Background: The functional role of the V-ATPase as a pH-sensing receptor remains unknown.
Results: N-terminal peptides from the a-subunit isoforms of the V-ATPase modulate the enzymatic GEF activity of cytohesin-2.
Conclusion: V-ATPase is a novel cytohesin-signaling receptor.
Significance: These data reveal an evolutionarily conserved signaling process between V-ATPase, cytohesin-2, and Arfs, which is crucial for understanding the cell biological functions of these proteins.
Keywords: Guanine Nucleotide Exchange Factor (GEF), Membrane Energetics, Nuclear Magnetic Resonance, Peptide Interactions, Vacuolar ATPase
Abstract
Previously, we reported an acidification-dependent interaction of the endosomal vacuolar H+-ATPase (V-ATPase) with cytohesin-2, a GDP/GTP exchange factor (GEF), suggesting that it functions as a pH-sensing receptor. Here, we have studied the molecular mechanism of signaling between the V-ATPase, cytohesin-2, and Arf GTP-binding proteins. We found that part of the N-terminal cytosolic tail of the V-ATPase a2-subunit (a2N), corresponding to its first 17 amino acids (a2N(1–17)), potently modulates the enzymatic GDP/GTP exchange activity of cytohesin-2. Moreover, this peptide strongly inhibits GEF activity via direct interaction with the Sec7 domain of cytohesin-2. The structure of a2N(1–17) and its amino acids Phe5, Met10, and Gln14 involved in interaction with Sec7 domain were determined by NMR spectroscopy analysis. In silico docking experiments revealed that part of the V-ATPase formed by its a2N(1–17) epitope competes with the switch 2 region of Arf1 and Arf6 for binding to the Sec7 domain of cytohesin-2. The amino acid sequence alignment and GEF activity studies also uncovered the conserved character of signaling between all four (a1–a4) a-subunit isoforms of mammalian V-ATPase and cytohesin-2. Moreover, the conserved character of this phenomenon was also confirmed in experiments showing binding of mammalian cytohesin-2 to the intact yeast V-ATPase holo-complex. Thus, here we have uncovered an evolutionarily conserved function of the V-ATPase as a novel cytohesin-signaling receptor.
Introduction
The vacuolar H+-ATPase (V-ATPase)6 is a multimeric complex that functions as a proton-pumping rotary nanomotor. The overall principle and structure of this nanomotor is conserved throughout evolution, which allows its universal function in proton pumping and acidification of the extracellular milieu as well as intracellular organelles in pro- and eukaryotic organisms (1, 2). The biochemistry, cell biology, and pathophysiology of V-ATPase were extensively reviewed by ourselves and others (1–4). Recently, the structure of the Saccharomyces cerevisiae eukaryotic holo-complex of intact V-ATPase was solved at 11 Å resolution (5). This structure explains intrasubunit interactions within the V-ATPase holo-complex and also allows studies of its interaction with regulatory proteins. Previously, we reported an acidification-dependent interaction of the endosomal V-ATPase with cytohesin-2 and Arf6 suggesting that it functions as a pH-sensing receptor (6–8). Cytohesins are Guanine nucleotide Exchange Factors (GEFs) for the ADP-ribosylation factor (Arf) family, which belongs to the Ras superfamily of GTP-binding proteins. Cytohesin-2 (also known as ARNO) is a member of the cytohesin family, which includes another three members as follows: cytohesin-1, cytohesin-3 (also called ARNO3 or GRP1), and cytohesin-4. These proteins share common structures, containing the following four domains: (i) an N-terminal coiled-coil; (ii) a central Sec7 domain; (iii) a pleckstrin homology domain; and (iv) a C-terminal polybasic motif region. The conserved Sec7 catalytic domain is responsible for Arf-GEF activity, which activates Arf GTP-binding proteins involved in signaling and regulation of the vesicular trafficking in both endocytic and exocytic pathways (9–13).
Both V-ATPase and cytohesins have been recently implicated as essential regulators of signaling and trafficking in the endosomal/lysosomal pathway upon activation of a broad range of plasma membrane receptors. In particular, V-ATPase itself and V-ATPase-driven acidification are required for Wnt/LRP6 (14) and Notch (15) receptor signaling, respectively. Moreover, the V-ATPase is involved in amino acid sensing, recruitment, and activation of the mammalian target of rapamycin complex on lysosomal membranes via its interaction with Rag (16). However, cytohesins have also been identified as crucial cytoplasmic activators of EGFR/ErbB family receptors that are involved in oncogenesis (17–19) and development of diabetic nephropathy (20–22). Cytohesins were also recently identified as crucial downstream effectors for the insulin-receptor signaling cascade (23, 24). These studies demonstrated that defective cytohesin signalings give rise to insulin resistance and consequent development of metabolic syndrome during diabetes (25).
Given the importance of both the V-ATPase and cytohesin to cellular signaling, we have been examining the potential relationship between the two proteins. In previous work from our laboratory, we found that the V-ATPase itself functions as a pH-sensing receptor in endosomes (6–8). In particular, it was shown that a2-subunit isoform containing V-ATPase directly interacts with cytohesin-2 and Arf family GTP-binding proteins, recruiting these proteins to early endosomes in an acidification-dependent manner. Moreover, our recent study demonstrated that cytohesin-2 directly interacts not only with the a2-subunit but with all four (a1–a4) a-subunit isoforms of the V-ATPase, indicating a widespread regulatory interaction between V-ATPase and Arf family GTP-binding proteins (26). However, the molecular mechanism and cell biological significance of this phenomenon remain obscure. During the course of this work, we identified two structural elements involved in specific and high affinity association of the V-ATPase a2-subunit isoform with cytohesin-2 as follows: (i) an N-terminal binding motif formed by the first 17 amino acids a2N(1–17) of the cytosolic N-terminal tail of a2-subunit (a2N), and (ii) an interaction pocket formed by the catalytic Sec7 and regulatory polybasic domains of cytohesin-2 (27). Biacore analysis revealed a very strong binding affinity between this a2N(1–17) peptide and the Sec7 domain of cytohesin-2, which was similar to the binding affinity between full-length cytohesin-2 and the complete a2N. We hypothesized that the a2N(1–17) peptide could be involved in the signaling and regulation of the enzymatic GEF activity of cytohesin-2 (27).
Here, we have examined the molecular mechanism of binding and signaling between the V-ATPase, cytohesin-2, and Arf GTP-binding proteins. We now report a novel function of the V-ATPase as a signaling receptor that modulates activity of cytohesin-2 and Arf GTP-binding proteins. Using a combination of NMR spectroscopy and in silico docking analysis, we identified the structural basis and molecular mechanism of this novel signaling phenomenon. Our data also suggest that the signaling between V-ATPase, cytohesin-2, and Arf family GTP-binding proteins is an evolutionarily conserved and a widespread phenomenon that may take place on a variety of membranes and organelles from yeast to mammalian cells.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
If not otherwise specified, all reagents were purchased from Sigma. All buffers, NU-PAGE gels, and Lipofectamine 2000 transfection reagent were from Invitrogen. Western Lightning chemiluminescence reagent plus was obtained from PerkinElmer Life Sciences. FluorSave reagent was from Calbiochem. Rabbit polyclonal V-ATPase a2-subunit-specific antibodies were previously described (6). Mouse monoclonal anti-GFP (B-2) and anti-His6 antibodies were purchased from Santa Cruz Biotechnology. Alexa 488-conjugated goat anti-mouse antibodies were obtained from Invitrogen. Horseradish peroxidase-conjugated sheep anti-mouse and anti-rabbit antibodies were purchased from GE Healthcare. Peptide synthesis resins and Fmoc-protected amino acids were purchased from EMD Chemicals. Dimethylformamide, N-methylpyrrolidone, dichloromethane, piperidine, 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate, 1-hydroxybenzotriazole, diisopropylethylamine, and trifluoroacetic acid were purchased from American Bioanalytical. Diisopropyl carbodiimide, phenol, thioanisole, thiophenol, ethanedithiol, 4,6-diamidino-2-phenylindole (DAPI), M2-Sepharose, and 3×FLAG peptide were from Sigma. Cytohesin inhibitor SecinH3 was a gift from Dr. Sylvain Bourgoin or from Calbiochem. A mini-extruder assembly was purchased from Avanti Polar Lipids. GTP, GDP, and GTPγS were from Sigma, and [35S]GTPγS was from PerkinElmer Life Sciences. Azolectin and phosphatidylglycerol were purchased from Sigma. Phosphatidylcholine was from Avanti Polar Lipids, and phosphatidylinositol 4, 5-bisphosphate (PIP2) was from Matreya and Cayman Chemical.
cDNA Constructs
GST-tagged cytohesin-2 (triglycine variant) and the Sec7 domain of human cytohesin-2 (61–252 amino acids) were cloned into a pGEX-6P-1 vector as described previously (26). Truncated cytohesin-2 lacking the coiled-coil domain (61–400 amino acids) (CTH2(61–400)) was cloned into a pET-28a(+) vector by restriction free cloning (28) including a His6 tag followed by a tobacco etch virus protease site. Truncated (Δ17)Arf1 and (Δ13)Arf6 proteins were cloned into a pET28b vector. cDNA encoding full-length wild-type mouse V-ATPase a2-isoform (g.i. 7363249) (29) was amplified using the PfuTurbo DNA polymerase (Stratagene) and subcloned into NheI/AgeI restriction sites of the pEGFP-N1 vector (Clontech) in-frame with the C-terminal enhanced green fluorescent protein (EGFP).
Peptides Synthesis, Labeling, and Purification
All peptides were synthesized, purified by HPLC, and analyzed by mass spectrometry in the MGH Peptide/Protein Core Facility as follows. Peptides were synthesized on an automatic peptide synthesizer (Applied Biosystems, Model 433A) by using the manufacturer's FastMoc chemistry cycles for Fmoc solid-phase synthesis (30). To render the V-ATPase-derived a2N(1–17) peptide soluble and cell penetrable, various versions of polyethylene glycol- or TAT (YGRKKRRQRRR sequence)-modified peptides were synthesized, and the final Fmoc group was removed. Peptides were further coupled at the N terminus (supplemental Table S1) with either biotin (in blue), (ii) fluorescein isothiocyanate (FITC) (in green), (iii) 5-carboxyfluorescein (in green), or 5-carboxytetramethylrhodamine (in red). The resin was then washed with dimethylformamide, dichloromethane, and methanol three times each and vacuum-dried. Peptides were cleaved from the solid support and de-protected using reagent K (TFA/phenol/thioanisole/water/ethanedithiol; 82.5:5.0:5.0:5.0:2.5 v/v) for 2.5 h at room temperature (31). Peptides were precipitated using cold methyl tertiary butyl ether. The precipitate was washed three times in methyl tertiary butyl ether, dissolved in a solvent (0.1% trifluoroacetic acid in 20% acetonitrile, 80% water) followed by freeze drying. Peptide purification was performed on a semi-preparative system (Waters Associates) using a Vydac C-18 reverse-phase column and water/acetonitrile gradient consisting of 0.1% trifluoroacetic acid giving rise to >95% purity. All purified peptides (see supplemental Table S1) were characterized by Ultra-high pressure liquid chromatography (UPLC) and matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS). FITC-TAT was also purchased from Anaspec. Peptide stock solutions were prepared in water at 5 mm concentration and stored at −20 °C.
Recombinant Protein Expression and Purification
GST-tagged human cytohesin-2 was expressed in BL21/DE3 cells (Stratagene) and purified as described previously (26). Cells were grown in LB at 37 °C. Expression of recombinant protein was induced by addition of 1 mm isopropyl β-d-thiogalactopyranoside. Cells were disrupted by sonication in 50 mm sodium phosphate buffer (pH 7.0), 300 mm NaCl, 1 mm MgCl2, 1 mm phenylmethylsulfonyl fluoride (PMSF), 0.5 mg/ml lysozyme, protease inhibitor mixture (Roche Applied Science). Lysate was centrifuged at 12,000 × g for 12 min. Supernatant was centrifuged at 20,000 × g for 60 min. Supernatant was applied to a PD10 column packed with glutathione-Sepharose 4B beads (GE Healthcare). The column was washed with 50 mm sodium phosphate buffer (pH 7.0), 300 mm NaCl. GST-tagged human cytohesin-2 was eluted with 50 mm Tris-HCl (pH 7.7) and 10 mm reduced glutathione and then further separated by SuperdexTM200 HR 10/30 pre-packed column (GE Healthcare) using an AKTA purifier system (GE Healthcare).
