Background: Ca2+ oscillations stimulate rhythmic proton signaling by Na+/H+ exchanger NHX-7 in C. elegans.
Results: The contribution of individual regulatory motifs to NHX-7 activity was defined by in vivo structure-function analysis.
Conclusion: NHX-7 activity is regulated by both Ca2+ and pH, leading to robust but transient signaling.
Significance: Understanding the mechanisms that distinguish proton signaling from pH regulation is critical for dual-function membrane transporters.
Keywords: C. elegans, Calcium Signaling, In vivo Imaging, pH Regulation, Sodium Proton Exchange, Proton Signaling
Abstract
Membrane proton transporters contribute to pH homeostasis but have also been shown to transmit information between cells in close proximity through regulated proton secretion. For example, the nematode intestinal Na+/H+ exchanger NHX-7 causes adjacent muscle cells to contract by transiently acidifying the extracellular space between the intestine and muscle. NHX-7 operates during a Ca2+-dependent rhythmic behavior and contains several conserved motifs for regulation by Ca2+ input, including motifs for calmodulin and phosphatidylinositol 4,5-bisphosphate binding, protein kinase C- and calmodulin-dependent protein kinase type II phosphorylation, and a binding site for calcineurin homologous protein. Here, we tested the idea that Ca2+ input differentiates proton signaling from pH housekeeping activity. Each of these motifs was mutated, and their contribution to NHX-7 function was assessed. These functions included pH recovery from acidification in cells in culture expressing recombinant NHX-7, extracellular acidification measured during behavior in live moving worms, and muscle contraction strength as a result of this acidification. Our data suggest that multiple levels of Ca2+ input regulate NHX-7, whose transport capacity normally exceeds the minimum necessary to cause muscle contraction. Furthermore, extracellular acidification limits NHX-7 proton transport through feedback inhibition, likely to prevent metabolic acidosis from occurring. Our findings are consistent with an integrated network whereby both Ca2+ and pH contribute to proton signaling. Finally, our results obtained by expressing rat NHE1 in Caenorhabditis elegans suggest that a conserved mechanism of regulation may contribute to cell-cell communication or proton signaling by Na+/H+ exchangers in mammals.
Introduction
pH homeostasis is vital for maintaining protein folding/function and cell volume and for facilitating the flow of ions and nutrients through various physiologically coupled mechanisms (for review, see Ref. 1). pH imbalance has been implicated in a number of human diseases, including renal tubular acidosis, osteoporosis, and mental retardation, as well as in the transformation and metastasis of cancer (2–4). Because pH regulation plays a ubiquitous role in cell physiology and is crucial to an organism's health, it is not surprising that a wide variety of acid-base transporters exist (1).
However, recent evidence suggests that protons themselves may have an additional role in signaling between cells. Several types of mammalian proton receptors have been identified, including a class of G-protein-coupled receptors (GPR4, OGR1, G2A, and TDAG8), adhesion the kinase Pyk2, and cation channels of the Na+ channel/degenerin superfamily (ASIC1/DRASIC) (5–9). These receptors are found in the kidney, brain, and central nervous system and have long been implicated in the perception of pain associated with tissue acidosis (10, 11). Reductions in extracellular pH arise during the course of ischemia, epileptic seizures, and electrical stimulation in the brain, suggesting a potential role for these proton receptors in human disease (12–16). Therefore, local regulation of proton availability may have important physiologic and pathophysiologic consequences through signal transmission processes. Membrane transporters that regulate pH homeostasis may also have a role in directing localized transient proton signals as part of an intercellular communication cascade. If so, it will be important to determine what particular features of such proteins distinguish their basal activity (which contributes to maintaining pH) from their stimulated acute activity during cell signaling.
Caenorhabditis elegans is a genetic model organism in which protons have been recently shown to be actively secreted as an intercellular signaling molecule during a rhythmic behavior (17, 18). Defecation is an ultradian behavior that occurs with a frequency of ∼45 s in well fed worms (19, 20). The defecation motor program (DMP)2 is characterized by three consecutive sets of muscle contractions (21). The first set is a contraction of the posterior body wall muscles (pBoc). This is closely followed by a contraction of the anterior body wall muscles and then expulsion of the luminal contents. Genetic analyses have shown that intestinal cell-autonomous oscillatory Ca2+ signaling is central to the defecation pacemaker (22). The first motor step of the DMP, pBoc, depends on Ca2+ signals initiating in the posterior cells of the intestine (17, 18), whereas a subsequent wave of Ca2+ that propagates posterior-to-anterior is necessary for the coupling and timing of the remaining motor steps (23–25).
A role for proton signaling during the DMP emerged from reports indicating that increased Ca2+ caused protons to move from the intestinal lumen into the cytoplasm, resulting in intracellular acidification, and that this in turn triggered proton extrusion across the basolateral membrane via the Na+/H+ exchanger NHX-7 (17, 18). Proton secretion was found to be both necessary and sufficient to trigger pBoc. Mechanistically, extracellular acidification resulting from NHX-7 activity was found to provide proton substrates that bound directly to and activated a ligand-gated cation channel, PBO-5/6 (pH50 ∼6.8), expressed on the muscle membrane. Activation of PBO-5/6 resulted in muscle depolarization and pBoc (17). This was the first example of protons and a Na+/H+ exchanger contributing to acute intercellular signaling.
Na+/H+ exchangers are a 12-transmembrane-spanning family of phosphoglycoproteins that contain a hydrophobic N-terminal region and a large cytoplasmic C-terminal tail. They exchange one extracellular Na+ for one intracellular H+ to regulate intracellular pH (pHi) and to maintain Na+ homeostasis (26). They facilitate adhesion, migration, proliferation, and volume regulation (27–32) and have been implicated in a number of human diseases, including epilepsy, mental retardation, and cancer metastasis, in addition to being a component of ischemia/reperfusion injury (2–4). Mammals have nine isoforms of Na+/H+ exchangers termed NHEs (33–40), whereas the nematode C. elegans likewise codes for nine isoforms termed NHXs (41). Although some of these isoforms are ubiquitously expressed (NHE1 (42) and NHX-4 (41)) in a “housekeeping” manner, others demonstrate a high degree of cell-specific expression, suggesting potentially unique modes of regulation and function.
The basal activity of these NHEs can be modulated via a cytoplasmic allosteric proton-binding site that enhances activity upon intracellular acidification (low pHi) (43) as well as by various protein interactions with regulatory motifs located in the C-terminal tail (26). This intracellular tail has also been shown to be the site of regulation by protein kinases, and these regulatory events can be a more powerful determinant of activity than the overall expression level of a given transporter (44). One ubiquitous and well studied regulatory mechanism is that of second messenger Ca2+ signaling. NHE1 and NHE3 both respond to changes in Ca2+ through a number of mechanisms, including phosphorylation by Ca2+/calmodulin-dependent protein kinase type II (CaMKII) and interactions with Ca2+-binding proteins such as calmodulin (CaM), calcineurin homologous protein (CHP), and tescalin (reviewed in Refs. 26) and 45). Thus, Ca2+ has the ability to both positively and negatively regulate Na+/H+ exchange and can do so in an isoform- and cell-specific manner.
Here, we present the results of a structure-function analysis focused on deciphering if and how Ca2+ signaling regulates NHX-7. We used dynamic fluorescent pH imaging to measure NHX-7 activity both in cells in culture and in live moving worms, where we could assess behavioral output. We hereby describe a role for CaM- and phosphatidylinositol 4,5-bisphosphate (PIP2)-binding motifs in activation of the exchanger. Although we were further able to demonstrate that CaM interacts directly with NHX-7, we also found that it impacts oscillatory Ca2+ signaling upstream of NHX-7. Finally, a potential role for CaMKII phosphorylation in limiting the extent of proton transport may relate to the observation that extracellular acidification provides feedback inhibition and likely prevents metabolic acidosis from occurring.
EXPERIMENTAL PROCEDURES
Strains
Standard nematode culture techniques were used (46). Strains containing pbo-4(ok583)X, pbo-5(n2303)V, and pha-1(e2123ts)III alleles were obtained from the Caenorhabditis Genetics Center (University of Minnesota) and the C. elegans Gene Knockout Consortium. Other strains used in this work are described in detail under supplemental Experimental Procedures.”
