Background: ARAP2 is an ArfGAP that regulates focal adhesions (FAs) by an unknown mechanism.
Results: ARAP2 activity controlled FA size and regulated cellular Arf6·GTP and Rac1·GTP. The effect of loss of ARAP2 on FAs was reversed by dominant negative Arf6 or Rac1.
Conclusion: ARAP2 functions in part by inhibiting the Arf6/Rac1 pathway.
Significance: We describe a mechanism by which an ArfGAP can regulate FAs.
Keywords: Adhesion, ARF, Cell Adhesion, G Proteins, Rac1, ARAP2, Arf6, GTP-binding Protein, GTPase-activating Protein, Focal Adhesion
Abstract
Focal adhesions (FAs) are dynamic structures that connect the actin cytoskeleton with the extracellular matrix. At least six ADP-ribosylation factor (Arf) GTPase-activating proteins (GAPs), including ARAP2 (an Arf6 GAP), are implicated in regulation of FAs but the mechanisms for most are not well defined. Although Rac1 has been reported to function downstream of Arf6 to control membrane ruffling and cell migration, this pathway has not been directly examined as a regulator of FAs. Here we test the hypothesis that ARAP2 promotes the growth of FAs by converting Arf6·GTP to Arf6·GDP thereby preventing the activation of the Rho family GTP-binding protein Rac1. Reduced expression of ARAP2 decreased the number and size of FAs in cells and increased cellular Arf6·GTP and Rac1·GTP levels. Overexpression of ARAP2 had the opposite effects. The effects of ARAP2 on FAs and Rac1 were dependent on a functional ArfGAP domain. Constitutively active Arf6 affected FAs in the same way as did reduced ARAP2 expression and dominant negative mutants of Arf6 and Rac1 reversed the effect of reduced ARAP2 expression. However, neither dominant negative Arf6 nor Rac1 had the same effect as ARAP2 overexpression. We conclude that changes in Arf6 and Rac1 activities are necessary but not sufficient for ARAP2 to promote the growth of FAs and we speculate that ARAP2 has additional functions that are effector in nature to promote or stabilize FAs.
Introduction
Focal adhesions (FAs)2 connect the actin cytoskeleton with the extracellular matrix. They are critical to cell survival, proliferation and movement (1–4). The function of FAs is closely related to their dynamic nature. Structural components such as integrin and paxillin cluster in the cell periphery forming the precursor structures, nascent adhesions, with an average size around 0.2 μm2 (5). Nascent adhesions can grow and mature into the intermediate sized focal complexes (0.5∼1 μm2) and then FAs that are typically larger than 1 μm2 (4). The formation and maturation of FAs is controlled by two subfamilies within the Ras superfamily. Rho family proteins have well described effects through a number of well established effectors (6). ADP-ribosylation factor (Arf) family GTP-binding proteins have also been implicated (7). The molecular basis for the function of Arfs in FAs is still being discovered but is thought to be related to the structurally diverse Arf GTPase-activating proteins (GAPs) that regulate Arfs and are found in FAs (8).
Over 20 genes encode Rho family GTP binding proteins in humans (9). Three have been studied extensively as regulators of the actin cytoskeleton. The activation of Rac1, i.e. formation of Rac1·GTP, drives the formation of actin rich membrane ruffles and focal complexes and opposes the formation of FAs. Similarly, Cdc42·GTP blocks the formation of FAs. RhoA·GTP induces the formation of FAs and actin stress fibers (10, 11). Many of the factors leading to the activation of Rho family proteins have been identified, and signaling pathways have been defined. Arf family GTP-binding proteins have been implicated as regulators of Rho family proteins, and the activity of the Arf and Rho family proteins are thought to be specifically coordinated (12–16).
Five genes in humans encode for Arfs, six in other mammals. The best studied Arf proteins are Arf1 and Arf6. Both have been implicated as regulators of the actin cytoskeleton and related structures. Activated Arf1 has been found to promote actin stress fiber formation and the recruitment of paxillin, a component of the cytoplasmic FA plaque, to FAs (7). Furthermore, activated Arf1 was recently shown to cooperate with Rac1 to stimulate WAVE-dependent actin polymerization (17). Activated Arf6-induced actin rich protrusions in HeLa cells (18). Subsequent work identified Arf6 function in cell migration, phagocytosis, and the disassembly of FAs (19–23). Rac1, a Rho family GTP-binding protein has been reported to function both upstream and downstream of Arf6 to control actin rich protrusions (12, 13, 24), but whether the inter-regulation between Arf6 and Rac1 plays any roles in FA formation has not been studied.
The function of Arf proteins relies on the binding and hydrolysis of GTP. Because Arfs have no detectable GTPase activity, their function is dependent on ArfGAPs, which catalyze the hydrolysis of GTP bound to Arfs. Thirty-one genes in humans encode ArfGAPs. At least nine ArfGAPs affect the actin cytoskeleton and six are in FAs (8, 24–32). Three of the FA associated ArfGAPs, ARAP2, GIT1, and GIT2, function with Arf6 (29, 33). The mechanisms by which these ArfGAPs affect FAs are incompletely described. Overexpression of GIT1 caused reduction in FAs independent of ArfGAP activity (34). Instead, the effect was due to a scaffolding function in which GIT1 binds to the Rac activator PIX and to P21-activated kinase, a Rac1 effector. On the other hand, GIT1 reduces membrane protrusions from cells, which presumably coincides with stabilizing FAs (5, 10, 11), by inactivating Arf6, which, in turn, reduces Rac1·GTP (24). ARAP2 has been reported to slow cell spreading and promote FAs dependent on its ArfGAP activity; however, the relationship to the Rho pathway has not been explored (29). Furthermore, with the complex domain structure of ARAP2 (Fig. 1A), an additional scaffolding function may contribute to the effect of ARAP2 on FAs.
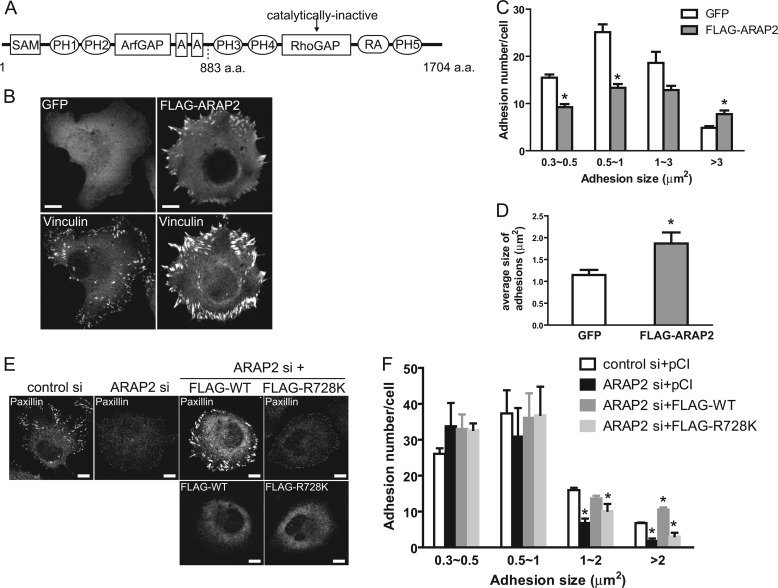
FIGURE 1.
Regulation of FA size by ARAP2 requires its ArfGAP activity. A, schematic representation of ARAP2 domain structure. SAM, sterile α-motif; PH, pleckstrin-homology domain; ArfGAP, ArfGTPase-activating protein; RhoGAP, RhoGTPase-activating protein; A, ankyrin repeat; RA, Ras-association domain. Numbers below the structure indicate corresponding amino acid position. B–D, effect of ARAP2 overexpression on FAs. HeLa cells were transfected with GFP or FLAG-ARAP2, plated onto fibronectin (FN)-coated (10 μg/ml) coverslips for 6 h, fixed and stained with anti-vinculin, anti-paxillin, and anti-FLAG antibodies to visualize FAs and FLAG-ARAP2. Representative images (B) and quantification of the adhesion number (C) and size (D) expressed as the mean ± S.E. of at least three experiments are shown. E and F, requirement for ArfGAP activity for effect of ARAP2. HeLa cells were cotransfected with siRNA and plasmids expressing FLAG-tagged wild type ARAP2 or [R728K]ARAP2 and processed as described for B–D. Representative images are shown in E and number of adhesions/cell (mean ± S.E.) for at least three experiments are shown in F. For all experiments presented in panels B–F, 30∼70 transfected cells were examined per construct. Bars, 10 μm. *, p < 0.05 compared with GFP control by Student's t test (C, D) or compared with control si+pCI using one way ANOVA with Dunnett's multiple comparison test (F).
Here, we examined the role of ARAP2 in the regulation of FAs, testing the hypothesis that the effects of ARAP2 on FAs are mediated by the ArfGAP activity resulting in reduced Arf6·GTP and consequent reduction in Rac1·GTP levels. We found that ARAP2, dependent on its ArfGAP activity, reduced both Arf6·GTP and Rac1·GTP levels. By quantification of FA size and number, we were also able to determine that the changes in Arf6 and Rac1 were necessary but not sufficient for the control of FAs by ARAP2. We speculate that in addition to the role as a negative regulator of Arf6, ARAP2 may have positive function as an effector.