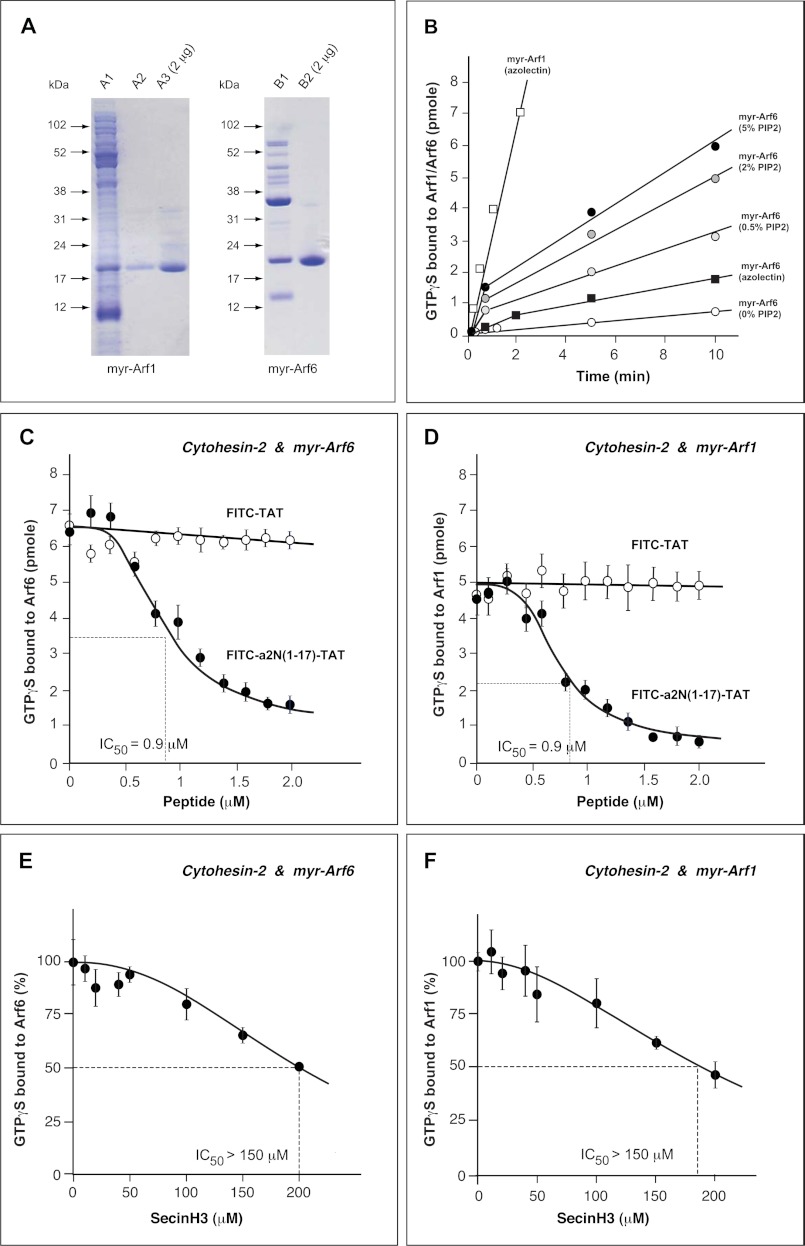
The Sec7 domain of human cytohesin-2 (61–252 amino acids) was expressed in BL21/DE3 cells and purified by sequential chromatography on TALON beads (Clontech) and glutathione-Sepharose 4B beads (GE Healthcare) as described previously (26). The GST tag was cleaved using PreScission ProteaseTM (GE Healthcare). Myristoylated Arf1 and Arf6 were prepared as described previously (32). Arf1 was expressed in BL21/DE3 together with yeast N-myristoyltransferase. Cells were disrupted by sonication in 20 mm Tris-HCl (pH 8.0), 100 mm NaCl, 1 mm MgCl2, 1 mm dithiothreitol (DTT), and 1 mg/ml lysozyme, protease inhibitor. The lysate was clarified by centrifugation at 100,000 × g for 60 min at 4 °C. The supernatant was loaded onto a 5-ml HiLoad Q column. Arf1 was eluted with 20 mm Tris-HCl (pH 8.0), 100 mm NaCl, 1 mm MgCl2, 1 mm DTT, 10%(v/v) glycerol. The fractions containing Arf1 were loaded onto a Sephacryl column. The fractions containing Arf1 were pooled. The protein solution was concentrated, and buffer was exchanged to 20 mm Tris-HCl (pH 8.0), 3 m NaCl, 1 mm MgCl2, 1 mm DTT by ultrafiltration. Myristoylated Arf1 (myr-Arf1) was isolated using a phenyl-Sepharose HP column. The column was developed in a gradient of 3 m to 100 mm NaCl. NU-PAGE analysis shows >95% purity of myr-Arf1 (Fig. 1A).
FIGURE 1.
Regulation of enzymatic GDP/GTP exchange activity of cytohesin-2 with myr-Arf6 and myr-Arf1 by V-ATPase-derived a2N(1–17) peptide on a membrane interface. A, preparation of recombinant myr-Arf1 and myr-Arf6 GTP-binding proteins. Human Arf1 was co-expressed with yeast N-myristoyltransferase in Escherichia coli BL21/DE3 strain. Recombinant myr-Arf1 was purified from the E. coli lysate (lane A1) using an AKTA purifier equipped with HiTrapQ, Superdex 200 columns (lane A2), and phenyl-Sepharose HP column (lane A3). Human Arf6 was also co-expressed with yeast N-myristoyltransferase in E. coli BL21/DE3 strain. Recombinant myr-Arf6 was extracted from the insoluble fraction using Triton X-100 (lane B1). The extract was precipitated by 35% ammonium sulfate and myr-Arf6 purified with HiTrapQ column (lane B2). B, time course of nucleotide exchange on myr-Arf1 and myr-Arf6 catalyzed by cytohesin-2. Exchange was followed by measuring the binding of [35S]GTPγS in an assay mixture that included PIP2-containing liposomes. C, dose-dependent inhibition of enzymatic GEF-activity of cytohesin-2 with myr-Arf6 by V-ATPase-derived a2N(1–17) peptide. Although FITC-a2N(1–17)-TAT peptide potently inhibits (IC50 = 0.9 μm), the GEF activity of cytohesin-2, the control FITC-TAT peptide has no effect on its activity. D, dose-dependent inhibition of enzymatic GEF activity of cytohesin-2 with myr-Arf1 by V-ATPase-derived a2N(1–17) peptide. Although FITC-a2N(1–17)-TAT peptide potently inhibits (IC50 = 0.9 μm), the GEF-activity of cytohesin-2, the control FITC-TAT peptide has no effect on its activity. E, dose-dependent inhibition by SecinH3 (IC50 >150 μm) of enzymatic GEF activity of cytohesin-2 with myr-Arf6. F, dose-dependent inhibition by SecinH3 (IC50 >150 μm) of enzymatic GEF-activity of cytohesin-2 with myr-Arf1.
Arf6 was expressed in BL21/DE3 together with yeast N-myristoyltransferase. Cells were disrupted by sonication in 20 mm Tris-HCl (pH 8.0 at 4 °C), 100 mm NaCl, 1 mm MgCl2, 1 mm DTT, 10% glycerol, 1 mg/ml lysozyme, protease inhibitor. The lysate was centrifuged at 100,000 × g for 60 min. The precipitate was resuspended in 20 mm Tris-HCl (pH 8.0 at 4 °C), 100 mm NaCl, 1 mm MgCl2, 1 mm DTT, 10% glycerol, 1% Triton X-100. The lysate was clarified by centrifugation at 100,000 × g for 60 min, and 10 μm GDP was added to the supernatant. Myristoylated Arf6 (myr-Arf6) was precipitated in 35% saturation ammonium sulfate, dissolved in 20 mm Tris-HCl (pH 8.0 at 4 °C), 25 mm NaCl, 1 mm MgCl2, 1 mm DTT, 1% (w/v) Triton X-100, 10% (v/v) glycerol, 10 μm GDP, and dialyzed against the same buffer. Myristoylated Arf6 was loaded on 5-ml HiTrapTM Q column and eluted with the same buffer. NU-PAGE analysis shows >95% purity of myr-Arf6 (Fig. 1A).
Truncated (Δ17)Arf1 and (Δ13)Arf6 proteins were expressed in BL21/DE3 cells grown in LB at 37 °C. Expression of recombinant proteins was induced by addition of 1 mm isopropyl β-d-thiogalactopyranoside. Cells were disrupted in 20 mm Tris-HCl (pH 8.0 at 4 °C), 100 mm NaCl, 1 mm MgCl2, 1 mm DTT, 10% glycerol, 1 mg/ml lysozyme, protease inhibitor. Lysates were centrifuged at 12,000 × g for 12 min. The supernatants were centrifuged at 100,000 × g for 60 min and were separated by gel filtration (HiLoad 16/600 Superdex 75 pg). Purified (Δ17)Arf1 and (Δ13)Arf6 proteins were >95% pure according to NU-PAGE analysis (data not shown). To load (Δ17)Arf1 and (Δ13)Arf6 with GDP, both recombinant proteins were incubated with 10-fold GDP and 2 mm EDTA. Excess GDP was removed using a column packed with Sephadex G-25M.
GEF Activity Assays
We used two assays to determine the GDP/GTP exchange activity of both full-length cytohesin-2 and Sec7 domain.
Radiolabel-based Assay
This assay allows the steady-state enzymatic GEF activity analysis of full-length cytohesin-2 or Sec7 domain with Arf1 and Arf6 in the presence of PIP2-containing liposomes (33). Phospholipid vesicles were prepared by the extrusion method (34). The lipids were dissolved in chloroform. A film of phospholipids was formed by evaporating chloroform and was resuspended in 50 mm Hepes (pH 7.5) and 1 mm DTT. The suspension was subjected to three freeze-thaw cycles and then passed through a 0.1-μm pore size polycarbonate filter (Millipore). For time course measurements, recombinant myristoylated myr-Arf1 or myr-Arf6 was diluted to 1 μm in a buffer containing 50 mm Hepes (pH 7.5), 1 mm MgCl2, 100 mm KCl, 1 mm DTT, and indicated lipid vesicles. Reactions were initiated by addition of 50 nm cytohesin-2 or Sec7 and 4 μm [35S]GTPγS and incubated at 37 °C. At the indicated times, aliquots (20 μl) were diluted into 2 ml of a buffer containing 20 mm Hepes (pH 7.5), 10 mm MgCl2, 100 mm KCl to stop the reaction. Proteins were trapped on nitrocellulose filters. The radioactivity was quantified after washing the filters three times with 2 ml of stop buffer. To test the effect of peptides, the 500 nm aliquots of cytohesin-2 or Sec7 were incubated with peptides (10-fold concentration shown on Figs. 1 and 6) for 10 min before the reactions were initiated. Reactions were continued for 2 min in the presence of 1.5 mg/ml azolectin with myr-Arf1. Alternatively, reactions were carried out for 30 min in the presence of 1 mg/ml liposomes containing 65% (w/w) phosphatidylcholine, 30% (w/w) phosphatidylserine, and 5%(w/w) PIP2 with myr-Arf6. To test the effects of SecinH3, the assays were performed as described above, and SecinH3 was diluted with 4% DMSO. Titration with SecinH3 was performed up to a 200 μm concentration, due to its limited solubility in GEF activity buffer.
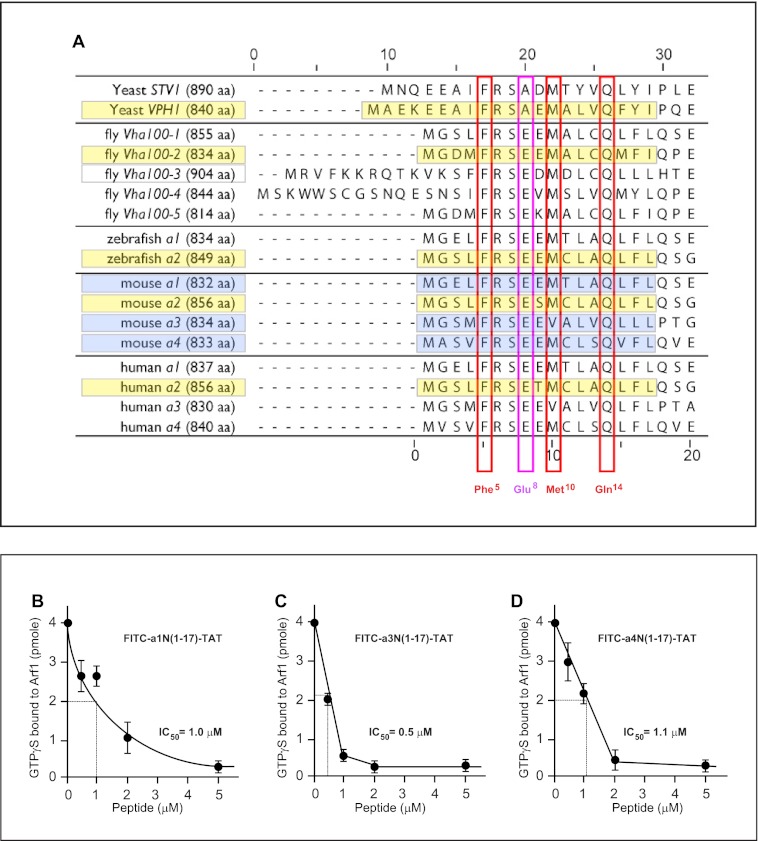
FIGURE 6.
Signaling of V-ATPase with cytohesin-2 and Arf GTP-binding proteins is evolutionarily conserved in eukaryotes among all four a-subunit isoforms. A, alignment of the first 20 amino acids of eukaryotic a-subunit isoforms of V-ATPase. The conserved character of binding surface-forming amino acids Phe5, Met10, and Gln14 (red boxes) and Glu8 (violet box) from yeast to humans. Although amino acids Phe5 and Gln14 are completely conserved in all eukaryotes, Met10 has one substitution to Val10 in the mouse and human a3-subunit V-ATPase (red boxes). The Glu8 residue is also conserved with only one substitution to Ala8 in yeast Stv1p and Vph1p (violet box). B–D, potent inhibition of enzymatic GEF activity of cytohesin-2 Sec7 domain by a1-, a3-, and a4-isoform-derived peptides. B, determination of IC50 = 1.0 μm for FITC-a1N(1–17)-TAT peptide. C, determination of IC50 = 0.5 μm for FITC-a3N(1–17)-TAT peptide. D, determination of IC50 = 1.1 μm for FITC-a4N(1–17)-TAT peptide.
