Background: Signals originated from mitochondria can affect other intracellular compartments.
Results: Decreased levels in mitochondrial cyclophilin D promote cell proliferation and cell motility via chemokine/STAT3 signaling.
Conclusion: Mitochondria regulate nuclear gene expression.
Significance: Interorganelle signaling affects cell proliferation and cell motility.
Keywords: Cell Invasion, Cell Migration, Cell Proliferation, Mitochondria, STAT3, Chemokines, Cyclophilin D
Abstract
Mitochondria control bioenergetics and cell fate decisions, but how they influence nuclear gene expression is understood poorly. Here, we show that deletion or reduction in the levels of cyclophilin D (CypD, also called Ppif), a mitochondrial matrix peptidyl prolyl isomerase and apoptosis regulator, results in increased cell proliferation and enhanced cell migration and invasion. These responses are associated with extensive transcriptional changes, modulation of a chemokine/chemokine receptor gene signature, and activation of the pleiotropic inflammatory mediator, STAT3. In the absence of CypD, active STAT3 enhances cell proliferation via accelerated entry into S-phase and stimulates autocrine/paracrine cell motility through Cxcl12-Cxcr4-directed chemotaxis. Therefore, CypD directs mitochondria-to-nuclei inflammatory gene expression in normal and tumor cells. This pathway may contribute to malignant traits under conditions of CypD modulation.
Introduction
Cyclophilin D (CypD, Ppif)2 is a mitochondrial matrix protein with cis-trans peptidyl prolyl isomerase activity (1) that participates in the regulation of organelle permeability transition and the initiation of apoptosis (2). Considerable effort has been devoted to elucidate a role of CypD in various aspects of mitochondrial cell death (3), map its molecular arrangement in an organelle permeability transition pore (4), and validate its therapeutic value as a target for improved cytoprotection (5) or, conversely, induction of apoptosis (6). Dynamically regulated by posttranslational modifications, including nitrosylation (7), deacetylation (8), and chaperone-directed folding (9), CypD has been implicated in additional mitochondrial functions, for instance stabilization of the glycolytic enzyme hexokinase II (10) at the outer membrane (11) and quality control of damaged organelles via modulation of autophagy (12). However, the possibility that CypD may connect to extramitochondrial signaling mechanism(s) and, in particular, changes in nuclear gene expression has not been investigated.
The concept of interorganelle signaling has been modeled on the cellular response to proteotoxic stress (13). Accordingly, defects in the protein folding environment in the endoplasmic reticulum initiates a complex gene expression response in the nucleus (14) aimed at restoring homeostasis while also dampening mitochondrial cell death pathways (15). There is also evidence that signals emanating from a mitochondrial unfolded protein response (16) may affect cellular homeostasis, participating in metabolic reprogramming, especially in tumor cells (9), and inducing secondary endoplasmic reticulum-dependent responses (17). Although some of these mitochondria → nuclei “retrograde” signaling mechanisms (18) are evolutionarily conserved and may affect broad homeostatic pathways, including aging, metabolism, and adaptive responses (19), their effector molecules in mitochondria have not been delineated clearly. In this study, we explored a potential role of CypD in mitochondria-to-nuclei interorganelle signaling.
EXPERIMENTAL PROCEDURES
Cell Lines
Human glioblastoma LN229 cells, monocytic THP-1 cells, breast adenocarcinoma MCF-7 cells, non-transformed NIH3T3 fibroblasts, and pancreatic adenocarcinoma MiaPACA cells were obtained from the ATCC and maintained in culture as recommended by the supplier. LN229 cells stably transfected with control, non-targeting shRNA or shRNA directed to CypD have been described (20). Clones of stable transfectants were maintained in the presence of 800 μg/ml of G418 sulfate (Sigma). Tumor cell types were maintained in high-glucose DMEM (except for THP-1 cells, which were propagated in RPMI 1640) with glutamine and supplemented with 10% FBS and 1% penicillin/streptomycin. WT or CypD−/− mouse embryonic fibroblasts (MEFs) were generously provided by Dr. Nika Danial, Harvard Medical School, and described previously (1).
Antibodies and Reagents
The following antibodies to Tyr-705-phosphorylated STAT3, Ser-727-phosphorylated STAT3, STAT3, Tyr-416-phosphorylated Src, Src, and Ser-15-phosphorylated p53 were obtained from Cell Signaling Technology, Inc. Antibodies to CypD and p53 were from Millipore. A neutralizing antibody to Cxcl12 or IL-6 was from Bethyl Laboratories, Inc. or Abcam, respectively. Oligofectamine, Lipofectamine, thymidine, phosphatase inhibitor mixtures 2 and 3, and antibodies to β-actin or β-tubulin were obtained from Sigma-Aldrich. Small molecule inhibitors of the STAT3 SH2 domain, Stattic (21) or Src, Dasatinib, were from Selleck Chemicals. EDTA-free protease inhibitor mixture and X-tremeGENE transfection reagent were from Roche. Luciferase assay kits were obtained from Promega. Cytokine arrays were from RayBiotech. Small molecule inhibitors to JAK2 (catalog nos. 420097 and TG101348) were obtained from EMD Chemicals and Chemie Tek, respectively. Acti-stain 488 fluorescent phalloidin was from Cytoskeleton.
