Background: Start codon recognition triggers eIF1 and Pi release from the preinitiation complex.
Results: The C-terminal tail of eIF1A moves closer to eIF5 upon start codon recognition, and this movement is required for Pi release.
Conclusion: eIF1 release and movement of the eIF1A C-terminal tail toward eIF5 are coupled processes.
Significance: Start codon recognition induces coordinated movements of initiation factors that trigger downstream events.
Keywords: Enzyme Kinetics, Fluorescence Resonance Energy Transfer (FRET), mRNA, Protein Synthesis, Protein-Nucleic Acid Interaction, Eukaryotic Translation Initiation, Protein Synthesis, Start Codon Recognition, eIF1A, eIF5
Abstract
Accurate recognition of the start codon in an mRNA by the eukaryotic translation preinitiation complex (PIC) is essential for proper gene expression. The process is mediated by eukaryotic translation initiation factors (eIFs) in conjunction with the 40 S ribosomal subunit and (initiator) tRNAi. Here, we provide evidence that the C-terminal tail (CTT) of eIF1A, which we previously implicated in start codon recognition, moves closer to the N-terminal domain of eIF5 when the PIC encounters an AUG codon. Importantly, this movement is coupled to dissociation of eIF1 from the PIC, a critical event in start codon recognition, and is dependent on the scanning enhancer elements in the eIF1A CTT. The data further indicate that eIF1 dissociation must be accompanied by the movement of the eIF1A CTT toward eIF5 in order to trigger release of phosphate from eIF2, which converts the latter to its GDP-bound state. Our results also suggest that release of eIF1 from the PIC and movement of the CTT of eIF1A are triggered by the same event, most likely accommodation of tRNAi in the P site of the 40 S subunit driven by base pairing between the start codon in the mRNA and the anticodon in tRNAi. Finally, we show that the C-terminal domain of eIF5 is responsible for the factor's activity in antagonizing eIF1 binding to the PIC. Together, our data provide a more complete picture of the chain of molecular events that is triggered when the scanning PIC encounters an AUG start codon in the mRNA.
Introduction
The initiation phase of translation in eukaryotes (1–3) begins with the assembly of a 43 S preinitiation complex (PIC).3 The PIC is formed when a ternary complex (TC) of eukaryotic initiation factor 2 (eIF2), GTP, and the methionyl initiator tRNA (Met-tRNAi) binds to the 40 S ribosomal subunit. Three initiation factors, eIF1, -1A, and -3, associate with the 40 S subunit and promote TC loading. The PIC then binds to the 5′-end of an mRNA with the assistance of the eIF4 factors, eIF3, and the poly(A)-binding protein and subsequently scans the mRNA in search of an initiation codon. Once the initiation codon is encountered, base pairing takes place between the anticodon of the initiator tRNA and the AUG in the mRNA, triggering a series of events that commit the complex to continuing the initiation process at that point on the message. These events include dissociation of eIF1 from the PIC and conversion of eIF2 to its GDP-bound form via gated phosphate (Pi) release, which requires the action of the GTPase-activating protein (GAP) eIF5. Subsequent dissociation of eIF5 and eIF2·GDP from the 40 S subunit clears the way for eIF5B·GTP-dependent joining of the 60 S ribosomal subunit to form an 80 S initiation complex. Subunit joining triggers GTP hydrolysis by eIF5B, releasing both it and eIF1A from the 80 S complex and allowing the first round of peptide bond formation to begin.
Start codon recognition by the PIC is a critical event for accurate gene expression. Over the last decade, significant progress has been made in our understanding of the molecular mechanics underlying the accurate recognition of an AUG codon in an mRNA by the translational machinery. In the current model for this process (1, 2), the PIC binds to the message in an “open” conformation that is competent to scan in search of a start codon (4). This open conformation is induced in Saccharomyces cerevisiae by eIF1 and eIF1A (5) and is probably also stabilized by eIF3 as well. Genetic and biochemical data (6, 7) have suggested that the tRNAi in the scanning PIC is not fully accommodated in the P site of the 40 S subunit and instead occupies a displaced state termed “Pout.” Biochemical and structural studies have shown that eIF1 binds adjacent to the P site and, in fact, sterically hinders full accommodation of the initiator tRNA (8, 9). The folded body of eIF1A binds in the A site of the 40 S subunit, but its long, unstructured N- and C-terminal tails (NTT and CTT, respectively) reach into the P site (10). The position of the CTT is also thought to sterically occlude full access of the tRNA to the P site.
We previously showed that when the PIC encounters the start codon, eIF1 is ejected from the complex (11). We also showed that start codon recognition induces a strong direct or indirect interaction between eIF1A and eIF5 (12). A variety of data indicate that base pairing between the anticodon of tRNAi and the start codon induces a transition from the open state to a closed one that is arrested on the mRNA (13). We have proposed that this codon-anticodon pairing drives the tRNAi fully into the P site (Pin state), which in turn ejects eIF1 and the CTT of eIF1A due to their steric clashes with the fully accommodated tRNA (2, 14). Release of eIF1 and interaction between eIF1A and eIF5 stabilize the closed conformation of the PIC. In addition, ejection of eIF1 was shown to set the rate of Pi release from eIF2, because eIF1 and Pi release occur with nearly the same rate constants, and mutations in eIF1 that speed up or slow down its ejection have the same effect on Pi release (15–17).
Additional studies have further defined the roles played by eIF1A and eIF5 in start codon recognition, although the mechanistic basis for these roles has remained obscure (18, 19). As noted above, a strong interaction between eIF1A and eIF5, dependent on the CTT of eIF1A, occurs upon start codon recognition. This interaction is sensitive to the codon in the P site and is strongly influenced by mutations in eIF5 and eIF1A that decrease (Sui− phenotype) or increase (Ssu− phenotype) the fidelity of start codon recognition in vivo (12, 20).
Recently, we identified elements in the CTT and NTT of eIF1A that play important roles in start codon recognition (6). Mutations in the two scanning enhancer (SE) elements in the CTT produce Sui− phenotypes, and our data suggested that these residues function to antagonize the closed state of the PIC and/or stabilize the Pout state of the tRNAi relative to the Pin state. Two scanning inhibitor (SI) elements were also identified, one in the NTT and the other in a helix adjacent to the CTT. Mutations in these SI elements increase the fidelity of start codon recognition (Ssu−), suggesting that the elements promote closed complex formation and/or transition to the Pin state of the tRNAi. One way to explain these opposing functions of the SE and SI elements is that the former binds in the P site of the 40 S subunit prior to start codon recognition, whereas the SI elements have affinity for a site(s) in the PIC that is incompatible with binding of the SE elements in the P site. In this model, mutation of the SE elements would facilitate removal of the CTT from the P site, stabilizing Pin relative to Pout, whereas mutation of the SI elements would have the opposite effect. Release of the eIF1A CTT from the P site upon start codon recognition presumably allows its direct or indirect interaction with eIF5, and this interaction might be the trigger for Pi release from eIF2.
There is also mounting evidence that eIF5 plays a direct role in start codon recognition in addition to its function as a GAP for eIF2. eIF5 consists of an N-terminal and a C-terminal domain (NTD and CTD), connected by a linker region (Fig. 1A) (21, 22). The NTD has an unstructured N-terminal tail, which contains the key arginine residue (Arg-15) required for GAP function and which presumably interacts directly with the GTP-binding pocket in eIF2γ. This tail is followed by a region with a fold similar to that of eIF1 and the α/β globular domain of archaeal homologs of eIF2β, which sits on top of a zinc-binding domain (ZBD) (22). The CTD of eIF5 is made up of a HEAT domain that has been shown to interact with eIF1, the unstructured NTT of eIF2β, and the eIF3c NTD, interactions that stabilize the yeast multifactor complex containing these eIFs (19, 21, 23–25). Mutations in the eIF5 NTD have been obtained that produce either Sui− or Ssu− phenotypes (26–28), whereas mutations in the CTD that disrupt interaction with eIF1 and eIF2β produce Ssu− phenotypes (25). One such Sui− substitution in the eIF5 NTD (G31R) was shown to alter the interaction of eIF5 with eIF1A in a manner that stabilizes the closed complex at UUG but destabilizes it at AUG, strongly implicating the eIF5 GAP domain in the conformational rearrangement of the PIC upon start codon recognition (12).
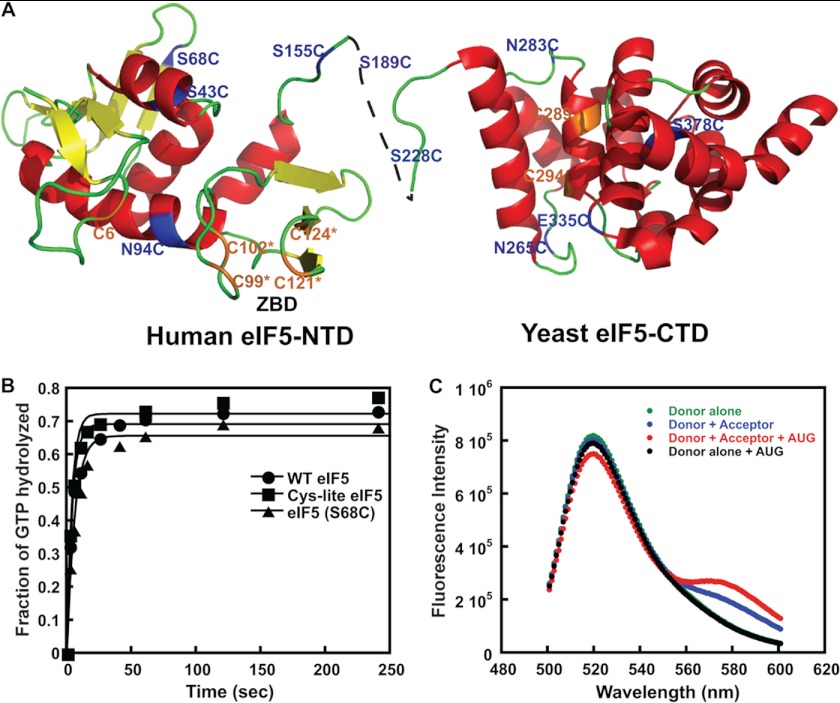
FIGURE 1.