Tryptophan Fluorescence-based GTPγS Binding Assay
In this assay, exchange was followed in real time. Truncated (Δ17)Arf1 and (Δ13)Arf6 were prepared as described (35) for the assay. These are soluble in both GDP- and GTP-bound forms (myr-Arf-GTP requires a hydrophobic surface for stability) allowing exchange to occur in the absence of lipid or detergent, which is desirable because lipid or detergent can scatter light and possibly confound interpretation of changes in fluorescence. To monitor tryptophan fluorescence, the fluorescent spectra of (Δ17)Arf1 in GDP-bound and GTPγS-bound forms were obtained at an excitation wavelength of 285 nm using a PTI spectrofluorimeter. The maximum difference was observed at 335 nm (Fig. 2A). Measurements were performed at 37 °C in 50 mm Hepes-KOH, 100 mm KCl, 1 mm MgCl2, 1 mm DTT, 10 μm GTPγS, 1 μm (Δ17)Arf1 or (Δ13)Arf6, and 10 nm Sec7 domain. All titration experiments were started by addition of mixture containing GTPγS, Sec7, and a2N(1–17) peptide or SecinH3 inhibitor (as shown on Fig. 2). Data were fitted by single exponential curve and analyzed using Microsoft Excel.
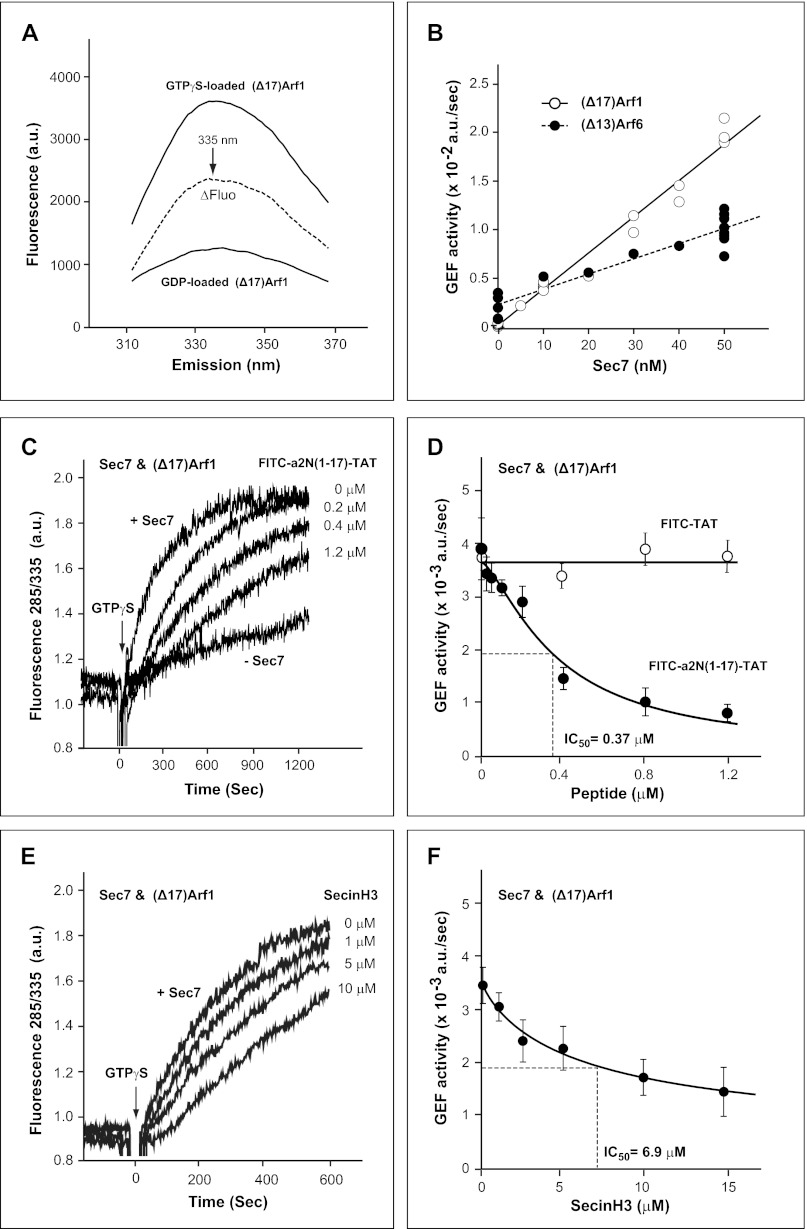
FIGURE 2.
Regulation of the enzymatic GDP/GTP-exchange activity of Sec7 domain using truncated (Δ17)Arf1 and (Δ13)Arf6 as substrates by V-ATPase derived a2N(1–17) peptide in solution. A, fluorescent spectra of (Δ17)Arf1 loaded with either GTPγS or GDP. Emission spectra were determined using an excitation wavelength of 285 nm, and the fluorescence difference (ΔFluo) between (Δ17)Arf1 loaded with GTPγS and GDP was calculated. B, dependence of GEF activity on concentration of Sec7 domain. GEF activity of Sec7 domain was determined with 1 μm of either (Δ17)Arf1 or (Δ13)Arf6 and 10 μm GTPγS. C, time course of enzymatic GEF activity of Sec7 with (Δ17)Arf1 and dose-dependent inhibition by V-ATPase-derived a2N(1–17) peptide. D, dose-dependent inhibition of enzymatic GEF activity of Sec7 with (Δ17)Arf1 by V-ATPase-derived a2N(1–17) peptide. Although FITC-a2N(1–17)-TAT peptide potently inhibits (IC50 = 0.37 μm) the GEF activity of Sec7, the control FITC-TAT peptide does not inhibit its activity. E, time course of enzymatic GEF activity of Sec7 with (Δ17)Arf1 and dose-dependent inhibition by SecinH3. F, dose-dependent inhibition (IC50 = 6.9 μm) of enzymatic GEF-activity of Sec7 with (Δ17)Arf1 by SecinH3. a.u., arbitrary units.
Circular Dichroism Spectroscopy and Secondary Structure Content of a2N(1–17)
For structure determination, the biotin-a2N(1–17)-TAT peptide (see supplemental Table S1) was dissolved in 25 mm phosphate buffer (pH 6.5) and 300 mm NaCl at a concentration of 1 mg/ml. The secondary structure of the a2N(1–17) peptide was determined from circular dichroism (CD) spectra. Steady-state CD spectra were measured in the far UV-light (190–260 nm) using a Chirascan spectropolarimeter (Applied Photophysics). Spectra were collected in a 60-μl quartz cell (Hellma) with a path length of 0.1 mm, at 20 °C, and a step resolution of 1 nm. The readings were an average of 2 s at each wavelength, and the recorded ellipticity values were the average of three determinations for each sample. The spectrum for the buffer was subtracted from the spectrum of the peptide. CD values were converted to mean residue ellipticity (Θ) in units of degree cm2 × dmol−1 using the software Chirascan Version 1.2 (Applied Photophysics). This base-line corrected spectrum was used as input for computer methods to obtain predictions of secondary structure. To analyze the CD spectrum, the following algorithms were used: Varselec (36), Selcon (37), Contin (38), and K2D (39). All methods were incorporated into the program Dicroprot (40) and Neural Net (41).
NMR Spectroscopy
All NMR experiments were performed on a Bruker DRX 600 MHz spectrometer equipped with a cryoprobe. The biotin-a2N(1–17)-TAT peptide was dissolved at a concentration of 2 mm in 25 mm phosphate buffer (pH 6.5) and 300 mm NaCl for determination of NMR structure in solution. Total correlation spectroscopy (TOCSY) and nuclear Overhauser effect spectroscopy (NOESY) of the peptide were recorded with mixing times of 80 and 300 ms, respectively, at a temperature of 25 °C. TopSpin (Bruker Biospin) and Sparky Suite (42) of programs were used for spectra processing, visualization, and peak picking. By using standard procedures for sequential assignment based on homonuclear TOCSY and NOESY experiments, all the residues of the peptide were assigned. Standard procedures based on spin-system identification and sequential assignment were adopted to identify the resonances (43). Interproton distances were obtained from the NOESY spectra. NOESY peaks were categorized as strong, medium, and weak based on the signal intensity and were translated into distance constraints as 3.0, 4.0, and 5.0 Å, respectively. Dihedral angle restraints as derived from torsion angle likelihood obtained from shift and sequence similarity (TALOS) (44) were employed to generate the three-dimensional structure of the peptide in the CYANA 2.1 package (45).
NMR Binding Studies
Interaction studies between Sec7 domain and a2N(1–17) as well as Sec7 and Vph1p(1–388) were performed using highly precise and reproducible NMR spectroscopy. Experiments were performed on a Bruker Avance 600 machine using Topspin for acquisition and processing of spectra. Respective spectra were overlapped to monitor chemical shift changes, and further analysis was done in SPARKY (42).
In Silico Sec7 and a2N(1–17) Docking Experiments
The AutoDock Version 4 and AutoDock 4 Tools programs (The Scripps Research Institute) were used for docking of a2N(1–17) peptide to the Sec7 domain (46, 47). Spatial structure of Sec7 domain was taken from the crystal structure of the complex of Sec7/Arf1 with brefeldin A as a stabilization agent (Protein Data Bank code 1S9D) (48), and all water molecules were removed from the Sec7 structure. The NMR structure of a2N(1–17) in solution was also used for these docking experiments (Fig. 3E). The Sec7 domain structure was rigid during docking. All backbone torsions in the helical part of a2N(1–17) were also treated as nonrotatable, although most of other torsions were rotatable. The Sec7 domain was covered by eight grids with 126 × 126 × 126 grid points and the default grid spacing of 0.375 Å. For each grid, the 1,000 runs of the Lamarckian genetic algorithm with default settings were performed. Thus, in total 8,000 blind and random in silico docking experiments have been done. All docking results were sorted by final docked energy. The contact areas for peptide residues were estimated with PyMOL Version 1.4 software (47) as a difference between solvent-accessible areas without Sec7 and with Sec7. Final docking complexes are shown for Arf1/Sec7/a2N(1–17) (see supplemental Movie S1) and Arf6/Sec7/a2N(1–17) (supplemental Movie S2).
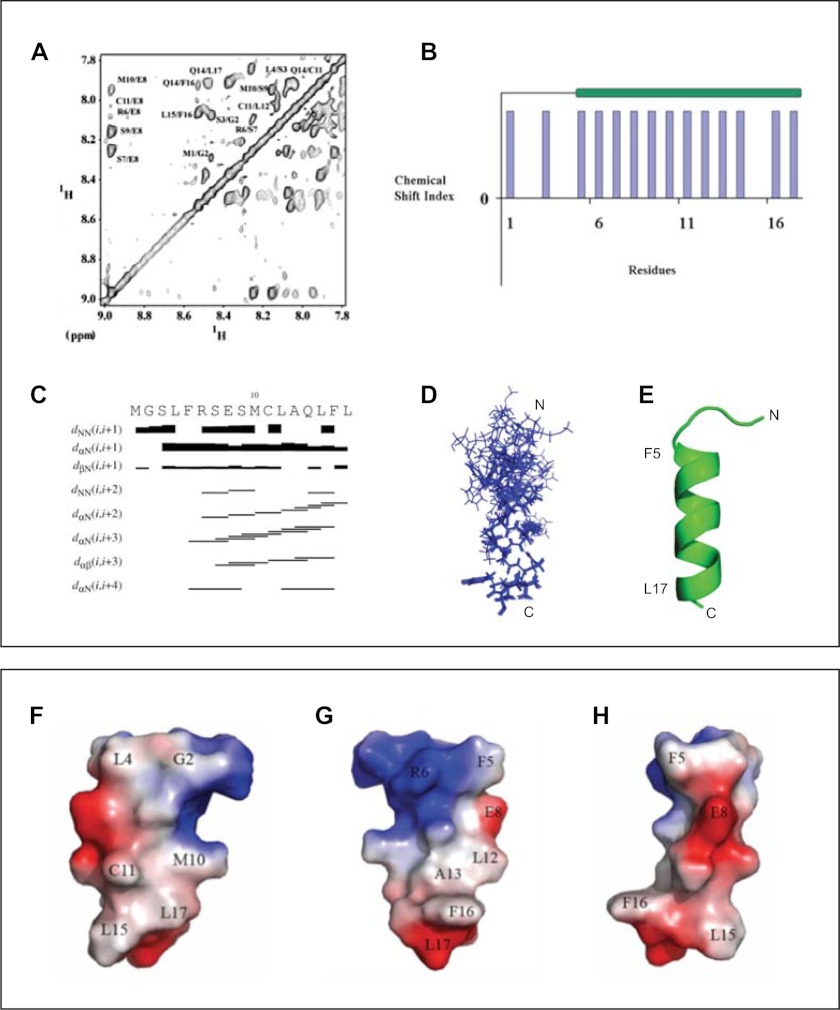
FIGURE 3.
