in the human brain. Am. J. Pathol. 155, 395–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Avril M., Tripathi A. K., Brazier A. J., Andisi C., Janes J. H., Soma V. L., Sullivan D. J., Jr., Bull P. C., Stins M. F., Smith J. D. (2012) A restricted subset of var genes mediates adherence of Plasmodium falciparum-infected erythrocytes to brain endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 109, E1782–E1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rask T. S., Hansen D. A., Theander T. G., Gorm Pedersen A., Lavstsen T. (2010) Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes. Divide and conquer. PLoS Comput. Biol. 6, e1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Howell D. P., Levin E. A., Springer A. L., Kraemer S. M., Phippard D. J., Schief W. R., Smith J. D. (2008) Mapping a common interaction site used by Plasmodium falciparum Duffy binding-like domains to bind diverse host receptors. Mol. Microbiol. 67, 78–87 [DOI] [PubMed] [Google Scholar]
- 18. Janes J. H., Wang C. P., Levin-Edens E., Vigan-Womas I., Guillotte M., Melcher M., Mercereau-Puijalon O., Smith J. D. (2011) Investigating the host binding signature on the Plasmodium falciparum PfEMP1 protein family. PLoS Pathog. 7, e1002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Khunrae P., Dahlbäck M., Nielsen M. A., Andersen G., Ditlev S. B., Resende M., Pinto V. V., Theander T. G., Higgins M. K., Salanti A. (2010) Full-length recombinant Plasmodium falciparum VAR2CSA binds specifically to CSPG and induces potent parasite adhesion-blocking antibodies. J. Mol. Biol. 397, 826–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Srivastava A., Gangnard S., Round A., Dechavanne S., Juillerat A., Raynal B., Faure G., Baron B., Ramboarina S., Singh S. K., Belrhali H., England P., Lewit-Bentley A., Scherf A., Bentley G. A., Gamain B. (2010) Full-length extracellular region of the var2CSA variant of PfEMP1 is required for specific, high-affinity binding to CSA. Proc. Natl. Acad. Sci. U.S.A. 107, 4884–4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Batchelor J. D., Zahm J. A., Tolia N. H. (2011) Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nat. Struct. Mol. Biol. 18, 908–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tolia N. H., Enemark E. J., Sim B. K., Joshua-Tor L. (2005) Structural basis for the EBA-175 erythrocyte invasion pathway of the malaria parasite Plasmodium falciparum. Cell 122, 183–193 [DOI] [PubMed] [Google Scholar]
- 23. Whitmore L., Wallace B. A. (2004) DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schuck P. (2003) On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal. Biochem. 320, 104–124 [DOI] [PubMed] [Google Scholar]
- 25. Laue T. M., Shah B. D., Ridgeway T. M., Pelletier S. L. (1992) Analytical Ultracentrifugation in Biochemistry and Polymer Science, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 26. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I. (2003) Windows-PC based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 27. Petoukhov M. V., Konarev P. V., Kikhney A. G., Svergun D. I. (2007) ATSAS 2.1. Towards automated and web-supported small-angle scattering data analysis. J. Appl. Crystallogr. 40, s223–s228 [Google Scholar]
- 28. Guinier A., Fournet G. (1955) in Small-angle Scattering of X-rays, John Wiley and Sons, New York [Google Scholar]
- 29. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 30. Porod G. (1982) in General Theory Small Angle X-ray Scattering, Academic Press, Inc., London [Google Scholar]
- 31. Svergun D. I. (2009) DAMMIF, a program for rapid ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Volkov V., Svergun D. I. (2006) ATSAS 2.1, a program package for small-angle scattering data analysis. J. Appl. Crystallogr. 39, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Birmanns S., Rusu M., Wriggers W. (2011) Using Sculptor and Situs for simultaneous assembly of atomic components into low-resolution shapes. J. Struct. Biol. 173, 428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kozin M. B., Svergun D. I. (2001) Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34, 33–41 [Google Scholar]
- 36. Ortega A., Amorós D., García de la Torre J. (2011) Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys. J. 101, 892–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Casasnovas J. M., Springer T. A. (1995) Kinetics and thermodynamics of virus binding to receptor. Studies with rhinovirus, intercellular adhesion molecule-1 (ICAM-1), and surface plasmon resonance. J. Biol. Chem. 270, 13216–13224 [DOI] [PubMed] [Google Scholar]
- 38. Owens R. M., Gu X., Shin M., Springer T. A., Jin M. M. (2010) Engineering of single Ig superfamily domain of ICAM-1 for native fold and function. J. Biol. Chem. 285, 15906–15915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Berendt A. R., McDowall A., Craig A. G., Bates P. A., Sternberg M. J., Marsh K., Newbold C. I., Hogg N. (1992) The binding site on ICAM-1 for Plasmodium falciparum-infected erythrocytes overlaps, but is distinct from, the LFA-1-binding site. Cell 68, 71–81 [DOI] [PubMed] [Google Scholar]
- 40. Tse M. T., Chakrabarti K., Gray C., Chitnis C. E., Craig A. (2004) Divergent binding sites on intercellular adhesion molecule-1 (ICAM-1) for variant Plasmodium falciparum isolates. Mol. Microbiol. 51, 1039–1049 [DOI] [PubMed] [Google Scholar]