Quantitative RT-PCR
Gene expression levels were assayed in first-strand cDNA samples using iQTM SYBR® Green Supermix (Bio-Rad) and standard protocols for real-time PCR and normalized to both pmp-3 and cdc-42 using the following primer pairs: nhx-7, 5′-CAGTACACTTTGCGAGTCGCTTAT-3′ and 5′-AGTTTGATCGTAGAACCCTGGATG-3′; pmp-3, 5′-GTTCCCGTGTTCATCACTCAT-3′ and 5′-ACACCGTCGAGAAGCTGTAGA-3′; and cdc-42, 5′-CTGCTGGACAGGAAGATTACG-3′ and 5′-CTCGGACATTCTCGAATGAAG-3′.
Molecular Biology
Plasmid constructs were created by conventional restriction site-mediated cloning methods. Site-directed mutagenesis was used to introduce point mutants, and inverse PCR was used to create deletions. Specific details as to the construction of the plasmids can be found under supplemental “Experimental Procedures.” A plasmid containing the extracellular pH sensor was a kind gift of Dr. L. Pablo Cid (Centro de Estudios Científicos, Valdivia, Chile) and has been described previously (47). All constructs were verified via DNA sequencing.
Contraction Measurements
pBoc strength and duration were determined post hoc from transmitted light videos by measuring the perimeter of late L4 worms in their maximally relaxed and contracted states. Worms in which the reference points were obscured by movement, in either assay, were excluded from analysis.
Imaging Fluorescent Biosensors in C. elegans
Transgenic nematodes were imaged live and unrestrained as described previously (18). Imaging was performed with a 10× Nikon Plan Apo objective. Illumination was provided by a Polychrome IV monochromator (TILL Photonics) rigged to a Nikon Eclipse TE2000-S inverted microscope equipped with a high-speed charge-coupled device camera (Cooke). TILLvisION software was used for analysis, data were compiled in Microsoft Excel, and graphs were generated using Origin.
Measurements of Na+/H+ Exchange Activity in AP-1 Cells
AP-1 cells were cultured as described (48) and transfected using Lipofectamine LTX reagent (Invitrogen). After 24 h, cells were made quiescent by placing them in low-serum medium for at least 1 h. The cells were then loaded with the pH-sensitive fluorescent dye 2′,7′-bi-(2-carboxyethyl)-5(6)-carboxyfluorescein acetoxymethyl ester (Molecular Probes) and superfused with physiologic saline on a Nikon/TILL Photonics fluorescent imaging rig (described above), and incubation with high K+/nigericin at varying pH values was used to calibrate the sensor (49). To measure Na+/H+ exchange activity, an ammonium prepulse was employed as described (48).
Confocal Microscopy
Confocal micrographs were obtained on an Olympus IX81 inverted laser scanning confocal microscope. The same parameters were used for all of the mutants when imaging across worm strains or across transfections.
Immunocytochemistry
Recombinant NHX-7 was detected using a mouse anti-V5 monoclonal antibody (1:2000; Invitrogen) and an Alexa Fluor 488-labeled goat anti-mouse secondary antibody (1:5000; Molecular Probes) using standard techniques.
CaM Binding Assay
In vitro transcription/translation (TnT; Promega) of linearized template DNA was used to label fusion proteins with [35S]methionine. Proteins associating with biotinylated bovine CaM (5 μm; Calbiochem) were precipitated with streptavidin-agarose. Equivalent fractions of the starting and bound material were resolved by SDS-PAGE and visualized using autoradiography.
RNAi Treatment and DMP Assay
Nematodes were grown on bacteria expressing double-stranded RNA as described (50). Execution of the DMP was visualized under a dissecting microscope, and timing data were collected using Etho software, a keystroke recorder (51).
RESULTS
Recombinant NHX-7 Is an Authentic Na+/H+ Exchanger
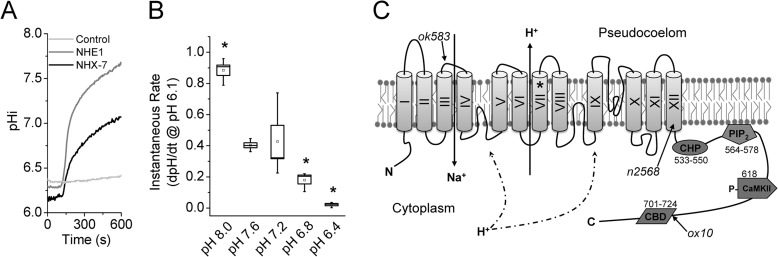
To analyze NHX-7 exchange activity, a cDNA was expressed in AP-1 cells, a mammalian cell line that lacks endogenous Na+/H+ exchanger activity (31). Antibody staining of a V5 epitope tag built into the C terminus of NHX-7 confirmed its expression at the plasma membrane (see Fig. 3F). As is commonly observed with overexpression, some of the protein was retained in the endoplasmic reticulum as well. To test whether or not the exchanger was active, an ammonium prepulse was used to acidify the cells (52), and the rate of Na+-dependent pH recovery was then measured. Eliciting NHX-7 activity from transfected cells required that they be starved of serum and that the measurements be performed within 24 h of transfection. Robust activity was observed, although the calculated exchange rate of NHX-7 was slower than that of the ubiquitous mammalian exchanger NHE1 (Fig. 1A). The NHX-7 exchange rate was sensitive to extracellular pH (pHe) (Fig. 1B) and Na+ (Km ∼ 35 mm). Surprisingly, agonists that raised intracellular Ca2+ levels had no significant effect on recombinant NHX-7 activity (data not shown).
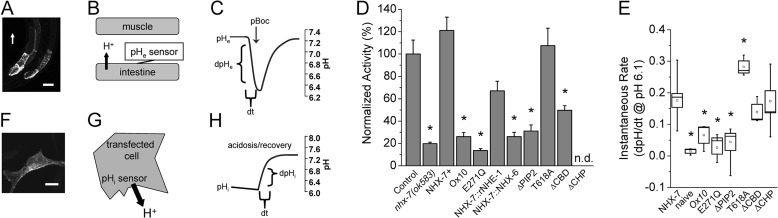
FIGURE 3.
Analysis of in vivo/in vitro exchanger activity. A genetically encoded fluorescent pH sensor was targeted to the extracellular side of the posterior intestinal basolateral membrane to measure regional changes in pseudocoelomic pH during defecation in transgenic strains expressing WT and mutant nhx-7 minigenes. To measure the basal activity of recombinant proteins, pHi was monitored in transfected AP-1 cells in culture, and the rate of Na+-dependent pH recovery following acidification was calculated. A, confocal micrograph of transgenic worms expressing the extracellular pH sensor at the basolateral membrane of intestinal rings 7–9. The white arrow denotes posterior-to-anterior orientation of the worm and is scaled to 50 μm. B, schematic of the extracellular pH sensor with protons moving from the intestine into the space between cells. C, representative trace of extracellular (pseudocoelomic) pH fluctuations that occur between the intestine and body wall muscle relative to pBoc. Dynamic fluorescent measurements of pHe in freely moving worms were used to calculate the initial rate of acidification (dpH/dt), which is a function of NHX-7 activity. D, extracellular acidification rates resulting from expression of the mutant constructs are represented as normalized to the control strain. The resting pH of the pseudocoelom was not significantly different between strains, and the overall extent of acidification was proportional to the initial rate of acidification. Error is S.E. for three independent trials, with more than five worms per trial. Asterisks indicate p < 0.05 compared with the wild-type control via analysis of variance. E, instantaneous Na+/H+ exchange rate data for mutant NHX-7 constructs expressed in AP-1 cells in vitro. To calculate instantaneous recovery rate data, a best line fit equation was calculated for a 1-min window following the re-addition of Na+ of a plot of dpHi/dt versus pH and extrapolated to pH 6.1. Values were obtained from between three and nine independent experiments, with >10 cells imaged per experiment. The mean and median are designated by the small interior box and horizontal line, respectively. Error (large box) is S.E., whiskers represent the maximum and minimum values, and asterisks indicate p < 0.05 compared with cells expressing the wild-type NHX-7 via analysis of variance. F, confocal micrograph of an AP-1 cell expressing recombinant V5 epitope-tagged NHX-7 that has been visualized using anti-V5 primary and fluorescent Alexa Fluor 488-conjugated anti-mouse secondary antibodies. Scale bar = 10 μm. G, schematic of pHi measurements in cell culture with protons moving out of the cell into the medium. H, representative trace of pHi during Na+-dependent pH recovery following acidification. The recovery rate was calculated based upon a plot of dpHi/dt versus pHi, with a best fit line extrapolated to pH 6.1.