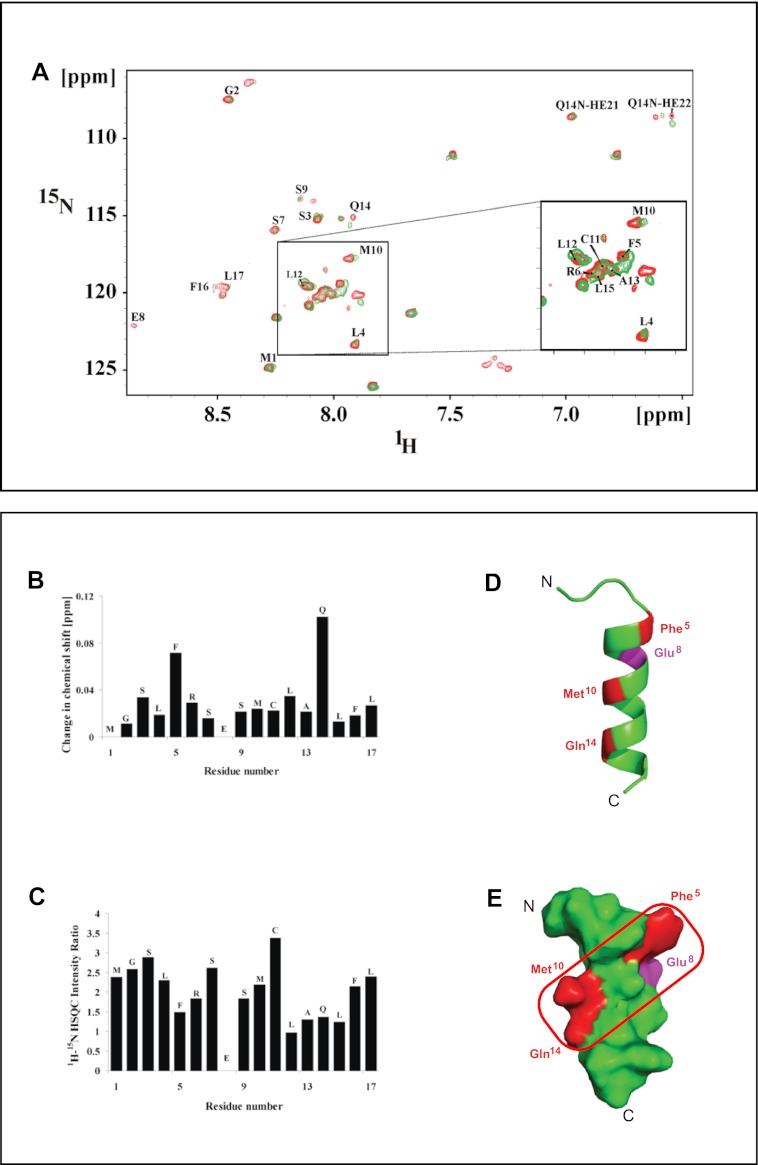
Determination of solution structure of V-ATPase-derived a2N(1–17) peptide by NMR. A, assignment of cross-peaks in the NOESY spectrum of a2N(1–17) in the HN-HN region of the spectrum. Peak picking was done by Sparky 3.1 software, and cross-peaks were identified based on the TOCSY and NOESY spectra. B, secondary structure prediction using the α-proton chemical shifts of a2N(1–17) by PREDITOR software. C, NOESY connectivity plot of peptide a2N(1–17). D, superimposition of the 10 lowest energy NMR structures of V-ATPase-derived a2N(1–17) peptide. E, NMR structure of the a2N(1–17) peptide in schematic representation. F–H, molecular surface electrostatic potential of peptide a2N(1–17) generated by PyMOL. The positive potentials are drawn in blue and negative potentials in red. The charge distribution of the helix reveals an amphiphilic surface. F, peptide has a hydrophobic surface on one side formed by residues Leu4, Met10, Cys11, Leu15, and Leu17. G, positively charged Arg6, the negative COOH group of Leu17, and the hydrophobic residues Leu12, Ala13, Phe16, and Leu17 form an amphiphilic surface potential on the opposite site. H, in a 90° view to the amphiphilic surface, the peptide is mainly negatively charged, formed by Glu8, and the carboxyl group of Leu17.
Purification of Organelles and V-ATPase Holo-complex
Isolation of rat and mouse kidney proximal tubules and early endosomes was performed as described previously (6, 49). Yeast vacuolar membranes were isolated, and the yeast V-ATPase holo-complex was purified by elution from the M2-Sepharose matrix with 3×FLAG peptide as described previously (5).
PAGE and Western Blot Analysis
Expression and integrity of endogenous full-length V-ATPase a2-subunit in purified kidney proximal tubules and early endosomes was analyzed by NU-PAGE and Western blot analysis as described previously (6). Expression and integrity of recombinant full-length a2-EGFP in HeLa cells was also analyzed by NU-PAGE and Western blotting. Briefly, overexpression of a2-EGFP was performed for 1 day using Lipofectamine 2000 transfection reagent. Total cell lysates were prepared, resolved by NU-PAGE, and analyzed by Western blotting using anti-GFP (B-2, 1:500) antibodies. Analysis of yeast V-ATPase integrity and its interaction with CTH2(61–400) was performed by SDS-PAGE and Western blot analysis as described previously (5). Briefly, yeast V-ATPase immobilized on the M2-Sepharose matrix was mixed with a large excess of recombinant CTH2(61–400) protein. Binding step was followed by four extensive washes to remove unbound CTH2(61–400) and contaminating proteins from the solubilized yeast membranes as described previously (6). Binding and co-purification of CTH2(61–400) with the V-ATPase holo-complex during elution with 3×FLAG peptide was analyzed by SDS-PAGE and confirmed by Western blotting with an anti-His6 antibody.
Immunocytochemistry and Confocal Microscopy Analysis
Overexpression of full-length mouse a2-EGFP was performed for 1 day in Madin-Darby canine kidney cells grown on premium glass coverslips in 6-well plates. Cells were rinsed in PBS and fixed in PBS containing 4% paraformaldehyde for 20 min. After three rinses in PBS, cells were permeabilized by treatment with 0.1% Triton X-100 in PBS for 10 min followed by blocking with 2% BSA in PBS for 1 h. To increase the fluorescence signal of the overexpressed recombinant protein, the a2-EGFP was then detected with mouse monoclonal anti-GFP (B-2) antibodies. Cells were incubated for 18 h at 4 °C with anti-GFP (B-2) antibodies (1:200 dilution in 2% BSA/PBS). After three PBS rinses, cells were incubated with Alexa 488-conjugated goat anti-mouse antibodies (1:1,000 dilution in 2% BSA/PBS). Nuclei were counterstained with 0.75 μm DAPI for 10 min. After three final PBS rinses, the coverslips were mounted with FluorSave reagent. Confocal microscopy was performed on a Nikon A1R laser scanning confocal microscope. Images were analyzed using Volocity Version 5 software.
Statistical Analysis
Data are presented as mean values, and error bars indicate the mean ± S.E. Statistical calculations were made using either Microsoft Excel or SigmaStatTM version 3.0 statistical software. The values of IC50 were calculated using a four-parameter logistic function with Microsoft Excel.
RESULTS
Signaling of a2-subunit V-ATPase with Cytohesin-2 and Arf GTP-binding Proteins
To test the potential signaling between V-ATPase and cytohesin-2/Arf GTP-binding proteins, we established a GEF activity assay for cytohesin-2(WT) and myr-Arf6(WT) and myr-Arf1(WT) GTP-binding proteins (Fig. 1A). This assay allows the steady-state enzymatic GEF activity analysis of full-length cytohesin-2 or Sec7 domain with myr-Arf1 and myr-Arf6 in the presence of PIP2-containing liposomes (Fig. 1B). We synthesized several soluble, cell-permeable, and fluorescently labeled a2N(1–17)-derived peptides, which were used in our current GEF activity and structural studies in vitro (see supplemental Table S1). After testing their solubility and cell permeability, the following V-ATPase-derived peptides were used: (i) FITC-a2N(1–17)-TAT; (ii) biotin-a2N(1–17)-TAT; and (iii) a control FITC-TAT peptide. Our GEF activity assays demonstrated that FITC-a2N(1–17)-TAT, but not FITC-TAT, potently inhibits the enzymatic activity of cytohesin-2 (IC50 = 0.9 μm) with both myr-Arf6 (Fig. 1C) and myr-Arf1 (Fig. 1D) as a substrate. In comparison, the SecinH3 small molecule, a known specific inhibitor of the cytohesin-2 Sec-7 domain, inhibited its activity with an IC50 >150 μm for myr-Arf6 (Fig. 1E) or IC50 >150 μm for myr-Arf1 (Fig. 1F). Thus, these studies show that a2N(1–17) peptide is a significantly more potent (up to 100 times) inhibitor of cytohesin-2 than SecinH3.
To analyze whether the a2N(1–17) peptide inhibits the GEF activity of cytohesin-2 via its direct and high affinity interaction with the Sec7 domain (27), a tryptophan fluorescence-based GEF activity assay was also used. This assay allows real time measurements of the GEF Sec7 domain-induced nucleotide exchange. (Δ17)Arf1 and (Δ13)Arf6, which are soluble in the GTP-bound form, were used as substrates, thereby eliminating the need for phospholipid or detergent in the assay. This approach eliminates the light scattering due to lipid or detergent that could confound interpretation of the fluorescence changes (50). Activation of (Δ17)Arf1 or (Δ13)Arf6 from the GDP-bound form to the GTPγS-bound form was monitored by tryptophan fluorescence (emission/excitation wavelengths of 285/335 nm) (Fig. 2A). Because recombinant (Δ17)Arf1 and (Δ13)Arf6 were tagged with His6 on the N terminus, we first determined whether the tag affected the enzymatic reaction. The concentration dependence of GEF activity of the Sec7 for both (Δ17)Arf1 and (Δ13)Arf6 demonstrated that the His6 tag did not interfere with the assay. Our data also revealed the preferential activity of Sec7 with (Δ17)Arf1 than (Δ13)Arf6 as reported previously (Fig. 2B) (50, 51). Using the purified Sec7 domain with truncated (Δ17)Arf1 as substrate, a direct and more potent inhibition of the Sec7 domain by the FITC-a2N(1–17)-TAT peptide was found (IC50 = 0.37 μm) (Fig. 2, C and D). In comparison, in this assay SecinH3 inhibits Sec-7/(Δ17)Arf1 with an IC50 = 6.9 μm (Fig. 2, E and F). Taken together, these experiments demonstrate that V-ATPase-derived a2N(1–17) peptide is a direct and potent inhibitor of the catalytic Sec7 domain of cytohesin-2.
Structure of a2-Subunit V-ATPase-derived a2N(1–17) Peptide
To understand the structural basis and molecular mechanism of signaling of the a2-subunit V-ATPase with cytohesin-2 and Arf GTP-binding proteins, we next studied the structure of the V-ATPase-derived a2N(1–17) peptide.
Circular Dichroism Spectroscopy
The secondary structure of biotin-a2N(1–17)-TAT was determined from circular dichroism spectra. The average secondary structure content was 70% α-helix and 30% random coil (data not shown). This result is consistent with secondary structure predictions based on subunit a2N(1–17) amino acid sequence.
NMR Structure of a2N(1–17) in Solution
The NMR structure of a2N(1–17) in solution was solved using standard procedures for sequential assignment based on homonuclear TOCSY and NOESY experiments as described under “Experimental Procedures.” Secondary structure prediction was done using the α-proton chemical shifts, which indicate the presence of α-helical structure at the C terminus of the peptide between residues Phe5 and Leu17 (Fig. 3A). Identified cross-peaks in the HN-HN region are also shown in (Fig. 3B). HN-HN, Hα–HN(i,i + 3), Hα–HN(i,i + 4), and Hα–Hβ(i,i + 3) connectivity were plotted from the assigned NOESY spectrum (Fig. 3C) and also support an α-helical content in the C terminus. Out of 100 structures generated, the 10 lowest energy structures were taken for further analysis. The ensemble of 10 calculated structures resulted in an overall root mean square deviation of 0.311 Å for the backbone atoms (Fig. 3D). All these structures have energies lower than −100 kcal/mol, no NOE violations greater than 0.3 Å, and no dihedral violations greater than 5°. The summary of the statistics for 10 structures is shown in supplemental Table S2. The calculated structure has a total length of 25.78 Å displaying an α-helical region between residues 5 and 17 (21.1 Å) and a flexible N-terminal region, formed by the amino acids 1–4, respectively (Fig. 3E). The molecular surface electrostatic potential of the peptide is presented in Fig. 3, F–H. At one side of the peptide, the residues Leu4, Met10, Cys11, Leu15, and Leu17 provide hydrophobic surface on a single face of the helix (Fig. 3F). The opposite side reflects an amphiphilic surface potential, formed by the positively charged Arg6, the negative COOH-group of Leu17, and the hydrophobic residues Leu12, Ala13, Phe16, and Leu17 (Fig. 3G). In a 90° view to the amphiphilic surface, the peptide is mainly negatively charged (Fig. 3H).
Interaction Interface between a2N(1–17) and Sec7 Domain of Cytohesin-2
We applied NMR titration experiments to determine the amino acid residues of a2N(1–17) that are involved in binding the Sec7 domain of cytohesin-2 as described under “Experimental Procedures.” This approach has been used successfully to study interaction interfaces between other peptides and truncated proteins (52). In our experiments the 1H-15N heteronuclear single quantum coherence (HSQC) spectrum of a2N(1–17) was used as starting point. The HSQC values of a2N(1–17) in the presence (green) and absence (red) of the Sec7 domain (22.1 kDa) (44) have been performed to assign the amino acid residues of a2N(1–17) peptide involved in the binding with Sec7 (Fig. 4A). Binding of Sec7 induces a structural change in a2N(1–17), which is indicated by changes in chemical shift, intensity, and/or the broadening in the resonances of residues Phe5, Met10, and Gln14 in the 1H-15N HSQC spectrum (Fig. 4, A–C). As can be seen for residue Glu8, the binding of equimolar amounts of Sec7 domain to a2N(1–17) caused a loss in signal intensity in the 1H-15N HSQC spectrum (Fig. 4, B and C). The loss of intensity of Glu8 may not be affected by direct amino acid interaction of Glu8 with a residue of the Sec7 domain of cytohesin-2 but rather be caused by the transformation of structural alterations at the actual binding site at Phe5, Met10, and Gln14 to Glu8. These four residues of a2N(1–17) are conserved according to the alignment with other mouse and human isoforms and yeast homologs (Fig. 6A). The binding region of a2N(1–17) is highlighted in the structure in red (Fig. 4, D and E).
