Background: Active DNA demethylation in plants is initiated by 5-meC DNA glycosylases of the ROS1/DME family and continued through a base excision repair pathway.
Results: XRCC1 stimulates ROS1-initiated demethylation and is required for efficient processing of post-excision intermediates.
Conclusion: XRCC1 functions at several stages during active DNA demethylation.
Significance: This study identifies a new component of the active DNA demethylation pathway in Arabidopsis.
Keywords: Base Excision Repair, DNA Enzymes, DNA Methylation, DNA Repair, Epigenetics
Abstract
DNA methylation patterns are the dynamic outcome of antagonist methylation and demethylation mechanisms, but the latter are still poorly understood. Active DNA demethylation in plants is mediated by a family of DNA glycosylases typified by Arabidopsis ROS1 (repressor of silencing 1). ROS1 and its homologs remove 5-methylcytosine and incise the sugar backbone at the abasic site, thus initiating a base excision repair pathway that finally inserts an unmethylated cytosine. The DNA 3′-phosphatase ZDP processes some of the incision products generated by ROS1, allowing subsequent DNA polymerization and ligation steps. In this work, we examined the possible role of plant XRCC1 (x-ray cross-complementing group protein 1) in DNA demethylation. We found that XRCC1 interacts in vitro with ROS1 and ZDP and stimulates the enzymatic activity of both proteins. Furthermore, extracts from xrcc1 mutant plants exhibit a reduced capacity to complete DNA demethylation initiated by ROS1. An anti-XRCC1 antibody inhibits removal of the blocking 3′-phosphate in the single-nucleotide gap generated during demethylation and reduces the capacity of Arabidopsis cell extracts to ligate a nicked DNA intermediate. Our results suggest that XRCC1 is a component of plant base excision repair and functions at several stages during active DNA demethylation in Arabidopsis.
Introduction
Cytosine methylation (5-meC)3 is an epigenetic mark associated with long term gene silencing that plays critical roles in genomic imprinting, suppression of transposable elements, and the establishment of correct gene expression patterns during development (1). Methylation landscapes are dynamically regulated by DNA methylation and demethylation processes (2, 3). In plants, active demethylation is performed by a family of bifunctional DNA glycosylases/lyases that remove 5-meC and initiate its replacement by unmethylated cytosine through a base excision repair (BER) process (2, 4, 5). In animals, no 5-meC DNA glycosylases have been unambiguously identified, and accumulative evidence indicates that demethylation involves excision of deaminated and/or oxidized derivatives of 5-meC (6, 7). Thus, BER is emerging as a central process for active DNA demethylation in both plants and animals.
Plant 5-meC DNA glycosylases are typified by Arabidopsis proteins ROS1 (repressor of silencing 1) (8), DME (Demeter), and DML2 and DML3 (Demeter-like proteins 2 and 3) (8–11). In vivo, DME demethylates the maternal allele of imprinted genes in the endosperm (12), whereas ROS1, DML2, and DML3 counteract excessive methylation at several hundred loci across the genome (10, 11, 13). All four proteins are bifunctional DNA glycosylases/lyases that cleave the phosphodiester backbone at the 5-meC removal site by β-elimination, creating a phospho-α,β-unsaturated aldehyde at the 3′-end of the strand break (10–12, 14, 15). A significant amount of β-elimination incisions proceed to β,δ-elimination, thus generating a single-nucleotide gap flanked by 3′-phosphate and 5′-phosphate termini (11, 14, 15). The DNA 3′-phosphatase ZDP functions downstream of ROS1 by removing the blocking 3′-phosphate present in β,δ-elimination products and allowing subsequent DNA polymerization and ligation steps (16). A deficiency in ZDP disrupts the methylation pathway and alters methylation control, leading to a hypermethylated state in many loci across the genome (16).
In an ongoing effort to identify additional components of the plant DNA demethylation pathway, we have turned our attention to XRCC1, which in mammalian cells plays a key role in BER and single-strand break repair as a scaffold protein (17). XRCC1 was originally identified by its capacity to restore DNA repair activity in CHO cell lines hypersensitive to DNA-damaging agents, including methyl methanesulfonate and ionizing radiation (18). XRCC1 lacks any detectable enzymatic activity, but interacts with many BER proteins to coordinate and enhance overall repair (17, 18). In mammalian cells, its interaction partners include several DNA glycosylases acting during lesion recognition/strand incision, such as hOGG1, MPG (3-methyladenine-DNA glycosylase), UNG2, hNTH1, hNEIL1, and hNEIL2 (19–22), gap-tailoring enzymes such as polynucleotide kinase and APE1 (23, 24), or proteins performing gap filling and ligation, such as DNA polymerase β and DNA ligase III (25, 26). Interestingly, the levels of chromatin-bound XRCC1 increase concurrently with DNA demethylation during reprogramming of the mouse germ line (27).
Plant XRCC1 homologs have been identified in Arabidopsis (28) and rice (29). Although XRCC1 proteins from mammals and other vertebrates possess an N-terminal domain (NTD) and two BRCT (BRCA1 C-terminal) domains (BRCT1 and BRCT2), plant XRCC1 only contains the BRCT1 domain, which displays a high degree of sequence conservation across all XRCC1 homologs (28, 29). Recently, it has been shown that Arabidopsis XRCC1 functions in a Ku-independent double-strand breakage repair pathway that involves nonhomologous end-joining (30). On the other hand, it has been reported in rice that XRCC1 interacts with ds- and ssDNA in vitro and with proliferating cell nuclear antigen in vitro and in vivo (29). However, the possible role of XRCC1 in plant BER, and specifically during active DNA demethylation, remains to be determined.
In this work, we report that Arabidopsis XRCC1 interacts with ROS1 and ZDP and stimulates their enzymatic activities in vitro. We found that cell extracts from xrcc1 mutant plants exhibit a reduced capability to complete DNA demethylation and that XRCC1 is required for efficient gap tailoring and ligation of DNA demethylation intermediates. Our results suggest that XRCC1 functions during active DNA demethylation in Arabidopsis.
EXPERIMENTAL PROCEDURES
Plant Material and Cell Extract Preparation
Arabidopsis thaliana wild-type plants were ecotype Columbia. The Arabidopsis mutant line SALK_027362 harboring a T-DNA insertion in the XRCC1 gene (30) was a kind gift from Charles White (Blaise Pascal University, France). Cells extracts were prepared from snap-frozen 15-day-old seedlings as described previously (31, 32).
Protein Expression and Purification
Expression and purification of His-ZDP, His-ROS1, and His-ROS1 deletion derivatives were performed as described previously (16). The full-length XRCC1 cDNA was obtained from the Arabidopsis Biological Resource Center (pZL1-XRCC1, clone 210J9) and subcloned into expression vectors pET30b (Novagen) and pMAL-c2X (New England Biolabs) to generate His-XRCC1 and MBP-XRCC1 fusion proteins, respectively. Expression was induced in Escherichia coli BL21 (DE3) dcm− Codon Plus cells (Stratagene). His-XRCC1 was purified by affinity chromatography on a Ni2+-nitrilotriacetic acid column (Amersham Biosciences), and MBP-XRCC1 was purified by amylose affinity chromatography (New England Biolabs).