FRET between fluorophores in the NTD of eIF5 and CTD of eIF1A in the PIC upon AUG recognition. A, ribbon representation of the human eIF5 NTD and yeast eIF5 CTD showing the single cysteine mutants generated in the Cys-lite background. Positions of native cysteines (C6, C289, and C294) that are not involved in the ZBD are shown in orange. Cysteines introduced at non-conserved surface residues are shown in blue. The cysteines involved in the ZBD (C99*, C102*, C121*, and C124*) are shown in orange. B, kinetics of GTP hydrolysis by eIF2, performed as described under “Experimental Procedures,” in the presence of native eIF5 (closed circles), Cys-lite eIF5 (closed squares), and Cys-lite eIF5(S68C) (closed triangles). Points are averages from two independent experiments. C, steady-state fluorescence measurements demonstrating FRET in the PIC upon the addition of mRNA(AUG) between the Cys-lite eIF5(S68C) derivative labeled with fluorescein and eIF1A labeled C-terminally with TAMRA. The following complexes were assembled, and their fluorescence was measured as a function of emission wavelength (excitation wavelength = 490 nm): Cys-lite eIF5(S68C)-Fl·eIF1A·eIF1·40 S·TC (Donor alone; green); eIF5(S68C)-Fl·eIF1A-TAMRA·eIF1·40 S·TC (Donor + Acceptor; blue); eIF5(S68C)-Fl·eIF1A-TAMRA·eIF1·40 S·TC·mRNA(AUG) (Donor + Acceptor + AUG; red); eIF5(S68C)-Fl·eIF1A·eIF1·40 S·TC·mRNA(AUG) (Donor alone + AUG; black). The FRET change can be seen as both a decrease in fluorescein (donor) fluorescence at 520 nm and increase in TAMRA (acceptor) fluorescence at 580 nm upon the addition of mRNA(AUG) to the donor + acceptor complex (red versus blue curves). The emission seen at 580 nm in the Donor + Acceptor curve in the absence of mRNA (blue) is due to weak excitation of TAMRA by the incident light. No change in donor fluorescence is observed in the absence of acceptor upon the addition of mRNA(AUG) (green versus black curves), demonstrating that the decrease in emission is due to FRET rather than a change in intrinsic fluorescence of the fluorescein moiety.
There is also evidence that eIF5 promotes AUG recognition by enhancing eIF1 dissociation from the PIC. We recently showed that high concentrations of eIF5 antagonize binding of eIF1 to the PIC in vitro (16). Consistent with this observation, overexpressing eIF5 in yeast cells elevates utilization of near-cognate UUG start codons (16), whereas overexpressing eIF1 has the opposite effect and suppresses UUG initiation (6, 29, 30). Similarly, it was shown that overexpression of eIF5 in mammalian cells increases use of near-cognate codons and AUG codons in suboptimal sequence contexts as start sites, and this effect is suppressed by co-overexpression of eIF1, consistent with the notion that high concentrations of eIF5 reduce the fidelity of start codon recognition in vivo by promoting release of eIF1 from the PIC (31).
Based on the available data, we proposed a model in which start codon recognition induces movement of the CTT of eIF1A out of the P site, allowing it to interact with eIF5 (12, 20). This interaction could be the trigger for Pi release from eIF2, particularly if it was coupled to dissociation of the eIF5 GAP domain from the GTP-binding pocket in eIF2γ. As noted above, displacement of the eIF1A CTT from the P site should also facilitate accommodation of tRNAi in the Pin state of the closed complex required for AUG recognition (6, 10). In addition, we proposed that upon dissociation of eIF1 from the PIC, one of the domains of eIF5 might move into the eIF1 binding site (e.g. a site on the 40 S subunit or in the unstructured NTT of eIF2β), preventing rebinding of eIF1 and promoting transition to downstream steps in the initiation process (16). This competition for the same binding site would explain the antagonism between eIF1 and eIF5. Recent results indicate that interaction of the eIF5 CTD with the eIF2β NTT is crucial for proper dissociation of eIF1 from the PIC and start codon recognition (25).
In this paper, we provide data that directly support and refine this model. Using fluorescence resonance energy transfer (FRET) between fluorophores on the C terminus of eIF1A and on eIF5, we show that the CTT of eIF1A moves closer to the NTD of eIF5 in response to start codon recognition in a manner controlled by the rate of eIF1 dissociation from the PIC and dependent on the SE elements in the eIF1A CTT. Remarkably, mutations in the SE elements uncouple eIF1 release from Pi release, dramatically impairing both Pi release and movement of the eIF1A CTT while minimally affecting eIF1 release. These findings demonstrate that eIF1 dissociation is not sufficient for Pi release and that movement of the eIF1A CTT toward the eIF5 NTD is additionally required for this key step in start codon recognition. The available data suggest that eIF1 release determines the timing of these events in WT PICs by setting the rate of accommodation of tRNAi into the P site, which in turn triggers movement of the eIF1A CTT toward the eIF5 NTD. Finally, we show that the CTD of eIF5 is responsible for the factor's antagonism with eIF1 in binding to the PIC, reinforcing the notion that interaction of the eIF5 CTD with the eIF2β NTT is critical for eIF1 release and stable TC binding with tRNAi fully accommodated in the P site. Our data indicate that a multistep series of movements of eIF1, eIF1A, and eIF5 takes place in response to start codon recognition and that these events are coupled to one another in WT PICs, most likely beginning with the movement of the initiator tRNA into the P site.
EXPERIMENTAL PROCEDURES
Buffers and Reagents
The reaction (“recon”) buffer was composed of 30 mm HEPES-KOH (pH 7.4), 100 mm KOAc (pH 7.4), 3 mm Mg(OAc)2, and 2 mm DTT. The enzyme storage buffer was composed of 20 mm HEPES-KOH (pH 7.4), 100 mm KoAC (pH 7.4), 2 mm DTT, and 10% glycerol. Purification of all components was performed as described previously (32). The model mRNAs used were of the sequence GGAA(UC)7UNNN(CU)10C, where NNN was either AUG or AUC (referred to as mRNA(AUG) and mRNA(AUC), respectively). The use of unstructured model mRNA obviates the need for mRNA recruitment and remodeling factors (e.g. eIF3 and eIF4F) as well as a 5′-cap and 3′-poly(A) tail.
Fluorescent Labeling of WT and Mutant Versions of eIF1 and eIF1A
WT eIF1, mutant G107K eIF1, WT eIF1A, and its mutants SE1*SE2* and SE2*, where SE1* designates 121FGFESDE127 → AAAAAAA and SE2* designates 131FEFGN135 → AAAAA (6), were labeled at their C termini with either Cys-Lys-ϵ-fluorescein dipeptide, or Cys-Lys-ϵ-TAMRA dipeptide using the Expressed Protein Ligation system as described previously (33).
Fluorescent Labeling of eIF5 Cysteine Mutants
Single cysteine mutants of eIF5 in the Cys-lite background (see below) were generated using QuikChange PCR (Stratagene). The eIF5 mutants were expressed in BL21(DE3) Codon Plus cells (Stratagene) as described (32). The purified proteins were then fluorescently labeled with fluorescein-maleimide (34).
Measurement of TC Binding Kinetics
Measurements of the kinetics of TC binding to 40 S subunits were carried out using the native gel assay as described (16). Component concentrations were 0.5–1 nm [35S]Met-tRNAi, 200 nm eIF2, 1 μm eIF1-G107K, 1 μm eIF1A, 200 nm 40 S subunits, and 1 mm GDPNP·Mg2+. The experiments were carried out in the absence of eIF5 and the presence of full-length eIF5 or the isolated eIF5 NTD or CTD. The concentration of eIF5 or its domains was saturating (2 μm) for effects on TC loading (data not shown). Data were fit with a first-order exponential equation to determine the observed pseudo-first-order rate constants for TC binding. The reactions were pseudo-first-order because the concentrations of 40 S subunits and initiation factors were much higher than the concentration of labeled Met-tRNAi.
Measurement of the Kinetics of eIF1 Dissociation from the PIC
The kinetics of eIF1 dissociation from the PIC in response to recognition of a start codon was measured on an SX.180MV-R stopped-flow fluorometer (Applied Photophysics) as described previously (16). Briefly, 43 S complex was made with 50 nm fluorescein-labeled WT eIF1 or eIF1-G107K (donor), 60 nm TAMRA-labeled WT or mutant eIF1A (acceptor), 100 nm 40 S subunits, and 450 nm TC (made with GDPNP). This was mixed rapidly with an equal volume of 20 μm mRNA(AUG) and 6 μm unlabeled eIF1 as chase. When full-length eIF5 or its domains were included, they were used at a final concentration of 2 μm. Loss of FRET between the two factors was observed as an increase in fluorescein fluorescence. The data were fit with a double exponential equation, with the first phase corresponding to a conformational change and the second to eIF1 dissociation.
Pi Release Kinetics
The kinetics of phosphate release from 43 S complexes in response to start codon recognition was measured by monitoring GTP hydrolysis using a rapid quench device (Kintek) as described previously (16). TC was formed at 4× concentration; 3.2 μm eIF2, 3.2 μm Met-tRNAi, and 250 pm [γ-32P]GTP were incubated in 1× recon buffer for 15 min at 26 °C. Ribosomal complex was also made at 4× concentration in 1× recon buffer using 800 nm 40 S subunits, 3.2 μm eIF1, and 3.2 μm eIF1A. Equal volumes of TC and ribosomal complex were mixed with 2 μm eIF5 and 20 μm mRNA(AUG) in a rapid quench. Reactions were quenched at different times with 100 mm EDTA. The samples were then on polyethyleneimine-cellulose TLC plates using 0.4 m Potassium phosphate buffer, pH 3.4, as the mobile phase, followed by PhosphorImager analysis to quantify the fraction of GTP hydrolyzed over time. The data were fit with a double exponential rate equation. The first phase corresponds to GTP hydrolysis, and the second phase corresponds to Pi release, which drives GTP hydrolysis forward (15).
Determination of Steady-state FRET Efficiencies between eIF5-Fluorescein and eIF1A-TAMRA
For each experiment, two identical samples were prepared. One contained the 43 S complex assembled with eIF5-Fl and unlabeled eIF1A. This was designated as a “donor alone” complex. The second complex was assembled with eIF5-Fl and eIF1A-TAMRA. This was designated as a “donor + acceptor” complex. In both cases, the concentrations of reagents were 100 nm eIF5-Fl, 200 nm eIF1A (labeled or unlabeled), 100 nm 40 S subunits, 1 μm eIF1, 200 nm TC, and 10 μm mRNA(AUG). After mixing all of the components, the fluorescein fluorescence was monitored as a function of time using λex = 490 nm and λem = 520 nm on a Spex Fluorolog-3 fluorometer. When equilibrium had been achieved (as indicated by stabilization of fluorescence intensity), samples were excited at 490 nm, and fluorescence emission was measured as a function of wavelength from 505 to 600 nm. Repeated measurements of the same sample gave identical results. FRET efficiency was calculated as 1 − IDA/ID, where IDA is the fluorescence intensity at 520 nm of the donor + acceptor sample, and ID is the intensity at 520 nm of the donor alone sample (35).
Measurement of the Kinetics of the Change in eIF1A-eIF5 FRET in Response to Start Codon Recognition by the PIC
Measurement of the change in eIF1A-eIF5 FRET in the PIC in response to start codon recognition was performed using an SX.180MV-R stopped-flow fluorometer (Applied Photophysics). Briefly, 43 S PICs were assembled using 400 nm WT or mutant eIF1A-TAMRA, 200 nm eIF5-Fl, 200 nm 40 S subunits, 400 nm TC (made with GDPNP), 800 nm eIF1 for 1 h at 26 °C in 1× recon buffer. This 2× complex was mixed with an equal volume of 20 μm mRNA(AUG or AUC). The increase in FRET between the dyes on the two labeled proteins with respect to time was measured as a decrease in fluorescein fluorescence. The data were fit with a double exponential equation.
General Kinetics
All kinetic experiments were repeated at least three times. The rate constants presented are averages, and the errors are mean deviations. Control experiments indicated that photobleaching was not significant on the time scale of any of the experiments.