MATERIALS AND METHODS
Plasmids
Mammalian expression vectors for N-terminal FLAG-tagged ARAP2 and [R728K]ARAP2 have been described (29). The bacterial expression vector for GST-fused VHSGAT of GGA3 has been described (13, 35). Plasmids for expressing C-terminal HA-tagged Arf mutants were kindly provided by Dr. Kazuhisa Nakayama (Kyoto University, Japan). pGEX-3X-PAK PBD plasmid was a generous gift from Dr. Silvio Gutkind (National Institutes of Health, Bethesda, MD). Mammalian expression constructs for N-terminal Myc-tagged [T17N]Rac1 and wild type Rac1(myc-Rac1) were purchased from Addgene (Addgene plasmids 12984 and 12985 deposited by Dr. Gary Bokoch, Cambridge, MA). An expression vector for GFP-[371-507]JIP3 LZII was a gift from Drs. Guillaume Montagnac and Philippe Chavrier (Institut Curie, France).
Antibodies and Reagents
Rabbit anti-ARAP2 antisera (antisera 1186) were raised against the same synthetic peptide RSRTLPKELQDEQILK as antisera 1185 described previously (29) (Covance Research, Princeton, NJ). 1186 antisera were affinity-purified using the peptides against which the antibody was raised with a purification kit from Thermo Scientific (Lafayette, CO). Affinity-purified 1186 antisera detected a band at 190 kDa by Western blotting. The band at 190 kDa was diminished in 3 cell lines treated with siRNA targeting ARAP2, providing evidence that this 190 kDa species was endogenous ARAP2. anti-FLAG polyclonal Ab, anti-Vinculin monoclonal Ab (hVIN-1), and fibronectin were purchased from Sigma-Aldrich. Anti-hemagglutinin (HA) rat monoclonal Ab (3F10) was from Roche Applied Science (Indianapolis, IN). Anti-Rac1 monoclonal Ab (23A8) was from Millipore (Temecula, CA). Anti-Paxillin monoclonal Ab (clone 349) was from BD Biosciences (Sparks, MD). Anti-AU5 monoclonal and anti-Myc polyclonal antibodies were from Covance and Abcam (Cambridge, MA). Anti-Arf6 serum were a generous gift from Dr. Julie Donaldson (National Institutes of Health, Bethesda, MD). Alexa Fluor-labeled secondary antibodies were from Invitrogen (Carlsbad, CA). Horseradish-peroxidase-conjugated anti-mouse and anti-rabbit IgG Abs were from Bio-Rad. All lipids were purchased from Avanti Polar Lipids (Alabaster, AL).
Cell Culture and Transfection
HeLa, T47D, and U118 cells were maintained at 37 °C in Dulbecco's modified Eagle's medium supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% fetal bovine serum (FBS). siRNA against ARAP2 (5′-GUAAGAAGACAUUGGGUUA-3′), and control siRNA (siCONTROL Non-Targeting siRNA#2) were purchased from Thermo Scientific (Lafayette, CO). Cells were transfected with 300 nm siRNA using Amaxa nucleofector system (Amaxa Inc., Gaithersburg, MD) or with 40 nm siRNA using Dharmafect (Thermo Scientific, Lafayette, CO) according to the manufacturer's instructions. Functional experiments with siRNA-transfected cells were performed 72 h following transfection when ARAP2 expression was reduced. For rescue experiments, 48 h after siRNA transfection, cells were transfected again with plasmids expressing epitope-tagged Arf mutants [T17N]Rac1 or GFP using Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA). 24 h later, focal adhesion morphology was assessed using confocal microscopy.
ArfGAP and RacGAP Assay
ArfGAP activity was determined by measuring hydrolysis of GTP bound to Arf using an in vitro assay (36). Myr-Arf1, myr-Arf6, and Rac1-His proteins were expressed and purified from bacteria as previously described (37–39). Myr-Arfs were loaded with [α-32P]GTP for 1 h at 30 °C as described in Refs. 37, 38 and Rac1-His was loaded with [α-32P]GTP under the same condition except for no MgCl2 in the buffer to reduce the basal GTP hydrolysis (39). Large Unilamellar Vesicles (LUVs) comprised of 40% phosphatidylcholine, 25% phosphatidylethanolamine, 15% phosphatidylserine, 7% PtdIns, 2.5% PtdIns (4,5)P2, 0.5% PtdIns (3,4,5)P3, and 10% cholesterol were prepared by extruding the lipid mixture through 1 μm pore polycarbonate membrane as described. Three sources of ARAP2 were used for the in vitro assay. Lysates of HeLa cells expressing FLAG-ARAP2 were prepared by scraping cells into 50 mm Tris-HCl pH 7.5, 100 mm NaCl, 2 mm MgCl2, 1 mm DTT, and 1% Triton X-100 supplemented with protease & phosphatase inhibitor mixture (Thermo Scientific, Lafayette, CO). Lysates were diluted with 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 2 mm MgCl2, and 1 mm DTT to decrease Triton X-100 concentration to 0.2%. Protein concentrations of the lysates were determined, and the amount of lysate indicated in the figure was included in the assay. The second source of ARAP2 was FA fractions from HeLa cells expressing FLAG-ARAP2 from plasmids. The third source of ARAP2 was His-[1–883]ARAP2 expressed and purified from bacteria using a His-Trap HP column followed by a PD-10 column to change buffer to PBS. Cell lysates, His-[1–883]ARAP2, or FA preparations were incubated with myr-Arf1, myr-Arf6, or Rac1-His preloaded with [α-32P]GTP in 25 mm HEPES, pH 7.5, 100 mm NaCl, 1 mm DTT, 2 mm MgCl2, 1 mm GTP, and 0.5 mm LUVs at 30 °C for 3 min (cell lysates and FA preparation) or for the times indicated (His-[1–883]ARAP2). The reaction was stopped by diluting the samples into ice-cold 10 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10 mm MgCl2, and 1 mm DTT. Protein-bound nucleotide was trapped on nitrocellulose filters, then released with formic acid, separated by thin layer chromatography on polyethyleneimine-cellulose plates and quantified using a Phosphor-Imager (GE Healthcare).
Preparation of Focal Adhesion Fraction
FA fraction was prepared following methods described previously with some modifications (40, 41). Briefly, cells were plated in 12-well plates at 120,000 cells/well. The next day, cells were transfected with plasmids as indicated. About 24 h later, cells were rinsed three times with ice-cold PBS, followed by incubation in hypotonic lysis buffer (2.5 mm Tris pH8, 2 mm MgCl2, 0.5 mm CaCl2, 1 mm DTT, 1 mm Na3VO4, 1 mm NaF, and protease inhibitors (Roche, Indianapolis, IN) for 5 min at 4 °C to promote osmotic swelling. Subsequently, cell bodies including nuclei, cytosol, and the bulk of the cytoskeleton were removed by strong trituration with 0.75 ml hypotonic lysis buffer using a repetitive pipette. The material remaining on the plate were referred as the “FA fraction” and used for adhesion site GAP assay.
Immunofluorescence Staining and Confocal Microscopy
Cells were trypsinized and resuspended in regular media. Subsequently, cells were rinsed once with plain Opti-MEM, reseeded at subconfluent density (3 × 104 cells/well of 24-well plate) on fibronectin-coated coverslips in plain Opti-MEM for 6 h and fixed for 20 min with 3.7% formaldehyde. Fixed cells were incubated in 15 mm glycine for 10 min, 50 mm NH4Cl for 2 × 10 min, then permeabilized and blocked with 0.2% saponin, 0.5% BSA, and 1% FBS in PBS for 20 min. An anti-vinculin monoclonal antibody (hVIN-1) or anti-paxillin monoclonal Ab was used to identify focal adhesions in cells. Cells were incubated in primary antibody for 1 h, followed by secondary antibodies for 1 h and mounted in DakoCytomation Fluorescent Mounting Medium (Dako, Carpinteria, CA). Images for fixed cells were taken on a Zeiss LSM 510 attached to a Zeiss Axiovert 100 m with a 63 × 1.4 numerical-aperture (NA) plan Neofluar oil immersion lens (Carl Zeiss, Thornwood, NY) and processed with Adobe Illustrator software.
Image Analysis and Statistics
Focal adhesion number per cell was analyzed using ImageJ (Rasband, W.S., ImageJ, NIH 1997-2010) as described previously (42). Images were high pass filtered. This method works well when the features (such as focal adhesions) are within a known size range and are high contrast from the background. Difference between multiple or two treatments were analyzed by one way ANOVA using Dunnett's multiple comparison test or Student's t test, with p < 0.05 considered to be significant. A lack of symbol means that it was not significant.
Pull-down Assays
Transfected cells were harvested 24 h after transfection. Arf6·GTP and Rac1·GTP levels were determined as described previously (43, 44). Briefly, cells were washed twice with ice-cold PBS and lysed in lysis buffer (25 mm Tris pH7.5, 150 mm NaCl, 10 mm MgCl2, 0.5 mm EDTA, 1% Triton X-100, and 1 mm Na3VO4) supplemented with protease & phosphatase inhibitor mixture (Thermo Scientific, Lafayette, CO). Immobilized GST-VHSGATGGA3 (for Arf6·GTP) or GST-PAK PBD (for Rac·GTP) was incubated with cleared cell lysates at 4 °C for 1.5 h on a rotator, washed twice with lysis buffer, and eluted with SDS sample buffer. The bound proteins were detected by Western blotting using antibodies against Arf6 or Rac.