Pulldown Assays
For maltose-binding protein (MBP) pulldown assays with ROS1 His-tagged protein, 50 pmol of purified MBP alone or MBP-XRCC1 in 100 μl of Column Buffer (20 mm Tris, pH 7.4, 200 mm NaCl, 1 mm EDTA, 10 mm β-mercaptoethanol) was added to 100 μl of amylose resin (New England Biolabs) and incubated for 1 h at 4 °C. The resin was washed twice with 600 μl of MBP Buffer (20 mm Tris, pH 7.4, 1 mm EDTA, 10 mm β-mercaptoethanol, 0.5% Triton X-100). Purified His-tagged ROS1 (12 pmol) was incubated at 4 °C for 1 h with either MBP or MBP-XRCC1 bound to resin. The resin was washed twice with MBP Buffer. Bound proteins were analyzed by Western blot using antibodies against His6 tag (Novagen).
For His tag pulldown assays with MBP-XRCC1 protein, 50 pmol of purified full-length ROS1, truncated ROS1 polypeptides, or ZDP fused to His tag in 400 μl of Sonication Buffer 2 (20 mm Tris, pH 8.0, 500 mm NaCl) was added to 100 μl of nickel-Sepharose resin (Amersham Biosciences) and incubated for 1 h at 4 °C. The resin was washed twice with MBP Buffer supplemented with 60 mm imidazole. Purified MBP or MBP-XRCC1 (12 pmol) was incubated at 4 °C for 1 h with FL-ROS1, truncated ROS1 polypeptides, or ZDP bound to resin. The resin was washed twice with MBP Buffer supplemented with 60 mm imidazole. Bound proteins were analyzed by Western blot using antibodies against MBP (Sigma).
DNA Substrates
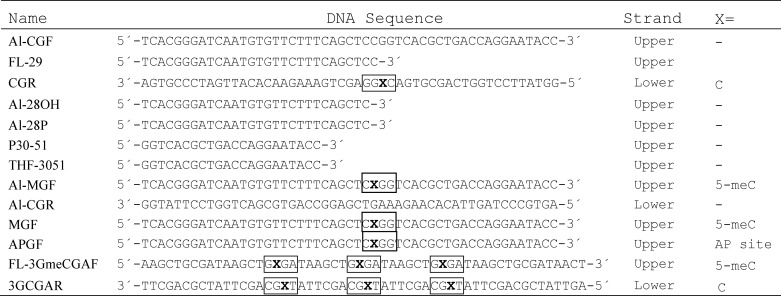
Oligonucleotides used as DNA substrates (see Table 1) were synthesized by Eurofins MWG Operon and/or Integrated DNA Technologies and purified by PAGE before use. Double-stranded DNA substrates were prepared by mixing a 5 μm solution of a 5′-fluorescein-labeled or 5′-Alexa Fluor-labeled oligonucleotide (upper strand) with a 10 μm solution of an unlabeled oligomer (lower strand), heating to 95 °C for 5 min, and slowly cooling to room temperature.
TABLE 1.
Oligonucleotides used as substrates
Relevant regions are boxed.
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was performed using Alexa Fluor-labeled duplex oligonucleotides prepared as described above. The labeled duplex substrate (100 nm) was incubated with different amounts of XRCC1 (0, 1, 2, 4, 6, and 8 μm) in DNA-binding reaction mixtures (15 μl) containing 10 mm Tris HCl, pH 8.0, 1 mm DTT, 10 μg/ml BSA, 1 mm EDTA. After a 60-min incubation at 25 °C, reactions were immediately loaded onto 0.2% agarose gels in 1× Tris acetate/EDTA. Electrophoresis was carried out in 1× Tris acetate/EDTA for 40 min at 80 V at room temperature. Alexa Fluor-labeled DNA was visualized in a FLA-5100 imager and analyzed using MultiGauge software (Fujifilm).
DNA 3′-Phosphatase Assay
Phosphatase reactions were performed as described previously (16).
DNA Glycosylase Assay
DNA glycosylase assays were performed as described previously (33).
DNA Repair/Demethylation Assay in Whole-cell Extracts
The DNA repair assay used to monitor gap filling and ligation during DNA demethylation was performed as described previously (31, 32).
RESULTS
Arabidopsis XRCC1 Interacts with ROS1, ZDP, and DNA
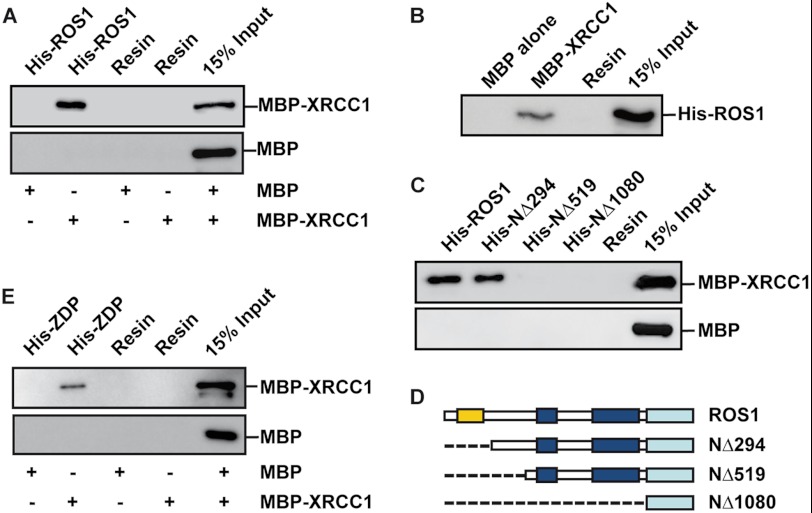
We used in vitro pulldown assays to test for a direct interaction between XRCC1 and ROS1. The fusion protein His-ROS1 was bound to a nickel-Sepharose resin and incubated with either purified MBP-XRCC1 or purified MBP. As shown in Fig. 1A, MBP-XRCC1, but not MBP alone, was bound by His-ROS1. We also confirmed that His-ROS1 was bound by MBP-XRCC1 immobilized on an amylose column (Fig. 1B). To define ROS1 regions implicated in the interaction, we generated a series of N- and C-terminal His-ROS1 deletion mutants, and the purified proteins were bound to a nickel-Sepharose resin and used to analyze their capacity to bind MBP-XRCC1 (Fig. 1, C and D). We found that ROS1 amino acids 295–519 are needed to interact with XRCC1. We also tested for possible interactions between XRCC1 and ZDP. As shown in Fig. 1E, MBP-XRCC1, but not MBP alone, interacts with the fusion protein His-ZDP bound to a nickel-Sepharose column. These results indicate that XRCC1 interacts both with the DNA glycosylase/lyase that incises DNA after removing the 5-meC residue and with the DNA 3′-phosphatase that performs cleaning of the ensuing 3′-blocking group.
FIGURE 1.
XRCC1 interacts with ROS1 and ZDP. A and C, purified full-length ROS1 or truncated ROS1 polypeptides fused to a His tag were fixed to a nickel-Sepharose column. The proteins bound to the column were incubated in the presence of either MBP-XRCC1 or MBP alone. After washes, the proteins associated to the resin were separated by SDS-PAGE, transferred to a membrane, and immunoblotted with antibodies against MBP. B, MBP either alone or fused to XRCC1 (MBP-XRCC1) was expressed in E. coli and fixed to an amylose resin. His-tagged ROS1 purified from E. coli (His-ROS1) was incubated with either MBP-XRCC1 or MBP bound to the resin. After washes, the proteins associated to the resin were separated by SDS-PAGE, transferred to a membrane, and immunoblotted with antibodies against His6 tag. D, schematic diagrams of full-length ROS1 and the different truncated ROS1 derivatives used in C: lysine-rich domain (yellow), DNA glycosylase domain (blue), and C-terminal domain (cyan). E, purified ZDP fused to His6 tag was fixed to a nickel-Sepharose column and incubated in the presence of either MBP-XRCC1 or MBP alone. After washes, the proteins associated to the resin were separated by SDS-PAGE, transferred to a membrane, and immunoblotted with antibodies against MBP.