SiRNA and Plasmid Vectors
Control, non-targeting siRNA pool (catalog no. D-001810) and CypD siRNA ON-Target SMARTpool (catalog no. L-009708-00-0005) were purchased from Dharmacon. Human (catalog no. sc-29493) and mouse (catalog no. sc-29494) STAT3-directed siRNA pools were obtained from Santa Cruz Biotechnology, Inc. A STAT3 reporter luciferase plasmid was from Panomics. A peptidyl prolyl isomerase-defective CypD mutant cDNA was described (9).
Western Blotting
Protein lysates were prepared from various cell types in extraction buffer containing 150 mm NaCl, 50 mm Tris (pH 8.0), and 1% Triton X-100 in the presence of protease and phosphatases inhibitors. 40 μg of protein was separated on SDS-polyacrylamide gels, transferred to PVDF membranes, and incubated with primary antibodies of various specificities for 16 h at 4 °C. After washes and incubation with secondary antibodies, immunoreactive bands were detected by chemiluminescence.
Fluorescence-activated Cell Sorting
Various cell types were fixed in glacial 70% ethanol for 24 h followed by incubation with propidium iodide (2.5 μg/ml) in the presence of RNAse A for an additional 16 h at 4 °C. Ten thousand events were acquired on a Calibur flow cytometer followed by analysis using Cell Quest Pro software (BD Biosciences). For cell cycle synchronization experiments, cells were arrested at the G1 phase of the cell cycle by double thymidine block and processed for analysis of DNA content at increasing time intervals (2–24 h) after release.
For determination of reactive oxygen species (ROS), cells were harvested, washed, and incubated in PBS (pH 7.4), in the presence of a carboxyl derivative of fluorescein (CM-H2DCFDA, Invitrogen) at a final concentration of 10 μm. After 30-min incubation, cells were centrifuged, washed, and suspended in PBS (pH 7.4) for flow cytometry analysis at excitation and emission wavelengths set at 488 nm and 575 nm, respectively.
Cell Viability Assay
Various cell types were seeded in 96-well plates at 5 × 104/ml (MEFs at 3 × 104) and incubated under the different experimental conditions for 24 h. Changes in cell viability were evaluated using a 3-(4,5-dimethyl-thyazoyl-2-yl)2,5 diphenyltetrazolium bromide colorimetric assay, and the production of formazan salts was quantified at 595 nm wavelength in a plate reader (DTX 880, Beckman Coulter). In some experiments, LN229 cells stably transfected with control shRNA or CypD-directed shRNA or, alternatively, WT or CypD−/− MEFs were incubated in the presence of increasing concentrations of the ROS scavenger N-acetyl cysteine and analyzed for cell proliferation by 3-(4,5-dimethyl-thyazoyl-2-yl)2,5 diphenyltetrazolium bromide assay.
Cell migration and Cell Invasion Assays
All migration experiments were carried out using uncoated transwell chambers (Becton Dickinson, catalog no. 353097) using NIH3T3-conditioned medium in the lower compartment as a chemoattractant. In the case of poorly invasive MCF-7 cells, transwell membranes were coated with collagen (15 μg/ml) before analysis of cell motility. All invasion experiments were carried out using Matrigel-coated transwell chambers (Becton Dickinson, catalog no. 354483) with NIH3T3-conditioned medium in the lower compartment as a chemoattractant.
Gene Expression Profiling
Total RNA was extracted using the TRI reagent (Ambion) method and treated with DNAse I (Fermentas). For microarray analysis, RNA quality was determined using the bioanalyzer (Agilent). Only samples with RNA Integrity numbers > 7.5 were used for further studies. Three WT (CypD+/+) and three CypD knockout (CypD−/−) samples were used. Equal amounts (400 ng) of total RNA were amplified under the different conditions and hybridized to the HumanWG-6 v2 whole genome bead arrays as recommended by Illumina.