FIGURE 4.
Determination of interaction-competent amino acids and the interaction surface plane of a2N(1–17) peptide involved in binding with Sec7 domain. A, NMR titration spectra of a2N(1–17) and Sec7 domain of cytohesin-2. Overlay of two-dimensional 1H-15N HSQC spectrum of a2N(1–17) alone (red) and a2N(1–17) with Sec7 domain of cytohesin-2 (green) in 25 mm sodium phosphate buffer (pH 6.5), 300 mm NaCl at 298 K. B, change in chemical shift; C, loss of 1H-15N HSQC peak intensity upon titration of unlabeled Sec7 domain to unlabeled a2N(1–17) at a molar ratio of 0.5. The stronger the decrease of the peak intensity compared with the spectrum without Sec7 domain, the higher the plotted intensity ratio. D, location of the four amino acids Phe5, Met10, Gln14 (red), and Glu8 (magenta) that were determined by NMR to be involved in interaction of a2N(1–17) peptide with Sec7 domain. E, identification of the interaction surface plane formed by amino acids Phe5, Met10, and Gln14 (shown in a red square) that is involved in binding of a2N(1–17) with Sec7 domain.
NMR Titration of the Sec7 Domain with a2N(1–17)
To further study the molecular site of interaction between Sec7 and a2N(1–17) peptide, the HSQC NMR spectra of the Sec7 domain (residues Arg61 to Arg252) of cytohesin-2 were also collected. The HSQC spectra confirm that the Sec7 domain is folded by the well dispersed peaks and their line shape (data not shown). The 1H-15N HSQC spectra were recorded at 298 K with a fixed concentration of 0.4 mm Sec7 domain, and the peptide a2N(1–17) was titrated to 0.6 mm (1:1.5) to the protein. These experiments revealed significant changes in the chemical shift of peaks of the Sec7 domain, indicating binding of the a2N(1–17) peptide to the Sec7 domain. Changes in 14 peaks were identified (data not shown). Nine of these peaks showed changes along the proton axis with decreases in ppm values, whereas three peaks showed change with increases in ppm. Along the nitrogen axis, two shifts were observed. One peak at 123.1/8.55 ppm 15N/1H had an increased ppm at the nitrogen axis (123.5/8.55 ppm 15N/1H), and peak 12 reflects a combined change in decreased ppm of nitrogen and proton from (127.1/8.32 ppm 15N/1H to 126.3/8.26 ppm 15N/1H) (data not shown).
NMR Titration of the Sec7 Domain with Vph1p(1–388)
We next tested whether the interaction of Sec7 also takes place with full-length V-ATPase holoprotein. The interaction of S. cerevisiae Vph1p(1–388) (the homolog of mammalian aN cytosolic tail) with Sec7 was also studied as recently described for Sca(104–363) (53). Vph1p(1–388) was purified according to Thaker et al. (54), and HSQC experiments were performed as described under “Experimental Procedures.” Respective spectra were overlapped to monitor chemical shift changes; further analysis was done in SPARKY (42). Sec7 was titrated with Vph1p(1–388) and 14 chemical shifts and losses of intensity of peaks were observed (data not shown). Importantly, 12 out of the 14 amino acids involved in this interaction were comparable with those identified in the a2N(1–17) peptide Sec7 domain titration experiments. These results strongly indicate that a2N(1–17) peptide and Vph1p(1–388) holo-protein share a common binding site on the Sec7 domain of cytohesin-2. The two additional changes in chemical shifts (peak 13 and 14) might be caused by the residues of Vph1p(1–388) that are not present in a2N(1–17).
Structural Basis of Binding and Signaling of V-ATPase Derived a2N(1–17) Peptide with Cytohesin-2 and Arf GTP-binding Proteins
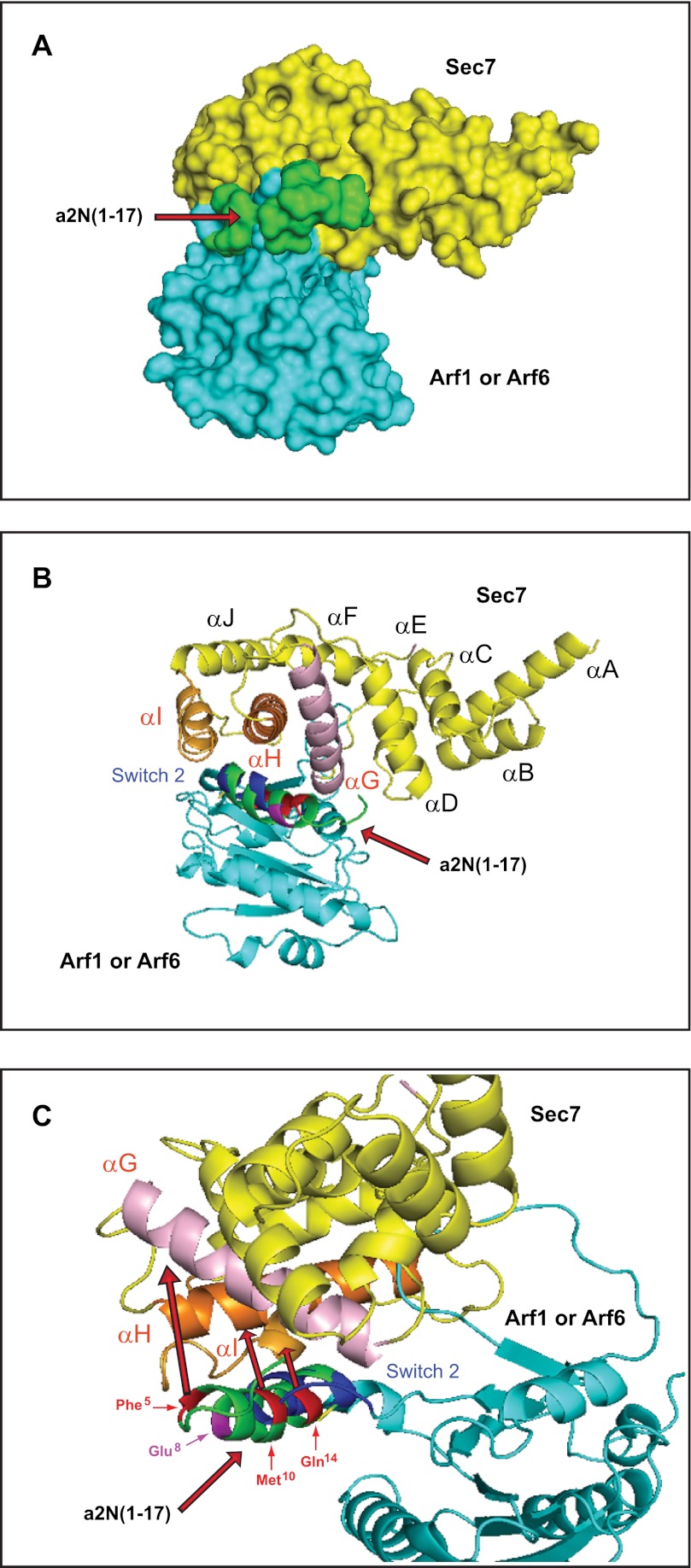
To map and characterize the molecular interface between peptide a2N(1–17) and the Sec7 domain in more detail, in silico docking experiments were performed as described under “Experimental Procedures.” In these studies, the lowest energy structure was found for docking the a2N(1–17) peptide on the interface of the binding between Sec7 domain and its substrates Arf1 or Arf6 (Fig. 5A). In particular, the helical part of the a2N(1–17) peptide, via its amino acids Phe5, Met10, and Gln14, situated at the interaction surface plane (Fig. 4E, red rectangle), binds to the αG, αH, and αI helices of the Sec7 domain (Fig. 5, B and C). More detailed analysis showed that the a2N(1–17)-binding site on the Sec7 domain overlaps with the interaction site of the regulatory switch 2 of Arf1 (Fig. 5, B and C) (see supplemental Movie S1) and Arf6 (see supplemental Movie S2).
FIGURE 5.
Structural basis and molecular mechanism of signaling between the V-ATPase a2-subunit and cytohesin-2/Arf-family GTP-binding proteins. A, in silico docking experiments revealed the binding site of a2N(1–17) peptide near the catalytic site of the Sec7 domain. B, binding of a2N(1–17) peptide on the Sec7 domain involves the αG, αH, and αI helixes, which are crucial for its catalytic activity with Arf1 and Arf6. C, molecular details of the interaction interfaces involved in binding of a2N(1–17) peptide to the catalytic site of the Sec7 domain with Arf1 (see supplemental Movie S1). Similar results were also shown for binding of a2N(1–17) peptide to the catalytic site of the Sec7 domain with Arf6 (see supplemental Movie S2).
Signaling between V-ATPase, Cytohesin-2, and Arf GTP-binding Proteins Is an Evolutionarily Conserved Phenomenon
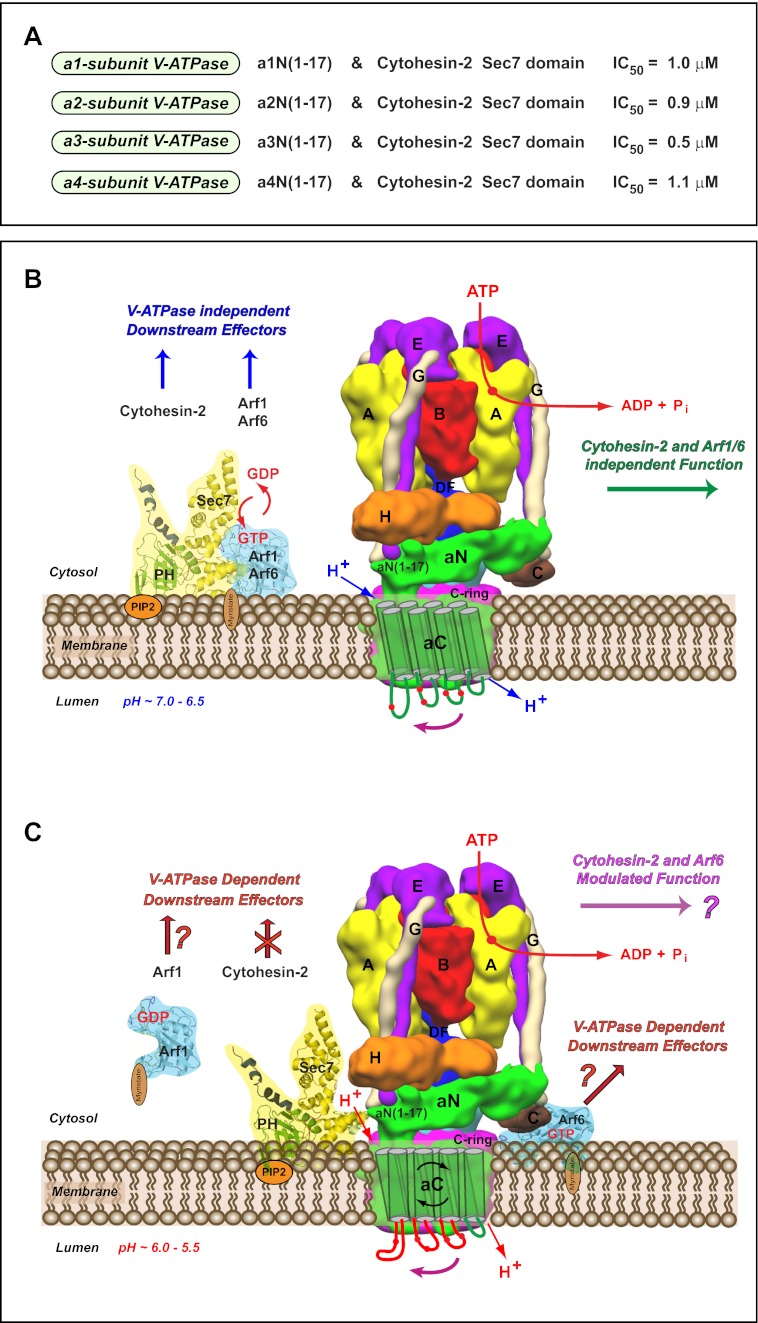
Alignment of the first 20 amino acids of the N-terminal tail of V-ATPase a-subunit isoforms revealed that the interface-forming amino acids Phe5 and Gln14 are completely conserved in all eukaryotes from yeast to humans (Fig. 6A, red boxes). The interface-forming amino acid Met10 is also conserved from yeast to humans with only one substitution to Val10, which is found in the mouse and human a3-subunit V-ATPase (Fig. 6A, red box). It is also noteworthy that although the Glu8 residue is highly conserved, it has substitution to Ala8 in yeast Stv1p and Vph1p (Fig. 6A, violet box). To test the potential signaling between other a-subunit (a1-, a3-, and a4-isoforms) of V-ATPase and cytohesin-2/Arf GTP-binding proteins, we have also synthesized the corresponding a-subunit isoform-derived peptides (Fig. 6A, highlighted in blue and supplemental Table S1). Additional enzymatic GEF activity experiments demonstrated that these peptides are also very potent inhibitors of GEF activity of cytohesin-2 Sec7 domain (Fig. 6, B–D). In these experiments, the following IC50 values were determined for a-isoform-specific peptides: (i) FITC-a1N(1–17)-TAT (IC50 = 1.0 μm) (Fig. 6B); (ii) FITC-a3N(1–17)-TAT (IC50 = 0.5 μm) (Fig. 6C); and (iii) FITC-a4N(1–17)-TAT (IC50 = 1.1 μm) (Fig. 6D). Thus, our data revealed the conserved character of signaling between all four a1-, a2-, a3-, and a4-subunit isoforms of V-ATPase with cytohesin-2 and Arf GTP-binding proteins.