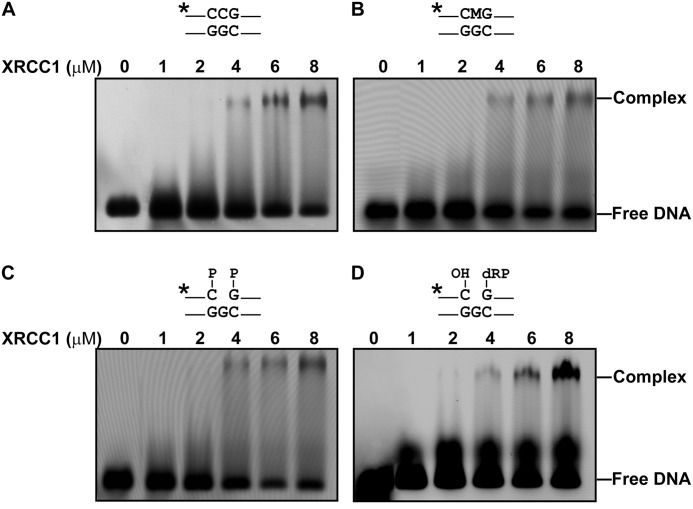
We also performed EMSA analysis to examine the capability of Arabidopsis XRCC1 to bind methylated DNA, as well as the gapped DNA repair intermediates generated during the reaction catalyzed by ROS1 (Fig. 2). We found that XRCC1 formed a complex with a 51-bp double-stranded DNA probe that contained a single 5-meC:G pair, but also bound with similar efficiency to an equivalent unmethylated probe or to a DNA containing a single-nucleotide gap flanked by 3′-phosphate and 5′-phosphate ends (Fig. 2, A–C). Analogous results were obtained when comparing XRCC1 binding to a DNA probe containing an incised abasic site (Fig. 2D). These results suggest that Arabidopsis XRCC1 has the capacity to bind a variety of DNA forms mimicking substrates, intermediates, and products of the demethylation process.
FIGURE 2.
Binding of Arabidopsis XRCC1 to DNA. A–D, increasing concentrations of XRCC1 (0, 1, 2, 4, 6, and 8 μm) were incubated with a fluorescently labeled (*) homoduplex DNA (A) or with an analogous duplex containing a single 5-meC:G pair (B), a single-nucleotide gap flanked by 3′-phosphate and 5′-phosphate termini (both indicated by P) (C), or a single-nucleotide gap flanked by 3′-OH and 5′-deoxyribose-5-phosphate (dRP) termini (D). After nondenaturing gel electrophoresis, protein-DNA complexes were identified by their retarded mobility when compared with that of free DNA, as indicated.
XRCC1 Stimulates the DNA Phosphatase Activity of ZDP
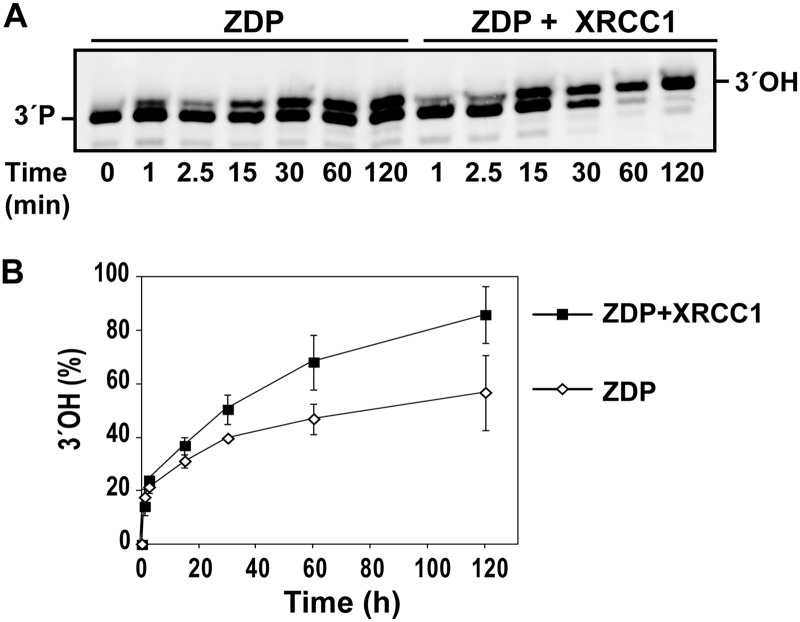
Because human XRCC1 stimulates the enzymatic activity of polynucleotide kinase (23), we asked whether plant XRCC1 may exert any effect on the activity of ZDP, the plant homolog of polynucleotide kinase. We incubated purified ZDP with a 5′-end-labeled single-nucleotide gapped substrate (Table 1), both in the absence and in the presence of XRCC1 (Fig. 3). We found that ZDP catalyzed the conversion of the 3′-phosphate end into the corresponding 3′-hydroxyl species in a time-dependent manner. As shown in Fig. 3, the DNA phosphatase activity was significantly increased in the presence of XRCC1. In the absence of ZDP, the XRCC1 protein showed no detectable activity on the gapped substrate (data not shown), confirming that the enhanced activity reflects stimulation of ZDP.
FIGURE 3.
XRCC1 stimulates the DNA phosphatase activity of ZDP. A, purified ZDP (0.09 nm) was incubated with a double-stranded oligonucleotide substrate (20 nm) containing a single-nucleotide gap flanked by 3′-phosphate (3′-P) and 5′-phosphate ends, in the absence or the presence of purified XRCC1 (0.9 nm). Reactions were stopped at the indicated times, and products were separated in a 15% denaturing polyacrylamide gel. B, conversion of 3′-phosphate termini to 3′-OH termini either in the absence (white diamonds) or in the presence (black squares) of XRCC1 was quantified by fluorescence scanning. Values are mean ± S.E. from two independent experiments.