RESULTS
ARAP2 Affects FAs and the Effect Is Dependent on ArfGAP Activity
The goal of our work was to determine the molecular basis for the control of FA morphology by ARAP2 that had been previously reported (29). As the growth or maturation of FAs is a continuous process and the stage of maturation is reflected by the size of adhesions (1, 4), we started our study by quantifying the effects of ARAP2 on the size distribution of all detectable adhesions in HeLa cells. To modulate the expression level of ARAP2 in HeLa cells, cells were either transiently transfected with a plasmid for the expression of ARAP2 with a FLAG epitope fused to the N terminus (FLAG-ARAP2) or with ARAP2 siRNA. The expression level of FLAG-ARAP2 was a least 100-fold of the endogenous ARAP2 and siRNA-mediated knockdown resulted in >90% reduction of endogenous ARAP2 (supplemental Fig. S1 and Fig. 2, 4, and 5). These cells were then plated onto fibronectin for 6 h before fixation and immunostained for vinculin or paxillin. Vinculin and paxillin were used as markers for FAs with similar results. As shown in Fig. 1, B–F, Fig. 4B, and Fig. 6B, although both overexpression and knockdown of ARAP2 caused a 25∼30% decrease in total adhesion number (all adhesions >0.3 μm2), they had opposite effects on the size distribution of adhesions. Overexpression of FLAG-ARAP2 resulted in more large adhesions (>3 μm2) and fewer small/medium size adhesions (0.3∼3 μm2) (Fig. 1, B and C). The average size of adhesions in FLAG-ARAP2-overexpressing cells was 1.9-fold larger than control cells (Fig. 1D). Reducing expression of endogenous ARAP2 using siRNA had the opposite result as overexpression: about 53% reduction for adhesions greater than 1 μm2 (typically considered as FAs) and no effects on the number of small adhesions (0.3∼0.5 μm2) (Fig. 1, E and F). This effect of ARAP2 siRNA was reproduced in two other cell lines, T47D breast cancer and U118 glioblastoma, indicating the regulation of FAs by ARAP2 is not restricted to HeLa cells (supplemental Fig. S2). Expressing wild type FLAG-ARAP2 from a plasmid reversed the effect of ARAP2 siRNA on large adhesions (Fig. 1, E and F), excluding that the FA phenotype is due to an off-target effect of ARAP2 siRNA. In contrast, a mutant of ARAP2 lacking ArfGAP activity (FLAG-[R728K]ARAP2) did not rescue the phenotype of reduced endogenous ARAP2. Based on these results, we conclude that ARAP2 promotes the growth of FAs and the effect is dependent on ArfGAP activity.
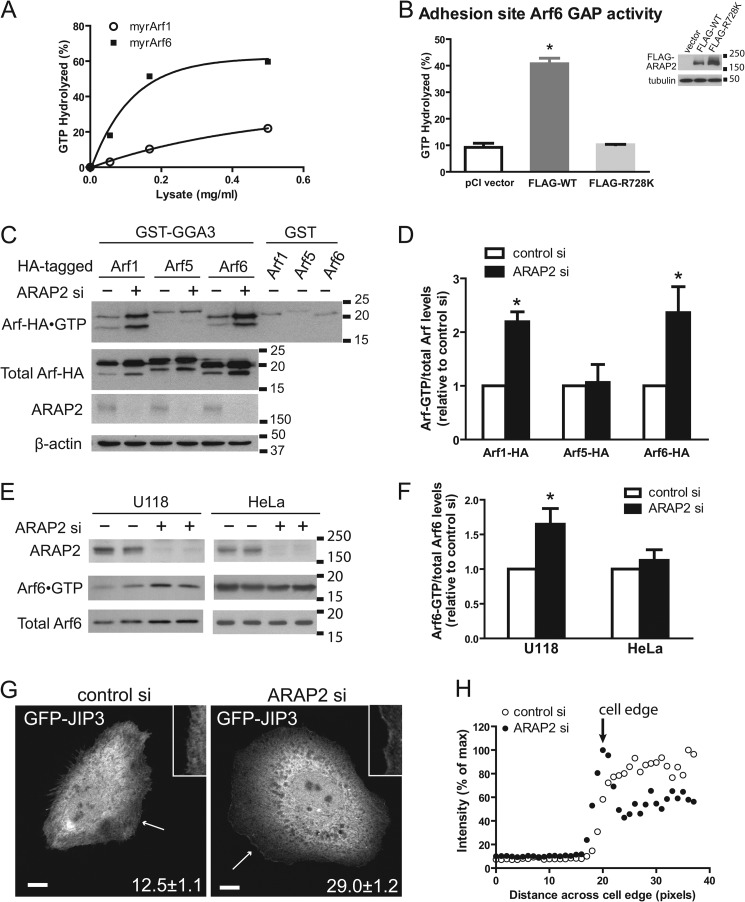
FIGURE 2.
ARAP2 uses Arf6 as substrates in vitro and in vivo. A, GAP activity in lysates of HeLa cells expressing ARAP2. The ArfGAP assay was performed as described in “Materials and Methods.” Different amount of lysates prepared from HeLa cells transfected with control GFP or FLAG-ARAP2 was incubated with purified myrArf1·GTP or myrArf6·GTP in the presence of large unilamellar vesicles (LUVs) consisting of 40% phosphatidylcholine, 25% phosphatidylethanolamine, 15% phosphatidylserine, 7% PtdIns, 2.5% PtdIns (4,5)-P2, 0.5% PtdIns (3,4,5)-P3, and 10% cholesterol. Activity was calculated by subtracting signal of GFP-transfected cell lysates from FLAG-ARAP2-transfected lysates. B, ArfGAP activity in FA preparations from HeLa cells expressing FLAG-ARAP2. ArfGAP assays were performed in triplicate using the FA fractions from the transfected HeLa cells using myrArf6·GTP as substrate. Insets, representative Western blots confirming the expression of FLAG-ARAP2. C and D, effect of reduced ARAP2 expression on Arf-HA·GTP levels. HeLa cells were transfected with control siRNA (−) or ARAP2 siRNA (+). 2 days later, cells were transfected again with HA-tagged Arf1, 5, or 6. Cells were lysed the next day and activated Arfs were co-precipitated with GST-VHSGATGGA3 beads. GST beads were included as a background control. Active and total Arf-HAs were detected by immunoblotting with antibodies against HA. Immunoblotting of ARAP2 and β-actin confirmed the down-regulation of ARAP2 and equal loading. Results shown are representative blots (C) and densitometric quantification of the relative Arf activity from three experiments (D). E and F, effect of reduced ARAP2 expression on levels of endogenous Arf6·GTP. Lysates from U118 or HeLa cells transfected with control siRNA or ARAP2 siRNA for 72 h were incubated with GST-VHSGATGGA3 beads. The beads were washed, and bound Arf6 (Arf6·GTP) was analyzed by Western blotting using anti-Arf6 antibody. Total Arf6 and the efficiency of ARAP2 protein knockdown were determined by Western blotting using anti-Arf6 and anti-ARAP2 antibody, respectively. Data shown are representative blots (E) and densitometric quantification (F) of the relative Arf6 activity (U118, n = 12; HeLa, n = 4, ± S.E.). For densitometric quantification, the Arf·GTP levels were normalized to the amount of total Arf and expressed as fold over control siRNA-treatment. G and H, cellular distribution of Arf6·GTP in HeLa cells with reduced ARAP2 expression. GFP-JIP3 leucine zipper II domain (GFP-JIP3) was transfected into control or ARAP2 siRNA-treated HeLa cells to monitor the localization of Arf6·GTP. Cells were then fixed and the intensity of GFP-JIP3 quantified across the cell border. Areas indicated by arrows were shown at higher magnification as insets in the images. Representative images of cells are shown in G, and representative plots of signal intensity versus distance across the cell edge are shown in H. ∼100 GFP-JIP3-transfected cells for each treatment were analyzed in each experiment. Numbers in the panels of the images in G indicate % of cells (the mean ± S.E. of three experiments) exhibiting higher GFP-JIP3 signal at the cell edge. Bars, 10 μm. * indicates significantly different from pCI using one way ANOVA with Dunnett's multiple comparison test, p < 0.05 (B) or control siRNA using Student's t test (D, F).
FIGURE 4.
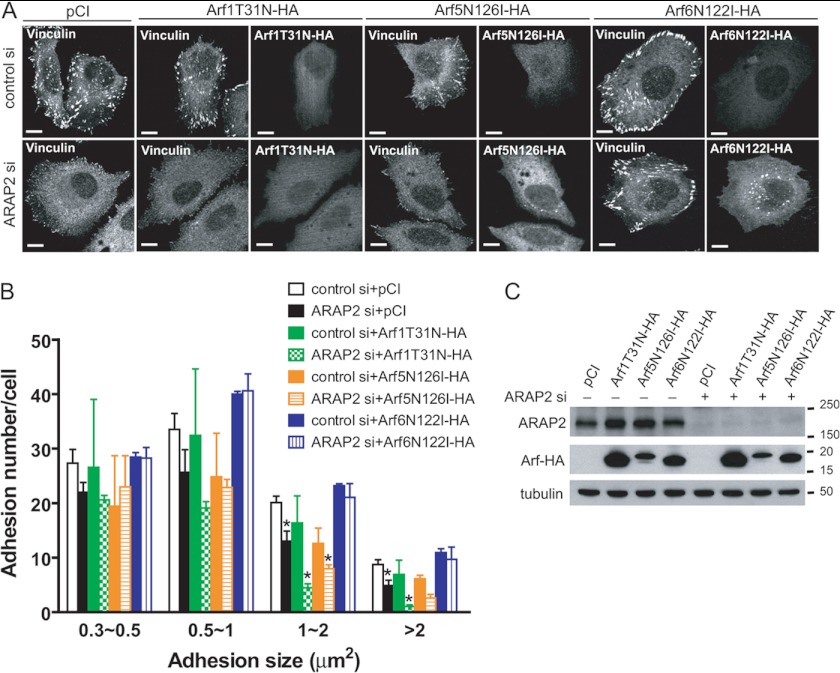
ARAP2 functions with Arf6 to regulate FAs. A—C, effect of dominant negative mutants of different Arfs on FA changes due to reduced ARAP2 expression. Control or ARAP2 siRNA-treated HeLa cells were transfected with plasmids directing the expression of either pCI, [T31N]Arf1-HA, [N126I]Arf5-HA, or [N122I]Arf6-HA and plated on FN-coated coverslips and processed as described for Fig. 3. A shows representative images and B the quantification for 40–90 transfected cells/treatment from three experiments. Mean number of adhesions ± S.E. is presented. C shows anti-ARAP2 and anti-HA immunoblots to determine the expression of ARAP2 and HA-tagged Arf mutants. Immunoblotting of tubulin was included as a loading control. *, p < 0.05 compared with control si+pCI using one way ANOVA with Dunnett's multiple comparison test.