For real-time PCR, cDNA was synthesized with polyT primers using the Maxima Universal first strand cDNA synthesis kit (Fermentas). Real-time PCR reactions were performed in triplicates using 1 μl of cDNA. Primers were obtained from IDT as follows (5′ → 3′), Spon2, TCACCTTCTCCTCCCCTAAC (forward) and AATGAATTTGCTGGGTGGCT (reverse); Mgp, AACACCTTTATGTCCCCTCAG (forward) and TCTCTGTTGATCTCGTAGGCA (reverse); Amigo2, TAACTCCCTAAACCTTCATCGT (forward) and TCTACTCAGCCATTCACATAACT (reverse); Col2a1, TTAGGGCAGAGAGAGAAGGG (forward) and CAGTGACTTGAGTGTAGCGT (reverse); Gjb3, GGGTGACGAGCAAAAAGACT (forward) and GGATGTTGGAGATGGGGAAG (reverse); GAPDH, AGGACACTGAGCAAGAGAGG (forward) and TGTTATTATGGGGGTCTGGGA (reverse); B2m, TTTCTGGTGCTTGTCTCACT (forward) and TTCAGTATGTTCGGCTTCCC (reverse); Hmbs, CATATCTGCCTTTCCCTCAGT (forward) and TGGTTTATTAGTGGTATTGGTTACA (reverse); IL-6, GATACCACTCCCAACAGACC (forward) and CAACTCTTTTCTCATTTCCACG (reverse); and Cxcl12, GCCCCTGCCGGTTCTTCGAG (forward) and GCCGTGCAACAATCTGAAGGGC (reverse). For profiling of mouse chemokine/chemokine receptor signatures and STAT3 gene signature, Qiagen RT2 Profiler PCR arrays were used (codes PAMM-022C-2 and PAMM-039ZC-2, respectively) according to the instructions of the manufacturer. Briefly, 500 ng of total RNA was retrotranscribed to cDNA, diluted 1:10 with the RT2 SYBR Green/ROX PCR Master Mix, and 25 μl of this final solution was added to each well of the array plate.
Transfections
Various cell types were seeded at 5 × 104/ml and transfected with control non-targeting siRNA or siRNA pool directed to the individual target gene(s) using Oligofectamine (25 pmol/ml). Plasmid DNA was transfected using Lipofectamine (0.5 μg/ml) in Opti-Mem, which was replaced with complete culture medium after 5-h incubation.
STAT3 Luciferase Reporter Assay
Cells were transfected with 0.25 μg of STAT3 reporter plasmid in the presence of 0.025 μg of Renilla plasmid as normalizer, using X-tremeGENE transfection reagent (2 ml). After 24 h, cells were lysed and analyzed for normalized fluorescence expression in a luminometer.
Chemokine Release
Cells (5 × 104 cells/ml) were serum-starved for 24 h, and medium was collected, centrifuged, and incubated onto membranes (catalog no. AAM-CYT-3-4, Raybiotech, Norcross, GA) spotted with antibodies against 62 different mouse cytokines. After washes, membranes were incubated with a mix of biotin-conjugated antibodies to the same cytokines, and immunoreactive spots were detected using HRP-conjugated streptavidin and chemiluminescence. Dots were quantified using ImageJ and normalized against positive controls.
Microarray Data Analysis
Illumina BeadStudio v.3.0 software was used to export expression levels and detection p values for each probe of each sample. Arrays were quartile-normalized between each other and filtered to remove non-informative probes (with detection p > 0.05). Differentially expressed genes between WT and CypD−/− groups were identified using two-tail unpaired Student's t test. The false discovery rate (FDR) was calculated according to the Storey procedure (22), and only genes with FDR < 5% were called significant. Heat maps for a list of genes were generated using two-way hierarchical clustering with a normalized Euclidean distance to cluster samples and a Spearman correlation distance to cluster the genes. Heat map color intensities were proportional to a value calculated as a ratio between the gene expression in a single sample and the geometric mean expression of the gene across all samples. A pathway enrichment analysis was carried out with Ingenuity Pathways Analysis software using Ingenuity Core Analysis (IPA 8.0, Ingenuity® Systems) with Benjamini-Hochberg correction for multiple testing and using p < 0.05 as a significance threshold. Enrichments of gene ontology terms and INTERPRO annotations in a gene list were done with DAVID (23) software. Results were filtered to satisfy criteria of FDR < 5% and fold enrichment > 4.
Statistical Analysis
Data were analyzed using GraphPad Prism 4.03 for Windows. Unless otherwise stated, data analyses were carried out using unpaired Student's t test. p < 0.05 was considered statistically significant.