RESULTS
The CTT of eIF1A and N-terminal Domain of eIF5 Move Closer to Each Other upon Start Codon Recognition
We showed previously that a direct or indirect interaction of the eIF1A CTT with eIF5 is crucial for stabilizing the closed, scanning-arrested conformation of the PIC (12). To further explore this interaction, we sought to observe FRET between fluorescently labeled positions in eIF5 and the C terminus of eIF1A. We reasoned that such a FRET signal could be used to monitor movements of eIF5 and eIF1A taking place within the PIC during the rearrangement from its open to closed conformations. To this end, we engineered a variety of eIF5 derivatives labeled site-specifically on single cysteine residues with fluorescein-maleimide and then tested for FRET in the PIC between these positions and eIF1A labeled C-terminally with TAMRA.
Yeast eIF5 has seven cysteines (Fig. 1A, shown in orange), four of which are part of a stable ZBD in the N-terminal domain (eIF5 NTD) (22) (Fig. 1A, orange ZBD residues). The three non-ZBD cysteines, Cys-6, Cys-289, and Cys-294, are located on the surface of the N- and C-terminal domains of the protein (Fig. 1A) and are not conserved in eukaryotic eIF5s, including those found in various plant, animal, and even other fungal species. We generated a “Cys-lite” derivative of eIF5 in which the three non-ZBD cysteines were changed to serines (Fig. 1A). The ZBD cysteines were not changed because they are important for maintaining the structure of eIF5 and are tightly coordinated to Zn2+ (22) and thus should be resistant to modification. Consistent with this last prediction, the Cys-lite protein is not modified by fluorescein-maleimide (data not shown). Cys-lite eIF5 was purified, and its ability to stimulate GTP hydrolysis by eIF2 in PICs was tested using a reconstituted S. cerevisiae translation initiation system. The results show that 2 μm Cys-lite eIF5 stimulates GTP hydrolysis to the same rate observed for the WT factor at the same concentration (Fig. 1B). The ability of the Cys-lite eIF5 to promote closed complex formation in conjunction with eIF1A upon start codon recognition was also tested by measuring rates of eIF1A dissociation from the PIC in the reconstituted system (12), and the mutant protein was found to behave indistinguishably from native eIF5 (data not shown). Next we generated various single cysteine mutants of Cys-lite eIF5 by introducing cysteines at non-conserved, surface-exposed positions in the NTD, CTD, and linker region using site-directed mutagenesis (Table 1). These mutants were then expressed in Escherichia coli and purified, and the cysteine residues were labeled with fluorescein-maleimide (34). We also labeled the three naturally occurring cysteines at positions 6, 289, and 294 in variants containing each one as the only surface-exposed cysteine (Table 1). We then screened all of our fluorescein-labeled Cys-lite eIF5 mutants for FRET with C-terminally TAMRA-labeled eIF1A in reconstituted PICs (also containing eIF1, TC, and 40 S subunits; TC contained GDPNP in place of GTP to prevent GTP hydrolysis and Pi release). The fluorescein dye was excited at 490 nm, and emission was monitored as a function of wavelength. Experiments were performed in the absence and presence of an unstructured model mRNA with a central AUG codon (mRNA(AUG)). Labels at two positions in the NTD of eIF5, S68C and N94C, showed FRET with the TAMRA on the C terminus of eIF1A only after the addition of the model mRNA, with the largest decrease in donor (fluorescein) emission at 520 nm. The FRET efficiency observed with the S68C (eIF5-S68C-Fl) mutant was ∼10% (Fig. 1C). A corresponding increase in acceptor (TAMRA) emission at 580 nm was also observed (compare red and blue curves in Fig. 1C). No change in emission intensity upon the addition of mRNA(AUG) was observed with PICs made with eIF5-S68C-Fl and unlabeled eIF1A (Fig. 1C, black curve, “donor alone + AUG”), indicating that the decreased fluorescein emission observed with complexes containing eIF5-S68C-Fl and TAMRA-labeled eIF1A is due to FRET rather than a change in intrinsic fluorescence of the fluorescein dye. Similar results were observed with the N94C mutant, although the extent of the FRET change was smaller (∼7%), and thus we chose to pursue the S68C mutant instead. No FRET was observed with dyes in the linker region or the CTD, suggesting that these domains are farther from the C terminus of eIF1A in the PIC after start codon recognition than is the NTD of eIF5. The decrease in fluorescein fluorescence (increase in FRET) after mRNA binding suggests that AUG recognition leads to movements within the PIC that bring the C terminus of eIF1A and the NTD of eIF5 closer together. The fluorescence anisotropy of eIF5-S68C-Fl bound to the PIC is 0.157, and that for eIF1A-TAMRA is 0.260. These values are considerably below the theoretical maximum of 0.4, indicating that the dyes in the PIC still have significant conformational flexibility and that changes in FRET are not due to changes in (fixed) orientations between the fluorophores (36). Thus, the increase in FRET reflects a decrease in distance between the two dyes.
TABLE 1.
Fluorescently labeled Cys-lite eIF5 variants
| eIF5 variant | Domain |
|---|---|
| Cys-6a | NTD |
| S43C | NTD |
| S68C | NTD |
| N94C | NTD |
| S155C | NTD |
| S189C | Linker |
| S228C | Linker |
| N265C | CTD |
| N283C | CTD |
| Cys-289a | CTD |
| Cys-294a | CTD |
| E335C | CTD |
| E378C | CTD |
a Native cysteines in eIF5.
To probe the events reported on by this change in FRET, we monitored the kinetics of the decrease in fluorescein fluorescence upon the addition of mRNA (Fig. 2A). PICs containing eIF1A-TAMRA, eIF5-S68C-Fl, eIF1, TC, and 40 S subunits were rapidly mixed with mRNA(AUG) in a stopped-flow fluorimeter, and the decrease in fluorescein fluorescence was monitored over time (λex = 490 nm, λem = 520 nm). The resulting curve (Fig. 2A) was biphasic; the first phase had a rate constant (k1) of 24 s−1 and a normalized amplitude of 0.5, and the second phase had a rate constant (k2) of 0.4 s−1 and an amplitude of 0.5. k2 is strikingly close to the rate constants for release of eIF1 and Pi upon start codon recognition (0.6 and 0.4 s−1, respectively). This observation suggests that these events are controlled by the same rate-limiting step in WT PICs.
FIGURE 2.
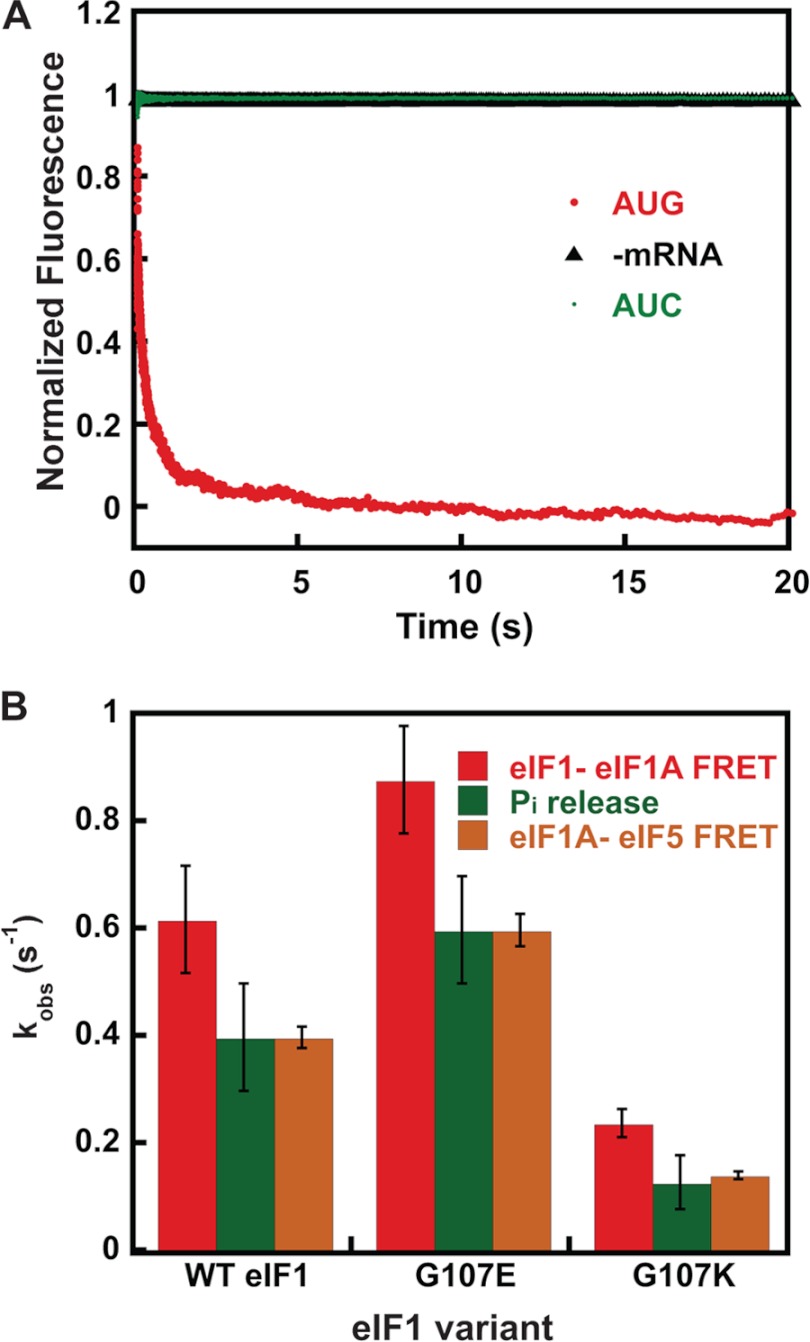
Increase in FRET between the eIF1A CTT and eIF5 NTD is dependent upon AUG codon recognition and is coupled to release of eIF1 from the PIC. A, kinetics of eIF1A-eIF5 FRET change upon binding of mRNAs to the PIC. Shown is the increase in FRET (decrease in fluorescein fluorescence) between Cys-lite eIF5(S68C)-Fl and eIF1A-TAMRA in the PIC after the addition of mRNA(AUG) (red). No FRET change was observed when buffer alone was mixed with labeled PICs (black; −mRNA) or when the same model mRNA with an AUC codon in place of the AUG codon was used (green), indicating that the FRET decrease is triggered by recognition of an AUG start codon. Curves were fit with a double exponential rate equation. Curves are the averages of three independent experiments. B, comparison of the kinetics of the eIF1-eIF1A FRET (red), eIF1A-eIF5 FRET change (orange), and Pi release (green) upon start codon recognition by PICs assembled with WT or mutant versions of eIF1. Values are averages from at least three independent experiments, and error bars represent average deviations (p < 0.05 for differences in rates between WT and mutants).
As for the eIF1 and Pi release steps (11, 15), the rate of the increase in FRET efficiency between fluorophores on the C terminus of eIF1A and the NTD of eIF5 depends on the identity of the start codon in the mRNA. Replacing the AUG codon with an AUC codon led to a complete loss of the increase in FRET, to the same extent as leaving out mRNA altogether and mixing the PICs with buffer alone (compare green AUC and black −mRNA curves in Fig. 2A). Thus, the change in FRET signal occurs in response to recognition of a start codon in the mRNA.