XRCC1 Stimulates the DNA Glycosylase/Lyase Activity of ROS1
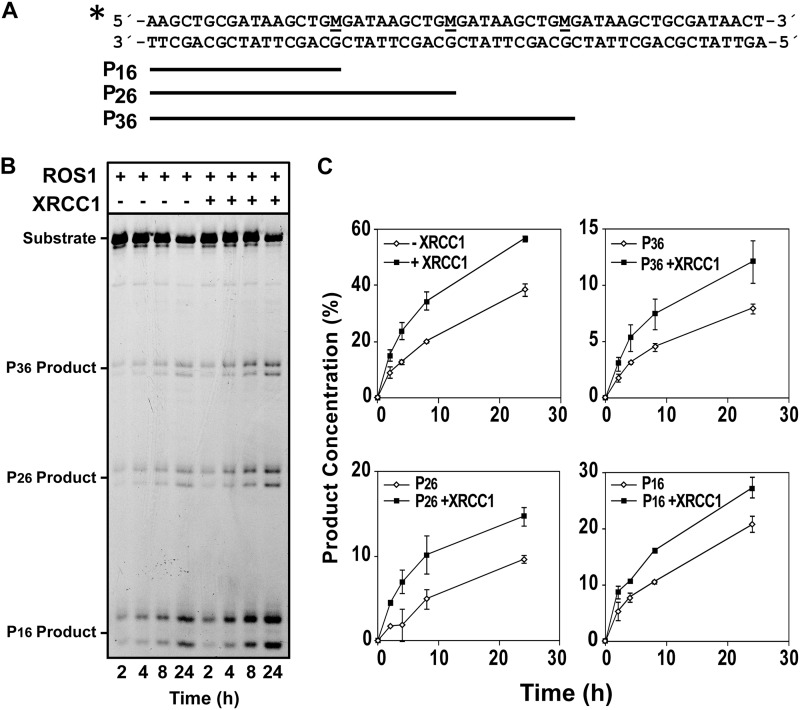
We also asked whether XRCC1 may modulate the DNA glycosylase activity of ROS1. Specifically, we decided to examine whether XRCC1 may exert any effect on the highly distributive behavior of ROS1, which does not exhibit significant processivity in vitro, due to strong binding to its reaction product (33, 34). We incubated ROS1, either in the absence or in the presence of XRCC1, with a double-stranded oligonucleotide substrate containing three 5-meC residues in the upper strand separated by 9 nt and located in the same sequence context (Fig. 4A). In this assay, a processive mechanism would rapidly convert the substrate to a final reaction product represented by a 16-nt labeled fragment, whereas a distributive mechanism would lead to the accumulation of partially processed reaction intermediates represented by 26- and 36-nt labeled fragments. Consistent with our previously reported observations (33), we found that ROS1 processed 5-meC in a highly distributive fashion, exhibiting a steady accumulation of the 16-nt fragment but also of significant amounts of partially processed substrates, even after long incubation times. As observed in Fig. 4, B and C, the addition of XRCC1 significantly increased the amount of the final reaction product (P16), but also augmented the accumulation of reaction intermediates (P26 and P36), which indicates that the low processivity of ROS1 is not significantly improved. These results suggest that the positive effect of XRCC1 on ROS1 is most likely not due to a displacement of the DNA glycosylase from its reaction product.
FIGURE 4.
XRCC1 stimulates ROS1 enzymatic activity. A, structure and length of the DNA substrates and products. M indicates 5-meC. B, purified ROS1 (18 nm) was incubated with a double-stranded oligonucleotide substrate (40 nm) containing three 5-meC:G pairs in the absence or presence of purified XRCC1 (18 nm). Reactions were stopped at the indicated times, and products were separated in a 12% denaturing polyacrylamide gel. C, product concentration either in the absence (diamonds) or in the presence (squares) of XRCC1 was quantified by fluorescence scanning. Values are mean ± S.E. from two independent experiments.
Extracts from Arabidopsis xrcc1−/− Mutants Exhibit a Reduced Capability to Complete DNA Demethylation
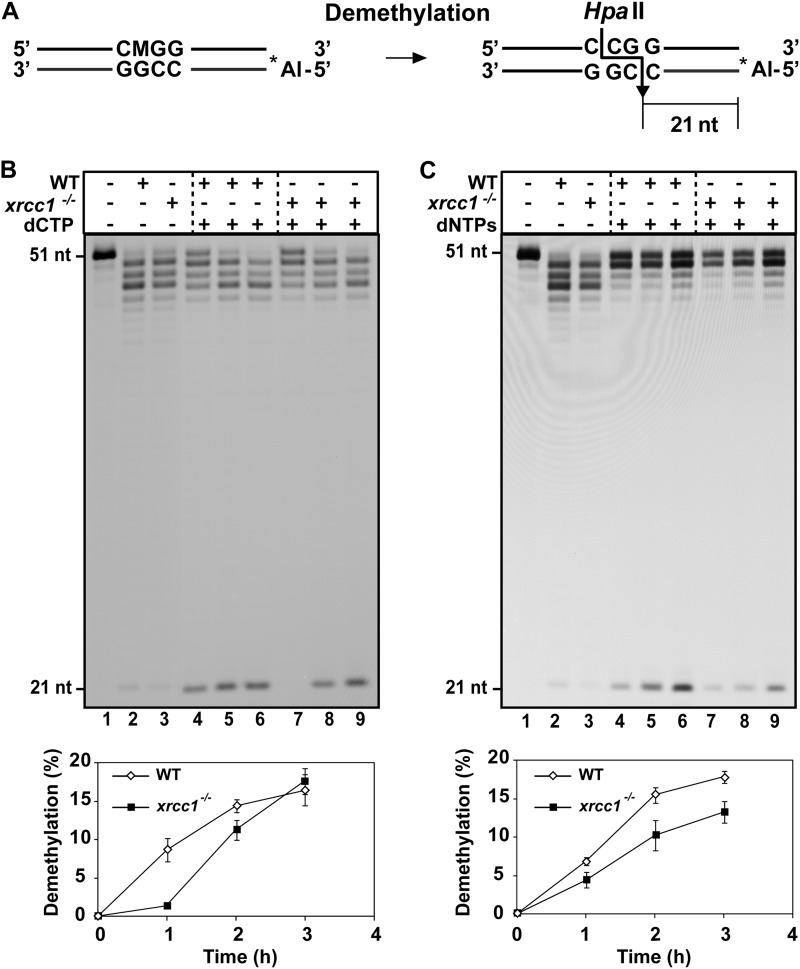
To further examine the possible role of XRCC1 in active DNA demethylation, we prepared cell extracts from an Arabidopsis xrcc1 mutant containing a T-DNA insertion in intron 5 (30) and analyzed their capacity to complete the DNA methylation process initiated by ROS1 (Fig. 5). Because ROS1 activity is too low to be detectable in Arabidopsis cell extracts,4 we preincubated purified recombinant ROS1 with a duplex oligonucleotide substrate that contained a 5-meC residue. ROS1 generated a mixture of β- and β,δ-elimination products that were purified and incubated with cell-free extracts from WT and xrcc1 plants. Incubations were performed in the presence of dCTP (Fig. 5B) or all four dNTPs (Fig. 5C), to distinguish demethylation completed via single-nucleotide insertion (SP-BER) or by long patch DNA synthesis (LP-BER) (32). In this assay, demethylation is detected by digestion with HpaII (Fig. 5A) so that fully demethylated products are visualized by the emergence of a 21-nt labeled fragment following denaturing PAGE (16, 32).
FIGURE 5.
Cell extracts from xrcc1−/− mutants show a reduced capacity to complete DNA demethylation. A, schematic diagram of molecules used as DNA substrates in the demethylation assay. Double-stranded oligonucleotides contained a 5-meC (indicated as M) at an HpaII site on the upper strand. The Alexa Fluor-labeled 5′-end is denoted by an asterisk. The size of the 5′-end-labeled fragment generated after HpaII digestion of the fully demethylated products is indicated. B, purified ROS1 (70 nm) was incubated at 30 °C for 8 h with a double-stranded oligonucleotide substrate (20 nm). Reaction products were purified and incubated with 35 μg of cell-free extract from WT and xrcc1−/− mutant plants at 30 °C in a reaction mixture that contained either dCTP (B) or all four dNTPs (C). Reactions were stopped at different times (1, 2, and 3 h), and products were separated in a 12% denaturing polyacrylamide gel and detected by fluorescence scanning. The lower panels show the percentage of DNA demethylation in reactions with WT (white diamonds) or xrcc1−/− extracts (black squares). Values are mean ± S.E. from two independent experiments.