RESULTS
CypD Regulation of Cell Cycle Progression
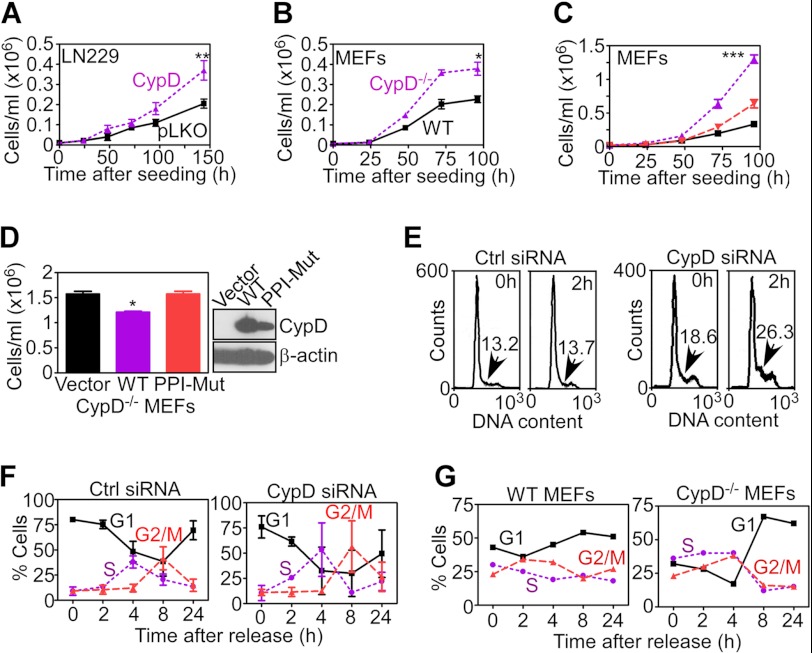
We began this study by examining a potential role of CypD in extramitochondrial signaling in normal or tumor cells representative of different genetic makeups. We found that glioblastoma LN229 cells with stable shRNA knockdown of CypD (20) exhibited increased cell proliferation compared with control cells transfected with non-targeting shRNA (Fig. 1A). Deletion of CypD (CypD−/−) in MEFs (1) also resulted in accelerated cell proliferation compared with WT MEFs (Fig. 1B). To test the specificity of this response, we next transfected CypD in CypD−/− MEFs and looked at changes in cell proliferation. Consistent with the data above, CypD−/− MEFs showed increased cell proliferation compared with WT cultures (Fig. 1C). Conversely, re-expression of CypD in CypD−/− cells reversed this effect and restored proliferation rates quantitatively comparable with WT MEFs (Fig. 1C). In contrast, transfection of a CypD mutant cDNA defective in peptidyl prolyl isomerase activity did not modulate proliferation of CypD−/− MEFs (Fig. 1D).
FIGURE 1.
CypD regulation of cell proliferation. A and B, LN229 cells stably transfected with non-targeting shRNA (pLKO), CypD-directed shRNA (A), or WT or CypD−/− MEFs (B) were analyzed for cell proliferation by direct cell counting at the indicated time intervals. Data are mean ± S.D. (n = 3). *, p < 0.05; **, p < 0.01. C, WT MEFs transfected with vector (black), CypD−/− MEFs expressing control vector (purple), or CypD cDNA (red) were analyzed for cell proliferation by direct cell counting. ***, p < 0.001. D, CypD−/− MEFs were transfected with vector (pcDNA), WT CypD, or a CypD mutant cDNA lacking peptidyl prolyl isomerase activity (PPI-Mut) and analyzed for cell proliferation by direct cell counting after 96 h. Data are mean ± S.D. (n = 3). *, p < 0.05. E, LN229 cells were transiently transfected with control non-targeting siRNA (Ctrl, left panel) or CypD-directed siRNA (right panel), arrested at the G1/S boundary by double thymidine block, and analyzed at the indicated time intervals after release for DNA content by propidium iodide staining and flow cytometry. The percentage of cells in the S-phase population is indicated. F and G, quantification of cell cycle transitions in thymidine-synchronized transfected LN229 cells (F) or WT or CypD−/− MEFs (G) by propidium iodide staining and flow cytometry at the indicated time intervals after release.
When analyzed in cell cycle-synchronized cultures, siRNA knockdown of CypD in LN229 (Fig. 1E) or breast adenocarcinoma MCF-7 cells (data not shown) resulted in faster and more sustained transition into S-phase, with accumulation of a larger fraction of cells with G2/M DNA content, compared with control transfectants (Fig. 1F). CypD−/− MEFs exhibited a similar phenotype with appearance of a larger S-phase population (data not shown) and further expansion of the G2/M fraction (Fig. 1G).
CypD Regulation of Cell Motility and Invasion
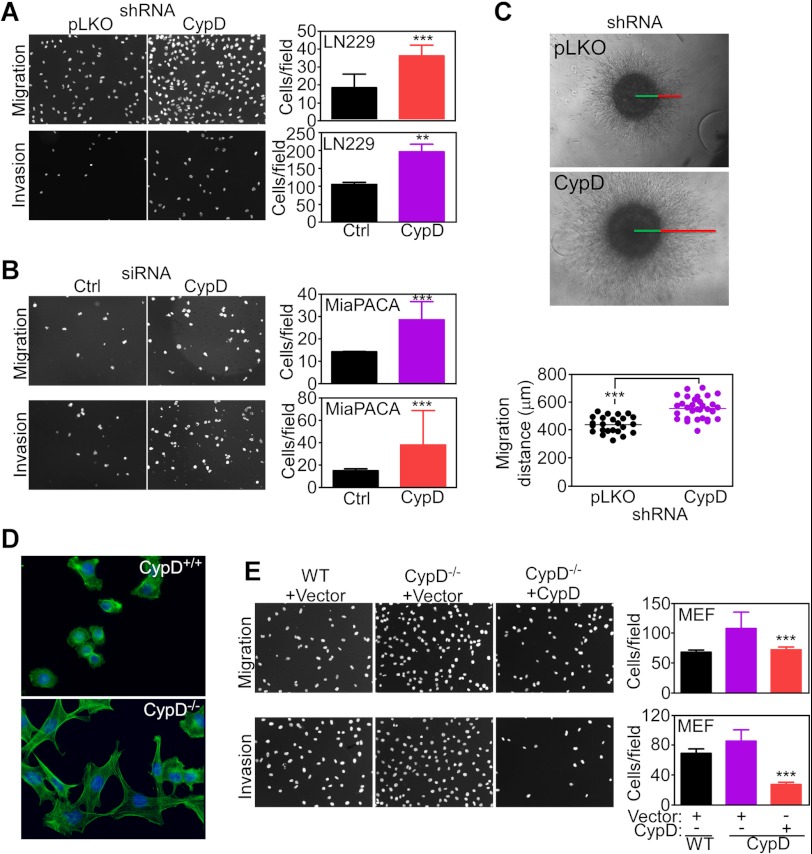
Stable shRNA depletion of CypD in LN229 cells (Fig. 2A) or acute knockdown of CypD in pancreatic adenocarcinoma MiaPACA cells (Fig. 2B) produced a second phenotype of increased cell migration and cell invasion compared with control transfectants (Fig. 2, A and B). Similar results were obtained in a more physiologic three-dimensional model of organotypic spheroids embedded in a collagen matrix where CypD knockdown considerably increased cell invasion compared with control transfectants (Fig. 2C). Consistent with modulation of cell motility, CypD−/− MEFs exhibited remodeling of the actin cytoskeleton with increased expression of cellular protrusions and lamellipodia formation compared with a rounded morphology of WT MEFs (Fig. 2D). In reconstitution experiments, transfection of a CypD cDNA reversed the increased cell migration and invasion of CypD−/− MEFs, whereas a control vector had no effect (Fig. 2E).