The Increase in FRET between the eIF1A CTT and eIF5 NTD Is Coupled to Release of eIF1 from the PIC
The fact that the newly identified eIF1A-eIF5 FRET signal is dependent on an AUG start codon (Fig. 2A) suggested that it might also be governed by eIF1 dissociation from the PIC. To address this possibility, we tested the effects of two mutations in eIF1 that were shown previously to speed up (G107E) or slow down (G107K) release of the factor from the PIC upon start codon recognition. In previous work, these mutations were shown to have corresponding effects on the rate constant for Pi release (16), and these findings were confirmed here for both mutants (Fig. 2B, eIF1 release and Pi release; in both cases, eIF5 was included in the PICs). The stopped-flow experiments monitoring FRET between eIF1A and eIF5 were repeated with PICs made with eIF1-G107E or eIF1-G107K. Remarkably, as with the rate constants for eIF1 release and Pi release, we found that G107E consistently increased the rate constant for the slow phase of eIF1A-eIF5 FRET by 1.5-fold (p < 0.05). Likewise, the G107K mutant decreased the rate constant for the slow phase by 4-fold, the same extent as it decreased the rate constants for eIF1 and Pi release. These data support the possibility that the rate of eIF1 release directly or indirectly governs the rate of the movement of eIF1A and/or eIF5 that produces the FRET change.
Mutations in the SE Elements in the CTT of eIF1A Uncouple Release of eIF1 and Pi from the PIC in Response to Start Codon Recognition
Having obtained evidence that the eIF1A CTT gets closer to the eIF5 NTD in the PIC upon AUG recognition, we wished to determine the role of the SE elements in the CTT of eIF1A in this movement. The SE elements (SE1 and SE2) are comprised of two loosely conserved tandem repeats of 9 or 10 residues located in the beginning of the unstructured CTT, each containing a pair of invariant Phe residues as critical constituents. We showed previously that the SE elements have partially overlapping functions that stabilize the open conformation of the PIC and promote scanning through near-cognate start codons and also support rapid TC loading in the Pout state. Replacing 7 of the 9 residues of SE1 with alanines (SE1*) and 5 of the 10 residues of SE2 with alanines (SE2*) is sufficient to inactivate each element, but combining these substitutions in a single mutant (SE1*,SE2*) is required to eliminate their overlapping contributions to TC recruitment and accurate start codon selection (6). Because SE2 is more potent than SE1, we chose to examine the SE2* single mutant and SE1*,SE2* double mutant in the experiments described below.
Versions of these two mutant factors were labeled with TAMRA on their C termini for use in FRET experiments. PICs were assembled with eIF5-S68C-Fl and either eIF1A-SE2*-TAMRA or eIF1A-SE1*,SE2*-TAMRA and then mixed rapidly in the stopped-flow device with mRNA(AUG). The rate constants of the first phase of the decrease in fluorescein fluorescence were unchanged by the mutations relative to the values observed with WT PICs (28 and 20 s−1 with the SE2* and SE1*,SE2* mutants, respectively versus 24 s−1 with WT eIF1A), although the amplitudes were decreased relative to the slow phases (Fig. 3A and Table 2). Remarkably, however, the rate constant for the slow phase was decreased 20–40-fold by the SE mutations relative to the value with the WT factor (Fig. 3A and Table 2). These findings indicate that the SE elements are required for rapid movement of eIF1A with respect to eIF5 upon start codon recognition.
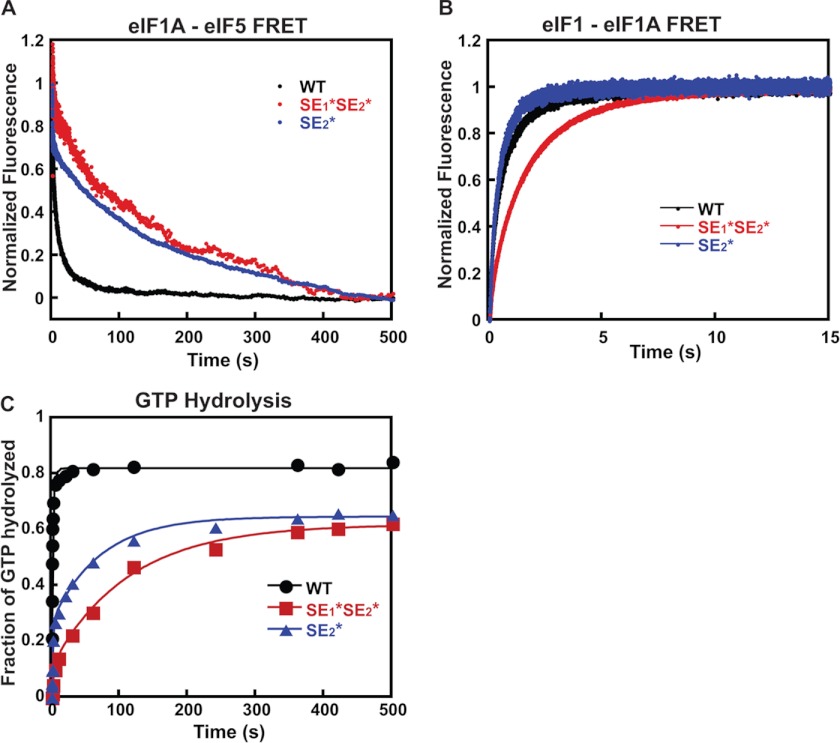
FIGURE 3.
Effect of mutations in the SE elements in the eIF1A CTT on the kinetics of the eIF1A-eIF5 FRET change, eIF1 release, and Pi release in response to AUG recognition. A, the kinetics of the increase in eIF1A-eIF5 FRET on AUG recognition was measured as a decrease in fluorescein (donor) fluorescence after rapid mixing with mRNA(AUG) in a stopped-flow fluorometer. Preinitiation complexes were assembled with eIF5(S68C)-Fl and either C-terminally TAMRA-labeled WT eIF1A (black), eIF1A-SE1*,SE2* (red), or eIF1A-SE2* (blue). Curves were fit with double exponential rate equations. The data shown are the averages of three experiments. B, the kinetics of the decrease in eIF1-eIF1A FRET in the PIC upon start codon recognition was monitored as an increase in fluorescein (donor) fluorescence between eIF1-Fl and eIF1A-TAMRA after rapid mixing of PICs and mRNA(AUG) in a stopped-flow fluorometer. Curves were fit with double exponential rate equations. The first phase corresponds to a conformational change in the PIC upon start codon recognition, and the second phase corresponds to eIF1 release. PICs were assembled with C-terminally labeled WT eIF1A (black), eIF1A-SE1*,SE2* (red), or eIF1A-SE2* (blue). 2 μm eIF5 was included for consistency with eIF1A-eIF5 FRET and GTPase experiments. The data shown are averages from three experiments. C, the kinetics of GTP hydrolysis and Pi release from 43 S PICs was measured after the addition of eIF5 and mRNA(AUG). Time points from reactions were stopped with 100 mm EDTA, and [γ-32P]GTP and [γ-32P]Pi were then separated using polyethyleneimine-cellulose TLC and quantified by PhosphorImager analysis. The fraction of GTP hydrolyzed versus time was plotted, and the data fit with a double exponential rate equation. The fast phase corresponds to GTP hydrolysis, and the slower phase corresponds to Pi release. PICs were assembled with WT eIF1A (black circles), eIF1A-SE1*,SE2* (red squares), or eIF1A-SE2* (blue triangles).
TABLE 2.
Kinetic parameters for the FRET change between eIF1A variants and eIF5 in PICs upon AUG recognition
| eIF1A variant | kobs | Amplitude (a) |
|---|---|---|
| s−1 | ||
| WT | k1 = 24 ± 4.0 | a1 = 0.5 ± 0.1 |
| k2 = 0.4 ± 0.1 | a2 = 0.5 ± 0.1 | |
| SE2* | k1 = 28 ± 6.0 | a1 = 0.3 ± 0.05 |
| k2 = 0.02 ± 0.1 | a2 = 0.7 ± 0.1 | |
| SE1*,SE2* | k1 = 20 ± 8.0 | a1 = 0.15 ± 0.05 |
| k2 = 0.01 ± 0.005 | a2 = 0.80 ± 0.05 |
We then asked whether the SE mutations evoke corresponding reductions in the rates of eIF1 and Pi release from reconstituted PICs. eIF1 release was monitored as a decrease in the efficiency of FRET between fluorescein-labeled eIF1 and the TAMRA-labeled eIF1A mutants. 2 μm eIF5 was included for consistency with the eIF1A-eIF5 and GTPase/Pi release assays. Using this assay with the WT factors, we previously showed that start codon recognition triggers a biphasic decrease in FRET (increase in fluorescein fluorescence) between the two fluorophores on the C termini of the respective factors (33). The first phase is thought to correspond to a conformational change within the PIC that moves the two fluorophores apart, and the second phase was shown to correspond to release of eIF1 from the complex. (For simplicity, we refer below to these two events as the fast and slow phases of eIF1 release.) With WT eIF1A in the PIC, the addition of a saturating concentration of mRNA(AUG) results in a biphasic increase in fluorescein fluorescence with rate constants of 4 and 0.6 s−1 for the first (k1) and second (k2) phases, respectively, and roughly equal amplitudes (Figs. 3 (B, black curve) and 4 (A and B) and Table 3, eIF1 release), consistent with previous studies (11, 15, 17). Despite its strong effect on the slow phase of eIF1A-eIF5 FRET, the SE2* mutation did not change the rate constants or amplitudes of either phase of eIF1 release (Figs. 3 (B, blue curve) and 4 (A and B) and Table 3), and the SE1*,SE2* mutation produced relatively small reductions in the rate constants of the fast and slow phases of eIF1 release of 4- and 2-fold, respectively (Figs. 3 (B, red curve) and 4 (A and B) and Table 3). These results indicate that eIF1 release can proceed normally in the absence of proper movement of eIF1A and eIF5 relative to one another.
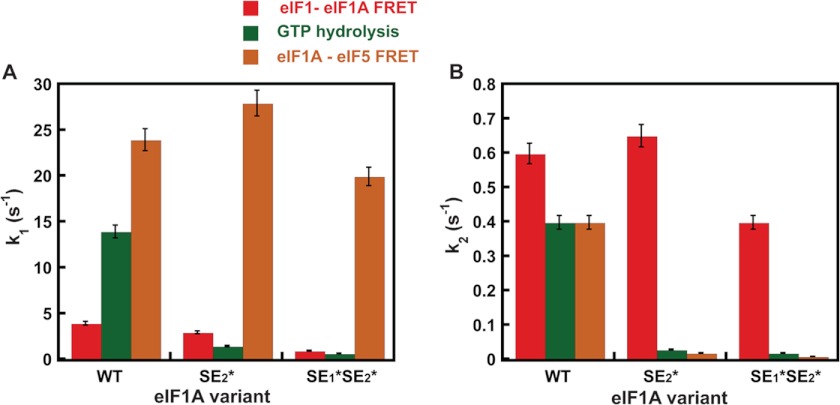
FIGURE 4.