- 41. Ockenhouse C. F., Betageri R., Springer T. A., Staunton D. E. (1992) Plasmodium falciparum-infected erythrocytes bind ICAM-1 at a site distinct from LFA-1, Mac-1, and human rhinovirus. Cell 68, 63–69 [DOI] [PubMed] [Google Scholar]
- 42. Gardner J. P., Pinches R. A., Roberts D. J., Newbold C. I. (1996) Variant antigens and endothelial receptor adhesion in Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 93, 3503–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Smith J. D., Craig A. G., Kriek N., Hudson-Taylor D., Kyes S., Fagan T., Fagen T., Pinches R., Baruch D. I., Newbold C. I., Miller L. H. (2000) Identification of a Plasmodium falciparum intercellular adhesion molecule-1 binding domain. A parasite adhesion trait implicated in cerebral malaria. Proc. Natl. Acad. Sci. U.S.A. 97, 1766–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Singh K., Gittis A. G., Nguyen P., Gowda D. C., Miller L. H., Garboczi D. N. (2008) Structure of the DBL3x domain of pregnancy-associated malaria protein VAR2CSA complexed with chondroitin sulfate A. Nat. Struct. Mol. Biol. 15, 932–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Higgins M. K. (2008) The structure of a chondroitin sulfate-binding domain important in placental malaria. J. Biol. Chem. 283, 21842–21846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Khunrae P., Philip J. M., Bull D. R., Higgins M. K. (2009) Structural comparison of two CSPG-binding DBL domains from the VAR2CSA protein important in malaria during pregnancy. J. Mol. Biol. 393, 202–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Juillerat A., Lewit-Bentley A., Guillotte M., Gangnard S., Hessel A., Baron B., Vigan-Womas I., England P., Mercereau-Puijalon O., Bentley G. A. (2011) Structure of a Plasmodium falciparum PfEMP1 rosetting domain reveals a role for the N-terminal segment in heparin-mediated rosette inhibition. Proc. Natl. Acad. Sci. U.S.A. 108, 5243–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miller J., Knorr R., Ferrone M., Houdei R., Carron C. P., Dustin M. L. (1995) Intercellular adhesion molecule-1 dimerization and its consequences for adhesion mediated by lymphocyte function associated-1. J. Exp. Med. 182, 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Staunton D. E., Dustin M. L., Erickson H. P., Springer T. A. (1990) The arrangement of the immunoglobulin-like domains of ICAM-1 and the binding sites for LFA-1 and rhinovirus. Cell 61, 243–254 [DOI] [PubMed] [Google Scholar]
- 50. Yang Y., Jun C.-D., Liu J.-H., Zhang R., Joachimiak A., Springer T. A., Wang J.-H. (2004) Structural basis for dimerization of ICAM-1 on the cell surface. Mol. Cell 14, 269–276 [DOI] [PubMed] [Google Scholar]
- 51. Bernadó P. (2010) Effect of interdomain dynamics on the structure determination of modular proteins by small-angle scattering. Eur. Biophys. J. 39, 769–780 [DOI] [PubMed] [Google Scholar]
- 52. Kolatkar P. R., Bella J., Olson N. H., Bator C. M., Baker T. S., Rossmann M. G. (1999) Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 18, 6249–6259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xiao C., Bator-Kelly C. M., Rieder E., Chipman P. R., Craig A., Kuhn R. J., Wimmer E., Rossmann M. G. (2005) The crystal structure of coxsackievirus A21 and its interaction with ICAM-1. Structure 13, 1019–1033 [DOI] [PubMed] [Google Scholar]
- 54. Singh S. K., Hora R., Belrhali H., Chitnis C. E., Sharma A. (2006) Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain. Nature 439, 741–744 [DOI] [PubMed] [Google Scholar]
- 55. Chattopadhyay R., Taneja T., Chakrabarti K., Pillai C. R., Chitnis C. E. (2004) Molecular analysis of the cytoadherence phenotype of a Plasmodium falciparum field isolate that binds intercellular adhesion molecule-1. Mol. Biochem. Parasitol 133, 255–265 [DOI] [PubMed] [Google Scholar]
- 56. Springer A. L., Smith L. M., Mackay D. Q., Nelson S. O., Smith J. D. (2004) Functional interdependence of the DBLβ domain and c2 region for binding of the Plasmodium falciparum variant antigen to ICAM-1. Mol. Biochem. Parasitol. 137, 55–64 [DOI] [PubMed] [Google Scholar]
- 57. Mayor A., Bir N., Sawhney R., Singh S., Pattnaik P., Singh S. K., Sharma A., Chitnis C. E. (2005) Receptor-binding residues lie in central regions of Duffy-binding-like domains involved in red cell invasion and cytoadherence by malaria parasites. Blood 105, 2557–2563 [DOI] [PubMed] [Google Scholar]
- 58. Bertonati C., Tramontano A. (2007) A model of the complex between the PfEMP1 malaria protein and the human ICAM-1 receptor. Proteins 69, 215–222 [DOI] [PubMed] [Google Scholar]
- 59. Hughes K. R., Biagini G. A., Craig A. G. (2010) Continued cytoadherence of Plasmodium falciparum infected red blood cells after antimalarial treatment. Mol. Biochem. Parasitol. 169, 71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Clausen T. M., Christoffersen S., Dahlbäck M., Langkilde A. E., Jensen K. E., Resende M., Agerbæk M. Ø., Andersen D., Berisha B., Ditlev S. B., Pinto V. V., Nielsen M. A., Theander T. G., Larsen S., Salanti A. (2012) Structural and functional insight into how the Plasmodium falciparum VAR2CSA protein mediates binding to chondroitin sulfate A in placental malaria. J. Biol. Chem. 287, 23332–23345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gruenberg J., Allred D. R., Sherman I. W. (1983) Scanning electron microscope-analysis of the protrusions (knobs) present on the surface of Plasmodium falciparum-infected erythrocytes. J. Cell Biol. 97, 795–802 [DOI] [PMC free article] [PubMed] [Google Scholar]