FIGURE 2.
Regulation of cell motility by mitochondrial CypD. A, LN229 cells were transfected with control shRNA (pLKO) or CypD-directed shRNA and analyzed for cell migration or cell invasion by DAPI staining of nuclei. Bar graphs show quantification of cell motility. Data are mean ± S.D. (n = 3). ***, p < 0.0001; **, p < 0.001. Magnification is ×10. B, pancreatic adenocarcinoma MiaPACA cells were transfected with control, non-targeting siRNA (Ctrl) or CypD-directed siRNA and analyzed for cell migration or cell invasion by DAPI staining of nuclei. Bar graphs show quantification of cell motility. Data are mean ± S.D. (n = 3). ***, p < 0.0001; **, p < 0.001. Magnification is ×10. C, stably transfected LN229 cells were embedded in a collagen matrix as three-dimensional organotypic spheroids and analyzed for cell invasion after 72 h. Bottom panel, quantification of migration distance. Each point corresponds to an independent determination of three-dimensional invasion. ***, p < 0.0001. D, WT or CypD−/− MEFs were analyzed for actin cytoskeleton assembly by FITC-conjugated phalloidin staining and fluorescence microscopy. Nuclei are stained by DAPI. Representative images are shown. Magnification is ×60. E, WT or CypD−/− MEFs were transfected with vector or CypD cDNA and analyzed for cell migration or invasion by DAPI staining of nuclei. Bar graphs show quantification of cell motility. Data are mean ± S.D. (n = 3). ***, p < 0.0001. Magnification is ×10.
CypD Deletion Results in Transcriptional Modulation of a Chemokine Gene Expression Signature
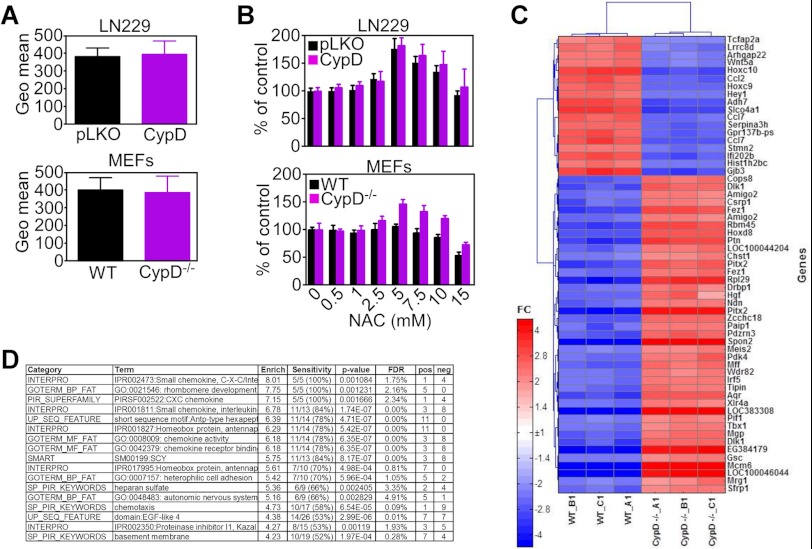
To begin elucidating how changes in CypD expression may affect cell behavior, we next looked at a potential differential mitochondrial production of ROS in control or CypD-depleted cells. In these experiments, ROS production was indistinguishable in control cultures or LN229 cells stably silenced for CypD or CypD−/− MEFs (Fig. 3A). Also, increasing concentrations of the ROS scavenger N-acetyl cysteine did not significantly affect proliferation of CypD−/− MEFs or CypD-silenced LN229 cells compared with control cultures (Fig. 3B).