Comparison of rate constants for eIF1-eIF1A FRET change, GTP hydrolysis, and eIF1A-eIF5 FRET change upon start codon recognition by the PIC. A, rate constants (k1) for the first phase of the decrease in eIF1-eIF1A FRET (red), GTP hydrolysis (green), and increase in eIF1A-eIF5 FRET (orange) upon start codon recognition by the PIC. For eIF1-eIF1A FRET, this phase corresponds to a first-order event, probably a conformational change. For the GTP hydrolysis reaction, this phase corresponds to a step or steps that limit the rate of cleavage of GTP to produce GDP and Pi. B, rate constants (k2) for the second phase of the decrease in eIF1-eIF1A FRET (red), GTP hydrolysis (green), and increase in eIF1A-eIF5 FRET (orange) upon start codon recognition by the PIC. For eIF1-eIF1A FRET, this phase corresponds to eIF1 release from the PIC, and for GTP hydrolysis, this phase corresponds to Pi release from eIF2, which drives the reaction to completion. PICs were assembled with WT eIF1A, eIF1A-SE1*SE2*, or eIF1A-SE2*. Data shown are averages of at least three experiments, and error bars represent average deviations.
TABLE 3.
Kinetic parameters for the eIF1-eIF1A FRET change and GTP hydrolysis in 43 S PICs
| eIF1A variant | eIF1 release kobs | GTP hydrolysis kobs |
|---|---|---|
| s−1 | s−1 | |
| WT | k1 = 4 ± 1 | k1 = 14 ± 4 |
| a1 = 0.6 ± 0.05 | a1 = 0.7 ± 0.1 | |
| k2 = 0.6 ± 0.1 | k2 = 0.4 ± 0.1 | |
| a2 = 0.4 ± 0.05 | a2 = 0.3 ± 0.1 | |
| k1 = 3 ± 1 | k1 = 1.4 ± 0.6 | |
| SE2* | a1 = 0.6 ± 0.1 | a1 = 0.4 ± 0.1 |
| k2 = 0.7 ± 0.1 | k2 = 0.03 ± 0.01 | |
| a2 = 0.4 ± 0.1 | a2 = 0.6 ± 0.1 | |
| k1 = 1 ± 0.5 | k1 = 0.7 ± 0.2 | |
| SE1*,SE2* | a1 = 0.5 ± 0.1 | a1 = 0.2 ± 0.1 |
| k2 = 0.4 ± 0.1 | k2 = 0.02 ± 0.005 | |
| a2 = 0.5 ± 0.1 | a2 = 0.8 ± 0.1 |
Finally, we measured the effect of these same mutations in eIF1A on Pi release from the PIC in response to start codon recognition. In these experiments, PICs containing [γ-32P]GTP are mixed in a rapid quench device with saturating mRNA(AUG) and eIF5, and the reactions are quenched after various times with EDTA. The amount of Pi generated over time is monitored using polyethyleneimine thin-layer chromatography followed by PhosphorImager analysis. Similar to eIF1 release, the rate of Pi formation is biphasic, with the first phase corresponding to eIF5-dependent GTP hydrolysis within the PIC and the second to Pi release, which drives GTP hydrolysis to completion (15, 16). PICs assembled with WT eIF1A hydrolyzed GTP with rate constants of 14 and 0.4 s−1 for the first (k1) and second (k2) phases, respectively (Figs. 3 (C, black curve) and 4 (A and B) and Table 3, GTP hydrolysis). These values are consistent with previous results and with the fact that eIF1 release limits the rate of Pi release (15–17). Remarkably, the SE2* substitution decreased the rates of both GTP hydrolysis (k1) and Pi release (k2) 10-fold (Figs. 3 (C, blue curve) and 4 (A and B) and Table 3), although it had no effect on the rate of eIF1 release (Fig. 4B). Likewise, the SE1*,SE2* substitution decreased the rate constants for GTP hydrolysis and Pi release by 20-fold (Figs. 3 (C, red curve) and 4 (A and B) and Table 3), whereas it had only a 2-fold effect on the rate constant for eIF1 release (Fig. 4B).
The fact that the rate constants for the fast phases of the eIF1-eIF1A FRET change, GTP hydrolysis/Pi release, and eIF1A-eIF5 FRET change cover a range from 4 to 23 s−1 in the WT system (Fig. 4A) suggests that they do not correspond to the same molecular event. This interpretation is further supported by the differing effects of the SE element mutations in the eIF1A CTT on these rate constants. For the eIF1-eIF1A FRET change, the SE2* mutation has no effect on k1, whereas the SE1*,SE2* mutation reduces k1 by 4-fold. In contrast, k1 for GTP hydrolysis/Pi release, which is thought to represent the rate-limiting step for GTP hydrolysis itself (15), is reduced ≥10-fold by both the SE2* and SE1*,SE2* mutations. Finally, k1 for the eIF1A-eIF5 FRET change is unaffected by either mutation. These data indicate that different molecular events correspond to each of the fast phases in the three assays.
On the other hand, the slow phases of the eIF1-eIF1A FRET change (eIF1 release), GTP hydrolysis (Pi release), and eIF1A-eIF5 FRET change occur with similar rate constants in the WT system (Fig. 4B), suggesting that they could correspond to the same molecular event. However, the fact that the SE mutations produce a much larger reduction in the rate of Pi release than in the rate of eIF1 release (Fig. 4B) indicates that ejection of eIF1 from the PIC does not directly trigger Pi release but requires the intermediary function of the eIF1A CTT. This proposal is consistent with our previous results demonstrating a functional interaction between the eIF1A CTT and eIF5 that is thought to stabilize the closed conformation of the PIC upon start codon recognition (12). Moreover, our finding that the SE mutations dramatically reduce the rate of the slow phase of the eIF1A-eIF5 FRET change, mirroring their effects on Pi release, is consistent with the possibility that the event corresponding to the slow phase in the eIF1A-eIF5 FRET assay is a prerequisite for Pi release. Supporting this proposal, the eIF1A-eIF5 FRET change occurs with the non-hydrolyzable GTP analog GDPNP in the PIC, indicating that Pi release is not required for the FRET change to occur.
Taken together, our results suggest that eIF1 release is required for and normally sets the rate of Pi release in WT PICs. eIF1 release is not sufficient for Pi release, however, which additionally requires the movement of the eIF1A CTT toward the eIF5 NTD manifested in the slow phase of the eIF1A-eIF5 FRET change. This movement of the two factors relative to each other is coupled to eIF1 release and is critically dependent on the SE elements in the eIF1A CTT. It is also noteworthy that both the SE2* and SE1*,SE2* mutants decrease the rate and amplitude of the fast kinetic phase of GTP hydrolysis in addition to their effects on the second phase. These data suggest that the CTT of eIF1A is involved in promoting full GTPase activation of eIF2, along with the GAP eIF5, in addition to its proposed role in triggering Pi release.
The CTD of eIF5 Promotes Displacement of eIF1 from the PIC
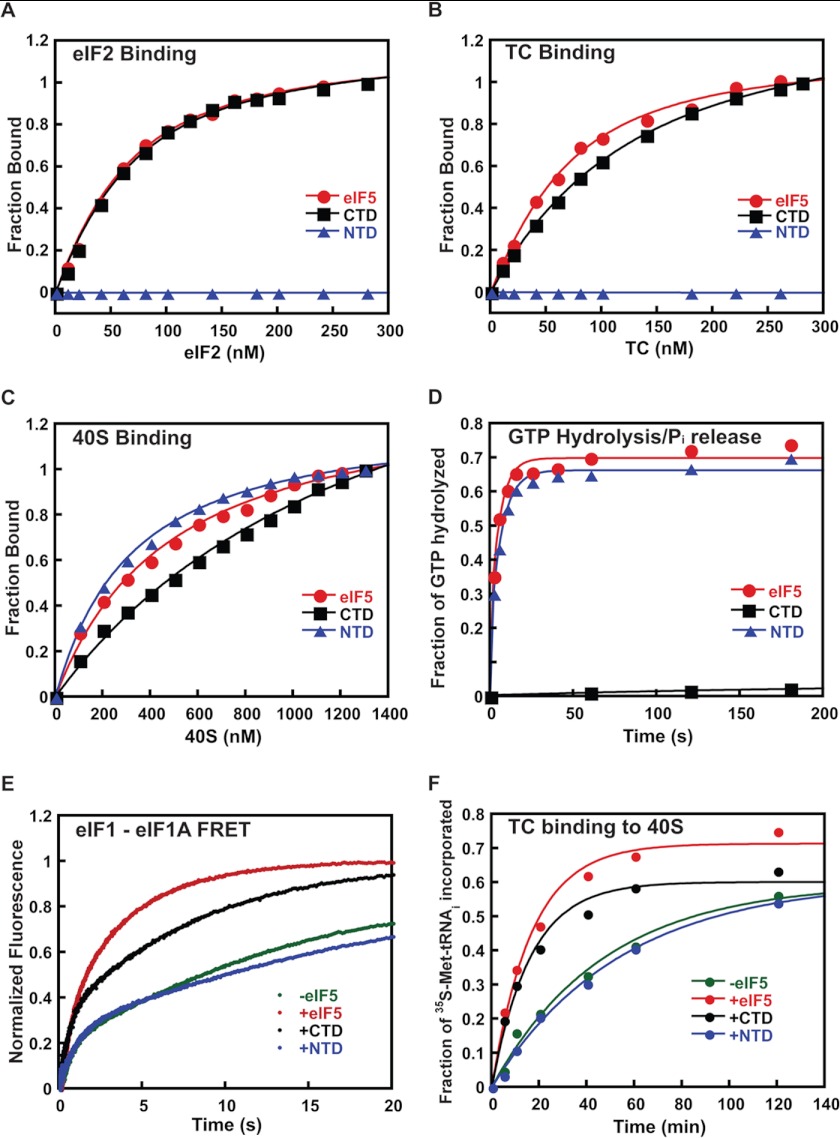
We showed previously that eIF5 antagonizes binding of eIF1 to the PIC and that overexpressing it in yeast promotes recognition of near-cognate UUG start codons, whereas overexpressing eIF1 has the opposite effect, suggesting that the ability of eIF5 to displace eIF1 from the PIC is part of its function in start codon recognition (16, 37). In this model, one of the domains of eIF5 would move into part of the binding site for eIF1 upon start codon recognition, promoting the latter factor's irreversible release from the PIC (16, 22). Consistent with an important role for competition between eIF5 and eIF1 in start codon recognition, recent work has shown that overexpression of eIF5 in mammalian cells also reduces the fidelity of start codon recognition, in a manner that can be suppressed by overexpression of eIF1 (31). To further explore the molecular basis of the interaction between these two factors and establish which domain of eIF5 is responsible for antagonizing eIF1 binding to the PIC, we expressed and purified the isolated NTD and CTD of eIF5 as separate proteins. Using C-terminally fluorescein-labeled versions of these protein domains, we first measured their affinity for eIF2 and TC by monitoring fluorescence anisotropy (Fig. 5, A and B). Full-length eIF5 and the CTD bound to eIF2 with identical affinity (Kd = 40 nm) (Fig. 5A). The NTD did not bind detectably at any concentration of eIF2 tested. Similar results were observed with the TC; full-length eIF5 and the CTD bound with nearly the same affinity (Kd values of 40 and 90 nm, respectively), whereas no binding of the NTD could be detected (Fig. 5B).