FIGURE 1.
Physiologic characterization of recombinant NHX-7 activity. AP-1 cells, a CHO cell derivative that lacks NHE activity, were used for transient expression and fluorescence-based pH measurements of recombinant Na+/H+ activity. A, representative traces of Na+-dependent pH recovery following acidification in cells expressing C. elegans NHX-7 or rat NHE1 as indicated. The negative control was the pcDNA3.1 vector alone. B, pHe dependence of NHX-7. To calculate instantaneous recovery rate data, a best line fit equation was calculated for a 1-min window following the re-addition of Na+ of a plot of dpHi/dt versus pH and extrapolated to pH 6.1. Each data point represents three to four replicate experiments (>10 cells per experiment). The mean and median are designated by the small interior box and horizontal line, respectively. Error (large box) is S.E., whiskers represent the maximum and minimum values, and asterisks indicate p < 0.05 versus pH 7.6 via analysis of variance. C, schematic of NHX-7 protein: domain organization and Ca2+ regulatory motifs. Transmembrane domains are labeled with Roman numerals. Relevant nhx-7 mutant alleles are listed in italics, with their positions denoted by arrows. ok583 is a deletion allele that acts as a null, whereas n2568 and ox10 result in truncated proteins. Motifs targeted for mutagenesis were based upon homology with rat NHE1 (supplemental Fig. S1) and are denoted as well. The E271Q pore mutation is indicated by an asterisk in transmembrane domain VII. Other targets are marked schematically and by amino acid location within the nhx-7 C-terminal coding sequence, including CHP, PIP2, and CBD motifs and the single potential CaMKII phosphorylation site at Thr-618. Fusions between the N terminus of NHX-7 and the C terminus of other Na+/H+ exchangers occur in a conserved sequence in transmembrane domain XII.
NHX-7 Is Regulated Similarly to Mammalian NHEs
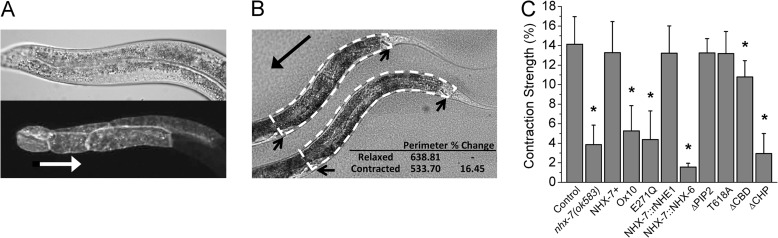
As an alternative approach to AP-1 cells, we turned to an in vivo model to test whether Ca2+ regulates NHX-7. First, the ability of recombinant NHX-7 to complement a C. elegans nhx-7(ok585) loss-of-function mutant was tested. An mCherry fusion to NHX-7 (Fig. 2A) rescued the pBoc deficit in the mutant (Fig. 2C and Table 1), consistent with previous observations regarding an NHX-7::GFP fusion protein (18). The specificity of rescue was suggested by two observations: recapitulating an nhx-7(ox10) loss-of-function mutation that arises from a deletion/frameshift starting at about A702 suppressed the ability to restore pBoc, as did mutation of a critical amino acid residue in the pore region (E271Q) that is essential for Na+/H+ exchange activity (Fig. 2C).
FIGURE 2.
In vivo structure-function analysis of NHX-7. A transgenic construct coding for an nhx-7 minigene fused to a cDNA for the red fluorescent protein mCherry was mutated at potential Ca2+ regulatory motifs or replaced with a homologous region from rNHE1 or NHX-6. The ability of these constructs to restore pBoc strength in an nhx-7(ok585) null mutant was assessed. A, confocal micrograph of pnhx-7::NHX-7::mCherry rescue construct expression in intestinal rings 7–9 (lower) and differential interference contrast (DIC) (upper). The white arrow points from posterior to anterior and is scaled to 50 μm. B, representative contraction assay using perimeter analysis. Values were calculated post hoc from a DIC video by measuring the perimeter (dashed lines) of the worm at both maximal relaxation (lower) and contraction (upper). Arrows indicate the vulva and anus, which were used as anterior and posterior reference points, respectively. An example calculation is shown in the inset. C, mutant pBoc strength. Transgenic constructs are denoted as follows: NHX-7+ is the WT transgene; ox10 recapitulates a loss-of-function mutation identified through a forward genetic screen (deletion/frameshift starting approximately A702); E271Q is a pore mutation; NHX-7::rNHE1 and NHX-7::NHX-6 are fusions between the transmembrane domain of NHX-7 and the cytoplasmic domain of the indicated NHEs; ΔPIP2 is a mutation of the PIP2-binding site (K569A/K570A); T618A is a mutation of a CaMKII recognition site; ΔCBD is a deletion of the CBD from amino acids 695 to 723; ΔCHP is a mutation of the PBO-1-binding site (amino acids 541–545, MVQHL to RRQHR). Error is S.D. for between 7 and 27 worms per strain. Asterisks denote p < 0.05 versus the wild-type control via analysis of variance.
TABLE 1.
Characteristics of in vivo NHX-7 mutagenesis
Shown below are the relative acidification rates and pBoc strength and duration in transgenic worms expressing mutant versions of NHX-7. pBoc duration values were derived post hoc from DIC videos. pbo-5 mutants lack pBoc, but NHX-7 activity is identical to wild-type controls. The nhx-7(ok583) mutant strains expressing rescue constructs also contained the pha-1 and him-5 mutant alleles. Error is S.D. for contraction strength and duration and S.E. for acidification rate. The n value for each trial is shown in parentheses. ND, not determined.
| Genetic background | Rescuing construct | Activity | Contraction strength | Contraction duration |
|---|---|---|---|---|
| % | % | s | ||
| pbo-5 (n2303) | 100 ± 10 (3) | ND | ND | |
| pha-1(2123ts)/him-5(e1490) | ND | 14.1 ± 2.8 (13) | 4.1 ± 0.8 (13) | |
| nhx-7(ok583) | 20 ± 1 (3)a | 3.9 ± 2.0 (17)a | 3.1 ± 0.8 (16)a | |
| nhx-7(ok583) | NHX-7+ | 120 ± 10 (3) | 13.3 ± 3.2 (7) | 5.0 ± 1.0 (7) |
| nhx-7(ok583) | ox10 | 26 ± 4 (3)a | 5.7 ± 4.2 (27)a | 3.3 ± 1.1 (24) |
| nhx-7(ok583) | E271Q | 14 ± 2 (3)a | 4.4 ± 2.9 (11)a | 2.6 ± 0.3 (9)a |
| nhx-7(ok583) | NHX-7::rNHE1 | 67 ± 9 (3) | 13.2 ± 2.8 (12) | 3.8 ± 0.7 (12) |
| nhx-7(ok583) | NHX-7::NHX-6 | 26 ± 4 (3)a | 1.6 ± 0.4 (8)a | 1.1 ± 0.2 (5)a |
| nhx-7(ok583) | ΔPIP2 | 31 ± 6 (3)a | 13.2 ± 1.5 (10) | 3.4 ± 0.4 (10) |
| nhx-7(ok583) | T618A | 110 ± 20 (3) | 13.3 ± 1.8 (11) | 5.7 ± 0.5 (11)a |
| nhx-7(ok583) | ΔCBD | 50 ± 4 (3)a | 10.8 ± 1.7 (8)a | 3.4 ± 0.3 (9) |
| nhx-7(ok583) | ΔCHP | ND | 3.0 ± 2.1 (10)a | 3.0 ± 0.3 (10)a |
| nhx-7(ok583) | rNHE1 | 30 ± 5 (3)a | 4.3 ± 1.7 (9)a | 3.9 ± 0.7 (4) |
a p < 0.05 versus the NHX-7+ control via analysis of variance.
NHX-7 has several motifs for input by Ca2+ signaling (Fig. 1C), which are shared with mammalian NHE1 in its C-terminal “regulatory domain” (supplemental Fig. S1). These shared motifs are likely to be functionally sufficient for coordinating upstream signaling, as the C-terminal coding sequence of rat NHE1 (rNHE1) effectively substituted for that of NHX-7 in the rescue construct (Fig. 2C). Conversely, a similar domain swap between NHX-7 and NHX-6, a worm exchanger that is coexpressed with NHX-7 in the intestine (41) but lacks notable homologous motifs for input by Ca2+ signaling (supplemental Fig. S1), did not rescue the mutant (Fig. 2C). However, NHE1 could not fully substitute for NHX-7, suggesting that there is some innate property of NHX-7 outside of the C terminus that strongly influences its ability to effectively function, through promoting proper localization, association with another protein(s), or transport activity itself.