FIGURE 3.
Microarray profiling of CypD deficiency. A, shRNA-transfected LN229 cell (top panel) or WT or CypD−/− MEFs (bottom panel) were mixed with the fluorescence dye H2DCFDA and analyzed for ROS production by flow cytometry. Data are expressed as mean fluorescence under the various conditions tested and are mean ± S.D. (n = 6). B, transfected LN229 cells (top panel) or WT or CypD−/− MEFs (bottom panel) were incubated with the indicated increasing concentrations of the ROS scavenger N-acetyl cysteine (NAC) and analyzed for cell proliferation by direct cell counting. Data are mean ± S.E. (n = 3). C, heat map of gene expression changes by microarray analysis of WT and CypD−/− MEFs. Only genes showing > 5-fold significant differences between the two cell lines are shown. D, list of the most enriched functional categories among genes exhibiting > 1.5-fold change between WT and CypD−/− MEFs. Enrich, enrichment of an annotation term; FDR, false discovery rate of the enrichment; N, number of genes in the analyzed list with the annotation; %, percent of all known genes with the annotation; FC, fold change CypD/WT.
We next subjected WT and CypD−/− MEFs to microarray profiling. We identified 1278 genes (FDR < 5%) that were modulated differentially (> 1.5-fold) between WT and CypD−/− MEFs and 454 genes that changed > 2-fold between the two cell types. A heat map of genes exhibiting a > 5-fold increased expression in CypD−/− MEFs compared with WT cultures is shown in Fig. 3C. Genes modulated under these conditions comprised multiple transcriptional regulators (Cops8, Hoxd8, Ndn, Meis2, Irf5, Pif1, Tbx1, Mcm6, and Sfrp1) and extracellular matrix proteins (Ptn, Spon2, and Mgp) (Fig. 3C). Analysis of overrepresented functions in the CypD−/− gene signature revealed a unique preponderance of chemokine activity (Fig. 3D) associated with inflammatory and cytokine-dependent transcriptional responses (STAT3 and NFκB) by pathway analysis (supplemental Fig. S1, A and B).
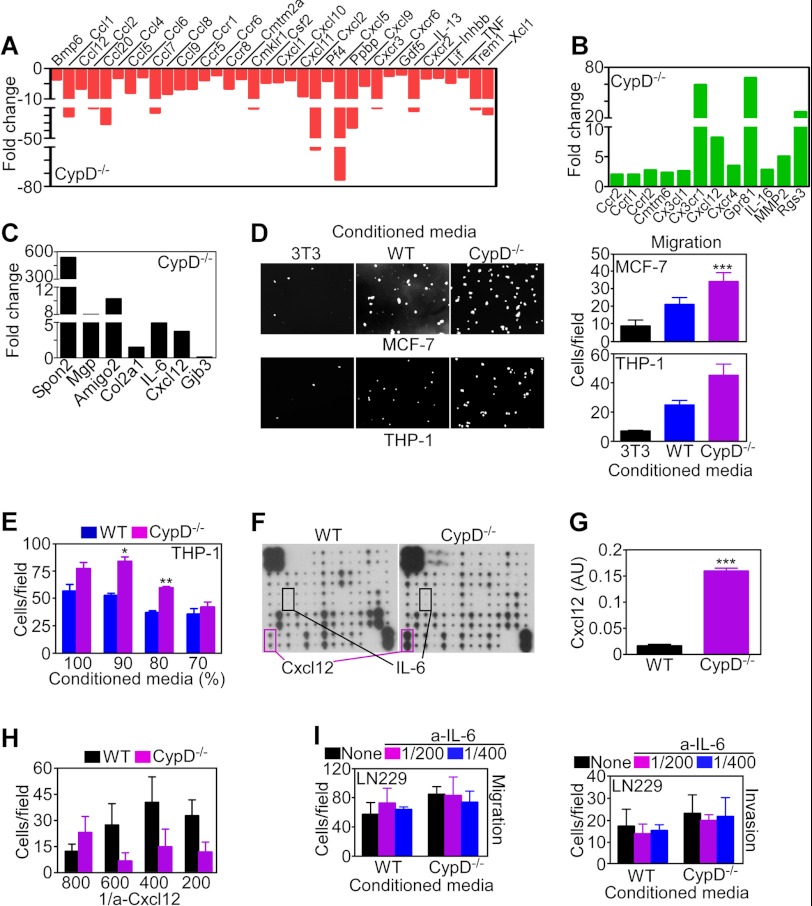
Consistent with these predictions, array studies identified extensive differential changes in chemokine/chemokine receptor expression in CypD−/− MEFs, including down-modulation (Fig. 4A) of Ccl2 (MCP-1, ↓ 10.6-fold), Ccl7 (MCP-3, ↓ 16.5-fold), Cmklr1 (chemokine receptor-1, ↓ 11-fold), Cxcr2 ligands, Cxcl2 (↓ 52-fold) and Cxcl5 (↓ 74.8-fold), platelet-basic protein (Ppbp, ↓ 36.1-fold), Cxcr3 (↓ 10.6-fold), and Xcl1 (↓ 18.4-fold). Compared with WT cultures, CypD−/− MEFs exhibited increased expression (Fig. 4B) of Cx3cl1 (fractalkine, ↑ 2.6-fold), Ccrl2 (↑ 2.7-fold), Cx3cr1 (↑ 59.3-fold), and the receptor-chemokine pair Cxcr4 (↑ 3.5-fold) and Cxcl12 (↑ 8.2-fold). In validation studies of RT-PCR amplification of selected gene products, CypD−/− MEFs expressed higher levels of the Wnt pathway regulator Spon2 (Spondin), vascular modulator Mgp (Matrix Gla protein), cell adhesion receptor AMIGO2, and the cytokine IL-6 (Fig. 4C).