Mammalian Cytohesin-2 Interacts with Yeast V-ATPase Holo-complex Containing the Intact a-Subunit Isoform
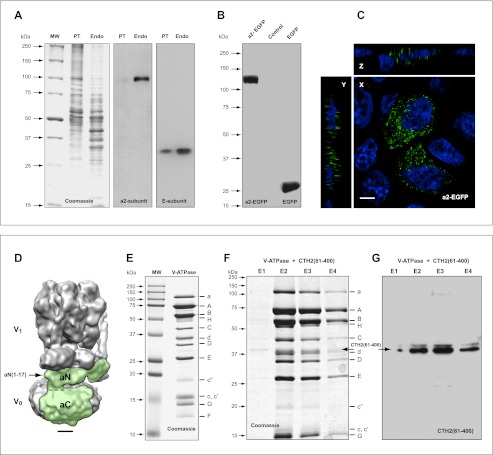
We next confirmed the conserved character of this signaling phenomenon in experiments showing binding of mammalian cytohesin-2 with the yeast V-ATPase holo-complex. However, first we verified that a-subunit isoforms, which are involved in signaling with cytohesin-2, remain intact and functional in both mammalian and yeast V-ATPases. We used the kidney endosomal/lysosomal experimental system that was also successfully employed in our previous studies (6, 55). Here, we demonstrated that under physiological conditions in situ, an endogenous a2-subunit isoform is not proteolytically processed and remains intact as a 100-kDa protein in kidney proximal tubules and endosomes (Fig. 7A). It is noteworthy that Western blot analysis did not reveal any additional low molecular weight proteins for either the a2-subunit (V0-sector) or E-subunit (V1-sector) of V-ATPase. These data indicate that in the mammalian kidney, the V-ATPase complex remains intact and competent for interaction and signaling with cytohesin-2 and Arf GTP-binding proteins. Indeed, our previous studies demonstrated that these intact proteins interact with the functional V-ATPase holo-complex in an acidification-dependent manner (6, 55). We also cloned EGFP-tagged full-length a2-subunit isoform and performed overexpression experiments. Our data demonstrate that recombinant a2-EGFP remains intact as a 130-kDa protein and does not undergo proteolytic cleavage (Fig. 7B). Importantly, overexpressed a2-EGFP is also correctly targeted to vesicular compartments in kidney cells (Fig. 7C).
FIGURE 7.
Structural integrity of a-subunit isoforms is crucially important for assembly, function, and evolutionarily conserved signaling of V-ATPase. A, in kidney experimental system, the V-ATPase complex remains intact and competent of acidification, interaction, and signaling with cytohesin-2 and Arf GTP-binding proteins. Under physiological conditions, in situ endogenous a2-subunit (V0 sector) and E-subunit (V1 sector) remain intact in kidney proximal tubules (PT) and endosomes (Endo). Note that to provide better visualization of the lack of proteolytic cleavage of these proteins, Western blots of a2- and E-isoforms are presented as complete gels, expanding the data that were previously published and showed only a small segment of these gel lanes (6). B and C, overexpressed recombinant full-length a2-EGFP-subunit isoform of V-ATPase remains intact (B) and competent to be targeted to vesicular compartment in kidney epithelial cells in culture (C). Scale bar, 4 μm. D, three-dimensional map shows holo-complex of S. cerevisiae eukaryotic V-ATPase solved at 11 Å resolution (5). Figure is showing well defined V0 and V1 sectors with almost all individual subunits resolved. The position of full-length intact a-subunit isoform (Vph1p homolog in yeast) in V-ATPase holo-complex is indicated in green. The positions of N-terminal cytosolic tail (aN) corresponding to amino acids of yeast Vph1p(1–404) and mouse a2N(1–402) is shown. The position of C-terminal transmembrane region (aC) corresponding to amino acids of yeast Vph1p(405–840) and mouse a2N(403–856) is also shown. The region of the epitope formed by the peptide aN(1–17) is indicated by arrow. Scale bar, 25 Å. E, isolation of S. cerevisiae V-ATPase by elution from the M2-Sepharose with the 3×FLAG peptide and characterization by SDS-PAGE analysis. This preparation of the V-ATPase contained 13 different subunits indicated on the right. It is noteworthy that the a-subunit isoform runs as the intact full-length 116-kDa protein. F and G, evolutionarily conserved interaction of intact yeast V-ATPase holo-complex with human cytohesin-2. Yeast V-ATPase was immobilized on M2-Sepharose, allowed to interact with CTH2(61–400), and purified as described under “Experimental Procedures.” SDS-PAGE (F) and Western blot analysis (G) demonstrated binding of human CTH2(61–400) and its elution with intact yeast V-ATPase holo-complex.
Recently, the structure of the S. cerevisiae eukaryotic V-ATPase was solved at 11 Å resolution by electron cryomicroscopy of protein particles in ice (5). This resolution was sufficient to resolve almost all of the individual subunits (Fig. 7D). In particular, the position of the intact a-subunit isoform (Vph1p homolog in yeast) in this V-ATPase (V1V0) holo-complex is shown in green and its epitope aN(1–17) is indicated by an arrow (Fig. 7D). This V-ATPase holo-complex assembles with intact subunits (Fig. 7E), and it allowed the study of its interaction with mammalian cytohesin-2. Human cytohesin-2 lacking its coiled-coil domain (CTH2(61–400)) was used in this study. As reported previously, this form of cytohesin-2 does not dimerize and strongly interacts with the intact full-length mammalian a2-subunit isoform (6). Indeed, here we show binding of human CTH2(61–400) with intact yeast V-ATPase immobilized on M2-Sepharose. Both CTH2(61–400) and V-ATPase holo-complexes are eluted together in an elution step proportional manner (Fig. 7, F and G). Thus, taken together, these data confirm the evolutionarily conserved interaction and signaling of intact V-ATPase holo-complex with cytohesin-2 and Arf GTP-binding proteins.
DISCUSSION
Besides being an ATP-driven proton-pumping nanomotor, the V-ATPase functions as a pH-sensing receptor by interacting with the Arf-GEF cytohesin-2 in an acidification-dependent manner (6–8). So far, the molecular mechanism underlying this cell biological event remains unknown. Here, we address this issue and describe the molecular details of a novel function of V-ATPase as an evolutionarily conserved cytohesin-signaling receptor (Fig. 8).
FIGURE 8.
V-ATPase nanomotor is a novel evolutionarily conserved cytohesin-signaling receptor. A, signaling of V-ATPase with cytohesin-2 and Arf GTP-binding proteins is evolutionarily conserved among all four a-subunit isoforms of the V-ATPase. Experimental IC50 values of GEF activity inhibition of the cytohesin-2 Sec7 domain by a-subunit isoform-specific peptides as follows: (i) a1N(1–17) (IC50 = 1.0 μm); (ii) a2N(1–17) (IC50 = 0.9 μm); (iii) a3N(1–17) (IC50 = 0.5 μm), and (iv) a4N(1–17) (IC50 = 1.1 μm). B and C, model of the novel role of V-ATPase nanomotor as a pH-sensing and cytohesin-signaling receptor. The molecular details of the interactions of V-ATPase with cytohesin-2 and Arf GTP-binding proteins at high (B) and low (C) levels of luminal acidification. B, schematic representation of the rotary proton-pumping eukaryotic V-ATPase that does not interact with cytohesin-2 and myristoylated Arf6/Arf1 GTP-binding proteins. Cytohesin-2 is anchored on the PIP2-containing membrane via its pleckstrin homology domain (PH). In this scenario cytohesin-2 and Arfs are signaling with their downstream effectors independently of V-ATPase. However, the function of V-ATPase does not affected by cytohesin-2 and Arf GTP-binding proteins. In this schematic, the recently solved structure of yeast V-ATPase was used according to Ref. 5. It shows some structural features of a-subunit isoform (green) and its binding partners within V-ATPase. The positions of the A (yellow), B (red), C (brown), E (purple), G (beige), H (orange), and DF (blue) subunits and c-ring (magenta) are also shown (5). The N-terminal cytosolic tail of V-ATPase a-subunit (aN) is situated in parallel to the membrane plane and could have contacts with membrane (66). The position of first 17-amino acid peptide-forming epitope (aN(1–17)) involved in interaction with the Sec-7 domain of cytohesin-2 is also indicated. The C-terminal transmembrane part of V-ATPase a-subunit (aC) is shown with the drawn eight transmembrane helices and four luminal loops as suggested previously for mouse a2-subunit isoform of V-ATPase (7). The location of corresponding pH-sensing histidine amino acid residues is indicated as red circles (7). This membrane topology was recently defined for S. cerevisiae Vph1p-subunit of V-ATPase (67) and identified by cryo-electron microscopy for Thermus thermophilus I-subunit of A-ATPase (68). C, after reaching certain low levels of pH, V-ATPase is sensing luminal acidification by its a-subunit isoform. We previously proposed that the transmembrane aC part of a-subunit senses pH by its histidine residues and signals through its conformational changes to the cytosolic aN part of the a-subunit (7). This results in interaction of cytohesin-2 with the cytosolic aN part of a-subunit and its recruitment to V-ATPase. The molecular details uncovered in this study revealed that this binding involves the N-terminal epitope of V-ATPase a-subunits (aN(1–17)) and Sec7 domain of cytohesin-2. Thus, during this cell biological event, V-ATPase serves as a “molecular sink” by recruiting cytohesin-2 and also inhibits its signaling. As a result of the competitive binding of V-ATPase with cytohesin-2, Arfs may be released from cytohesin-2, and Arf6 could interact with the c-subunit ring of V-ATPase in a pH-dependent manner (6, 7). Thus, under this scenario both Arf1 and Arf6 may continue signaling with different GEFs and modulate the function of a different set of downstream effectors. Finally, the interaction of V-ATPase with cytohesin-2 and Arf6 also may be involved in modulation of the function of V-ATPase itself (1).
Previously, we identified the N-terminal epitope of the cytosolic tail of the a2-subunit of V-ATPase as a major interacting site with cytohesin-2 (27). This epitope corresponds to the peptide a2N(1–17), which is formed by the first 17 amino acids (MGSLFRSESMCLAQLFL) of the a2-subunit isoform (27). Thus, here we hypothesized that this V-ATPase epitope-forming a2N(1–17) peptide is crucial for signaling between V-ATPase and cytohesin-2. Indeed, our studies revealed that a2N(1–17) peptide is a potent inhibitor of the enzymatic GDP/GTP exchange activity of cytohesin-2 with both Arf1 and Arf6 as substrates. Moreover, we demonstrated that this peptide strongly inhibits GEF activity via its direct interaction with the catalytic Sec7 domain. The α-helical structure of a2N(1–17) and its residues Phe5, Met10, and Gln14 binding with the Sec7 domain were also identified. Moreover, in silico docking experiments revealed that a2N(1–17) peptide binds to the αG, αH, and αI helices of the Sec7 domain and thus competes with Arf1 and Arf6 GTP-binding proteins for a binding site on the switch 2 region (48, 56). Together, our data reveal the structural basis and molecular mechanism of the inhibitory action of the V-ATPase derived peptide a2N(1–17) on GEF activity. Therefore, we have uncovered a novel mechanism of signaling involving interaction of the V-ATPase holo-complex with cytohesin-2 and Arf family GTP-binding proteins (Fig. 8).
It is generally accepted that the two yeast a-subunit isoforms (Vph1p and Stv1p) and four mammalian a-subunit isoforms (a1, a2, a3, and a4) have a central role in assembly, differential targeting, and function of V-ATPase (1, 3, 4). In humans, the importance of the structural integrity of full-length a-subunit isoforms in V-ATPase function is strongly supported by the existence of genetic diseases associated with mutations in these proteins (57–59). For example, loss-of-function mutations in the a2-subunit were identified as cause of autosomal recessive cutis laxa (ARCL) type II (Debre type) or wrinkly skin syndrome (59). To date, a total of 41 mutations were identified, of which 18 missense and frameshift mutations were located on the N-terminal cytosolic tail of a2-subunit (a2N) (60, 61). Importantly, one of the frameshift mutations described in these studies is involved in modification of structural integrity of the a2N(1–17) epitope described here. At the molecular and cellular level, these mutations give rise to loss of integrity of the a2-subunit, loss of V-ATPase function, impaired vesicular trafficking, and cell survival (59–61). These genetic studies strongly argue in favor of the importance of the structural integrity of full-length a-subunit isoforms in V-ATPase function in situ. Similarly, our previous (6) and current (Fig. 7) studies also show that an intact a-subunit containing V-ATPase holo-complex is involved in interaction and signaling with cytohesin-2 and Arf GTP-binding proteins (Fig. 8).