FIGURE 5.
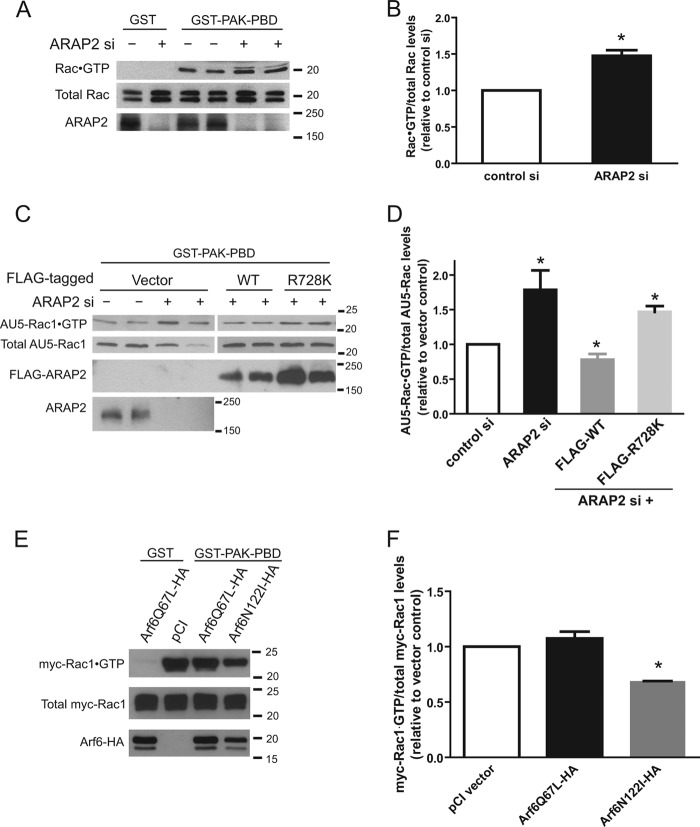
ARAP2 regulation of Rac1·GTP. A and B, reduction in ARAP2 results in increased Rac1·GTP. HeLa cells were transfected with control (−) or ARAP2 siRNA (+) and lysed 72 h later. Rac·GTP was co-precipitated with GST-PAK PBD beads. GTP-bound and total Rac were detected by anti-Rac immunoblots. Representative Western blots (A) and densitometric quantification (B, n = 3, ± S.E.) are shown. C and D, ARAP2 reduces AU5-Rac1·GTP levels dependent on ArfGAP activity of ARAP2. siRNA-treated HeLa cells were transfected with wild-type FLAG-ARAP2 (WT) or FLAG-[R728K]ARAP2 together with AU5-Rac1. AU5-Rac1·GTP was measured as in A except that an anti-AU5 antibody was used. Representative Western blots (C) and quantification (D) are shown (n ≥5). E and F, inhibition of Arf6 activity reduces Rac1 activity. pCI, [Q67L]Arf6-HA or [N122I]Arf6-HA was cotransfected with myc-Rac1 into HeLa cells. Myc-Rac1·GTP was measured as in A except that an anti-Myc antibody was used. Representative Western blots (E) and quantification (F) are shown (n ≥2). *, p < 0.05 compared with control siRNA or pCI using Student's t test (B) and one way ANOVA with Dunnett's multiple comparison test (D and F).
FIGURE 6.
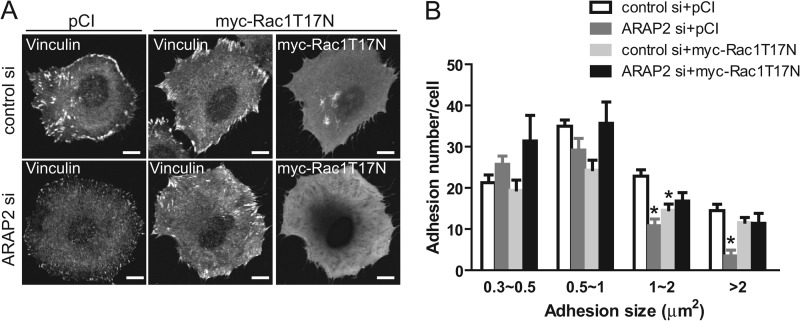
Effect of reduced ARAP2 expression on FAs depends on activation of Rac1. siRNA-treated HeLa cells were transfected with pCI vector or myc-[T17N]Rac1, then immunostained with anti-vinculin and anti-Myc antibodies. The number of FAs was quantified as described in Fig. 1 and “Materials and Methods.” A shows representative images. B is the quantification of three experiments. 45–60 transfected cells were examined in each experiment. Bars, 10 μm. *, p < 0.05 compared with control si+pCI using one way ANOVA with Dunnett's multiple comparison test.
Regulation of Arf6 Is Required for the Effects of ARAP2 on FAs: ARAP2 Has Arf6 GAP Activity in Vitro and in Vivo
Since ArfGAP activity of ARAP2 is required for its effect on FAs, we next determined whether regulation of Arf6 by ARAP2 is also essential. To test this idea, we began by confirming the function of ARAP2 as an Arf6 GAP. Previous work used nonmyristoylated Arfs as substrate (29). Because native Arfs are covalently modified on the N-terminal glycine with a myristate, we reassessed Arf specificity in vitro using the myristoylated Arfs. First, we determined activity in lysates of HeLa cells expressing FLAG-ARAP2 from a plasmid. By titrating the cell lysates, we determined that mammalian-expressed FLAG-ARAP2 hydrolyzed myr-Arf6·GTP about 10-fold more efficiently than myr-Arf1·GTP (Fig. 2A). To examine whether ARAP2 contributed Arf6 GAP activity associated with FAs, we performed the in vitro ArfGAP assay using a crude FA fraction that contains cellular ventral surface membranes and FA proteins (45). The crude FA fraction was prepared following a method described by Kuo et al. (40) with minor modifications. Briefly, HeLa cells were hypotonically lysed with shearing force generated by a repetitive pipette. Microscopic examination confirmed the loss of nuclei and the existence of FA proteins including paxillin and vinculin by immunostaining (supplemental Fig. S3). Myr-Arf6·[α-32P]GTP was added to the FA fraction on the plate. [α-32P]GTP hydrolysis as a reflection of ArfGAP activity was measured as described under “Materials and Methods.” Consistent with the association of ARAP2 with FAs (Fig. 1) (29), the FA fraction from cells overexpressing FLAG-ARAP2 contained more Arf6 GAP activity than the FA fraction from control cells (Fig. 2B). Expression of the GAP dead FLAG-[R728K]ARAP2 did not affect ArfGAP activity associated with the FA fraction. To determine the effect of ARAP2 on the in vivo levels of activated Arfs, HA-tagged Arf1, -5, and -6 (Arf1-HA, Arf5-HA, and Arf6-HA) were expressed in the control and ARAP2 siRNA-transfected HeLa cells. Activated Arf (i.e. Arf·GTP) was co-precipitated with GST-GGA and Arf-HA in the precipitate was detected by anti-HA immunoblotting (44). Reducing ARAP2 expression with siRNA increased Arf1-HA·GTP (2.2-fold) and Arf6-HA·GTP (2.4-fold) levels. It had no effect on Arf5-HA·GTP levels (Fig. 2, C and D). We also determined if reduced ARAP2 affects endogenous Arf6·GTP levels in U118 glioblastoma and HeLa cells. In U118 cells, endogenous Arf6·GTP level was elevated 1.6-fold when ARAP2 expression was reduced, similar to the results with exogenously expressed Arf6-HA. In HeLa cells, reduced ARAP2 expression did not affect Arf6·GTP levels globally (Fig. 2, E and F).
The lack of effect of reduced ARAP2 expression on global Arf6·GTP levels in HeLa cells could be a consequence of FA-associated Arf6 representing a smaller pool of total cellular Arf6 as compared with the situation in U118 cells (consequently, the fractional change in total cellular Arf6·GTP would be less in the HeLa cells). Given this possibility, we used a GFP-tagged JIP3 leucine zipper domain (GFP-JIP3) as a probe to detect localized changes in Arf6·GTP. JIP3 is an Arf6-specific effector with a leucine zipper domain that binds to Arf6·GTP (46). In HeLa cells with reduced ARAP2 consequent to siRNA treatment, GFP-JIP3 was enriched at the edge of membrane protrusions (Fig. 2, G and H), supporting the idea that reduced ARAP2 increased Arf6·GTP locally in HeLa cells. Together with the results of the in vitro assay and the GGA pull down, these data are consistent with the idea that ARAP2 functions as an Arf6 GAP in vivo.