Mutation of Putative Motifs for Ca2+ Regulation of NHX-7
Conserved potential Ca2+ regulatory motifs in NHX-7 (Fig. 1C) were individually mutated, and the mutant constructs were tested for their ability to complement the loss-of-function mutant. These motifs included 1) a CaM-binding site of the 1-5-8-14 class, 2) a site for PIP2 binding, 3) a CaMKII phosphorylation site, and 4) a binding site for the C. elegans CHP ortholog PBO-1 (54). Although there are other potential sites for Ca2+ regulation, these four sites represented the most likely candidates for the following reasons. First, the nonfunctional protein coded for by the nhx-7(ox10) allele has been shown to be truncated immediately prior to the start of the potential CaM-binding domain (CBD), suggesting that CaM binding might be import for NHX-7 function (17). Second, PIP2 is a necessary cofactor for a variety of plasma membrane transporters, including NHEs (55), and during defecation, reductions in PIP2 have been shown to activate TRPM channels, hence coordinating their activity with phospholipase C-mediated inositol trisphosphate production (56). Third, CHP is a cofactor for NHEs that has been suggested to mediate Ca2+ regulation of the exchanger (57, 58), and a pbo-1 loss-of-function mutant lacks pBoc (54). Finally, phosphoregulation by CaMKII of a conserved site in NHE1 increases its activity in response to Ca2+ signaling (59, 60).
We were initially quite surprised to find that all of the mutant constructs supported robust pBoc (data not shown), at least until we considered that the mutant transgenes were being overexpressed and that overexpression could conceivably circumvent endogenous regulatory mechanisms. Hence, we established strains that approximated the wild-type expression levels of nhx-7 as judged by quantitative RT-PCR (data not shown). Within this context, the PIP2-binding site (ΔPIP2) and CaMKII (T618A) mutants still rescued pBoc completely, but deletion of the CBD (ΔCBD) or CHP-binding sites (ΔCHP) reduced pBoc strength (Fig. 2C). The ΔCBD mutation resulted in a small but significant decrease in pBoc strength, whereas the ΔCHP mutation suppressed the ability of the transgene to rescue pBoc entirely (Table 1). Confocal microscopy was used to establish the abundance and membrane localization of the fluorescent mCherry-tagged NHX-7 mutant proteins, and whereas most of the mutants were indistinguishable from wild-type NHX-7::mCherry (data not shown), the ΔCHP protein accumulated at intracellular locations rather than at the basolateral membrane.3
We next considered the idea that the strength of pBoc may not directly reflect NHX-7 activity. pBoc is triggered by activation of proton receptors, whose pKa is ∼6.8 (17), on the body wall muscle cells, but proton extrusion, receptor activation, muscle response, and proton clearance mechanisms all likely impact the overall signaling output. Therefore, it seemed possible that reduction of NHX-7 activity could result in compensation from these other mechanisms or perhaps that NHX-7 normally extrudes an abundance of protons above and beyond that necessary for a normal behavioral response. If that were the case, then reductions in NHX-7 activity might be tolerated without a corresponding reduction in contraction strength.
We surmised that measuring changes in pHe directly might be more informative regarding the mutations' effects on NHX-7 activity. To accomplish this, an extracellular pH biosensor (47) was expressed in the posterior intestinal cells and directed to the basolateral membrane (Fig. 3, A and B). In agreement with previous results (18), we were able to detect a transient robust acidification of the extracellular space between the posterior intestine and the overlying body wall muscle during pBoc in live moving worms (Fig. 3C). The observed acidification was largely absent in nhx-7(ok583) mutant worms, and measurements made in a pbo-5(n2303) background, which lacks the proton receptor and pBoc, suggested that the signal was not influenced by muscle contractility (Table 1). We then assessed the effect of each mutation on pHe fluctuations and extended this analysis to include strains expressing the chimeric fusions with NHX-6 and rNHE1 described above (Fig. 3D and Table 1).
Our results suggest that three distinct categories of mutant phenotypes exist. There are mutants that support normal pH oscillations and rescue pBoc, mutants that have reduced exchange activity but still rescue pBoc, and mutants with little or no exchange activity and do not rescue pBoc. This last class mimics the nhx-7(ok583) null mutant. These results are consistent with our hypothesis that a reduction in NHX-7 activity can be tolerated without causing a visible phenotype. For example, the ΔCBD, ΔPIP2, and chimeric rNHE1 proteins all rescued pBoc at least to some extent, but pHe measurements suggested that they are less active than wild-type NHX-7 (Fig. 3D). In contrast, the NHX-6 chimera, ΔCHP mutant, ox10 mutant, and E271Q pore mutant resembled the null mutant and exhibited low-amplitude pH oscillations that were unable to invoke contractions. The residual oscillations observed in the null mutant are likely due to the activities of other pH homeostatic mechanisms responding to intracellular acidification and causing some low level of proton efflux into the pseudocoelom. Finally, we note that the T618A mutation of the putative CaMKII phosphorylation site appears to have increased the activity of NHX-7, resulting in a strong contraction that was extended in duration (Table 1).
To directly assess whether the reduced activity observed with several of the mutants might be due to a reduction in basal levels of transport rather than disruption of Ca2+ signaling input, these mutants were expressed in AP-1 cells, and their transport rates were measured under quiescent conditions (Fig. 3, E–H). Approximately equal levels of NHX-7 expression and residence at the plasma membrane for each of the mutant proteins were suggested by immunolabeling and confocal microscopy (data not shown). Surprisingly, unlike our single-copy gene worm model, we found that enough of the ΔCHP mutant was trafficked to the cell surface in AP-1 cells to permit Na+/H+ exchange activity at similar levels as wild-type NHX-7 (Fig. 3E). Thus, the inability of the ΔCHP mutant to trigger pBoc was not due to it lacking transport activity. Similarly, the reduced activity of the ΔCBD mutant observed in worms was not a secondary consequence of reduced basal transport capacity (Fig. 3E). On the other hand, the enhanced activity of the T618A mutant in worms was recapitulated in AP-1 cells (Fig. 3E), suggesting that phosphorylation at Thr-618 by CaMKII may normally suppress basal activity. We also found that the ΔPIP2 mutant, which had greatly reduced activity in worms but was able to support a nearly normal pBoc, did not function at all in AP-1 cells (Fig. 3E). This could conceivably be due to higher levels of PIP2 in worms versus cells in culture compensating for reduced PIP2 binding affinity in the mutated exchanger, but it is also possible that Ca2+ activation in worms overcomes the observed reduced basal activity. Regardless, this result is inconsistent with the idea that reductions in PIP2 might activate NHX-7 such as they do TRPM channels during defecation (56).