FIGURE 5.
The C-terminal domain of eIF5 antagonizes binding of eIF1 to the PIC. Full-length WT eIF5 (red curves, circle) and its isolated CTD (black curves, square) and NTD (blue curves, triangle) were expressed and purified as described under “Experimental Procedures.” In all cases, data are the averages of at least two independent experiments. A, binding of C-terminally fluorescein-labeled eIF5 derivatives (50 nm) to eIF2 was assessed by monitoring fluorescence anisotropy (λex = 497 nm; λem = 520 nm) as a function of the concentration of eIF2. Kd values were determined by fitting the resulting data with a hyperbolic binding equation. The measured Kd values were 40 ± 3 nm for full-length eIF5 and 45 ± 5 nm for the eIF5 CTD; the eIF5 NTD did not detectably bind to eIF2, and thus no Kd was determined. B, the binding of the eIF5 derivatives to ternary complex (eIF2·Met-tRNAi·GDPNP) was measured using fluorescence anisotropy as in A. The measured Kd values were 45 ± 5 nm for full-length eIF5 and 90 ± 5 nm for the eIF5 CTD. No binding was detected to the eIF5 NTD. C, the binding of eIF5 derivatives to the 40 S ribosomal subunit was determined by monitoring the change in fluorescence anisotropy of C-terminally fluorescein-labeled full-length eIF5 or the NTD or CTD (50 nm) as a function of concentration of 40 S subunits. The measured Kd values were 300 ± 50 nm for full-length eIF5, 250 ± 30 nm for the NTD, and 1500 ± 500 nm for the CTD. D, kinetics of GTP hydrolysis by eIF2 within the PIC stimulated by full-length eIF5 or the NTD or CTD (2 μm). Curves were fit with double-exponential rate equations to determine rate constants: full-length eIF5 (red circles); eIF5 CTD (black squares); and eIF5 NTD (blue triangles). E, the effect of eIF5 and its domains on release of eIF1-G107K-Fl from the PIC in response to start codon recognition. The decrease in FRET between G107K-Fl and eIF1A-TAMRA within the PIC after rapid mixing with mRNA(AUG) without or with 2 μm full-length eIF5 or the NTD or CTD was measured as an increase in fluorescein (donor) fluorescence: −eIF5 (green); +eIF5 (red); eIF5 CTD (black); eIF5 NTD (blue). Curves were fit with double-exponential rate equations to determine rate constants. F, the effects of eIF5 NTD and CTD on the kinetics of TC recruitment to the PIC. TC containing [35S]Met-tRNAi and GDPNP was mixed with 40S subunits, eIF1-G107K, and eIF1A in the absence or presence of eIF5 or the NTD or CTD (2 μm). Time points were loaded directly on a running native gel, and the fraction of TC bound over time was analyzed by phosphorimaging: −eIF5 (green diamonds); +eIF5 (red circles); eIF5 CTD (black squares); eIF5 NTD (blue triangles). Curves were fit with single-exponential equations to determine observed pseudo-first-order rate constants.
We next tested whether eIF5 and its domains bind to the 40 S subunit itself (Fig. 5C). Using the same C-terminally labeled derivatives of full-length eIF5 and its domains, we monitored fluorescence anisotropy as a function of 40 S subunit concentration. In this case, the full-length factor and the NTD bound with similar affinities (Kd values of 400 and 300 nm, respectively). The CTD also bound but with a 3-fold higher Kd. Thus, both domains of eIF5 are capable of binding directly to the 40 S ribosomal subunit.
The NTD of eIF5 contains the Arg-15 residue essential for the factor's GAP function. This domain was previously reported to be sufficient to promote GTP hydrolysis by eIF2 (22, 38, 39). Consistent with these results, we found that the NTD was capable of promoting full GTPase activity within the PIC, whereas the CTD did not stimulate GTP hydrolysis detectably (Fig. 5D). These data indicate that although the NTD does not interact detectably with eIF2 or the TC free in solution, it must be able to do so in the context of the PIC, consistent with previous work showing an interaction between the NTD of eIF5 and the isolated γ-subunit of eIF2 (39).
To determine which domain of eIF5 is responsible for promoting release of eIF1 from the PIC, we assembled 43 S complexes using eIF1-G107K labeled on its C terminus with fluorescein, eIF1A labeled on its C terminus with TAMRA, TC, and 40 S subunits. Start codon recognition was initiated by rapid mixing in a stopped-flow device of mRNA(AUG) alone or mRNA(AUG) together with either full-length eIF5 or the isolated NTD or CTD (2 μm). Loss of FRET between the fluorescein and TAMRA labels was monitored (increase in fluorescein fluorescence) to follow the kinetics of eIF1 release from the complex. The G107K mutant of eIF1 was used because its release from the complex is impaired, allowing the effect of eIF5 to be seen more readily in response to an AUG start codon; release of WT eIF1 is so facile on a cognate AUG codon that the addition of eIF5 has only a small effect (16). In the absence of eIF5, eIF1-G107K was released with a rate constant of 0.08 s−1 (Fig. 5E and Table 4), which is 5-fold lower than that observed for WT eIF1 in the absence of eIF5 (11). The addition of 2 μm eIF5 NTD had no effect on the rate constant for eIF1-G107K release, whereas the addition of the same concentration of full-length eIF5 or the CTD increased the observed rate constant ≥2-fold to 0.25 and 0.17 s−1, respectively. These data indicate that the CTD of eIF5 is responsible for promoting release of eIF1 from the PIC.
TABLE 4.
Rate constants for eIF1 dissociation from and TC recruitment to PICs
| eIF5 variant | eIF1-G107K dissociation kobs | TC recruitment to PIC kobs |
|---|---|---|
| s−1 | min−1 | |
| −eIF5 | 0.05 ± 0.01 | 0.02 ± 0.01 |
| +eIF5 | 0.25 ± 0.05 | 0.06 ± 0.01 |
| +CTD | 0.17 ± 0.03 | 0.05 ± 0.005 |
| +NTD | 0.05 ± 0.01 | 0.02 ± 0.01 |
We previously presented data indicating that in the reconstituted yeast translation initiation system, binding of TC to the 40 S subunit (in the presence of eIF1, eIF1A, and model mRNA) occurs in two steps: an initial encounter that is not codon-dependent and cannot be detected in our native gel-based assay, followed by a start codon-dependent conformational change that locks the complex into a stable state that is detected in the native gel assay (13, 16). We showed that release of eIF1 is required for the transition to this stable state and that high concentrations of eIF5 accelerate the apparent rate of TC binding by enhancing eIF1 release from the PIC and thus conversion to the stable, closed state that is detected in the gel-based assay (16). As a further test of the functions of the domains of eIF5 in promoting eIF1 release, we also measured their effects on the kinetics of TC binding to 40 S subunits in the presence of eIF1-G107K and eIF1A but in the absence of mRNA, using the native gel assay. At the concentration of 40 S subunits used (200 nm) in the absence of eIF5, the observed pseudo-first-order rate constant (kobs) for detectable TC binding was 0.02 min−1. The addition of 2 μm full-length eIF5 increased kobs 3-fold to 0.06 min−1. As in the eIF1 release experiments above, a 2 μm concentration of the eIF5 NTD had no effect on the rate of TC binding, but the same concentration of the CTD increased the rate nearly as much as the full-length factor did (kobs = 0.05 min−1). Thus, these experiments strongly support the conclusion that the CTD of eIF5 is responsible for the factor's ability to promote eIF1 release from the PIC with attendant enhancement of TC binding.
DISCUSSION
Previous work has elucidated a number of key events taking place within the PIC when it encounters a start codon. These events trigger downstream steps and commit the complex to continuing initiation at the selected position on the mRNA. The data presented herein have significantly refined and strengthened the model for the events surrounding start codon recognition by providing evidence for new initiation codon-dependent movements of eIF1A and eIF5 within the PIC and elucidating the connections between Pi release from eIF2 and conformational changes in the initiation factors.
Coupling of Start Codon-dependent Movements of eIF1, eIF1A, and eIF5 to Pi Release by eIF2
We have found a FRET signal between fluorophores on the C terminus of eIF1A and the folded N-terminal domain of eIF5 that occurs upon start codon recognition, indicating that the CTT of eIF1A and the NTD of eIF5 move closer to each other after the AUG is encountered. Importantly, the rate of this rearrangement is strongly dependent on the SE elements in the eIF1A CTT. These results are consistent with our previous data showing a strong interaction, either direct or indirect, between the CTT of eIF1A and eIF5 that takes place upon start codon recognition (12) and consistent with more recent hydroxyl radical footprinting results indicating that the CTT of eIF1A must move out of the P site of the 40 S subunit when the initiator tRNA is fully engaged (10).
In addition, we have shown that mutations in the SE elements in the eIF1A CTT decouple the release of eIF1 from the release of Pi in response to start codon recognition. The SE2* and SE1*SE2* mutations have ≤2-fold effects on the rate of eIF1 release but evoke 13–20-fold reductions in the rate of Pi release. These data indicate that, although eIF1 dissociation is necessary for Pi release, it is not sufficient and that the SE elements are additionally required for rapid Pi release from eIF2 on AUG recognition. The fact that the SE mutations also reduce the rate of GTP hydrolysis itself indicates that the CTT of eIF1A has a previously unrecognized function, working along with eIF5 to promote the GTPase activity of eIF2 in the PIC.
Taken together, our data suggest that release of eIF1 and movement of the CTT of eIF1A out of the P site toward the NTD of eIF5 are triggered separately by the same event. We consider the most likely candidate for this event to be accommodation of tRNAi fully into the P site because it is expected to produce steric clashes with both factors (9, 10) and because tRNAi binds more tightly to the PIC in the absence of eIF1 (5). In this model, mutations in eIF1 that speed or slow release of the factor exert their influence on Pi release by facilitating or impeding accommodation of the tRNA, respectively. This altered rate of tRNA accommodation would then directly affect the rate of movement of the CTT of eIF1A out of the P site, which in turn would affect the rate of Pi release. Indeed, this is exactly what we observed with the G107E and G107K mutants of eIF1 (Fig. 2B). The fact that the rate of the slow kinetic phase of the eIF1A-eIF5 FRET is always the same (within error) as the rate of Pi release (Fig. 4B; k2 values in Tables 2 and 3) supports the proposal that movement of the CTT is a key step for triggering Pi release required in addition to dissociation of eIF1.
The fast kinetic phase in the loss of eIF1-eIF1A FRET reflects a conformational change upon AUG recognition that increases the distance between eIF1 and the eIF1A CTT (11). As described above, our data indicate that this rearrangement and the fast kinetic phases of GTP hydrolysis and the eIF1A-eIF5 FRET change do not reflect the same molecular event. This conclusion argues against the possibility that the fast phases of either of the changes in FRET correspond to movement of the CTT of eIF1A because one of the fluorophores in each case is on the C terminus of eIF1A, and movement of the CTT that affected FRET with one partner (e.g. eIF5) would most likely affect FRET with the other partner (e.g. eIF1) as well. Instead, we suggest that the fast phases of the eIF1-eIF1A and eIF1A-eIF5 FRET changes correspond to movement of eIF1 and eIF5, respectively (see below). Overall, these data support the model in which start codon recognition and attendant accommodation of the initiator tRNA into the P site of the 40 S subunit cause movement of eIF1 away from its initial binding site in the PIC and subsequent movement of the CTT of eIF1A toward the NTD of eIF5, which in turn triggers Pi release from eIF2.