Regulation of Arf6 Is Required for the Effects of ARAP2 on FAs: Effect of Arf Mutants on FA Morphology
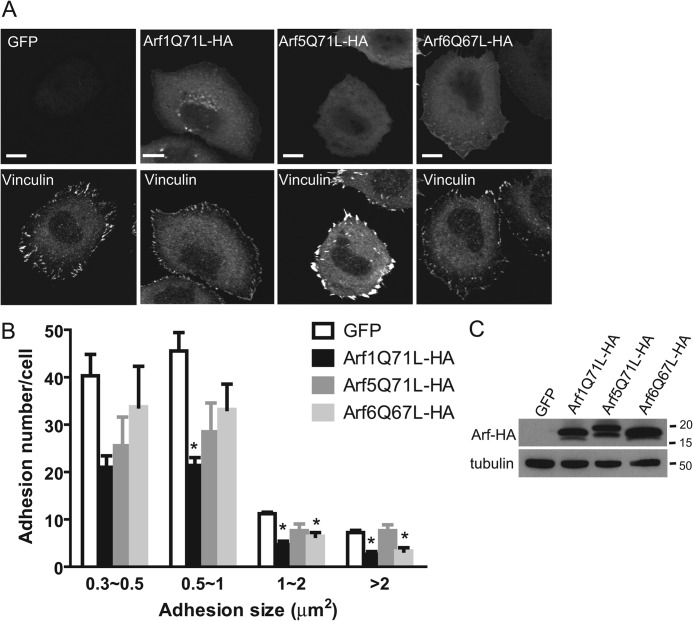
As one test of the idea that the effects of ARAP2 on FAs were mediated by Arf6, we determined the effect of different classes of Arf mutants on FAs. We examined HA-tagged Arf1, -5, and -6 as representatives of class I, II, and III Arfs. The prediction was that if the effect of ARAP2 was, at least in part, due to reduction of Arf6·GTP levels, then a constitutively active Arf6 ([Q67L]Arf6-HA) should have a similar effect as ARAP2 knockdown. Consistent with this prediction, transient expression of [Q67L]Arf6-HA in HeLa cells decreased total adhesion number by about 29%. More importantly, while [Q67L]Arf6-HA expressing cells did not affect the number of small adhesions, they had 50% fewer FAs (>1 μm2) than did controls (Fig. 3, A and B), which mimicked the size distribution of adhesions in ARAP2 siRNA cells. Interestingly, expression of the equivalent mutant of Arf1 ([Q71L]Arf1-HA) in HeLa cells had similar effects except that [Q71L]Arf1-HA also caused a reduction in medium sized adhesions (0.5∼1 μm2). In contrast, expression of the constitutively active [Q71L]Arf5-HA in HeLa cells had no effect on the number of FAs (>1 μm2) and increased the average size of adhesions by 22%. The expression level of all three Arf mutants was similar (Fig. 3C). These data indicate that different classes of Arfs had distinct effects on FA morphology and their effects could be divided into two groups: activated Arf1 and Arf6 inhibit the growth of FAs whereas activated Arf5 increases FA size and/or number.
FIGURE 3.
Effects of different classes of Arfs on FAs. HeLa cells were transiently transfected with control GFP or various HA-tagged mutants of Arf1, Arf5, and Arf6. 14∼18 h later, cells were replated onto fibronectin (FN)-coated (10 μg/ml) coverslips for 6 h, then fixed and stained with anti-vinculin and anti-HA antibodies to visualize FAs and Arf-HAs, respectively. Results shown are representative images (A) and quantification (B) expressed as the mean ± S.E. of four experiments with examination of 30∼70 transfected cells per construct in each experiment. C, representative Western blots showing expression levels of different Arf-HAs. Tubulin was included as a loading control. Bars, 10 μm. *, p < 0.05 indicates significantly different from GFP control by one way ANOVA using Dunnett's multiple comparison test.
If the reduction in FAs in ARAP2 knockdown cells is a result of increased Arf6·GTP levels, blocking the change in Arf6 activity by expressing a dominant negative variant of Arf6 ([N122I]Arf6-HA) should rescue the FA phenotype in ARAP2 knockdown cells. For these experiments, dominant negative mutants of different Arfs ([T31N]Arf1-HA, [N126I]Arf5-HA, and [N122I]Arf6-HA) were expressed in HeLa cells treated with control or ARAP2 siRNA (immunoblots to determine protein expression levels are shown in Fig. 4C). The cells were stained for vinculin as a FA marker, and the number of FAs was quantified from the micrographs (Fig. 4, A and B). As described above, reduction of ARAP2 (ARAP2 si) reduced the number of FAs. Consistent with the prediction that this was the result of increased Arf6·GTP levels, expression of [N122I]Arf6-HA, but not [T31N]Arf1-HA or [N126I]Arf5-HA, rescued the effect of ARAP2 knockdown, with greater number of FAs (>1 μm2) than in cells with ARAP2 knockdown alone. However, unlike ARAP2 overexpression, [N122I]Arf6-HA by itself had little effect on FA size and number (compare Fig. 4, A and B to Fig. 1, B and C). Together with the result that the effect of the ArfGAP dead mutant on FAs (Fig. 1, E and F), we conclude that regulation of Arf6 activity by ARAP2 is necessary but not sufficient to mediate the effects of ARAP2 on FAs.
Regulation of Rac1 Is Also Necessary for the Effect of ARAP2 on FAs
Expression of constitutively active Rac1 has been reported to affect substrate adhesions in a manner similar to ARAP2 knockdown (10, 11). Furthermore, Arf6·GTP has been reported to facilitate the formation of Rac1·GTP (13, 14, 19). Based on these considerations, we hypothesized that ARAP2 may control FAs by regulating Rac1 downstream of Arf6. To test this idea, we first determined if reduction of ARAP2 expression affects Rac1·GTP levels in cells. In these experiments, HeLa cells with reduced ARAP2 consequent to siRNA treatment were lysed, and endogenous Rac1·GTP was precipitated using the binding domain of PAK1, a Rac1 effector, fused to GST (GST-PAK-PBD) and bound to glutathione beads. The precipitated Rac1 was detected by immunoblots. Consistent with the hypothesis, Rac1·GTP levels were elevated 1.5-fold in cells with reduced ARAP2 (Fig. 5, A and B). We next determined if the change in Rac1·GTP levels requires ArfGAP activity of ARAP2. In these experiments, FLAG-tagged wild type ARAP2 or mutant ARAP2 lacking ArfGAP activity, FLAG-[R728K]ARAP2, was coexpressed with AU5-tagged Rac1 at a ratio of 7 to 1 in order to specifically detect changes in Rac1·GTP levels in FLAG-ARAP2-expressing cells. Replacing the endogenous ARAP2 with wild type FLAG-ARAP2 expressed from a plasmid not only reversed the knockdown effect but further decreased AU5-Rac1·GTP levels (Fig. 5, C and D). In contrast, expressing the ArfGAP dead mutant, FLAG-[R728K]ARAP2, did not rescue the increased AU5-Rac1·GTP in ARAP2 knockdown cells (Fig. 5, C and D), suggesting that the effect of ARAP2 on Rac1·GTP levels was mediated by the changes in Arf6·GTP levels.
It is possible that the ArfGAP domain of FLAG-ARAP2 functions as both a RacGAP and an ArfGAP. In this case, FLAG-[R728K]ARAP2, which is a mutation in the catalytic arginine in the ArfGAP domain, would fail to rescue changes in Rac1 activity caused by ARAP2 knockdown because this mutant abolishes both RacGAP activity and ArfGAP activity. To exclude this possibility, we determined if ARAP2 has RacGAP activity in vitro using bacterially purified proteins. Myr-Arf6 or His-Rac1 was loaded with [α-32P]GTP and incubated with bacterially purified His-[1–883]ARAP2 (Fig. 1A) and [α-32P]GTP hydrolysis was measured as described under “Materials and Methods.” At a concentration of 80 nm, His-[1–883]ARAP2 effectively stimulated hydrolysis of GTP bound to myr-Arf6 but had no detectable effect on GTP hydrolysis by His-Rac1 (supplemental Fig. S4). To further test the hypothesis that ARAP2 affects Arf6 activity leading to changes in Rac1·GTP levels, we examined the effects of Arf6 mutants on Rac1·GTP levels in cells. HeLa cells were cotransfected with myc-Rac1 and one of the HA-tagged Arf6 mutants or pCI vector as controls. Myc-Rac1·GTP levels were measured using the GST-PAK pull down assay as described above. As shown in Fig. 5, E and F, under the normal growth condition with serum in the culture media, [Q67L]Arf6-HA had no effect on myc-Rac1·GTP levels, possibly because Rac1 was already maximally activated in the presence of serum. On the other hand, decreasing Arf6·GTP levels by expression of [N122I]Arf6-HA reduced myc-Rac1·GTP levels by 31%. This result indicates that changes in Arf6 activity can affect Rac activity. Together, these data support the idea that ARAP2 regulates Rac1 activity through control of Arf6 activity.
To test the idea that the effects of ARAP2 on FAs were, at least in part, mediated by regulation of Rac1, we examined the effect of dominant negative Rac1 (myc-[T17N]Rac1) on changes in FAs due to reduced ARAP2 expression. Myc-[T17N]Rac1 was expressed in HeLa cells with reduced expression of ARAP2 consequent to siRNA treatment. As described above, FAs were visualized by staining with vinculin and adhesion size and number were quantified (Fig. 6, A and B). As previously described, cells with reduced ARAP2 expression had fewer FAs. If the increased Rac1 activity accounts for the effect of ARAP2 knockdown on FAs, preventing Rac1 activation by expressing myc-[T17N]Rac1 should reverse FA phenotype caused by ARAP2 knockdown. While in control siRNA cells, expression of myc-[T17N]Rac1 caused no change in FAs (>1 μm2), it prevented the reduction of FAs (>1 μm2) in ARAP2 knockdown cells. Together, these data support the conclusion that negative regulation of Rac1, like the negative regulation of Arf6, is necessary but not sufficient for the effects of ARAP2 on FAs.