NHX-7/Calmodulin Binding and Knockdown of cmd-1, a Nematode Calmodulin Gene
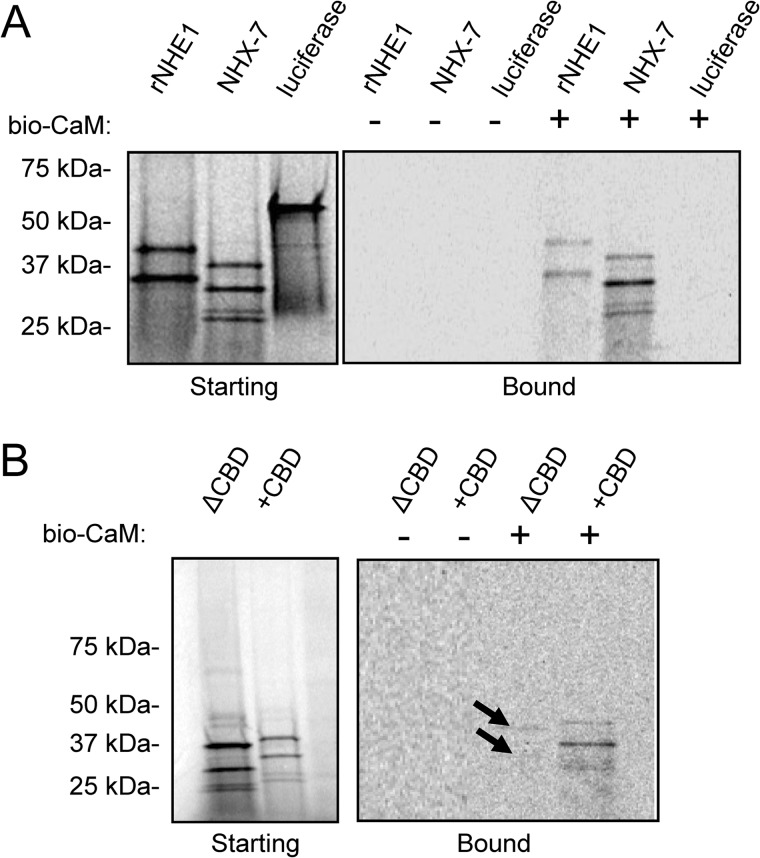
The results of our mutagenesis suggested that CaM binding contributes to NHX-7 proton signaling during defecation. To test this more directly, an NHX-7 C-terminal protein fragment was labeled with [35S]Met via in vitro transcription/translation, and the ability of the labeled protein to associate with biotinylated CaM was assessed by coprecipitation. NHE1, which is known to bind CaM (61), was used as a positive control to validate the assay (Fig. 4A). CaM associated with the soluble cytoplasmic C terminus of NHX-7 but did not associate with luciferase, which was used as a negative control (Fig. 4A). We also found that the Kd for CaM was similar to that for NHE1 (∼8 nm) (data not shown). To confirm the specificity of this interaction, we demonstrated that the ΔCBD mutation significantly reduced CaM binding to NHX-7 (Fig. 4B).
FIGURE 4.
Analysis of in vitro calmodulin binding by NHX-7. The cytoplasmic C termini of both NHX-7 and rNHE1 were expressed in vitro as radiolabeled proteins, and their ability to bind to biotinylated bovine calmodulin (bio-CaM; 98% identical to worm CMD-1) was assessed by streptavidin fractionation, gel electrophoresis of the starting and bound fractions, and autoradiographic detection. A, autoradiograms of gel-fractionated 35S-labeled proteins. The starting products from in vitro transcription/translation reactions (left) and bound fractions (right) are shown. An equivalent fraction of each was analyzed. Luciferase was used as a negative control. Multiple products are likely due to the use of PCR-amplified templates for in vitro transcription and do not interfere with the conclusion. Predicted molecular masses are as follows: rNHE1, ∼36 kDa; NHX-7, ∼31 kDa; and luciferase, 61 kDa. B, the CBD (identical to ΔCBD) was deleted, and the ability of the mutant ΔCBD protein to bind CaM was assessed as described above. Arrows denote slight residual CaM binding. The predicted molecular mass of ΔCBD is ∼28 kDa.
Having confirmed CaM binding and its physiologic relevance to NHX-7 function, we next asked whether reducing CaM levels in vivo would phenocopy the effect of the ΔCBD mutant on pBoc. The C. elegans genome contains five genes that encode Ca2+-binding CaM-like proteins. The cmd-1 gene codes for a nematode CaM ortholog that is 100% conserved with human CALM3. A transcriptional fusion between the cmd-1 promoter and the red fluorescent protein mCherry coding sequence was broadly expressed in multiple cells and cell types, including body wall muscle, hypodermal cells, neurons, and, most relevant to our work, posterior intestinal cells (Fig. 5A). Therefore, the functional role of cmd-1 was queried using RNAi-mediated knockdown. Unfortunately, global RNAi screens have shown that cmd-1 is necessary for viability, and worms treated with cmd-1 RNAi for several days died (data not shown). To circumvent this limitation, we turned to cell-specific RNAi. The RDE-1 protein is required for effective RNAi to occur, and an rde-1 mutant strain can be complemented with a transgene expressing recombinant RDE-1 in a limited set of cells such as the intestine (25).
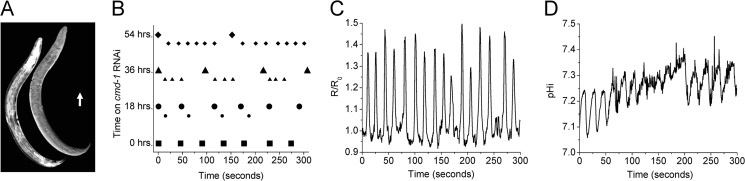
FIGURE 5.
The calmodulin gene cmd-1 is required for normal defecation signaling. RNAi was used to reduce cmd-1 expression specifically in the intestine. Worms were placed onto cmd-1 or control RNAi plates as L3 larva, and measurements were made following 0, 18, 36, and 54 h of RNAi. The fluorescent biosensors D3cpv and pHluorin were used to follow intestinal Ca2+ and pHi oscillations, respectively, as cmd-1 expression was reduced over time. A, composite image of a transgenic worm expressing a transcriptional fusion of the cmd-1 promoter and the fluorescent protein mCherry (left) and the corresponding DIC image (right). The arrow indicates posterior-to-anterior orientation and is scaled to 50 μm. B, dot plots representing pBoc timing in individual worms following exposure to cmd-1 RNAi for the time periods indicated (0, 18, 36, and 54 h). Strong contractions are plotted as symbols, whereas shadows or weak reiterative contractions are plotted as smaller symbols of like type placed lower on the y axis. C, representative Ca2+ oscillations in a cmd-1 RNAi worm at 54 h. The oscillatory period shown here is similar to that of the behavioral reiterations shown in B. D, representative intestinal pHi oscillations in a cmd-1 RNAi worm at 54 h. Note that this trace represents fluctuations in pHi rather than pHe and that the period is similar to that of both Ca2+ oscillations and pBoc.
Here, we found that intestinal RNAi of cmd-1 resulted in a progressive phenotype in which the cumulative loss of cmd-1 expression altered defecation frequency (Fig. 5B). In general, when young adult worms were subjected to cmd-1 RNAi for 18 h, they began to exhibit weak reiterations of the DMP. This has been observed previously in unc-43 CaMKII loss-of-function mutants (20, 25), where these reiterations were referred to as “shadows” or “echos.” The interval between the principal, or strong, DMP and the shadows was much shorter (∼16–18 s) in cmd-1 RNAi worms than the average defecation period (∼45 s), consistent with cmd-1 normally functioning to suppress inappropriate intracycle execution of the DMP. Surprisingly, however, an increased time of exposure to cmd-1 RNAi resulted in not just a single shadow occurring after the principal DMP, but multiple shadows occurring (Fig. 5B). The period between principal executions of the DMP increased with loss of cmd-1 primarily due to an increased number of shadows occurring between cycles, whereas the period between shadows, which was ∼18 s, did not increase correspondingly. Eventually, the weaker shadows became nearly indistinguishable from the principal DMP. At this point, the worms were essentially defecating every ∼18 s (Fig. 5). Continued exposure to cmd-1 RNAi led to the cessation of defecation entirely, followed by death of the animal (data not shown).
Given that the DMP is initiated by oscillatory Ca2+ signaling and cyclic acidification of the intestine, we used fluorescent biosensors to measure Ca2+ and pH in the intestines of live moving worms subjected to cmd-1 RNAi. Following 54 h, we observed oscillations in both of these second messengers occurring every ∼18 s, mirroring the observed changes to the behavioral period (Fig. 5, C and D). We conclude that CaM directly influences intestinal oscillatory Ca2+ signaling upstream of acidification, and hence, its loss has effects that preclude analysis of its role in regulating NHX-7 directly. Nevertheless, these results are interesting in the context of signaling pathways that regulate the periodicity of Ca2+ oscillations and emphasize the integration between Ca2+ and proton signaling during defecation in worms.
DISCUSSION
Na+/H+ exchangers are recognized to contribute to pHi homeostasis and electrolyte and water balance (for review, see Ref. 62). Recently, a new role for Na+/H+ exchangers has emerged from work in C. elegans showing that NHX-7 allows adjacent cells to communicate through direct proton signaling (17, 18). This event is coordinated by oscillatory Ca2+ signaling, which in turn leads to repetitive cellular acidification and recovery. Hence, defecation represents a behavior whose physiologic output is integrated by cross-talk between Ca2+ and pH signals. Counterintuitively, apart from its role in signaling, NHX-7 contributes little to re-establishing pH homeostasis following defecation. Because Na+/H+ exchangers are activated by allosteric proton binding and have been shown to be regulated by Ca2+, it is reasonable to ask whether regulation of NHX-7 by Ca2+ is a determining factor in its ability to function as a signaling protein as opposed to a “pH housekeeper.”