Consistent with our previously reported observations (16), we found that WT Arabidopsis extracts are capable of completing DNA demethylation both in the presence of dCTP (Fig. 5B) and in the presence of all four dNTPs (Fig. 5C), thus suggesting that active DNA demethylation may proceed via both SP and LP-BER. However, the levels of fully demethylated products in reactions with xrcc1 extracts were significantly reduced when compared with those with WT plants (Fig. 5, B and C). The diminished demethylation capacity of xrcc1 extracts was noticeable after 2- and 3-h incubation times in LP-BER conditions, whereas it was only detected at shorter incubation times in SP-BER conditions (Fig. 5, B and C). In parallel control reactions with a DNA substrate containing an AP site, we found that the levels of incision activity were not significantly different between WT and xrcc1 extracts (supplemental Fig. S1). Altogether, these results indicate that xrcc1 extracts show a reduced capacity to complete DNA demethylation.
Anti-OsXRCC1 Inhibits 3′-End Cleaning and Nick Ligation of DNA Demethylation Intermediates
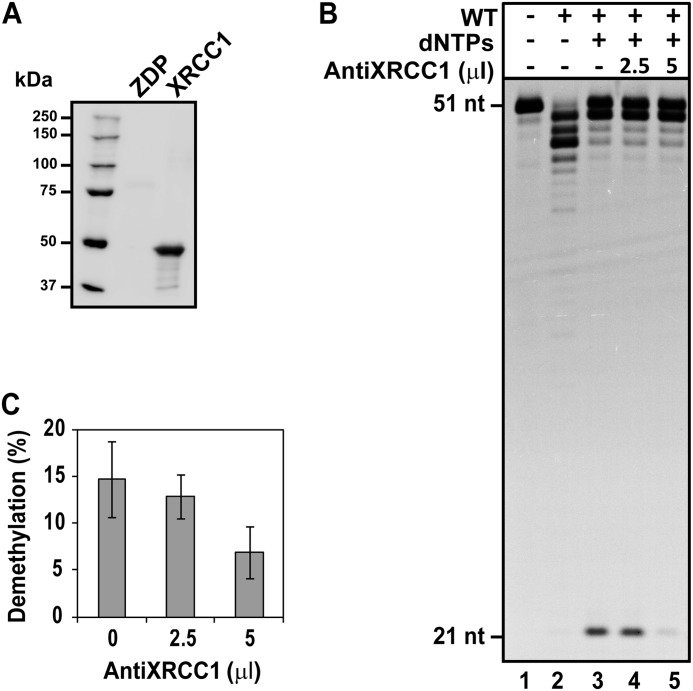
We looked for additional evidence of XRCC1 function during demethylation by using antiserum generated against recombinant rice XRCC1 (anti-OsXRCC1) (29) (Fig. 6). Immunoblotting analysis confirmed that anti-OsXRCC1 specifically recognizes XRCC1 (Fig. 6A). We found that when reactions were performed in the presence of increasing amounts of anti-OsXRCC1, the capacity of WT cell extracts to complete DNA demethylation was inhibited (Fig. 6, B and C). In contrast, the antibody did not affect the reduced DNA demethylation activity observed in xrcc1−/− extracts (supplemental Fig. S2). These results suggest that DNA demethylation is dependent on XRCC1.
FIGURE 6.
Anti-XRCC1 serum inhibits completion of DNA demethylation in cell extracts. A, detection of recombinant Arabidopsis XRCC1 protein (0.5 μg) by Western blotting using serum against OsXRCC1. Recombinant ZDP protein (0.5 μg) was used as a control. B, purified ROS1 (70 nm) was incubated at 30 °C for 8 h with a double-stranded oligonucleotide substrate (20 nm). Reaction products were purified and incubated with 35 μg of cell-free extract from WT plants at 30 °C for 3 h in a reaction mixture that contained increasing amounts of anti-OsXRCC1 serum (0, 2.5, and 5 μl) and either no dNTPs or all four dNTPs, as indicated. Reactions were stopped, and products were digested with HpaII and separated in a 12% denaturing polyacrylamide gel for detection with fluorescence scanning. C, the percentage of fully demethylated products. Values are mean ± S.E. from two independent experiments.
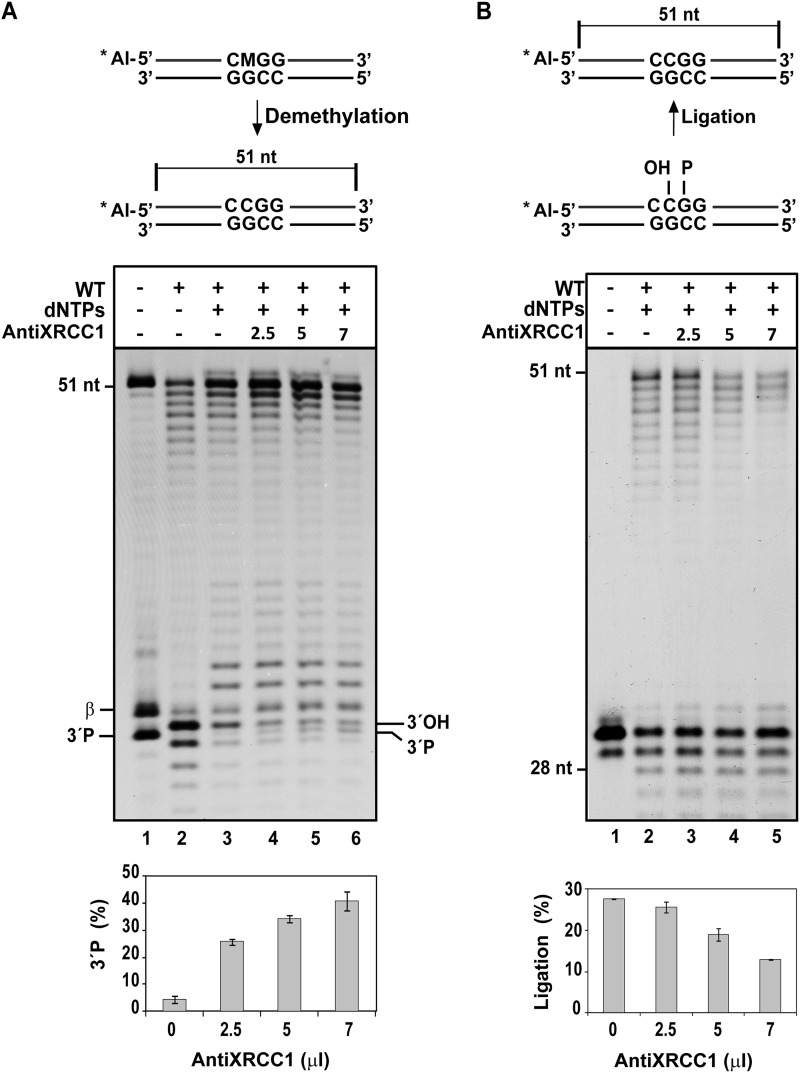
To identify which DNA demethylation steps are dependent on XRCC1, we performed reactions with a DNA substrate labeled at the 5′-end of the upper strand (Fig. 7A). As expected, the incision performed by the DNA glycosylase/lyase activity of ROS1 generated a mixture of β- and β,δ-elimination products, which contained either an unsaturated aldehyde or a phosphate group at their 3′-ends, respectively (Fig. 7A, lane 1). Consistent with our previously reported observations (16), we found that WT Arabidopsis extracts efficiently removed both the unsaturated aldehyde and the 3′-phosphate from the 3′ terminus of the gap to generate free 3′-OH ends (Fig. 7A, lane 2), plus smaller fragments probably arising by exonucleolytic degradation. When deoxynucleotides were added to the reaction, the 3′-OH ends were used for gap filling, allowing insertion of up to 3 nt (Fig. 7A, lane 3). However, the addition of increasing amounts of anti-OsXRCC1 led to the accumulation of unprocessed 3′-phosphate ends (Fig. 7A, lanes 4–6 and lower panel). Such accumulation was not observed when a control unrelated antibody (anti-Con7, raised in rabbits against a transcription factor of the fungus Fusarium oxysporum) was added to the reaction mixture (supplemental Fig. S3A). Because ZDP is the only detectable 3′-DNA phosphatase in Arabidopsis cell extracts (16), these results strongly suggest that ZDP function during DNA demethylation is partially dependent on XRCC1. Furthermore, they support the idea that ZDP and XRCC1 interact with each other during processing of 3′-blocked ends.