DISCUSSION
Multiple ArfGAPs associate with FAs but little is known about the signaling pathways by which most affect FAs. Here we examined ARAP2 as a potential negative regulator of the Arf6/Rac1 pathway in the control of FAs. We found that ARAP2 inhibits Rac1 activity consequent to its negative regulation of Arf6 and that negative regulation of both Arf6 and Rac1 was critical for the effects of ARAP2 on FAs. To our knowledge, this is the first study on the role of Arf6/Rac1 pathway in the control of FAs. Furthermore, the quantification of the effects caused by mutants of Arf6 and Rac1 also revealed that negative regulation, by itself, could not fully mimic the effects of ARAP2, which has led us to speculate about additional mechanisms of ARAP2 action.
The finding that Arf6 together with Rac1 regulates FAs is consistent with previous reports focusing on membrane protrusions driven by actin polymerization. The function of Arf6 with Rac1 to control the actin cytoskeleton was first identified in work examining Arf6 induced actin-rich protrusions and membrane ruffles (12). The pathway was further defined by examining the Arf exchange factor ARNO. ARNO activation of Arf6 and consequent activation of Rac1 induced lamellipodia (13, 14). The same pathway was implicated in the action of an ArfGAP, GIT1 (14, 24). In the case of GIT1, recruitment of GIT1 to the lateral and rear plasma membrane resulted in reduced Arf6·GTP. Rac1·GTP levels were consequently reduced, which prevented membrane protrusion. The assembly of adhesions is coupled to the rate of membrane protrusion. Nascent adhesions form at the leading front of lamellipodia and begin to grow into mature FAs when protrusions pause (5). Therefore, our data link the function of Arf6/Rac1 pathway from regulating membrane protrusions to the growth of FAs.
The idea that Arfs may regulate FAs is based mostly on studies examining the effects of Arf6 on actin cytoskeleton, membrane protrusion, and cell migration. Direct examination of effects of Arfs on FAs has been limited. In one report, bacterially expressed recombinant Arf1, added to permeabilized cells or microinjected into cells, mediated recruitment of paxillin to FAs (7). Although these approaches allowed for the examination of the acute effect of Arf1, the bacterially expressed Arf is not properly modified, which affects the activity of the protein (47, 48). Both Arf1 and 6 were detected in FAs in a proteomic study (40), suggesting Arfs may regulate FAs. We compared three Arf isoforms and found that active Arf1 and Arf6 inhibit the growth of FAs, opposite from what was found with the bacterially expressed Arf1. On the other hand, active Arf5 induced large FAs. The difference between our result with Arf1, and the previous report using the bacterially expressed Arf1 could be due to different cell types or the differences in Arf expressed in bacterial and mammalian cells. We cannot exclude indirect effects because of relatively long term expression of the constitutively active proteins. Nevertheless, the results provide the first evidence of specific and opposing effects of Arf isoforms in the regulation of FAs.
Several Arf regulatory proteins, ArfGEFs and ArfGAPs, had been shown to affect cell adhesion and/or migration (13, 26, 30, 49–52). However, in most cases, the requirement for regulation of Arf·GTP levels for their respective effects was not examined. Brag2, an Arf exchange factor, was reported to cause the disassembly of FAs secondary to the activation of Arf6 (53). This result will have to be reconciled with recent work identifying Arf5 as the substrate for Brag2 in vivo (54). Our work, complementing the study of the Arf exchange factors including Brag2 and ARNO, is the first report examining the effects of the ArfGAP regulation of the Arf6/Rac1 pathway on FAs. ARAP2, by inactivating Arf6 and, consequently Rac1, would stabilize FAs and oppose the activity of Brag2 or ARNO.
We have not previously observed the changes in Rac1 due to ARAP2 (29). In those experiments, the effect of overexpression of ARAP2 was examined. In the current paper, we found reduced expression of ARAP2 resulted in an easily detectable increase in total Rac1·GTP whereas overexpression induced small decreases in total Rac1·GTP. The small effect could be because the changes were in a pool of Rac1 associated with a specific site and constituted only a small fraction of total Rac1. We may not have detected the effect of overexpression in previous work either because changes were small or because there were cell line differences. For similar reasons, we may not have detected an increase in Rac1·GTP upon expression of constitutively active Arf6 ([Q67L]Arf6-HA) in this study. Whereas ARAP2 had site specific effects, expression of [Q67L]Arf6-HA results in global activation of Arf6. Effects of Arf6 at other sites within the cell could result in opposing effects on Rac1. In addition, previous work in which [Q67L]Arf6 was reported to stimulate Rac1·GTP was done in a serum-free condition (19) whereas we measured Rac1 activity in the presence of serum, in which Rac1·GTP was already maximally elevated. On the other hand, because the basal Rac1 activity was high in our experiments, a reduction of Rac1·GTP levels was readily detectable when dominant negative Arf6 ([N122I]Arf6-HA) was expressed. The mechanism by which regulation of Arf6 by ARAP2 leads to a change in Rac1 activity remains elusive but likely involves proteins including DOCK180/Elmo or IQGAP1 that have been shown to mediate Arf6-dependent Rac1 activation under different circumstances (14, 19).
The size of adhesions is determined by the assembly and disassembly rate. One way ARAP2 could affect FAs through the Arf6/Rac1 pathway could be by preventing Rac1·GTP-induced disassembly. Targeting of FAs by microtubules (MTs) coincides with their disassembly (55, 56). Rac, the Rac GEF Tiam2, and the Rac effector PAK have been found to facilitate FA disassembly by promoting the growth or targeting of MTs to FAs (57–63). Based on these considerations, a plausible hypothesis is that ARAP2 reduces Arf6·GTP leading to lower Rac1·GTP levels in cells, which impairs targeting of MTs to FAs consequently stabilizing the FAs.
Although dominant negative Rac1 blocked the effect of reduced ARAP2 expression, it did not fully mimic the effect of ARAP2. It is possible that the effects require strict site-specific regulation of Rac1. Other plausible explanations are that changes in Arf6 have a parallel effect or that ARAP2 affects additional regulatory pathways independently of Arf6/Rac1. A number of mechanisms involving Arf6 not directly related to Rac1 may function in parallel with Rac1 to affect membrane traffic and actin resulting in changes in FAs. For instance, Arf6 activates the β isoform of phosphatidylinositol 4-phosphate 5-kinase (PIP5-kinase) but not PIP5Kγ (64). PIP5Kβ but not PIP5Kγ has been found to regulate FA disassembly by promoting endocytosis of β1-integrin (65). It is possible that the effects of ARAP2 on FAs are in part the result of specific control of Arf6/PIP5Kβ-dependent endocytosis of integrin. Arf6 also regulates endosomal recycling of β1-integrin back to the plasma membrane (49, 66, 67). Through regulation of Arf6, ARAP2 may control the recycling of integrins or other adhesion components, leading to a change in the adhesion turnover.
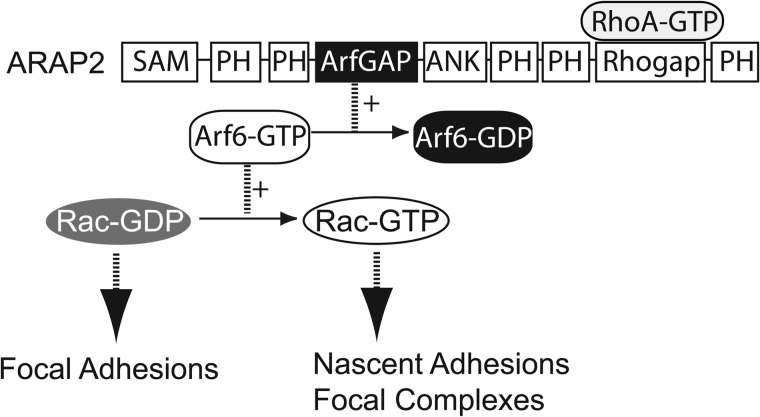
The other domains of ARAP2 may have functions both related to and independent of Arf6. The fact that dominant negative Arf6 failed to recapitulate the effect of ARAP2 on FAs supports this idea. For instance, site-specific targeting of ARAP2 that may provide localized changes in Arf is likely mediated by a domain outside of the ArfGAP domain. A PH domain may be involved, although they were not found to be important for PIP3-dependent targeting of the related ARAP1 to plasma membranes (68). RhoA·GTP binds to the RhoGAP domain, which could also have a role in targeting or other functions of ARAP2. Two-hybrid screens for binding partners of ARAP1 and ARAP3 have identified Cin85, Ship-2, and β-catenin as potential binding partners.3 Cin85 and Ship-2 are experimentally confirmed binding partners (69, 70). Rap1 has been reported to bind to ARAP3 through the RA domain (71). These or similar binding partners of ARAP2 could explain how ARAP2, with similar biochemical effects on Arf6 and Rac1 activity as GIT1 (24), has an opposite effect on FAs than does GIT1, which has been reported to promote FA disassembly (34). In conclusion, the data presented support a model where ARAP2 regulates Arf6·GTP and consequently, Rac1·GTP levels to control FAs (Fig. 7).
FIGURE 7.