Apart from a relative insensitivity to the amiloride derivative 5-(N-ethyl-N-isopropyl)amiloride, NHX-7 functions much like mammalian NHEs and can facilitate recovery from induced acidosis when transfected into cells in culture (Fig. 1). However, initial attempts to discern the signaling pathway through which Ca2+ might stimulate NHX-7 were hampered by the fact that recombinant NHX-7 activity was not responsive to pharmacologic manipulations that increased intracellular Ca2+ levels. Although we considered the possibility that NHX-7 is not stimulated by Ca2+ signaling, we feel that it is more likely either that the relevant signaling pathway is not present in AP-1 cells or that it does not recognize the worm signaling motifs. Similarly, it is possible that overexpression in cell culture could mask Ca2+ regulatory effects on NHX-7.
Nevertheless, these simple observations helped to inform us as to how NHX-7 may contribute to the integrative physiology of defecation. In the worm, protons are an important means of performing work, and a proton gradient formed between the intestinal lumen and cell is essential for nutrient uptake across the apical membrane (50, 63). Maintenance of this gradient is critical and presents some physiologic challenges given that a significant portion of the intestine's luminal contents are expulsed every 45 s. To prevent their loss during expulsion, protons enter the cell from the lumen during defecation. These protons must then be quickly returned to the lumen immediately following defecation to drive nutrient uptake. Viewed in this context, sustained H+ efflux through NHX-7 on the basolateral membrane could be catastrophic, leading to both caloric deprivation and systemic acidosis.
Our data demonstrate one mechanism for ensuring that proton efflux to the pseudocoelomic space via NHX-7 is acute. A brief pulse of activity is sufficient to rapidly acidify the small pseudocoelom to a nadir value at which we found NHX-7 was virtually inactive (Fig. 1B). Therefore, the inherent pHe sensitivity of NHX-7 appears to provide a failsafe method of ensuring that H+ signaling is turned off following stimulation of the muscle proton receptor.
A transgenic worm model allowed us to perform structure-function mutagenesis of NHX-7 and complementation of a null mutant to assess the role of potential Ca2+ regulatory motifs. Our results from both behavioral output and in vivo pH measurements taken together suggest that 1) PIP2 binding stimulates basal NHX-7 activity, 2) CaM binding contributes to Ca2+ activation of NHX-7, and 3) stimulated NHX-7 activity exceeds what is necessary to trigger a “normal” pBoc. Binding of the CHP ortholog PBO-1 appears to affect NHX-7 trafficking and/or membrane residence through an unexplored mechanism (data not shown); hence, we were unable to define its role in Ca2+ activation, although given its EF-hand Ca2+-binding domains, we hypothesize that it may serve dual roles in transport and signaling. We also found that a T618A mutation of a potential CaMKII phosphorylation site increased the duration of pBoc and the period of extracellular acidification during pBoc (Fig. 3 and Table 1). Although the effect of the T618A mutation was relatively modest, phosphorylation by CaMKII at this site may provide a “cellular memory” of Ca2+ signaling, such as occurs during long-term potentiation in neurons (64), hence contributing to feedback inhibition of NHE activity.
Interestingly, crystal structure data from bacterial NhaA and from a CHP/NHE1 fragment suggest a possible mechanism of action for CHP and PIP2 whereby their binding near helical segment 11, which has been shown to be important for exchange activity, could have direct implications in the transport of ions (65, 66). Moreover, Ca2+ regulation of NHX-7 appears to be uncoupled from the primary regulation by H+, consistent with the pH sensor responsible for H+ recognition being located at the beginning of helix 9 on the opposite side of the unwound helix 11 (65).
Regulation by CaM is more difficult to interpret mechanistically, as its binding occurs farther downstream in the C-terminal tail, and the binding site(s) in NHX-7 and NHE1 are not precisely conserved at the linear sequence level (supplemental Fig. S1). Although CaM itself is not well studied in C. elegans, CaMKII has been studied extensively, and genetic approaches have shown that mutations in unc-43, the worm CaMKII ortholog, cause the DMP to be reiterated, albeit weakly (20). This ability of CaMKII to normally suppress inappropriate Ca2+ oscillations between cycles is consistent with cellular memory. We discovered that loss of CaM itself resulted in a qualitatively similar result (Fig. 5). However, we also discovered that this phenotype was progressive and that the strength and occurrence of these reiterations became more pronounced over time. Astoundingly, after several days of exposure to cmd-1 RNAi, the worms were executing the entire DMP every ∼18 s, and shortly thereafter, they expired.
To understand the physiologic alteration accompanying these shadow contractions, we also went on to show that late-stage cmd-1 RNAi worms exhibited Ca2+ and pH oscillations at nearly three times the rate of normal worms (Fig. 5). At this rate, intestinal pH recovery processes, most likely those of the energy-dependent second-phase V-ATPase (63), can not accommodate the cyclic acidification that occurs, resulting in sustained cellular acidosis well below that of the normal resting pH at ∼7.4 (Fig. 5D). We speculate that cmd-1 may contribute to regulating the inositol 1,4,5-trisphosphate receptor ITR-1, which is the molecular pacemaker for defecation. Although we found these results to be noteworthy and to deserve reporting, the fact that cmd-1 contributed to Ca2+ pacemaking and that its loss induced massive cellular acidification derailed our attempts to confirm CaM regulation of NHX-7 through cmd-1 RNAi.
Considered as a whole, our data support a model in which Ca2+ activation of NHX-7 is not essential for its function but may contribute to proton conservation. Moreover, we have shown that PIP2 binding is required for basal function of the exchanger and suggested that Ca2+ may also play a role in suppressing the activity of NHX-7 following its activation. Finally, although it appears likely that CaM helps to coordinate Ca2+ activation of NHX-7, it also regulates oscillatory Ca2+ signaling frequency upstream of NHX-7 activity.
In conclusion, the apparent sensitivity of NHX-7 within the integrative context of defecation to changes in both Ca2+ and pH is consistent with the importance that protons play in intestinal physiology in the worm but may also provide mechanistic insights into the regulation of NHE activity in mammals. For example, Na+/H+ exchange has been implicated in the regulation of synaptic transmission at glutamatergic, GABAergic, and dopaminergic synapses (67–69), and NHE1 has been shown to contribute to synaptic acidification and the creation of an acidic microdomain at the synaptic cleft (70). It is likely not coincidental that synaptic vesicle release is a Ca2+-mediated event. Alternatively, NHE5 is credited with supporting neuronal activity-dependent dendritic spine growth through a novel pH-mediated negative feedback mechanism (53). These results suggest that acute control of pH and proton transport across the membrane has profound consequences for mammalian neurophysiology and may potentially affect communication between adjacent cells in a variety of contexts, including, as we have shown here, during signaling between intestinal and muscle cells in C. elegans.
Supplementary Material
Acknowledgments
We thank Teresa Sherman for technical assistance, Dr. L. Pablo Cid for the extracellular pH sensor, and Dr. Maureen Peters for critical comments. Some strains used in this work were provided by the C. elegans Gene Knockout Consortium and the Caenorhabditis Genetics Center, which is supported by National Institutes of Health Office of Research Infrastructure Program P40 OD010440.
This work was supported by Ruth L. Kirschstein National Research Service Institutional Award GM068411 from the National Institutes of Health (to E. A.). This work was also supported by National Science Foundation Grant IOS0919848 (to K. N.).

This article contains supplemental “Experimental Procedures,” Fig. S1, and additional references.
E. Allman and K. Nehrke, unpublished data.
- DMP
- defecation motor program
- pBoc
- posterior body wall muscle contraction
- NHE
- mammalian Na+/H+ exchanger
- NHX
- C. elegans Na+/H+ exchanger
- CaMKII
- Ca2+/calmodulin-dependent protein kinase type II
- CaM
- calmodulin
- CHP
- calcineurin homologous protein
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- rNHE1
- rat NHE1
- CBD
- CaM-binding domain
- DIC
- differential interference contrast.