FIGURE 7.
Anti-OsXRCC1 serum inhibits 3′-phosphate end processing and ligation of DNA demethylation intermediates catalyzed by cell extracts. A, analysis of DNA incision products generated on the upper strand during DNA demethylation. Purified ROS1 (70 nm) was incubated at 30 °C for 8 h with a 51-mer double-stranded oligonucleotide substrate (20 nm) containing a single 5-meC:G pair. Reaction products, which contained a mixture of β- and β,δ-elimination products (lane 1), were purified and incubated with 35 μg of cell-free extract from WT plants at 30 °C for 3 h in a reaction mixture that contained increasing amounts of anti-OsXRCC1 serum (0, 2.5, 5, and 7 μl) and all four dNTPs, except when indicated. Reactions were stopped, and products were separated in a 12% denaturing polyacrylamide gel and detected by fluorescence scanning. The lower panel shows the percentage of 3′-phosphate (3′-P) not converted into 3′-OH in two independent experiments. B, DNA ligation assay. DNA duplexes containing a nick in the upper, 5′-Alexa Fluor-labeled strand were incubated with 35 μg of extract from WT plants at 30 °C for 3 h in a reaction mixture supplemented with increasing amounts of anti-OsXRCC1 serum (0, 2.5, 5, and 7 μl) and all four dNTPs, except when indicated. Reaction products were separated using a 12% denaturing polyacrylamide gel and detected by fluorescence scanning. The lower panel shows the percentage of fully ligated products. Values are mean ± S.E. from two independent experiments.
In addition, we tested whether XRCC1 may have also a role during the ligation step of the DNA demethylation process. We incubated WT cell extracts with a substrate mimicking a nicked DNA intermediate and found that increasing amounts of anti-OsXRCC1 strongly inhibited the levels of ligated products (Fig. 7B). Such inhibition was not observed when the control unrelated antibody anti-Con7 was added to the reaction mixture (supplemental Fig. S3B). These results indicate that XRCC1 modulates the capacity to seal DNA nicks in Arabidopsis extracts and suggest that the protein may also play a role during the ligation step of DNA demethylation.
DISCUSSION
Active DNA demethylation in Arabidopsis is initiated by DNA glycosylases of the ROS1/DME family and presumably continued by proteins participating in the plant BER pathway. In mammalian cells, XRCC1 performs a central role by coordinating the activities of several enzymes functioning in both single-strand break repair and BER. In this work, we have aimed to determine the biochemical role of the plant XRCC1 homolog during active DNA demethylation in Arabidopsis. Our results suggest that plant XRCC1 functions both at the initial stages of demethylation, by stimulating ROS1 activity, and also at post-excision steps, by favoring 3′-end cleaning and DNA ligation.
Similarly to its mammalian counterpart, Arabidopsis XRCC1 might function as a scaffold protein that helps to coordinate the several stages of the base excision process. However, there may be significant differences. Mammalian XRCC1 binds undamaged DNA (35), but specifically interacts with gapped or nicked DNA through its N-terminal domain (35–37), which is not conserved between animal and plant homologs (28). In contrast, Arabidopsis XRCC1 binds DNA with low affinity and exhibits no discrimination for DNA demethylation intermediates. Our results argue that it is likely that the interaction with ROS1 and ZDP increases the affinity of XRCC1 for the target sites where demethylation is taking place. Thus, XRCC1 might be recruited to the DNA by its interaction with ROS1 and might later on be maintained at the target site both by its ability to interact with DNA in a nonspecific way and through specific interactions with both ROS1 and ZDP.
It has been previously reported that mammalian XRCC1 interacts with several DNA glycosylases and stimulates the activity of at least three of them, hOGG1, MPG, and hNTH1, although the underlying mechanism is unknown (21, 22). We have found that the processivity of ROS1 is not significantly increased in the presence of XRCC1, thus suggesting that the stimulating effect is not due to facilitated product release and increased turnover. It is therefore unclear how the interaction with XRCC1 leads to a higher ROS1 activity. ROS1 binds nonspecifically to DNA through its N-terminal domain, and we have recently shown that such nonspecific binding provides the protein with the capacity to slide along DNA in the search for 5-meC (34, 38). Given the DNA binding capacity of XRCC1, one possibility is that it helps to stabilize a productive complex between ROS1 and DNA once the target base has been located. However, our attempts to detect a putative ROS1-XRCC1-DNA ternary complex have been unsuccessful. Therefore, the exact nature of the mechanism underlying the stimulating effect of XRCC1 on ROS1 activity remains to be determined.
XRCC1 not only modulates the activity of ROS1, but also may help to recruit proteins that function downstream in the demethylation pathway, such as ZDP. We have recently reported that ROS1 and ZDP interact in vitro and co-localize in vivo in nucleoplasmic foci (16). We report here that XRCC1 interacts with ZDP and stimulates its DNA phosphatase activity in vitro. Furthermore, the activity of ZDP in Arabidopsis extracts becomes rate-limiting when an anti-XRCC1 antibody is present in the reaction mixture. Altogether, our results suggest that the interaction of ZDP with ROS1 is required for optimal processing of blocked 3′-ends during active demethylation in Arabidopsis. Such a positive effect is similar to the stimulation of polynucleotide kinase by interaction with mammalian XRCC1 (23). This interaction is facilitated through phosphorylation of XRCC1 by casein kinase 2 (CK2) at a segment that binds specifically to the Forkhead-associated (FHA) domain of polynucleotide kinase (39, 40). Although Arabidopsis XRCC1 partially conserves this segment and is also phosphorylated in vitro by CK2 (39), its interaction partner ZDP lacks an identifiable Forkhead-associated domain. Therefore, it is unclear whether phosphorylation plays a role in the interaction between Arabidopsis XRCC1 and ZDP.
Our results indicate that XRCC1 is also required to reach high levels of strand rejoining by DNA ligation. In mammalian BER, XRCC1 exerts a positive effect on ligation by interaction with DNA ligase III isoform α (Lig3α) (26, 41). The interaction between XRCC1 and Lig3α is mediated by their C termini, each of which contains a BRCT domain (41–43). In plants, there is no DNA ligase III homolog, and XRCC1 only possesses the BRCT1 domain and lacks the C-terminal BRCT2 domain (28, 29). It is therefore tempting to speculate that XRCC1 facilitates DNA ligation in Arabidopsis extracts through interaction with DNA ligase I (LIG1). LIG1 is an ortholog of mammalian Lig1 (44, 45). Disruption of the LIG1 gene is lethal in Arabidopsis (46), and RNAi lines with reduced levels of LIG1 display severe growth defects and show slower repair of both single-strand and double-strand DNA breaks (47). We have recently reported that LIG1 is essential for BER of uracil and abasic sites in Arabidopsis (48), and there is genetic evidence that LIG1 acts downstream of DME during maternal allele demethylation and gene imprinting in the endosperm (49). LIG1 appears to supply the major, perhaps the only, DNA ligase activity to seal DNA nicks in plant cell extracts, and other DNA ligases cannot substitute for its nick-closing functions (48). It is therefore likely that most BER events in Arabidopsis cells are completed by LIG1.