FIGURE 4.
Chemokine regulation of CypD-directed cell motility. A and B, array profiling of fold changes in expression of chemokines/inflammatory mediators in CypD−/− MEFs compared with WT MEFs. A, decreased expression. B, increased expression. C, expression of selected gene products in the CypD−/− MEF signature by quantitative PCR. Data are expressed as fold changes compared with WT MEFs. D, MCF-7 or THP-1 cells were incubated in the presence of conditioned media collected from NIH3T3 fibroblasts (3T3), WT, or CypD−/− MEFs and analyzed for cell migration after 16 h by DAPI staining of nuclei. Bar graphs show quantification of cell migration. Data are mean ± S.D. (n = 3). Magnification is ×10. ***, p < 0.001. E, THP-1 cells were incubated with the indicated dilutions of conditioned media collected from WT or CypD−/− MEFs and analyzed for cell migration after 16 h. Data are mean ± S.D. (n = 3). *, p < 0.05; **, p < 0.01. F, supernatants from WT or CypD−/− MEFs were analyzed for changes in chemokine secretion by array profiling. The levels of Cxcl12 or IL-6 in the supernatants are indicated. G, quantification of Cxcl12 release in supernatants of WT or CypD−/− MEFs. Data are mean ± S.D. (n = 3). ***, p < 0.001. H, LN229 cells transfected with CypD-directed shRNA were incubated with conditioned media from WT or CypD−/− MEFs and analyzed for cell invasion after 6 h in the presence of the indicated dilutions of a neutralizing antibody to Cxcl12. Data are mean ± S.D. (n = 10). I, LN229 cells were incubated with WT or CypD−/− conditioned media, mixed with the indicated dilutions of a neutralizing antibody to IL-6 (a-IL-6), and analyzed for cell migration (left panel) or cell invasion (right panel). Data are mean ± S.D. (n = 5).
On the basis of these results, we next asked whether CypD−/− MEFs secreted active chemokine(s) in their conditioned medium, potentially supporting autocrine/paracrine cell motility. Accordingly, exposure of MCF-7 or monocyte THP-1 cells to CypD−/− conditioned media resulted in increased cell migration compared with control cultures incubated with NIH3T3 or WT MEF-conditioned media (Fig. 4D). The enhancing effect on cell motility was concentration-dependent for increasing doses of CypD−/−-conditioned media (Fig. 4E). To identify which secreted chemokine was potentially responsible for the enhanced migratory phenotype induced by CypD loss, array profiling studies were carried out next. In these experiments, supernatants from CypD−/− MEFs exhibited a dramatic increase in Cxcl12 compared with WT cells (Fig. 4F). In contrast, IL-6 levels were increased less prominently, and other chemokines on the array did not change significantly between the two cell types (Fig. 4F). Consistent with these data, supernatants from CypD−/− MEFs contained high levels of Cxcl12 compared with WT cultures (Fig. 4G). Functionally, a neutralizing antibody to Cxcl12 inhibited cell migration mediated by CypD−/−-conditioned media (Fig. 4H), whereas a non-binding IgG had no effect (not shown). Conversely, a neutralizing antibody to IL-6 did not significantly affect tumor cell migration or invasion in the presence of CypD−/−-conditioned medium (Fig. 4I).