However, an unconventional role of a truncated form of the a2-subunit in lymphoid and trophoblast cells was also reported previously (62). These cells express 45-, 52-, and 70-kDa homologs of the a2-isoform, which was formerly known as regeneration and tolerance factor (RTF or TJ6) (63–65). In particular, it was reported that upon cell activation, the 70-kDa RTF/TJ6 is relocated to plasma membrane and proteolytically cleaved to generate a so-called a2NTD peptide, a 20-kDa protein that corresponds to amino acids a2N(142–344) of the a2N cytosolic tail (62). However, although the effects of extracellular a2NTD in cell-cell communication were intensively studied, its intracellular generation and relevance to the physiological function of V-ATPase as a proton pump are currently unknown. Moreover, it is also unknown if RTF/TJ6 is ever a part of an intact and functional V-ATPase. Indeed, a recent model proposes that RTF/TJ6 traffics and functions as a distinct and separate protein (62). Therefore, because RTF/TJ6 lacks the major critical part of its N-terminal cytosolic tail and functions as a separate protein, it is most likely that it is not relevant to the cell biological events described in our study.
However, we previously demonstrated that interaction with cytohesin-2 is not only restricted to the a2-subunit isoform but also occurs with the three other a-subunit isoforms (a1, a3, and a4) of the V-ATPase (26). It is noteworthy that these mammalian isoforms as well as two yeast a-subunit isoforms (Vph1p and Stv1p) function as full-length proteins and do not undergo proteolytic cleavage. Sequence alignment of the N-terminal epitope of a-subunit isoforms shows that the V-ATPase amino acids involved in interaction with Sec7 domain are highly conserved in all eukaryotes from yeast to humans. Accordingly, our additional enzymatic GEF activity studies revealed that N-terminal peptides derived from another three a-subunit isoforms (a1, a3, and a4) of the V-ATPase are potent inhibitors of cytohesin-2. Thus, these data reveal the conserved character of signaling between all four a1-, a2-, a3-, and a4-subunit isoforms of mammalian V-ATPase and cytohesin-2 (Fig. 8A). Our present findings also indicate that signaling between V-ATPase and cytohesin-2 is a general cell biological phenomenon. We suggest that, because a-isoforms are involved in targeting the V-ATPase to different cellular compartments (1), the signaling between intact V-ATPase and cytohesin-2, Arf GTP-binding proteins could take place on endosomal, lysosomal, Golgi, or plasma membranes among others (1).
What are the downstream effectors of the signaling between V-ATPase and cytohesin-2/Arf GTP-binding proteins? There are various intracellular downstream effectors and pathways that may be modulated by the signaling between V-ATPase and cytohesin/Arf GTP-binding proteins. First, we have shown that pH-dependent interaction between cytohesin-2 and a2-isoform containing holo-complexes of V-ATPase is crucial for the function of early endosomes both in vitro (55) and in vivo (Fig. 8, B and C) (6). Our studies indicate the accessibility of endogenous cytohesin-2 to intact and functional V-ATPase complexes in vivo. Recently, the structure of intact eukaryotic holo-complex of S. cerevisiae V-ATPase was solved at 11 Å resolution (5). This resolution is sufficient to resolve almost all of the subunits in the V-ATPase holo-complex and to reveal the binding interfaces with its interacting partners. A three-dimensional map of V-ATPase with the a-subunit (Vph1p homolog) structure highlighted is shown in Figs. 7D and 8, B and C. Our data clearly show the accessibility of the aN(1–17) forming an epitope for binding with cytohesin-2 (Figs. 7D and 8, B and C) and its interaction with an intact V-ATPase holo-complex (Fig. 7, F and G). Second, we have previously proposed that interaction of V-ATPase with cytohesin-2 and Arf6 could be an integral part of their role as “molecular switches.” In this scenario, cytohesin-2 and Arf6 may be involved in self-regulation of the V-ATPase function and, thus turning “on/off” this remarkable nanomotor (Fig. 8, B and C) (1). Third, both cytohesin-2 and Arf GTP-binding proteins are also important for modulation of the following: (i) membrane coat recruitment; (ii) phospholipid modification; (iii) microtubule-dependent vesicular movement (13), and (iv) remodeling of the actin cytoskeleton (9–12, 26). Fourth, cytohesin-2 recently emerged as an essential modulator of the insulin receptor (23, 24) and EGFR/ErbB-receptors (18) signaling at the plasma membrane and endosomal/lysosomal pathway. Thus, the intracellular interaction and signaling of the V-ATPase with cytohesin-2/Arf GTP-binding proteins could potentially modulate these multiple downstream effectors as well. Importantly, it is noteworthy that under this scenario V-ATPase, cytohesin-2/Arf GTP-binding proteins, and their downstream effectors should be located at the same membrane microenvironment (Fig. 8, B and C).
Here, we identified and characterized four novel a-isoform-specific V-ATPase-derived peptides (a1N(1–17), a2N(1–17), a3N(1–17), and a4N(1–17)) that are potent inhibitors of cytohesin-2 GEF activity in vitro. Importantly, we determined the IC50 values for all four a-subunit isoform-specific peptides (Fig. 8A). The interaction-competent amino acids (Phe5 and Gln14) are completely conserved in all eukaryotic a1-, a2-, a3-, and a4-isoforms, and their corresponding specific peptides show similar IC50 ∼1.0 μm values. Interestingly, a single substitution in one of the interaction competent amino acids of the a3-isoform (Met10 to Val10) doubles (IC50 ∼0.5 μm) the anti-cytohesin activity of the a3-isoform-specific peptide. Thus, these data open up the possibility of designing novel peptides with differential anti-cytohesin activities. In conclusion, the structural interface of signaling between the V-ATPase and cytohesin-2 reported here provides the impetus for further development of novel and more potent anti-cytohesin pharmaceuticals/peptides. It also helps to direct future screening and/or the rational design of compounds targeting cytohesins. In particular, these novel isoform-specific pharmaceuticals and drugs could be used to prevent the cytohesin-2-dependent activation of EGFR/ErbB receptors to treat a variety of cancers (17–19) and to prevent development of diabetic nephropathy in type I diabetes (20–22). Moreover, the cytohesin/insulin-receptor signaling is inhibited during type II diabetes. Therefore, design of the peptides or drugs able to activate cytohesins and prevent this inhibition might be useful in treating type II diabetes (23, 24).
In summary, we have discovered an evolutionarily conserved function of the V-ATPase as a novel cytohesin-signaling receptor. Our data reveal that during its function as a pH-sensing receptor, V-ATPase is not simply a scaffold to attach cytohesin-2 and Arfs to their target membranes, but it also modulates signaling of cytohesin-2 and Arf GTP-binding proteins. These cell biological events could consequently modulate function of V-ATPase itself as well as downstream effectors, including EGFR/ErbB and insulin receptors among others (Fig. 8, B and C). Finally, our data also underline the evolutionarily conserved character of signaling between V-ATPase and cytohesins/Arfs, which may take place in different intracellular organelles and plasma membranes of eukaryotic cells from yeast to humans.
Supplementary Material
Acknowledgments
We thank Dr. Dennis Brown for critical reading and constructive suggestions during the preparation of this manuscript. We are grateful to Dr. Masamitsu Futai and Dr. James Casanova for providing cDNA encoding V-ATPase a2-isoform and cytohesin-2, respectively. We are also grateful to Dr. Sylvain Bourgoin for the generous gift of SecinH3 inhibitor. The Microscopy Core Facility of the Program in Membrane Biology received additional support from National Institutes of Health Grant DK57521 to BADERC and Grant DK43351 to Center for the Study of Inflammatory Bowel Disease.
This work was supported, in whole or in part, by National Institutes of Health Grant DK038452. This work was also supported by Boston Area Diabetes Endocrinology Research Center Grant DK057521-08 (to V. M.).

This article contains supplemental Tables S1 and S2 and Movies S1 and S2.
The atomic coordinates and structure factors (code 2LX4) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- V-ATPase
- vacuolar H+-ATPase
- GEF
- GDP/GTP-exchange factor
- GTPγS
- guanosine 5′-O-[γ-thio]triphosphate
- PG
- phosphatidylglycerol
- PIP2
- phosphatidylinositol-4,5-bisphosphate; (PIP2)
- EGFP
- enhanced green fluorescent protein
- TOCSY
- total correlation spectroscopy
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- myr
- myristoylated
- HSQC
- heteronuclear single quantum coherence
- RTF
- regeneration and tolerance factor
- EGFR
- EGF receptor.
REFERENCES
- 1. Marshansky V., Futai M. (2008) The V-type H+-ATPase in vesicular trafficking: targeting, regulation, and function. Curr. Opin. Cell Biol. 20, 415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grüber G., Marshansky V. (2008) New insights into structure-function relationships between archaeal ATP synthase (A1A0) and vacuolar type ATPase (V1V0). BioEssays 30, 1096–1109 [DOI] [PubMed] [Google Scholar]
- 3. Nishi T., Forgac M. (2002) The vacuolar H+-ATPases–nature's most versatile proton pumps. Nat. Rev. Mol. Cell Biol. 3, 94–103 [DOI] [PubMed] [Google Scholar]
- 4. Forgac M. (2007) Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8, 917–929 [DOI] [PubMed] [Google Scholar]
- 5. Benlekbir S., Bueler S. A., Rubinstein J. L. (2012) Structure of the vacuolar-type ATPase from Saccharomyces cerevisiae at 11-Å resolution. Nat. Struct. Mol. Biol. 19, 1356–1362 [DOI] [PubMed] [Google Scholar]
- 6. Hurtado-Lorenzo A., Skinner M., El Annan J., Futai M., Sun-Wada G. H., Bourgoin S., Casanova J., Wildeman A., Bechoua S., Ausiello D. A., Brown D., Marshansky V. (2006) V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat. Cell Biol. 8, 124–136 [DOI] [PubMed] [Google Scholar]
- 7. Marshansky V. (2007) The V-ATPase a2-subunit as a putative endosomal pH-sensor. Biochem. Soc. Trans. 35, 1092–1099 [DOI] [PubMed] [Google Scholar]
- 8. Recchi C., Chavrier P. (2006) V-ATPase: a potential pH sensor. Nat Cell Biol. 8, 107–109 [DOI] [PubMed] [Google Scholar]
- 9. Donaldson J. G., Jackson C. L. (2000) Regulators and effectors of the ARF GTPases. Curr. Opin. Cell Biol. 12, 475–482 [DOI] [PubMed] [Google Scholar]
- 10. D'Souza-Schorey C., Chavrier P. (2006) ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7, 347–358 [DOI] [PubMed] [Google Scholar]
- 11. Casanova J. E. (2007) Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic 8, 1476–1485 [DOI] [PubMed] [Google Scholar]
- 12. Randazzo P. A., Nie Z., Miura K., Hsu V. W. (2000) Molecular aspects of the cellular activities of ADP-ribosylation factors. Sci. STKE 2000, re1 [DOI] [PubMed] [Google Scholar]
- 13. Montagnac G., de Forges H., Smythe E., Gueudry C., Romao M., Salamero J., Chavrier P. (2011) Decoupling of activation and effector binding underlies ARF6 priming of fast endocytic recycling. Curr. Biol. 21, 574–579 [DOI] [PubMed] [Google Scholar]
- 14. Cruciat C. M., Ohkawara B., Acebron S. P., Karaulanov E., Reinhard C., Ingelfinger D., Boutros M., Niehrs C. (2010) Requirement of prorenin receptor and vacuolar H+-ATPase-mediated acidification for Wnt signaling. Science 327, 459–463 [DOI] [PubMed] [Google Scholar]
- 15. Vaccari T., Duchi S., Cortese K., Tacchetti C., Bilder D. (2010) The vacuolar ATPase is required for physiological as well as pathological activation of the Notch receptor. Development 137, 1825–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., Sabatini D. M. (2011) mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H-ATPase. Science 334, 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hafner M., Vianini E., Albertoni B., Marchetti L., Grüne I., Gloeckner C., Famulok M. (2008) Displacement of protein-bound aptamers with small molecules screened by fluorescence polarization. Nat. Protoc. 3, 579–587 [DOI] [PubMed] [Google Scholar]
- 18. Bill A., Schmitz A., Albertoni B., Song J. N., Heukamp L. C., Walrafen D., Thorwirth F., Verveer P. J., Zimmer S., Meffert L., Schreiber A., Chatterjee S., Thomas R. K., Ullrich R. T., Lang T., Famulok M. (2010) Cytohesins are cytoplasmic ErbB receptor activators. Cell 143, 201–211 [DOI] [PubMed] [Google Scholar]