XRCC1 deficiency apparently has more drastic consequences in mammals than in plants. XRCC1 knock-out mice show increased DNA breakage and apoptosis before death at embryonic day 6.5 (18, 50). In contrast, Arabidopsis xrcc1 mutants develop normally and are fully fertile, although they show hypersensitivity to γ rays (30). There is evidence that plants tolerate genome maintenance deficiencies that trigger lethal apoptotic responses in mammals (51). However, an alternative explanation is that xrcc1 plant mutants may express a protein retaining some functionality. Although the T-DNA insertion in xrcc1 plants prevents production of a full-length WT transcript (30), we cannot exclude the possibility that mutants express a C-terminal truncated protein retaining the BRCT1 domain. We tried to test this hypothesis by immunoblotting, but the levels of AtXRCC1 are too low to be detectable in cell extracts, at least with the anti-OsXRCC1 antibody. In any case, the generation of XRCC1 null alleles or RNAi-silenced lines will facilitate a full understanding of the physiological consequences of XRCC1 deficiency in plants.
In summary, our data indicate a biochemical role for XRCC1 during BER-mediated DNA demethylation in plants, during both excision and post-excision steps. These results suggest that the central role of XRCC1 in coordinating DNA repair pathways involving single-strand breaks, including DNA demethylation, is conserved in both animals and plants.
Supplementary Material
Acknowledgments
We thank Dr. Charles I. White (CNRS-University-INSERM unit at Université Blaise Pascal, France) for the kind gift of xrcc1−/− seeds. We are also very grateful to Dr. Kengo Sakaguchi (Tokyo University of Science, Japan) for generously providing anti-OsXRCC1 serum and to Dr. Carmen Ruiz-Roldán (University of Córdoba, Spain) for kindly providing anti-Con7 serum. We thank members of our laboratory for helpful discussions and advice.
This work was supported by the Spanish Ministry of Economy and Competitiveness and the European Regional Development Fund (Grant BFU2010-18838) and by the Junta de Andalucía (Grant P07-CVI-02770).

This article contains supplemental Figs. S1–S3.
M. I. Martínez-Macías, D. Córdoba-Cañero, R. R. Ariza, and T. Roldán-Arjona, unpublished observations.
- 5-meC
- cytosine methylation
- ZDP
- zinc finger DNA 3′-phosphoesterase
- BER
- base excision repair
- SP-BER
- single-nucleotide insertion BER
- LP-BER
- long patch DNA synthesis BER
- ROS1
- repressor of silencing 1
- XRCC1
- x-ray cross-complementing group protein 1
- BRCT
- BRCA1 C-terminal
- MBP
- maltose-binding protein
- nt
- nucleotide(s)
- h
- human
- OsXRCC1
- Oryza sativa XRCC1
- LIG1
- DNA ligase I.
REFERENCES
- 1. Law J. A., Jacobsen S. E. (2010) Establishing, maintaining, and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roldan-Arjona T., Ariza R. R. (2009) DNA demethylation. in DNA and RNA Modification Enzymes: Comparative Structure, Mechanism, Functions, Cellular Interactions and Evolution (Grosjean H., ed) pp. 149–161, Landes Bioscience, Austin, TX [Google Scholar]
- 3. Wu S. C., Zhang Y. (2010) Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 11, 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhu J. K. (2009) Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 43, 143–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gehring M., Reik W., Henikoff S. (2009) DNA demethylation by DNA repair. Trends Genet. 25, 82–90 [DOI] [PubMed] [Google Scholar]
- 6. Bhutani N., Burns D. M., Blau H. M. (2011) DNA demethylation dynamics. Cell 146, 866–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gong Z., Zhu J. K. (2011) Active DNA demethylation by oxidation and repair. Cell Res. 21, 1649–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gong Z., Morales-Ruiz T., Ariza R. R., Roldán-Arjona T., David L., Zhu J. K. (2002) ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111, 803–814 [DOI] [PubMed] [Google Scholar]
- 9. Choi Y., Gehring M., Johnson L., Hannon M., Harada J. J., Goldberg R. B., Jacobsen S. E., Fischer R. L. (2002) DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110, 33–42 [DOI] [PubMed] [Google Scholar]
- 10. Penterman J., Zilberman D., Huh J. H., Ballinger T., Henikoff S., Fischer R. L. (2007) DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. U.S.A. 104, 6752–6757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ortega-Galisteo A. P., Morales-Ruiz T., Ariza R. R., Roldán-Arjona T. (2008) Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 67, 671–681 [DOI] [PubMed] [Google Scholar]
- 12. Gehring M., Huh J. H., Hsieh T. F., Penterman J., Choi Y., Harada J. J., Goldberg R. B., Fischer R. L. (2006) DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124, 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu J., Kapoor A., Sridhar V. V., Agius F., Zhu J. K. (2007) The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr. Biol. 17, 54–59 [DOI] [PubMed] [Google Scholar]
- 14. Morales-Ruiz T., Ortega-Galisteo A. P., Ponferrada-Marín M. I., Martínez-Macías M. I., Ariza R. R., Roldán-Arjona T. (2006) DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc. Natl. Acad. Sci. U.S.A. 103, 6853–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Agius F., Kapoor A., Zhu J. K. (2006) Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc. Natl. Acad. Sci. U.S.A. 103, 11796–11801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Martínez-Macías M. I., Qian W., Miki D., Pontes O., Liu Y., Tang K., Liu R., Morales-Ruiz T., Ariza R. R., Roldán-Arjona T., Zhu J. K. (2012) A DNA 3′ phosphatase functions in active DNA demethylation in Arabidopsis. Mol. Cell 45, 357–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Caldecott K. W. (2003) XRCC1 and DNA strand break repair. DNA Repair 2, 955–969 [DOI] [PubMed] [Google Scholar]
- 18. Thompson L. H., West M. G. (2000) XRCC1 keeps DNA from getting stranded. Mutat. Res. 459, 1–18 [DOI] [PubMed] [Google Scholar]
- 19. Wiederhold L., Leppard J. B., Kedar P., Karimi-Busheri F., Rasouli-Nia A., Weinfeld M., Tomkinson A. E., Izumi T., Prasad R., Wilson S. H., Mitra S., Hazra T. K. (2004) AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell 15, 209–220 [DOI] [PubMed] [Google Scholar]
- 20. Akbari M., Solvang-Garten K., Hanssen-Bauer A., Lieske N. V., Pettersen H. S., Pettersen G. K., Wilson D. M., 3rd, Krokan H. E., Otterlei M. (2010) Direct interaction between XRCC1 and UNG2 facilitates rapid repair of uracil in DNA by XRCC1 complexes. DNA Repair 9, 785–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marsin S., Vidal A. E., Sossou M., Ménissier-de Murcia J., Le Page F., Boiteux S., de Murcia G., Radicella J. P. (2003) Role of XRCC1 in the coordination and stimulation of oxidative DNA damage repair initiated by the DNA glycosylase hOGG1. J. Biol. Chem. 278, 44068–44074 [DOI] [PubMed] [Google Scholar]