REFERENCES
- 1. Casey J. R., Grinstein S., Orlowski J. (2010) Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 11, 50–61 [DOI] [PubMed] [Google Scholar]
- 2. Alper S. L. (2002) Genetic diseases of acid-base transporters. Annu. Rev. Physiol. 64, 899–923 [DOI] [PubMed] [Google Scholar]
- 3. Garbern J. Y., Neumann M., Trojanowski J. Q., Lee V. M., Feldman G., Norris J. W., Friez M. J., Schwartz C. E., Stevenson R., Sima A. A. (2010) A mutation affecting the sodium/proton exchanger, SLC9A6, causes mental retardation with tau deposition. Brain 133, 1391–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Martínez-Zaguilán R., Seftor E. A., Seftor R. E., Chu Y. W., Gillies R. J., Hendrix M. J. (1996) Acidic pH enhances the invasive behavior of human melanoma cells. Clin. Exp. Metastasis 14, 176–186 [DOI] [PubMed] [Google Scholar]
- 5. Ludwig M. G., Vanek M., Guerini D., Gasser J. A., Jones C. E., Junker U., Hofstetter H., Wolf R. M., Seuwen K. (2003) Proton-sensing G-protein-coupled receptors. Nature 425, 93–98 [DOI] [PubMed] [Google Scholar]
- 6. Li S., Sato S., Yang X., Preisig P. A., Alpern R. J. (2004) Pyk2 activation is integral to acid stimulation of sodium/hydrogen exchanger 3. J. Clin. Invest. 114, 1782–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Waldmann R., Champigny G., Bassilana F., Heurteaux C., Lazdunski M. (1997) A proton-gated cation channel involved in acid-sensing. Nature 386, 173–177 [DOI] [PubMed] [Google Scholar]
- 8. Ishii S., Kihara Y., Shimizu T. (2005) Identification of T cell death-associated gene 8 (TDAG8) as a novel acid sensing G-protein-coupled receptor. J. Biol. Chem. 280, 9083–9087 [DOI] [PubMed] [Google Scholar]
- 9. Tomura H., Mogi C., Sato K., Okajima F. (2005) Proton-sensing and lysolipid-sensitive G-protein-coupled receptors: a novel type of multi-functional receptors. Cell. Signal. 17, 1466–1476 [DOI] [PubMed] [Google Scholar]
- 10. Wemmie J. A., Price M. P., Welsh M. J. (2006) Acid-sensing ion channels: advances, questions and therapeutic opportunities. Trends Neurosci. 29, 578–586 [DOI] [PubMed] [Google Scholar]
- 11. Dray A. (1995) Inflammatory mediators of pain. Br. J. Anaesth. 75, 125–131 [DOI] [PubMed] [Google Scholar]
- 12. Issberner U., Reeh P. W., Steen K. H. (1996) Pain due to tissue acidosis: a mechanism for inflammatory and ischemic myalgia? Neurosci. Lett. 208, 191–194 [DOI] [PubMed] [Google Scholar]
- 13. Cobbe S. M., Poole-Wilson P. A. (1980) The time of onset and severity of acidosis in myocardial ischaemia. J. Mol. Cell. Cardiol. 12, 745–760 [DOI] [PubMed] [Google Scholar]
- 14. Chesler M., Kaila K. (1992) Modulation of pH by neuronal activity. Trends Neurosci. 15, 396–402 [DOI] [PubMed] [Google Scholar]
- 15. Krishtal O. A., Osipchuk Y. V., Shelest T. N., Smirnoff S. V. (1987) Rapid extracellular pH transients related to synaptic transmission in rat hippocampal slices. Brain Res. 436, 352–356 [DOI] [PubMed] [Google Scholar]
- 16. Vessey J. P., Stratis A. K., Daniels B. A., Da Silva N., Jonz M. G., Lalonde M. R., Baldridge W. H., Barnes S. (2005) Proton-mediated feedback inhibition of presynaptic calcium channels at the cone photoreceptor synapse. J. Neurosci. 25, 4108–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Beg A. A., Ernstrom G. G., Nix P., Davis M. W., Jorgensen E. M. (2008) Protons act as a transmitter for muscle contraction in C. elegans. Cell 132, 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pfeiffer J., Johnson D., Nehrke K. (2008) Oscillatory transepithelial H+ flux regulates a rhythmic behavior in C. elegans. Curr. Biol. 18, 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thomas J. H. (1990) Genetic analysis of defecation in Caenorhabditis elegans. Genetics 124, 855–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu D. W., Thomas J. H. (1994) Regulation of a periodic motor program in C. elegans. J. Neurosci. 14, 1953–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Croll N. A., Smith J. M. (1978) Integrated behaviour in the feeding phase of Caenorhabditis elegans (Nematoda). J. Zool. 184, 507–517 [Google Scholar]
- 22. Dal Santo P., Logan M. A., Chisholm A. D., Jorgensen E. M. (1999) The inositol triphosphate receptor regulates a 50-second behavioral rhythm in C. elegans. Cell 98, 757–767 [DOI] [PubMed] [Google Scholar]
- 23. Espelt M. V., Estevez A. Y., Yin X., Strange K. (2005) Oscillatory Ca2+ signaling in the isolated Caenorhabditis elegans intestine: role of the inositol-1,4,5-trisphosphate receptor and phospholipases C β and γ. J. Gen. Physiol. 126, 379–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Teramoto T., Iwasaki K. (2006) Intestinal calcium waves coordinate a behavioral motor program in C. elegans. Cell Calcium 40, 319–327 [DOI] [PubMed] [Google Scholar]
- 25. Nehrke K., Denton J., Mowrey W. (2008) Intestinal Ca2+ wave dynamics in freely moving C. elegans coordinate execution of a rhythmic motor program. Am. J. Physiol. Cell Physiol. 294, C333–C344 [DOI] [PubMed] [Google Scholar]
- 26. Slepkov E. R., Rainey J. K., Sykes B. D., Fliegel L. (2007) Structural and functional analysis of the Na+/H+ exchanger. Biochem. J. 401, 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Denker S. P., Barber D. L. (2002) Cell migration requires both ion translocation and cytoskeletal anchoring by the Na-H exchanger NHE1. J. Cell Biol. 159, 1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pouysségur J., Sardet C., Franchi A., L'Allemain G., Paris S. (1984) A specific mutation abolishing Na+/H+ antiport activity in hamster fibroblasts precludes growth at neutral and acidic pH. Proc. Natl. Acad. Sci. U.S.A. 81, 4833–4837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Putney L. K., Barber D. L. (2003) Na-H exchange-dependent increase in intracellular pH times G2/M entry and transition. J. Biol. Chem. 278, 44645–44649 [DOI] [PubMed] [Google Scholar]
- 30. Tominaga T., Barber D. L. (1998) Na-H exchange acts downstream of RhoA to regulate integrin-induced cell adhesion and spreading. Mol. Biol. Cell 9, 2287–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rotin D., Grinstein S. (1989) Impaired cell volume regulation in Na+-H+ exchange-deficient mutants. Am. J. Physiol. 257, C1158–C1165 [DOI] [PubMed] [Google Scholar]
- 32. Rotin D., Steele-Norwood D., Grinstein S., Tannock I. (1989) Requirement of the Na+/H+ exchanger for tumor growth. Cancer Res. 49, 205–211 [PubMed] [Google Scholar]
- 33. Sardet C., Franchi A., Pouysségur J. (1989) Molecular cloning, primary structure, and expression of the human growth factor-activatable Na+/H+ antiporter. Cell 56, 271–280 [DOI] [PubMed] [Google Scholar]
- 34. Orlowski J., Kandasamy R. A., Shull G. E. (1992) Molecular cloning of putative members of the Na/H exchanger gene family. cDNA cloning, deduced amino acid sequence, and mRNA tissue expression of the rat Na/H exchanger NHE-1 and two structurally related proteins. J. Biol. Chem. 267, 9331–9339 [PubMed] [Google Scholar]
- 35. Wang Z., Orlowski J., Shull G. E. (1993) Primary structure and functional expression of a novel gastrointestinal isoform of the rat Na/H exchanger. J. Biol. Chem. 268, 11925–11928 [PubMed] [Google Scholar]
- 36. Numata M., Petrecca K., Lake N., Orlowski J. (1998) Identification of a mitochondrial Na+/H+ exchanger. J. Biol. Chem. 273, 6951–6959 [DOI] [PubMed] [Google Scholar]
- 37. Attaphitaya S., Park K., Melvin J. E. (1999) Molecular cloning and functional expression of a rat Na+/H+ exchanger (NHE5) highly expressed in brain. J. Biol. Chem. 274, 4383–4388 [DOI] [PubMed] [Google Scholar]