- 19. Bill A., Schmitz A., König K., Heukamp L. C., Hannam J. S., Famulok M. (2012) Anti-proliferative effect of cytohesin inhibition in gefitinib-resistant lung cancer cells. PLoS ONE 7, e41179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sandholm N., Salem R. M., McKnight A. J., Brennan E. P., Forsblom C., Isakova T., McKay G. J., Williams W. W., Sadlier D. M., Mäkinen V. P., Swan E. J., Palmer C., Boright A. P., Ahlqvist E., Deshmukh H. A., Keller B. J., Huang H., Ahola A. J., Fagerholm E., Gordin D., Harjutsalo V., He B., Heikkilä O., Hietala K., Kytö J., Lahermo P., Lehto M., Lithovius R., Osterholm A. M., Parkkonen M., Pitkäniemi J., Rosengård-Bärlund M., Saraheimo M., Sarti C., Söderlund J., Soro-Paavonen A., Syreeni A., Thorn L. M., Tikkanen H., Tolonen N., Tryggvason K., Tuomilehto J., Wadén J., Gill G. V., Prior S., Guiducci C., Mirel D. B., Taylor A., Hosseini S. M., DCCT/EDIC Research Group, Parving H. H., Rossing P., Tarnow L., Ladenvall C., Alhenc-Gelas F., Lefebvre P., Rigalleau V., Roussel R., Tregouet D. A., Maestroni A., Maestroni S., Falhammar H., Gu T., Möllsten A., Cimponeriu D., Ioana M., Mota M., Mota E., Serafinceanu C., Stavarachi M., Hanson R. L., Nelson R. G., Kretzler M., Colhoun H. M., Panduru N. M., Gu H. F., Brismar K., Zerbini G., Hadjadj S., Marre M., Groop L., Lajer M., Bull S. B., Waggott D., Paterson A. D., Savage D. A., Bain S. C., Martin F., Hirschhorn J. N., Godson C., Florez J. C., Groop P. H., Maxwell A. P. (2012) New susceptibility Loci associated with kidney disease in type 1 diabetes. PLoS Genet. 8, e1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Böger C. A., Sedor J. R. (2012) GWAS of diabetic nephropathy. Is the GENIE out of the bottle? PLoS Genet 8, e1002989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Panchapakesan U., Pollock C., Saad S. (2011) Renal epidermal growth factor receptor: its role in sodium and water homeostasis in diabetic nephropathy. Clin. Exp. Pharmacol. Physiol. 38, 84–88 [DOI] [PubMed] [Google Scholar]
- 23. Hafner M., Schmitz A., Grüne I., Srivatsan S. G., Paul B., Kolanus W., Quast T., Kremmer E., Bauer I., Famulok M. (2006) Inhibition of cytohesins by SecinH3 leads to hepatic insulin resistance. Nature 444, 941–944 [DOI] [PubMed] [Google Scholar]
- 24. Fuss B., Becker T., Zinke I., Hoch M. (2006) The cytohesin Steppke is essential for insulin signaling in Drosophila. Nature 444, 945–948 [DOI] [PubMed] [Google Scholar]
- 25. Jackson C. (2006) Diabetes: kicking off the insulin cascade. Nature 444, 833–834 [DOI] [PubMed] [Google Scholar]
- 26. Merkulova M., Hurtado-Lorenzo A., Hosokawa H., Zhuang Z., Brown D., Ausiello D. A., Marshansky V. (2011) Aldolase directly interacts with ARNO and modulates cell morphology and acidic vesicle distribution. Am. J. Physiol. Cell Physiol. 300, C1442–C1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Merkulova M., Bakulina A., Thaker Y. R., Grüber G., Marshansky V. (2010) Specific motifs of the V-ATPase a2-subunit isoform interact with catalytic and regulatory domains of ARNO. Biochim. Biophys. Acta 1797, 1398–1409 [DOI] [PubMed] [Google Scholar]
- 28. van den Ent F., Löwe J. (2006) RF cloning: a restriction-free method for inserting target genes into plasmids. J Biochem. Biophys. Methods 67, 67–74 [DOI] [PubMed] [Google Scholar]
- 29. Toyomura T., Oka T., Yamaguchi C., Wada Y., Futai M. (2000) Three subunit a isoforms of mouse vacuolar H+-ATPase. Preferential expression of the a3 isoform during osteoclast differentiation. J. Biol. Chem. 275, 8760–8765 [DOI] [PubMed] [Google Scholar]
- 30. Fields C. G., Lloyd D. H., Macdonald R. L., Otteson K. M., Noble R. L. (1991) HBTU activation for automated Fmoc solid-phase peptide synthesis. Pept. Res. 4, 95–101 [PubMed] [Google Scholar]
- 31. King D. S., Fields C. G., Fields G. B. (1990) A cleavage method which minimizes side reactions following Fmoc solid phase peptide synthesis. Int. J. Pept. Protein Res. 36, 255–266 [DOI] [PubMed] [Google Scholar]
- 32. Ha V. L., Thomas G. M., Stauffer S., Randazzo P. A. (2005) Preparation of myristoylated Arf1 and Arf6. Methods Enzymol. 404, 164–174 [DOI] [PubMed] [Google Scholar]
- 33. Santy L. C., Frank S. R., Hatfield J. C., Casanova J. E. (1999) Regulation of ARNO nucleotide exchange by a PH domain electrostatic switch. Curr. Biol. 9, 1173–1176 [DOI] [PubMed] [Google Scholar]
- 34. Macia E., Paris S., Chabre M. (2000) Binding of the PH and polybasic C-terminal domains of ARNO to phosphoinositides and to acidic lipids. Biochemistry 39, 5893–5901 [DOI] [PubMed] [Google Scholar]
- 35. Viaud J., Zeghouf M., Barelli H., Zeeh J. C., Padilla A., Guibert B., Chardin P., Royer C. A., Cherfils J., Chavanieu A. (2007) Structure-based discovery of an inhibitor of Arf activation by Sec7 domains through targeting of protein-protein complexes. Proc. Natl. Acad. Sci. U.S.A. 104, 10370–10375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Manavalan P., Johnson W. C., Jr. (1987) Variable selection method improves the prediction of protein secondary structure from circular dichroism spectra. Anal. Biochem. 167, 76–85 [DOI] [PubMed] [Google Scholar]
- 37. Sreerama N., Woody R. W. (1993) A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 209, 32–44 [DOI] [PubMed] [Google Scholar]
- 38. Provencher S. W. (1982) A constrained regularization method for inverting data represented by linear algebraic or integral equations. Comput. Physics Commun. 27, 213–227 [Google Scholar]
- 39. Andrade M. A., Chacón P., Merelo J. J., Morán F. (1993) Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 6, 383–390 [DOI] [PubMed] [Google Scholar]
- 40. Deléage G., Geourjon C. (1993) An interactive graphic program for calculating the secondary structure content of proteins from circular dichroism spectrum. Comput. Appl. Biosci. 9, 197–199 [DOI] [PubMed] [Google Scholar]
- 41. Böhm G., Muhr R., Jaenicke R. (1992) Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 5, 191–195 [DOI] [PubMed] [Google Scholar]
- 42. Goddard T. D., Kneller D. G. (1997) SPARKY Version 3.110, University of California, San Francisco [Google Scholar]
- 43. Wüthrich K. (1986) NMR of Proteins and Nucleic Acids, pp. 146–154, Wiley Interscience, New York [Google Scholar]
- 44. Cornilescu G., Delaglio F., Bax A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 45. Güntert P., Mumenthaler C., Wüthrich K. (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 46. Huey R., Morris G. M., Olson A. J., Goodsell D. S. (2007) A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 28, 1145–1152 [DOI] [PubMed] [Google Scholar]
- 47. Seeliger D., de Groot B. L. (2010) Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 24, 417–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Renault L., Guibert B., Cherfils J. (2003) Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature 426, 525–530 [DOI] [PubMed] [Google Scholar]
- 49. Liu Z., Segawa H., Aydin C., Reyes M., Erben R. G., Weinstein L. S., Chen M., Marshansky V., Fröhlich L. F., Bastepe M. (2011) Transgenic overexpression of the extra-large Gsα variant XLαs enhances Gsα-mediated responses in the mouse renal proximal tubule in vivo. Endocrinology 152, 1222–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Macia E., Chabre M., Franco M. (2001) Specificities for the small G proteins ARF1 and ARF6 of the guanine nucleotide exchange factors ARNO and EFA6. J. Biol. Chem. 276, 24925–24930 [DOI] [PubMed] [Google Scholar]
- 51. Béraud-Dufour S., Robineau S., Chardin P., Paris S., Chabre M., Cherfils J., Antonny B. (1998) A glutamic finger in the guanine nucleotide exchange factor ARNO displaces Mg2+ and the β-phosphate to destabilize GDP on ARF1. EMBO J. 17, 3651–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rishikesan S., Thaker Y. R., Priya R., Gayen S., Manimekalai M. S., Hunke C., Grüber G. (2008) Spectroscopical identification of residues of subunit G of the yeast V-ATPase in its connection with subunit E. Mol. Membr. Biol. 25, 400–410 [DOI] [PubMed] [Google Scholar]
- 53. Dip P. V., Saw W. G., Roessle M., Marshansky V., Grüber G. (2012) Solution structure of subunit a, a(104–363), of the Saccharomyces cerevisiae V-ATPase and the importance of its C-terminus in structure formation. J. Bioenerg. Biomembr. 44, 341–350 [DOI] [PubMed] [Google Scholar]
- 54. Thaker Y. R., Hunke C., Yau Y. H., Shochat S. G., Li Y., Grüber G. (2009) Association of the eukaryotic V1VO ATPase subunits a with d and d with A. FEBS Lett. 583, 1090–1095 [DOI] [PubMed] [Google Scholar]
- 55. Maranda B., Brown D., Bourgoin S., Casanova J. E., Vinay P., Ausiello D. A., Marshansky V. (2001) Intra-endosomal pH-sensitive recruitment of the Arf-nucleotide exchange factor ARNO and Arf6 from cytoplasm to proximal tubule endosomes. J. Biol. Chem. 276, 18540–18550 [DOI] [PubMed] [Google Scholar]
- 56. Mossessova E., Gulbis J. M., Goldberg J. (1998) Structure of the guanine nucleotide exchange factor Sec7 domain of human ARNO and analysis of the interaction with ARF GTPase. Cell 92, 415–423 [DOI] [PubMed] [Google Scholar]
- 57. Frattini A., Orchard P. J., Sobacchi C., Giliani S., Abinun M., Mattsson J. P., Keeling D. J., Andersson A. K., Wallbrandt P., Zecca L., Notarangelo L. D., Vezzoni P., Villa A. (2000) Defects in TCIRG1 subunit of the vacuolar proton pump are responsible for a subset of human autosomal recessive osteopetrosis. Nat. Genet. 25, 343–346 [DOI] [PubMed] [Google Scholar]
- 58. Smith A. N., Skaug J., Choate K. A., Nayir A., Bakkaloglu A., Ozen S., Hulton S. A., Sanjad S. A., Al-Sabban E. A., Lifton R. P., Scherer S. W., Karet F. E. (2000) Mutations in ATP6N1B, encoding a new kidney vacuolar proton pump 116-kD subunit, cause recessive distal renal tubular acidosis with preserved hearing. Nat. Genet. 26, 71–75 [DOI] [PubMed] [Google Scholar]
- 59. Kornak U., Reynders E., Dimopoulou A., van Reeuwijk J., Fischer B., Rajab A., Budde B., Nürnberg P., Foulquier F., ARCL Debré-type Study Group, Lefeber D., Urban Z., Gruenewald S., Annaert W., Brunner H. G., van Bokhoven H., Wevers R., Morava E., Matthijs G., Van Maldergem L., Mundlos S. (2008) Impaired glycosylation and cutis laxa caused by mutations in the vesicular H+-ATPase subunit ATP6V0A2. Nat. Genet. 40, 32–34 [DOI] [PubMed] [Google Scholar]
- 60. Hucthagowder V., Morava E., Kornak U., Lefeber D. J., Fischer B., Dimopoulou A., Aldinger A., Choi J., Davis E. C., Abuelo D. N., Adamowicz M., Al-Aama J., Basel-Vanagaite L., Fernandez B., Greally M. T., Gillessen-Kaesbach G., Kayserili H., Lemyre E., Tekin M., Türkmen S., Tuysuz B., Yüksel-Konuk B., Mundlos S., Van Maldergem L., Wevers R. A., Urban Z. (2009) Loss-of-function mutations in ATP6V0A2 impair vesicular trafficking, tropoelastin secretion, and cell survival. Hum. Mol. Genet. 18, 2149–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fischer B., Dimopoulou A., Egerer J., Gardeitchik T., Kidd A., Jost D., Kayserili H., Alanay Y., Tantcheva-Poor I., Mangold E., Daumer-Haas C., Phadke S., Peirano R. I., Heusel J., Desphande C., Gupta N., Nanda A., Felix E., Berry-Kravis E., Kabra M., Wevers R. A., van Maldergem L., Mundlos S., Morava E., Kornak U. (2012) Further characterization of ATP6V0A2-related autosomal recessive cutis laxa. Hum. Genet. 131, 1761–1773 [DOI] [PubMed] [Google Scholar]
- 62. Ntrivalas E., Levine R., Kwong C., Gilman-Sachs A., Beaman K. (2010) The a2 isoform of vacuolar ATPase is a modulator of implantation and feto-maternal immune tolerance in early pregnancy. J. Reprod. Immunol. 85, 106–111 [DOI] [PubMed] [Google Scholar]
- 63. Lee C., Ghoshal K., Beaman K. D. (1990) Cloning of a cDNA for a T cell produced molecule with a putative immune regulatory role. Mol. Immunol. 27, 1137–1144 [DOI] [PubMed] [Google Scholar]
- 64. Nichols T. C., Kang J. A., Angkachatchai V., Beer A. E., Beaman K. D. (1994) Expression of a membrane form of the pregnancy-associated protein TJ6 on lymphocytes. Cell. Immunol. 155, 219–229 [DOI] [PubMed] [Google Scholar]
- 65. Beaman K., Angkachatchai V., Gilman-Sachs A. (1996) TJ6: the pregnancy-associated cytokine. Am. J. Reprod. Immunol. 35, 338–341 [DOI] [PubMed] [Google Scholar]
- 66. Merkulova M., McKee M., Dip P. V., Grüber G., Marshansky V. (2010) N-terminal domain of the V-ATPase a2-subunit displays integral membrane protein properties. Protein Sci. 19, 1850–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Toei M., Toei S., Forgac M. (2011) Definition of membrane topology and identification of residues important for transport in subunit a of the vacuolar ATPase. J. Biol. Chem. 286, 35176–35186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lau W. C., Rubinstein J. L. (2011) Subnanometre-resolution structure of the intact Thermus thermophilus H+-driven ATP synthase. Nature 481, 214–218 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.