- 22. Campalans A., Marsin S., Nakabeppu Y., O'connor T. R., Boiteux S., Radicella J. P. (2005) XRCC1 interactions with multiple DNA glycosylases: a model for its recruitment to base excision repair. DNA Repair 4, 826–835 [DOI] [PubMed] [Google Scholar]
- 23. Whitehouse C. J., Taylor R. M., Thistlethwaite A., Zhang H., Karimi-Busheri F., Lasko D. D., Weinfeld M., Caldecott K. W. (2001) XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell 104, 107–117 [DOI] [PubMed] [Google Scholar]
- 24. Vidal A. E., Boiteux S., Hickson I. D., Radicella J. P. (2001) XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J. 20, 6530–6539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kubota Y., Nash R. A., Klungland A., Schär P., Barnes D. E., Lindahl T. (1996) Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase β and the XRCC1 protein. EMBO J. 15, 6662–6670 [PMC free article] [PubMed] [Google Scholar]
- 26. Caldecott K. W., McKeown C. K., Tucker J. D., Ljungquist S., Thompson L. H. (1994) An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell Biol. 14, 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hajkova P., Jeffries S. J., Lee C., Miller N., Jackson S. P., Surani M. A. (2010) Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 329, 78–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Taylor R. M., Thistlethwaite A., Caldecott K. W. (2002) Central role for the XRCC1 BRCT I domain in mammalian DNA single-strand break repair. Mol. Cell Biol. 22, 2556–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Uchiyama Y., Suzuki Y., Sakaguchi K. (2008) Characterization of plant XRCC1 and its interaction with proliferating cell nuclear antigen. Planta 227, 1233–1241 [DOI] [PubMed] [Google Scholar]
- 30. Charbonnel C., Gallego M. E., White C. I. (2010) Xrcc1-dependent and Ku-dependent DNA double-strand break repair kinetics in Arabidopsis plants. Plant J. 64, 280–290 [DOI] [PubMed] [Google Scholar]
- 31. Córdoba-Cañero D., Roldán-Arjona T., Ariza R. R. (2012) Using Arabidopsis cell extracts to monitor repair of DNA base damage in vitro. Methods Mol. Biol. 920, 263–277 [DOI] [PubMed] [Google Scholar]
- 32. Córdoba-Cañero D., Morales-Ruiz T., Roldán-Arjona T., Ariza R. R. (2009) Single-nucleotide and long-patch base excision repair of DNA damage in plants. Plant J. 60, 716–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ponferrada-Marín M. I., Roldán-Arjona T., Ariza R. R. (2009) ROS1 5-methylcytosine DNA glycosylase is a slow-turnover catalyst that initiates DNA demethylation in a distributive fashion. Nucleic Acids Res. 37, 4264–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ponferrada-Marín M. I., Martínez-Macías M. I., Morales-Ruiz T., Roldán-Arjona T., Ariza R. R. (2010) Methylation-independent DNA binding modulates specificity of repressor of silencing 1 (ROS1) and facilitates demethylation in long substrates. J. Biol. Chem. 285, 23032–23039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mani R. S., Karimi-Busheri F., Fanta M., Caldecott K. W., Cass C. E., Weinfeld M. (2004) Biophysical characterization of human XRCC1 and its binding to damaged and undamaged DNA. Biochemistry 43, 16505–16514 [DOI] [PubMed] [Google Scholar]
- 36. Marintchev A., Mullen M. A., Maciejewski M. W., Pan B., Gryk M. R., Mullen G. P. (1999) Solution structure of the single-strand break repair protein XRCC1 N-terminal domain. Nat. Struct. Biol. 6, 884–893 [DOI] [PubMed] [Google Scholar]
- 37. Nazarkina Z. K., Khodyreva S. N., Marsin S., Lavrik O. I., Radicella J. P. (2007) XRCC1 interactions with base excision repair DNA intermediates. DNA Repair 6, 254–264 [DOI] [PubMed] [Google Scholar]
- 38. Ponferrada-Marín M. I., Roldán-Arjona T., Ariza R. R. (2012) Demethylation initiated by ROS1 glycosylase involves random sliding along DNA. Nucleic Acids Res. 40, 11554–11562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Loizou J. I., El-Khamisy S. F., Zlatanou A., Moore D. J., Chan D. W., Qin J., Sarno S., Meggio F., Pinna L. A., Caldecott K. W. (2004) The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell 117, 17–28 [DOI] [PubMed] [Google Scholar]
- 40. Ali A. A., Jukes R. M., Pearl L. H., Oliver A. W. (2009) Specific recognition of a multiply phosphorylated motif in the DNA repair scaffold XRCC1 by the FHA domain of human PNK. Nucleic Acids Res. 37, 1701–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nash R. A., Caldecott K. W., Barnes D. E., Lindahl T. (1997) XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry 36, 5207–5211 [DOI] [PubMed] [Google Scholar]
- 42. Wei Y. F., Robins P., Carter K., Caldecott K., Pappin D. J., Yu G. L., Wang R. P., Shell B. K., Nash R. A., Schär P. (1995) Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA ligase III, an enzyme active in DNA repair and recombination. Mol. Cell Biol. 15, 3206–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mackey Z. B., Ramos W., Levin D. S., Walter C. A., McCarrey J. R., Tomkinson A. E. (1997) An alternative splicing event which occurs in mouse pachytene spermatocytes generates a form of DNA ligase III with distinct biochemical properties that may function in meiotic recombination. Mol. Cell Biol. 17, 989–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Taylor R. M., Hamer M. J., Rosamond J., Bray C. M. (1998) Molecular cloning and functional analysis of the Arabidopsis thaliana DNA ligase I homologue. Plant J. 14, 75–81 [DOI] [PubMed] [Google Scholar]
- 45. Wu Y. Q., Hohn B., Ziemienowic A. (2001) Characterization of an ATP-dependent type I DNA ligase from Arabidopsis thaliana. Plant Mol. Biol. 46, 161–170 [DOI] [PubMed] [Google Scholar]
- 46. Babiychuk E., Cottrill P. B., Storozhenko S., Fuangthong M., Chen Y., O'Farrell M. K., Van Montagu M., Inzé D., Kushnir S. (1998) Higher plants possess two structurally different poly(ADP-ribose) polymerases. Plant J. 15, 635–645 [DOI] [PubMed] [Google Scholar]
- 47. Waterworth W. M., Kozak J., Provost C. M., Bray C. M., Angelis K. J., West C. E. (2009) DNA ligase 1 deficient plants display severe growth defects and delayed repair of both DNA single and double strand breaks. BMC Plant Biol. 9, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Córdoba-Cañero D., Roldán-Arjona T., Ariza R. R. (2011) Arabidopsis ARP endonuclease functions in a branched base excision DNA repair pathway completed by LIG1. Plant J. 68, 693–702 [DOI] [PubMed] [Google Scholar]
- 49. Andreuzza S., Li J., Guitton A. E., Faure J. E., Casanova S., Park J. S., Choi Y., Chen Z., Berger F. (2010) DNA LIGASE I exerts a maternal effect on seed development in Arabidopsis thaliana. Development 137, 73–81 [DOI] [PubMed] [Google Scholar]
- 50. Tebbs R. S., Flannery M. L., Meneses J. J., Hartmann A., Tucker J. D., Thompson L. H., Cleaver J. E., Pedersen R. A. (1999) Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev. Biol. 208, 513–529 [DOI] [PubMed] [Google Scholar]
- 51. Hays J. B. (2002) Arabidopsis thaliana, a versatile model system for study of eukaryotic genome-maintenance functions. DNA Repair 1, 579–600 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.