- 38. Baird N. R., Orlowski J., Szabó E. Z., Zaun H. C., Schultheis P. J., Menon A. G., Shull G. E. (1999) Molecular cloning, genomic organization, and functional expression of Na+/H+ exchanger isoform 5 (NHE5) from human brain. J. Biol. Chem. 274, 4377–4382 [DOI] [PubMed] [Google Scholar]
- 39. Goyal S., Vanden Heuvel G., Aronson P. S. (2003) Renal expression of novel Na+/H+ exchanger isoform NHE8. Am. J. Physiol. Renal Physiol. 284, F467–E473 [DOI] [PubMed] [Google Scholar]
- 40. Nakamura N., Tanaka S., Teko Y., Mitsui K., Kanazawa H. (2005) Four Na+/H+ exchanger isoforms are distributed to Golgi and post-Golgi compartments and are involved in organelle pH regulation. J. Biol. Chem. 280, 1561–1572 [DOI] [PubMed] [Google Scholar]
- 41. Nehrke K., Melvin J. E. (2002) The NHX family of Na+-H+ exchangers in Caenorhabditis elegans. J. Biol. Chem. 277, 29036–29044 [DOI] [PubMed] [Google Scholar]
- 42. Noël J., Pouysségur J. (1995) Hormonal regulation, pharmacology, and membrane sorting of vertebrate Na+/H+ exchanger isoforms. Am. J. Physiol. 268, C283–C296 [DOI] [PubMed] [Google Scholar]
- 43. Aronson P. S., Nee J., Suhm M. A. (1982) Modifier role of internal H+ in activating the Na+-H+ exchanger in renal microvillus membrane vesicles. Nature 299, 161–163 [DOI] [PubMed] [Google Scholar]
- 44. Imahashi K., Mraiche F., Steenbergen C., Murphy E., Fliegel L. (2007) Overexpression of the Na+/H+ exchanger and ischemia-reperfusion injury in the myocardium. Am. J. Physiol. Heart Circ. Physiol. 292, H2237–H2247 [DOI] [PubMed] [Google Scholar]
- 45. He P., Yun C. C. (2010) Mechanisms of the regulation of the intestinal Na+/H+ exchanger NHE3. J. Biomed. Biotechnol. 2010, 238080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Urra J., Sandoval M., Cornejo I., Barros L. F., Sepúlveda F. V., Cid L. P. (2008) A genetically encoded ratiometric sensor to measure extracellular pH in microdomains bounded by basolateral membranes of epithelial cells. Pflugers Arch. 457, 233–242 [DOI] [PubMed] [Google Scholar]
- 48. Johnson D., Allman E., Nehrke K. (2012) Regulation of acid-base transporters by reactive oxygen species following mitochondrial fragmentation. Am. J. Physiol. Cell Physiol. 302, C1045–C1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thomas J. A., Buchsbaum R. N., Zimniak A., Racker E. (1979) Intracellular pH measurements in Ehrlich ascites tumor cells utilizing spectroscopic probes generated in situ. Biochemistry 18, 2210–2218 [DOI] [PubMed] [Google Scholar]
- 50. Nehrke K. (2003) A reduction in intestinal cell pHi due to loss of the Caenorhabditis elegans Na+/H+ exchanger NHX-2 increases life span. J. Biol. Chem. 278, 44657–44666 [DOI] [PubMed] [Google Scholar]
- 51. Iwasaki K., Liu D. W., Thomas J. H. (1995) Genes that control a temperature-compensated ultradian clock in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 92, 10317–10321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Boron W. F., De Weer P. (1976) Intracellular pH transients in squid giant axons caused by CO2, NH3, and metabolic inhibitors. J. Gen. Physiol. 67, 91–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Diering G. H., Mills F., Bamji S. X., Numata M. (2011) Regulation of dendritic spine growth through activity-dependent recruitment of the brain-enriched Na+/H+ exchanger NHE5. Mol. Biol. Cell 22, 2246–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wagner J., Allman E., Taylor A., Ulmschneider K., Kovanda T., Ulmschneider B., Nehrke K., Peters M. A. (2011) A calcineurin homologous protein is required for sodium-proton exchange events in the C. elegans intestine. Am. J. Physiol. Cell Physiol. 301, C1389–C1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Aharonovitz O., Zaun H. C., Balla T., York J. D., Orlowski J., Grinstein S. (2000) Intracellular pH regulation by Na+/H+ exchange requires phosphatidylinositol 4,5-bisphosphate. J. Cell Biol. 150, 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xing J., Strange K. (2010) Phosphatidylinositol 4,5-bisphosphate and loss of PLCγ activity inhibit TRPM channels required for oscillatory Ca2+ signaling. Am. J. Physiol. Cell Physiol. 298, C274–C282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pang T., Su X., Wakabayashi S., Shigekawa M. (2001) Calcineurin homologous protein as an essential cofactor for Na+/H+ exchangers. J. Biol. Chem. 276, 17367–17372 [DOI] [PubMed] [Google Scholar]
- 58. Lin X., Barber D. L. (1996) A calcineurin homologous protein inhibits GTPase-stimulated Na-H exchange. Proc. Natl. Acad. Sci. U.S.A. 93, 12631–12636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fliegel L., Walsh M. P., Singh D., Wong C., Barr A. (1992) Phosphorylation of the C-terminal domain of the Na+/H+ exchanger by Ca2+/calmodulin-dependent protein kinase II. Biochem. J. 282, 139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vila-Petroff M., Mundiña-Weilenmann C., Lezcano N., Snabaitis A. K., Huergo M. A., Valverde C. A., Avkiran M., Mattiazzi A. (2010) Ca2+/calmodulin-dependent protein kinase II contributes to intracellular pH recovery from acidosis via Na+/H+ exchanger activation. J. Mol. Cell. Cardiol. 49, 106–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bertrand B., Wakabayashi S., Ikeda T., Pouysségur J., Shigekawa M. (1994) The Na+/H+ exchanger isoform 1 (NHE1) is a novel member of the calmodulin-binding proteins. Identification and characterization of calmodulin-binding sites. J. Biol. Chem. 269, 13703–13709 [PubMed] [Google Scholar]
- 62. Putney L. K., Denker S. P., Barber D. L. (2002) The changing face of the Na+/H+ exchanger, NHE1: structure, regulation, and cellular actions. Annu. Rev. Pharmacol. Toxicol. 42, 527–552 [DOI] [PubMed] [Google Scholar]
- 63. Allman E., Johnson D., Nehrke K. (2009) Loss of the apical V-ATPase a-subunit VHA-6 prevents acidification of the intestinal lumen during a rhythmic behavior in C. elegans. Am. J. Physiol. Cell Physiol. 297, C1071–C1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lisman J., Yasuda R., Raghavachari S. (2012) Mechanisms of CaMKII action in long-term potentiation. Nat. Rev. Neurosci. 13, 169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hunte C., Screpanti E., Venturi M., Rimon A., Padan E., Michel H. (2005) Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature 435, 1197–1202 [DOI] [PubMed] [Google Scholar]
- 66. Mishima M., Wakabayashi S., Kojima C. (2007) Solution structure of the cytoplasmic region of Na+/H+ exchanger 1 complexed with essential cofactor calcineurin B homologous protein 1. J. Biol. Chem. 282, 2741–2751 [DOI] [PubMed] [Google Scholar]
- 67. Trudeau L. E., Parpura V., Haydon P. G. (1999) Activation of neurotransmitter release in hippocampal nerve terminals during recovery from intracellular acidification. J. Neurophysiol. 81, 2627–2635 [DOI] [PubMed] [Google Scholar]
- 68. Jang I. S., Brodwick M. S., Wang Z. M., Jeong H. J., Choi B. J., Akaike N. (2006) The Na+/H+ exchanger is a major pH regulator in GABAergic presynaptic nerve terminals synapsing onto rat CA3 pyramidal neurons. J. Neurochem. 99, 1224–1236 [DOI] [PubMed] [Google Scholar]
- 69. Rocha M. A., Crockett D. P., Wong L. Y., Richardson J. R., Sonsalla P. K. (2008) Na+/H+ exchanger inhibition modifies dopamine neurotransmission during normal and metabolic stress conditions. J. Neurochem. 106, 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dietrich C. J., Morad M. (2010) Synaptic acidification enhances GABAA signaling. J. Neurosci. 30, 16044–16052 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.