One seeming paradox is the fact that mutations in the SE elements increase utilization of near-cognate UUG codons relative to AUG codons as start sites in vivo, yet they slow movement of the eIF1A CTT and Pi release from eIF2, steps that are thought to be important for commitment of the PIC to proceeding with initiation upon start codon recognition. A possible explanation to resolve this seeming paradox is that substituting the critical phenylalanine residues in the SE elements with alanines leads to elimination of the CTT from the P site and also impairs the interaction with the eIF5 NTD that is required to trigger dissociation of Pi from eIF2. Because the CTT with SE mutations is not in the P site, accommodation of the initiator tRNA is less hindered and can take place more readily in response to near-cognate codons. This enhanced transit of the tRNAi from the Pout to Pin state would correspondingly increase the rate of complex closure and arrest scanning PICs on near-cognate codons long enough to proceed with the remaining steps of initiation with increased frequency relative to WT complexes at the same sites (in this model, the SE mutations cannot enhance transit to Pin to a level faster than that which already occurs on AUG codons in WT complexes). This proposal is consistent with our previous data suggesting that the SE mutations stabilize the closed/Pin state of the PIC relative to the open one at near-cognate codons (6).
This model can also explain why the SE element mutations do not slow release of eIF1 to the same degree they slow movement of the eIF1A CTT and Pi release; because the CTT of eIF1A with the SE mutations is not in the P site, its hindered movement does not impede accommodation of the initiator tRNA into the P site or subsequent displacement of eIF1 upon start codon recognition. In addition, the proposal can explain why the SE mutations slow TC binding to the PIC (6) because proper positioning of the eIF1A CTT within the complex could be required to directly stabilize binding of TC to the 40 S subunit as well as to promote the open state of the complex to which TC initially binds.
The Domains of eIF5 Play Multiple Roles in Start Codon Recognition
Previous studies have indicated that eIF5 plays multiple roles in start codon recognition in addition to its function as a GAP for eIF2. To better understand the various activities of eIF5, we tested the ability of the isolated NTD and CTD of the factor to interact with other components of the system and to promote release of eIF1 from the PIC. The isolated eIF5 CTD bound to eIF2 and TC with the same affinity as the full-length factor, whereas the eIF5 NTD did not bind detectably to either. In contrast, previous studies showed that the eIF5 NTD binds directly to the isolated eIF2γ subunit in solution (39). Together, these data suggest that the binding site for the eIF5 NTD on eIF2γ is occluded in free eIF2 and TC in solution and raise the possibility that a conformational change occurs in the scanning PIC to open this binding site and allow the eIF5 NTD to interact with eIF2γ. One appealing possibility is that this switch involves displacement of eIF2β from eIF2γ by the eIF5 NTD because the NTD of eIF5 and eIF2β share a common fold, and both have a ZBD. Consistent with this idea, the crystal structure of an archaeal counterpart of eIF2 (aIF2) (40), reveals that the ZBD interacts with the γ subunit and places residues corresponding to those altered by Sui− substitutions in yeast eIF2β in proximity to the GTP binding pocket of aIF2γ, including a Sui− mutation thought to elevate the latent GTPase activity of eIF2. This and other findings support the notion that the WT eIF2β ZBD blocks the GTPase activity of eIF2γ in a manner relieved by eIF5 in the PIC (7, 26, 40, 41). We suggest that the eIF2β ZBD is displaced from eIF2γ by the homologous ZBD domain of eIF5 in the scanning PIC, allowing the unstructured NTT of eIF5 (containing the key GTPase activating residue Arg-15) to stimulate GTP hydrolysis by eIF2γ and suggest that the eIF5 NTT must be withdrawn from the GTP-binding pocket of eIF2γ to enable Pi release upon AUG recognition (see below).
In addition to binding to eIF2, our data indicate that both domains of eIF5 can interact with the 40 S subunit, with the NTD and full-length factor binding 3–4-fold more tightly than the CTD (Fig. 5C). The mechanistic significance of these interactions is unclear, but they suggest the possibility that eIF5 could mediate communication between the ribosome and eIF2.
Importantly, we found that the eIF5 CTD is the domain responsible for promoting release of eIF1 from the PIC because it functions nearly as well as the WT factor to promote release of eIF1-G107K from the PIC, whereas the isolated NTD has no detectable effect. Consistent with this, the eIF5 CTD promotes stable TC binding to PICs reconstituted with eIF1-G107K as effectively as does full-length eIF5, whereas the eIF5 NTD, again, has no detectable effect despite its ability to activate GTP hydrolysis.
A Complex Series of Molecular Rearrangements Underlies the Response to Start Codon Recognition by the PIC
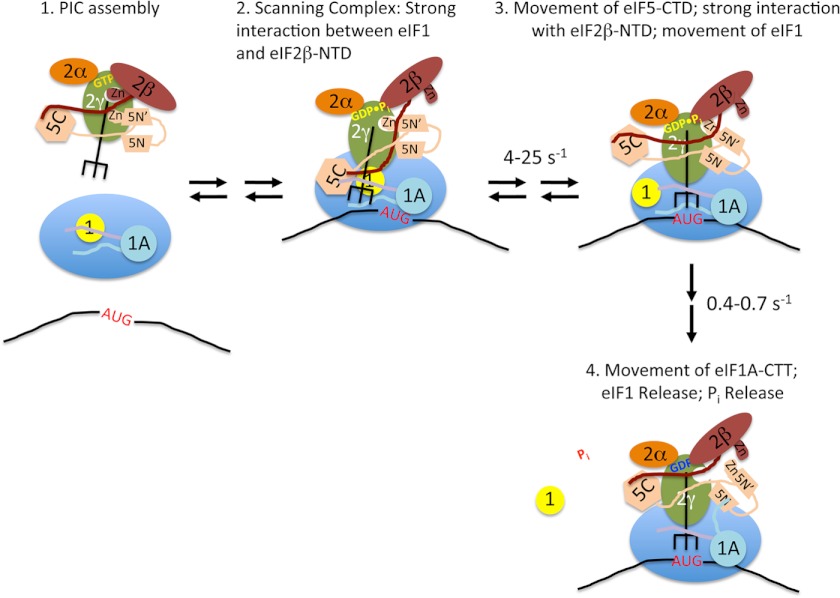
Based on all of the available data, we suggest the following model for the events surrounding start codon recognition by the PIC (Fig. 6), which synthesizes and builds off of several previously proposed models (6, 16, 20, 25). Prior to encountering a start codon, the PIC is in an open conformation, with tRNAi in the Pout state, not fully engaged with the P site of the 40 S subunit (Fig. 6, stage 2). eIF1 and the CTT of eIF1A partially occupy the P site, inhibiting full accommodation of the tRNAi (6, 8–10). Binding of eIF1 to the open conformation of the PIC is stabilized by its interaction with the NTT of eIF2β (42) and possibly also its weak interaction with the CTD of eIF5, accounting for the previous finding that eIF1 substitutions that weaken these contacts reduce eIF1·PIC association and elevate UUG initiation (a Sui− phenotype) (19). Having demonstrated here 40 S binding by the eIF5 CTD, we propose that this domain occupies a site adjacent to the eIF1 binding site on the 40 S platform, where it may interact weakly with both the eIF2β NTT and eIF1 (25, 42). We speculate that at this stage, the ZBD of eIF2β has been displaced by the ZBD of eIF5, allowing the unstructured NTT of eIF5 (containing Arg-R15) to stimulate GTP hydrolysis by eIF2γ; however, the position of the eIF5 NTT on eIF2γ prevents Pi release from the scanning complex. Entry of an AUG codon into the P site (Fig. 6, stage 3) drives formation of the codon-anticodon helix, which pulls the initiator tRNA more fully into the P site (Pin state), ejecting the CTT of eIF1A and displacing eIF1 to a second, lower affinity binding site on the 40 S platform. Movement of the tRNA and eIF1 allows strengthened interaction between the eIF5 CTD and the eIF2β NTT, resulting in displacement of eIF1 from eIF2β by the eIF5 CTD. Loss of this interaction between eIF1 and eIF2β in turn enhances the rate of eIF1 release from the PIC. High concentrations of full-length eIF5 or its CTD can exogenously compete with eIF1 for binding to the NTT of eIF2β, thus weakening binding of eIF1 to the PIC and promoting its release at near-cognate codons. This start codon-dependent switch in strong binding partners for the eIF2β NTT between eIF1 and the eIF5 CTD, which explains the observed effects of high concentrations of eIF5 and its CTD, is the main difference between our model and the one previously proposed by Luna et al. (25), in which the NTT of eIF2β did not interact with eIF1.
FIGURE 6.
Model for the events taking place within the PIC upon start codon recognition. Stage 1, TC·eIF5 complex binds to the 40S subunit. eIF1 occupies a site on the platform of the 40S adjacent to the P site, and the body of eIF1A binds in the A site, with its NTT (purple) and CTT (light blue) binding in the P site. Stage 2, the scanning PIC is in an open conformation with the tRNAi in the Pout state. eIF1 binding is stabilized by a strong interaction with the NTT of eIF2β. Stage 3, entry of the start codon into the P site allows formation of the codon-anticodon helix between the mRNA and tRNAi, which drives the tRNA into the Pin state. This displaces eIF1 to a second, weaker binding site on the 40S subunit, breaking its interaction with the eIF2β NTT, which in turn binds strongly to the eIF5 CTD. Movements of the tRNA and/or eIF5 CTD result in changes in the orientation of the eIF5 NTD. Stage 4, eIF1 dissociates from the complex, which, along with accommodation of tRNAi into the P site, causes the CTT of eIF1A to move and interact with the eIF5 NTD. This interaction triggers Pi release from eIF2, possibly by moving the unstructured NTT of eIF5 (not shown for clarity). The resulting complex is in a closed, scanning-arrested state.
Consistent with the notion that the previously demonstrated interaction between eIF1 and the NTT of eIF2β has a role in stabilizing binding of eIF1 to the PIC, mutations in eIF1 that disrupt the interface between the two factors produce Sui− phenotypes (19). In contrast, mutations that destabilize the interaction between the eIF5-CTD and eIF2β-NTT result in an Ssu− phenotype and decrease the ability of eIF5 to promote eIF1 release and closed complex formation upon AUG recognition (25, 43), consistent with the proposal that this interaction occurs after the start codon is encountered and competes with the interaction between the eIF2β NTT and eIF1 to promote eIF1 release from the PIC.
At this stage (Fig. 6, stage 4), having been ejected from the P site, the eIF1A CTT now engages with the eIF5 NTD, dependent on the critical Phe residues of the SE elements, and this new interaction helps to displace the eIF5 NTT from the G domain of eIF2γ, triggering Pi release. These last features of the model are based on our discovery of eIF1A-eIF5 FRET upon start codon recognition and the fact that the slow kinetic phase of this FRET change and the rate of Pi release are the same and are reduced coordinately by the SE mutations in the CTT of eIF1A.