Model for signaling function of ARAP2 in control of FA growth. As a RhoA·GTP-binding Arf6GAP, ARAP2 stimulates the conversion of Arf6·GTP to Arf6·GDP, leading to lower Rac1·GTP levels near FAs, allowing for stabilization/maturation of FAs from nascent adhesions/focal complexes.
Supplementary Material
Acknowledgments
We thank Drs. James Casanova, Martin Schwartz, and Clare Waterman for discussions, Dr. Ruibai Luo for critically reading the manuscript and Drs. Guillaume Montagnac and Philippe Chavrier for JIP3 constructs.
This work was supported, in whole or in part, by the intramural program of the NCI, National Institutes of Health (Project BC 007365).

This article contains supplemental Figs. S1—S4.
P.A. Randazzo, unpublished data.
- FA
- focal adhesions
- Arf
- ADP-ribosylation factor
- GAP
- GTPase-activating protein
- MT
- microtubule
- FN
- fibronectin.
REFERENCES
- 1. Gardel M. L., Schneider I. C., Aratyn-Schaus Y., Waterman C. M. (2010) Mechanical integration of actin and adhesion dynamics in cell migration. Annu. Rev. Cell Dev. Biol. 26, 315–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Geiger B., Spatz J. P., Bershadsky A. D. (2009) Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 10, 21–33 [DOI] [PubMed] [Google Scholar]
- 3. Geiger B., Yamada K. M. (2011) Molecular architecture and function of matrix adhesions. Cold Spring Harb. Perspect. Biol. 3, doi: 10.1101/cshperspect.a005033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Parsons J. T., Horwitz A. R., Schwartz M. A. (2010) Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11, 633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choi C. K., Vicente-Manzanares M., Zareno J., Whitmore L. A., Mogilner A., Horwitz A. R. (2008) Actin and α-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 10, 1039–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ridley A. J. (2006) Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 16, 522–529 [DOI] [PubMed] [Google Scholar]
- 7. Norman J. C., Jones D., Barry S. T., Holt M. R., Cockcroft S., Critchley D. R. (1998) ARF1 mediates paxillin recruitment to focal adhesions and potentiates Rho-stimulated stress fiber formation in intact and permeabilized Swiss 3T3 fibroblasts. J. Cell Biol. 143, 1981–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Randazzo P. A., Inoue H., Bharti S. (2007) Arf GAPs as regulators of the actin cytoskeleton. Biol. Cell 99, 583–600 [DOI] [PubMed] [Google Scholar]
- 9. Boureux A., Vignal E., Faure S., Fort P. (2007) Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol. Biol. Evol. 24, 203–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hall A. (1998) Rho GTPases and the actin cytoskeleton. Science 279, 509–514 [DOI] [PubMed] [Google Scholar]
- 11. Nobes C. D., Hall A. (1995) Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81, 53–62 [DOI] [PubMed] [Google Scholar]
- 12. Radhakrishna H., Al-Awar O., Khachikian Z., Donaldson J. G. (1999) ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. J. Cell Sci. 112, 855–866 [DOI] [PubMed] [Google Scholar]
- 13. Santy L. C., Casanova J. E. (2001) Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J. Cell Biol. 154, 599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Santy L. C., Ravichandran K. S., Casanova J. E. (2005) The DOCK180/Elmo complex couples ARNO-mediated Arf6 activation to the downstream activation of Rac1. Curr. Biol. 15, 1749–1754 [DOI] [PubMed] [Google Scholar]
- 15. Koo T. H., Eipper B. A., Donaldson J. G. (2007) Arf6 recruits the Rac GEF Kalirin to the plasma membrane facilitating Rac activation. BMC Cell Biol. 8, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tarricone C., Xiao B., Justin N., Walker P. A., Rittinger K., Gamblin S. J., Smerdon S. J. (2001) The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways. Nature 411, 215–219 [DOI] [PubMed] [Google Scholar]
- 17. Koronakis V., Hume P. J., Humphreys D., Liu T., Hørning O., Jensen O. N., McGhie E. J. (2011) WAVE regulatory complex activation by cooperating GTPases Arf and Rac1. Proc. Natl. Acad. Sci. U.S.A. 108, 14449–14454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Radhakrishna H., Klausner R. D., Donaldson J. G. (1996) Aluminum fluoride stimulates surface protrusions in cells overexpressing the ARF6 GTPase. J. Cell Biol. 134, 935–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu B., Shi B., Jarzynka M. J., Yiin J. J., D'Souza-Schorey C., Cheng S. Y. (2009) ADP-ribosylation factor 6 regulates glioma cell invasion through the IQ-domain GTPase-activating protein 1-Rac1-mediated pathway. Cancer Res. 69, 794–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sakurai A., Gavard J., Annas-Linhares Y., Basile J. R., Amornphimoltham P., Palmby T. R., Yagi H., Zhang F., Randazzo P. A., Li X., Weigert R., Gutkind J. S. (2010) Semaphorin 3E initiates antiangiogenic signaling through plexin D1 by regulating Arf6 and R-Ras. Mol. Cell Biol. 30, 3086–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Torii T., Miyamoto Y., Sanbe A., Nishimura K., Yamauchi J., Tanoue A. (2010) Cytohesin-2/ARNO, through its interaction with focal adhesion adaptor protein paxillin, regulates preadipocyte migration via the downstream activation of Arf6. J. Biol. Chem. 285, 24270–24281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cotton M., Boulay P. L., Houndolo T., Vitale N., Pitcher J. A., Claing A. (2007) Endogenous ARF6 interacts with Rac1 upon angiotensin II stimulation to regulate membrane ruffling and cell migration. Mol. Biol. Cell 18, 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Daher Z., Noël J., Claing A. (2008) Endothelin-1 promotes migration of endothelial cells through the activation of ARF6 and the regulation of FAK activity. Cell Signal. 20, 2256–2265 [DOI] [PubMed] [Google Scholar]
- 24. Nishiya N., Kiosses W. B., Han J., Ginsberg M. H. (2005) An α4 integrin-paxillin-Arf-GAP complex restricts Rac activation to the leading edge of migrating cells. Nat. Cell Biol. 7, 343–352 [DOI] [PubMed] [Google Scholar]
- 25. Ha V. L., Bharti S., Inoue H., Vass W. C., Campa F., Nie Z., de Gramont A., Ward Y., Randazzo P. A. (2008) ASAP3 is a focal adhesion-associated Arf GAP that functions in cell migration and invasion. J. Biol. Chem. 283, 14915–14926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Manabe R., Kovalenko M., Webb D. J., Horwitz A. R. (2002) GIT1 functions in a motile, multi-molecular signaling complex that regulates protrusive activity and cell migration. J. Cell Sci. 115, 1497–1510 [DOI] [PubMed] [Google Scholar]
- 27. Mazaki Y., Hashimoto S., Okawa K., Tsubouchi A., Nakamura K., Yagi R., Yano H., Kondo A., Iwamatsu A., Mizoguchi A., Sabe H. (2001) An ADP-ribosylation factor GTPase-activating protein Git2-short/KIAA0148 is involved in subcellular localization of paxillin and actin cytoskeletal organization. Mol. Biol. Cell 12, 645–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Randazzo P. A., Andrade J., Miura K., Brown M. T., Long Y. Q., Stauffer S., Roller P., Cooper J. A. (2000) The Arf GTPase-activating protein ASAP1 regulates the actin cytoskeleton. Proc. Natl. Acad. Sci. U.S.A. 97, 4011–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yoon H. Y., Miura K., Cuthbert E. J., Davis K. K., Ahvazi B., Casanova J. E., Randazzo P. A. (2006) ARAP2 effects on the actin cytoskeleton are dependent on Arf6-specific GTPase-activating protein activity and binding to RhoA-GTP. J. Cell Sci. 119, 4650–4666 [DOI] [PubMed] [Google Scholar]
- 30. Zhu Y., Wu Y., Kim J. I., Wang Z., Daaka Y., Nie Z. (2009) Arf GTPase-activating protein AGAP2 regulates focal adhesion kinase activity and focal adhesion remodeling. J. Biol. Chem. 284, 13489–13496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hasegawa J., Tsujita K., Takenawa T., Itoh T. (2012) ARAP1 regulates the ring size of circular dorsal ruffles through Arf1 and Arf5. Mol. Biol. Cell 23, 2481–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nie Z., Stanley K. T., Stauffer S., Jacques K. M., Hirsch D. S., Takei J., Randazzo P. A. (2002) AGAP1, an endosome-associated, phosphoinositide-dependent ADP-ribosylation factor GTPase-activating protein that affects actin cytoskeleton. J. Biol. Chem. 277, 48965–48975 [DOI] [PubMed] [Google Scholar]
- 33. Vitale N., Patton W. A., Moss J., Vaughan M., Lefkowitz R. J., Premont R. T. (2000) GIT proteins, A novel family of phosphatidylinositol 3,4, 5-trisphosphate-stimulated GTPase-activating proteins for ARF6. J. Biol. Chem. 275, 13901–13906 [DOI] [PubMed] [Google Scholar]
- 34. Zhao Z. S., Manser E., Loo T. H., Lim L. (2000) Coupling of PAK-interacting exchange factor PIX to GIT1 promotes focal complex disassembly. Mol. Cell Biol. 20, 6354–6363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Puertollano R., Randazzo P. A., Presley J. F., Hartnell L. M., Bonifacino J. S. (2001) The GGAs promote ARF-dependent recruitment of clathrin to the TGN. Cell 105, 93–102 [DOI] [PubMed] [Google Scholar]