In this model, the fast phase of the change in eIF1-eIF1A FRET corresponds to the initial movement of eIF1 away from the P site in response to accommodation of the initiator tRNA (stage 2 to stage 3). This rapid movement would be followed by slower dissociation of the factor from the PIC (stage 3 to stage 4). The fast phase of the increase in eIF1A-eIF5 FRET, which is >5-fold faster than the first phase of the decrease in eIF1-eIF1A FRET, could correspond to a conformational change in eIF5 induced by movements of the initiator tRNA and/or interaction between the NTT of eIF2β and the CTD of eIF5 (stage 2 to stage 3). Movement of the eIF1A CTT out of the P site would then correspond to the second, slower phase of the change in eIF1A-eIF5 FRET. This latter event may also involve breaking of the interaction between eIF1 and eIF1A (33) upon dissociation of eIF1 from the PIC.
Although this is just one possible model that is consistent with currently available data, it should serve as a useful framework to plan and interpret future experiments aimed at developing a complete understanding of the molecular mechanics of start codon recognition during eukaryotic translation initiation.
Acknowledgments
We thank the members of our laboratories for comments and suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grants GM 62128 (to J. R. L.) and by the National Institutes of Health Intramural Research Program (to A. G. H.).
- PIC
- preinitiation complex
- TC
- ternary complex
- GAP
- GTPase-activating protein
- GDPNP
- Guanosine 5′ β, γ imido triphosphate
- NTT
- N-terminal tail
- CTT
- C-terminal tail
- SE
- scanning enhancer
- SI
- scanning inhibitor
- NTD
- N-terminal domain
- CTD
- C-terminal domain
- ZBD
- zinc-binding domain.
REFERENCES
- 1. Hinnebusch A. G., Lorsch J. R. (2012) The mechanism of eukaryotic translation initiation. New insights and challenges. Cold Spring Harb. Perspect. Biol. 4, 1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aitken C. E., Lorsch J. R. (2012) A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 19, 568–576 [DOI] [PubMed] [Google Scholar]
- 3. Jackson R. J., Hellen C. U., Pestova T. V. (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pestova T. V., Borukhov S. I., Hellen C. U. (1998) Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394, 854–859 [DOI] [PubMed] [Google Scholar]
- 5. Passmore L. A., Schmeing T. M., Maag D., Applefield D. J., Acker M. G., Algire M. A., Lorsch J. R., Ramakrishnan V. (2007) The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell 26, 41–50 [DOI] [PubMed] [Google Scholar]
- 6. Saini A. K., Nanda J. S., Lorsch J. R., Hinnebusch A. G. (2010) Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNAiMet binding to the ribosome. Genes Dev. 24, 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hinnebusch A. G. (2011) Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 75, 434–467, first page of table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lomakin I. B., Kolupaeva V. G., Marintchev A., Wagner G., Pestova T. V. (2003) Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 17, 2786–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rabl J., Leibundgut M., Ataide S. F., Haag A., Ban N. (2011) Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331, 730–736 [DOI] [PubMed] [Google Scholar]
- 10. Yu Y., Marintchev A., Kolupaeva V. G., Unbehaun A., Veryasova T., Lai S. C., Hong P., Wagner G., Hellen C. U., Pestova T. V. (2009) Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Res. 37, 5167–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maag D., Fekete C. A., Gryczynski Z., Lorsch J. R. (2005) A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol. Cell 17, 265–275 [DOI] [PubMed] [Google Scholar]
- 12. Maag D., Algire M. A., Lorsch J. R. (2006) Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal pre-initiation complex plays a role in start site selection. J. Mol. Biol. 356, 724–737 [DOI] [PubMed] [Google Scholar]
- 13. Kolitz S. E., Takacs J. E., Lorsch J. R. (2009) Kinetic and thermodynamic analysis of the role of start codon/anticodon base pairing during eukaryotic translation initiation. RNA 15, 138–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lorsch J. R., Dever T. E. (2010) Molecular view of 43 S complex formation and start site selection in eukaryotic translation initiation. J. Biol. Chem. 285, 21203–21207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Algire M. A., Maag D., Lorsch J. R. (2005) Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell 20, 251–262 [DOI] [PubMed] [Google Scholar]
- 16. Nanda J. S., Cheung Y. N., Takacs J. E., Martin-Marcos P., Saini A. K., Hinnebusch A. G., Lorsch J. R. (2009) eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J. Mol. Biol. 394, 268–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheung Y. N., Maag D., Mitchell S. F., Fekete C. A., Algire M. A., Takacs J. E., Shirokikh N., Pestova T., Lorsch J. R., Hinnebusch A. G. (2007) Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 21, 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fekete C. A., Applefield D. J., Blakely S. A., Shirokikh N., Pestova T., Lorsch J. R., Hinnebusch A. G. (2005) The eIF1A C-terminal domain promotes initiation complex assembly, scanning and AUG selection in vivo. EMBO J. 24, 3588–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reibarkh M., Yamamoto Y., Singh C. R., del Rio F., Fahmy A., Lee B., Luna R. E., Ii M., Wagner G., Asano K. (2008) Eukaryotic initiation factor (eIF) 1 carries two distinct eIF5-binding faces important for multifactor assembly and AUG selection. J. Biol. Chem. 283, 1094–1103 [DOI] [PubMed] [Google Scholar]
- 20. Fekete C. A., Mitchell S. F., Cherkasova V. A., Applefield D., Algire M. A., Maag D., Saini A. K., Lorsch J. R., Hinnebusch A. G. (2007) N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. EMBO J. 26, 1602–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wei Z., Xue Y., Xu H., Gong W. (2006) Crystal structure of the C-terminal domain of S. cerevisiae eIF5. J. Mol. Biol. 359, 1–9 [DOI] [PubMed] [Google Scholar]
- 22. Conte M. R., Kelly G., Babon J., Sanfelice D., Youell J., Smerdon S. J., Proud C. G. (2006) Structure of the eukaryotic initiation factor (eIF) 5 reveals a fold common to several translation factors. Biochemistry 45, 4550–4558 [DOI] [PubMed] [Google Scholar]
- 23. Asano K., Clayton J., Shalev A., Hinnebusch A. G. (2000) A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNA(Met) is an important translation initiation intermediate in vivo. Genes Dev. 14, 2534–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yamamoto Y., Singh C. R., Marintchev A., Hall N. S., Hannig E. M., Wagner G., Asano K. (2005) The eukaryotic initiation factor (eIF) 5 HEAT domain mediates multifactor assembly and scanning with distinct interfaces to eIF1, eIF2, eIF3, and eIF4G. Proc. Natl. Acad. Sci. U.S.A. 102, 16164–16169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luna R. E., Arthanari H., Hiraishi H., Nanda J., Martin-Marcos P., Markus M. A., Akabayov B., Milbradt A. G., Luna L. E., Seo H. C., Hyberts S. G., Fahmy A., Reibarkh M., Miles D., Hagner P. R., O'Day E. M., Yi T., Marintchev A., Hinnebusch A. G., Lorsch J. R., Asano K., Wagner G. (2012) The C-terminal domain of eukaryotic initiation factor 5 promotes start codon recognition by its dynamic interplay with eIF1 and eIF2β. Cell Rep. 1, 689–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang H. K., Yoon H., Hannig E. M., Donahue T. F. (1997) GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev. 11, 2396–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Asano K., Shalev A., Phan L., Nielsen K., Clayton J., Valássek L., Donahue T. F., Hinnebusch A. G. (2001) Multiple roles for the C-terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J. 20, 2326–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Singh C. R., Curtis C., Yamamoto Y., Hall N. S., Kruse D. S., He H., Hannig E. M., Asano K. (2005) Eukaryotic translation initiation factor 5 is critical for integrity of the scanning preinitiation complex and accurate control of GCN4 translation. Mol. Cell Biol. 25, 5480–5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Valásek L., Nielsen K. H., Zhang F., Fekete C. A., Hinnebusch A. G. (2004) Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol. Cell Biol. 24, 9437–9455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alone P. V., Cao C., Dever T. E. (2008) Translation initiation factor 2γ mutant alters start codon selection independent of Met-tRNA binding. Mol. Cell Biol. 28, 6877–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Loughran G., Sachs M. S., Atkins J. F., Ivanov I. P. (2012) Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res. 40, 2898–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Acker M. G., Kolitz S. E., Mitchell S. F., Nanda J. S., Lorsch J. R. (2007) Reconstitution of yeast translation initiation. Methods Enzymol. 430, 111–145 [DOI] [PubMed] [Google Scholar]
- 33. Maag D., Lorsch J. R. (2003) Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. J. Mol. Biol. 330, 917–924 [DOI] [PubMed] [Google Scholar]
- 34. Nanda J., Lorsch J. (2011) Labeling of proteins with fluorophores using maleimide derivitization. in Methods Navigator, 1st Ed (Lorsch J. R., ed) Elsevier, Amsterdam: [DOI] [PubMed] [Google Scholar]
- 35. Shih W. M., Gryczynski Z., Lakowicz J. R., Spudich J. A. (2000) A FRET-based sensor reveals large ATP hydrolysis-induced conformational changes and three distinct states of the molecular motor myosin. Cell 102, 683–694 [DOI] [PubMed] [Google Scholar]
- 36. Lakowicz J. R. (1999) Principles of Fluorescence Spectroscopy, pp. 353–381, Kluwer Academic/Plenum Publishers, New York, NY [Google Scholar]
- 37. Martin-Marcos P., Cheung Y. N., Hinnebusch A. G. (2011) Functional elements in initiation factors 1, 1A, and 2β discriminate against poor AUG context and non-AUG start codons. Mol. Cell Biol. 31, 4814–4831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Das S., Ghosh R., Maitra U. (2001) Eukaryotic translation initiation factor 5 functions as a GTPase-activating protein. J. Biol. Chem. 276, 6720–6726 [DOI] [PubMed] [Google Scholar]
- 39. Alone P. V., Dever T. E. (2006) Direct binding of translation initiation factor eIF2γ-G domain to its GTPase-activating and GDP-GTP exchange factors eIF5 and eIF2B ϵ. J. Biol. Chem. 281, 12636–12644 [DOI] [PubMed] [Google Scholar]
- 40. Yatime L., Mechulam Y., Blanquet S., Schmitt E. (2007) Structure of an archaeal heterotrimeric initiation factor 2 reveals a nucleotide state between the GTP and the GDP states. Proc. Natl. Acad. Sci. U.S.A. 104, 18445–18450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hashimoto N. N., Carnevalli L. S., Castilho B. A. (2002) Translation initiation at non-AUG codons mediated by weakened association of eukaryotic initiation factor (eIF) 2 subunits. Biochem. J. 367, 359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Singh C. R., He H., Ii M., Yamamoto Y., Asano K. (2004) Efficient incorporation of eukaryotic initiation factor 1 into the multifactor complex is critical for formation of functional ribosomal preinitiation complexes in vivo. J. Biol. Chem. 279, 31910–31920 [DOI] [PubMed] [Google Scholar]
- 43. Laurino J. P., Thompson G. M., Pacheco E., Castilho B. A. (1999) The β subunit of eukaryotic translation initiation factor 2 binds mRNA through the lysine repeats and a region comprising the C2-C2 motif. Mol. Cell Biol. 19, 173–181 [DOI] [PMC free article] [PubMed] [Google Scholar]