- 36. Randazzo P. A., Kahn R. A. (1994) GTP hydrolysis by ADP-ribosylation factor is dependent on both an ADP-ribosylation factor GTPase-activating protein and acid phospholipids. J. Biol. Chem. 269, 10758–10763 [PubMed] [Google Scholar]
- 37. Luo R., Ahvazi B., Amariei D., Shroder D., Burrola B., Losert W., Randazzo P. A. (2007) Kinetic analysis of GTP hydrolysis catalysed by the Arf1-GTP-ASAP1 complex. Biochem. J. 402, 439–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ha V. L., Thomas G. M., Stauffer S., Randazzo P. A. (2005) Preparation of myristoylated Arf1 and Arf6. Methods Enzymol. 404, 164–174 [DOI] [PubMed] [Google Scholar]
- 39. Miura K., Jacques K. M., Stauffer S., Kubosaki A., Zhu K., Hirsch D. S., Resau J., Zheng Y., Randazzo P. A. (2002) ARAP1: a point of convergence for Arf and Rho signaling. Mol. Cell 9, 109–119 [DOI] [PubMed] [Google Scholar]
- 40. Kuo J. C., Han X., Hsiao C. T., Yates J. R., 3rd, Waterman C. M. (2011) Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for beta-Pix in negative regulation of focal adhesion maturation. Nat. Cell Biol. 13, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tong W. Y., Liang Y. M., Tam V., Yip H. K., Kao Y. T., Cheung K. M., Yeung K. W., Lam Y. W. (2010) Biochemical characterization of the cell-biomaterial interface by quantitative proteomics. Mol. Cell Proteomics 9, 2089–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen P. W., Kroog G. S. (2010) Leupaxin is similar to paxillin in focal adhesion targeting and tyrosine phosphorylation but has distinct roles in cell adhesion and spreading. Cell Adh. Migr. 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Benard V., Bokoch G. M. (2002) Assay of Cdc42, Rac, and Rho GTPase activation by affinity methods. Methods Enzymol. 345, 349–359 [DOI] [PubMed] [Google Scholar]
- 44. Yoon H. Y., Bonifacino J. S., Randazzo P. A. (2005) In vitro assays of Arf1 interaction with GGA proteins. Methods Enzymol. 404, 316–332 [DOI] [PubMed] [Google Scholar]
- 45. Chen P. W., Jian X., Luo R., Randazzo P. A. (2012) Approaches to Studying Arf GAPs in Cells: In Vitro Assay with Isolated Focal Adhesions. Curr. Protocols Cell Biol. Chapter 17, Unit17 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Montagnac G., Sibarita J. B., Loubéry S., Daviet L., Romao M., Raposo G., Chavrier P. (2009) ARF6 Interacts with JIP4 to control a motor switch mechanism regulating endosome traffic in cytokinesis. Curr. Biol. 19, 184–195 [DOI] [PubMed] [Google Scholar]
- 47. Haun R. S., Tsai S. C., Adamik R., Moss J., Vaughan M. (1993) Effect of myristoylation on GTP-dependent binding of ADP-ribosylation factor to Golgi. J. Biol. Chem. 268, 7064–7068 [PubMed] [Google Scholar]
- 48. Randazzo P. A., Terui T., Sturch S., Fales H. M., Ferrige A. G., Kahn R. A. (1995) The myristoylated amino terminus of ADP-ribosylation factor 1 is a phospholipid- and GTP-sensitive switch. J. Biol. Chem. 270, 14809–14815 [DOI] [PubMed] [Google Scholar]
- 49. Dunphy J. L., Moravec R., Ly K., Lasell T. K., Melancon P., Casanova J. E. (2006) The Arf6 GEF GEP100/BRAG2 regulates cell adhesion by controlling endocytosis of β1 integrins. Curr. Biol. 16, 315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Geiger C., Nagel W., Boehm T., van Kooyk Y., Figdor C. G., Kremmer E., Hogg N., Zeitlmann L., Dierks H., Weber K. S., Kolanus W. (2000) Cytohesin-1 regulates β2 integrin-mediated adhesion through both ARF-GEF function and interaction with LFA-1. EMBO J. 19, 2525–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Quast T., Tappertzhofen B., Schild C., Grell J., Czeloth N., Förster R., Alon R., Fraemohs L., Dreck K., Weber C., Lämmermann T., Sixt M., Kolanus W. (2009) Cytohesin-1 controls the activation of RhoA and modulates integrin-dependent adhesion and migration of dendritic cells. Blood 113, 5801–5810 [DOI] [PubMed] [Google Scholar]
- 52. Gambardella L., Anderson K. E., Nussbaum C., Segonds-Pichon A., Margarido T., Norton L., Ludwig T., Sperandio M., Hawkins P. T., Stephens L., Vermeren S. (2011) The GTPase-activating protein ARAP3 regulates chemotaxis and adhesion-dependent processes in neutrophils. Blood 118, 1087–1098 [DOI] [PubMed] [Google Scholar]
- 53. Sakurai A., Jian X., Lee C. J., Manavski Y., Chavakis E., Donaldson J., Randazzo P. A., Gutkind J. S. (2011) Phosphatidylinositol-4-phosphate 5-kinase and GEP100/Brag2 protein mediate antiangiogenic signaling by semaphorin 3E-plexin-D1 through Arf6 protein. J. Biol. Chem. 286, 34335–34345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Moravec R., Conger K. K., D'Souza R., Allison A. B., Casanova J. E. (2012) BRAG2/GEP100/IQSec1 Interacts with Clathrin and Regulates α5β1 Integrin Endocytosis through Activation of Arf5. J. Biol. Chem. 287, 31138–31147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kaverina I., Krylyshkina O., Small J. V. (1999) Microtubule targeting of substrate contacts promotes their relaxation and dissociation. J. Cell Biol. 146, 1033–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kaverina I., Rottner K., Small J. V. (1998) Targeting, capture, and stabilization of microtubules at early focal adhesions. J. Cell Biol. 142, 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wittmann T., Bokoch G. M., Waterman-Storer C. M. (2003) Regulation of leading edge microtubule and actin dynamics downstream of Rac1. J. Cell Biol. 161, 845–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wittmann T., Waterman-Storer C. M. (2005) Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3β in migrating epithelial cells. J. Cell Biol. 169, 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rooney C., White G., Nazgiewicz A., Woodcock S. A., Anderson K. I., Ballestrem C., Malliri A. (2010) The Rac activator STEF (Tiam2) regulates cell migration by microtubule-mediated focal adhesion disassembly. EMBO Rep. 11, 292–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nayal A., Webb D. J., Brown C. M., Schaefer E. M., Vicente-Manzanares M., Horwitz A. R. (2006) Paxillin phosphorylation at Ser273 localizes a GIT1-PIX-PAK complex and regulates adhesion and protrusion dynamics. J. Cell Biol. 173, 587–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Frost J. A., Khokhlatchev A., Stippec S., White M. A., Cobb M. H. (1998) Differential effects of PAK1-activating mutations reveal activity-dependent and -independent effects on cytoskeletal regulation. J. Biol. Chem. 273, 28191–28198 [DOI] [PubMed] [Google Scholar]
- 62. Manser E., Huang H. Y., Loo T. H., Chen X. Q., Dong J. M., Leung T., Lim L. (1997) Expression of constitutively active α-PAK reveals effects of the kinase on actin and focal complexes. Mol. Cell Biol. 17, 1129–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wittmann T., Bokoch G. M., Waterman-Storer C. M. (2004) Regulation of microtubule destabilizing activity of Op18/stathmin downstream of Rac1. J. Biol. Chem. 279, 6196–6203 [DOI] [PubMed] [Google Scholar]
- 64. Funakoshi Y., Hasegawa H., Kanaho Y. (2010) Activation mechanisms of PIP5K isozymes by the small GTPase ARF6. Adv. Enzyme Regulat. 50, 72–80 [DOI] [PubMed] [Google Scholar]
- 65. Chao W. T., Ashcroft F., Daquinag A. C., Vadakkan T., Wei Z., Zhang P., Dickinson M. E., Kunz J. (2010) Type I phosphatidylinositol phosphate kinase β regulates focal adhesion disassembly by promoting β1 integrin endocytosis. Mol. Cell Biol. 30, 4463–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Powelka A. M., Sun J., Li J., Gao M., Shaw L. M., Sonnenberg A., Hsu V. W. (2004) Stimulation-dependent recycling of integrin β1 regulated by ARF6 and Rab11. Traffic 5, 20–36 [DOI] [PubMed] [Google Scholar]
- 67. Radhakrishna H., Donaldson J. G. (1997) ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J. Cell Biol. 139, 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Campa F., Yoon H. Y., Ha V. L., Szentpetery Z., Balla T., Randazzo P. A. (2009) A PH domain in the Arf GTPase-activating protein (GAP) ARAP1 binds phosphatidylinositol 3,4,5-trisphosphate and regulates Arf GAP activity independently of recruitment to the plasma membranes. J. Biol. Chem. 284, 28069–28083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Raaijmakers J. H., Deneubourg L., Rehmann H., de Koning J., Zhang Z., Krugmann S., Erneux C., Bos J. L. (2007) The PI3K effector Arap3 interacts with the PI(3,4,5)P3 phosphatase SHIP2 in a SAM domain-dependent manner. Cell Signal 19, 1249–1257 [DOI] [PubMed] [Google Scholar]
- 70. Yoon H. Y., Kales S. C., Luo R., Lipkowitz S., Randazzo P. A. (2011) ARAP1 association with CIN85 affects epidermal growth factor receptor endocytic trafficking. Biol. Cell 103, 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Krugmann S., Williams R., Stephens L., Hawkins P. T. (2004) ARAP3 is a PI3K- and rap-regulated GAP for RhoA. Curr. Biol. 14, 1380–1384 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.