Signaling Requirements of CypD-directed Cellular Responses
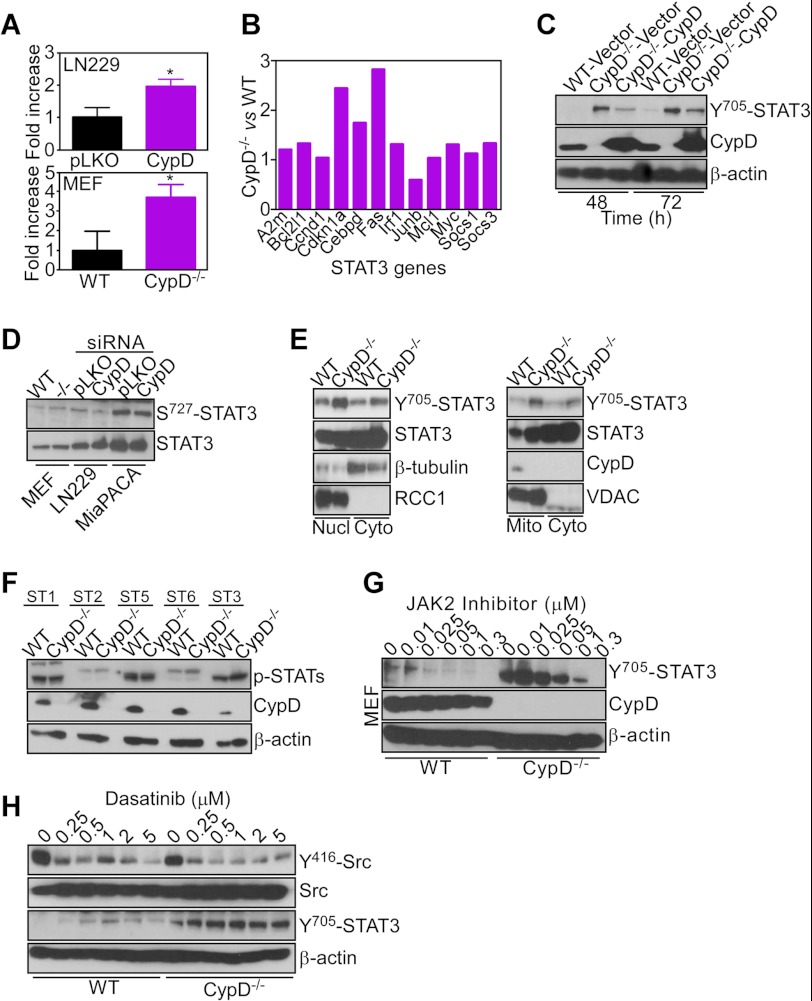
Analysis of transcriptional changes in CypD−/− MEFs suggested a potential involvement of STAT3 (supplemental Fig. S1) (24). Consistent with these predictions, CypD knockdown in LN229 cells or deletion of CypD in MEFs resulted in increased STAT3 luciferase promoter activity compared with control cultures (Fig. 5A). In addition, CypD−/− MEFs exhibited increased mRNA expression of a panel of STAT3 target genes, compared with WT MEFs, by quantitative PCR amplification (Fig. 5B). Biochemically, this was associated with increased phosphorylation of STAT3 on Tyr-705, a marker of transcriptional activity (Fig. 5C). In reconstitution experiments, transfection of a CypD cDNA attenuated STAT3 phosphorylation in CypD−/− MEFs, whereas a control vector had no effect (Fig. 5C). Confirming the specificity of this response, CypD targeting in different cell types did not affect STAT3 phosphorylation on Ser-727 (Fig. 5D), an activation marker of mitochondria-localized STAT3 (25, 26).
FIGURE 5.
Selective STAT3 activation induced by CypD loss. A, LN229 transfected with control (Ctrl), CypD-directed shRNA (top panel), or WT or CypD−/− MEFs (bottom panel) were transfected with a STAT3 luciferase reporter construct and analyzed for promoter activity in a luminometer after 24 h. Data are mean ± S.D. (n = 3). B, RNA extracted from WT or CypD−/− MEFs was amplified with primers specific for the indicated STAT3-regulated gene products by PCR. Data are expressed as fold differences in mRNA expression normalized to WT MEFs. C, WT or CypD−/− MEFs were transfected with the indicated plasmids and analyzed by Western blotting. D, the indicated cell types were analyzed by Western blotting. E, WT or CypD−/− MEFs were fractionated in the indicated subcellular compartments and analyzed by Western blotting. Nucl, nuclei; Cyto, cytosol; Mito, mitochondria. VDAC or RCC1 were used as mitochondrial or nuclear markers, respectively. F, WT or CypD−/− MEFs were analyzed for changes in expression of various STAT molecules by Western blotting. G, WT or CypD−/− MEFs were incubated with the indicated increasing concentrations of a JAK2 inhibitor and analyzed by Western blotting. H, WT or CypD−/− MEFs were treated with the indicated increasing concentrations of Dasatinib and analyzed by Western blotting. The phosphorylation sites in Src or STAT3 are indicated.
Consistent with STAT3 transcriptional activity, subcellular fractionation experiments identified Tyr-705-phosphorylated STAT3 in the nuclei of CypD−/− MEFs, with less pronounced changes observed in mitochondria (Fig. 5E). Conversely, phosphorylation of STAT1, STAT2, STAT5, or STAT6 was unchanged in WT or CypD−/− MEFs (Fig. 5F). Finally, incubation with a small molecule inhibitor of JAK, a cytokine-regulated STAT3 activator (24), dose-dependently suppressed STAT3 phosphorylation on Tyr-705 in CypD−/− MEFs (Fig. 5G) or CypD-silenced LN229 transfectants (data not shown). In control experiments, treatment of CypD−/− MEFs with Dasatinib, a small molecule inhibitor of the Src kinase that also activates STAT3 (24), abrogated Src phosphorylation on Tyr-416 but had no effect on Tyr-705 phosphorylation of STAT3 (Fig. 5H).
STAT3 Regulation of CypD-directed Cell Motility and Cell Proliferation
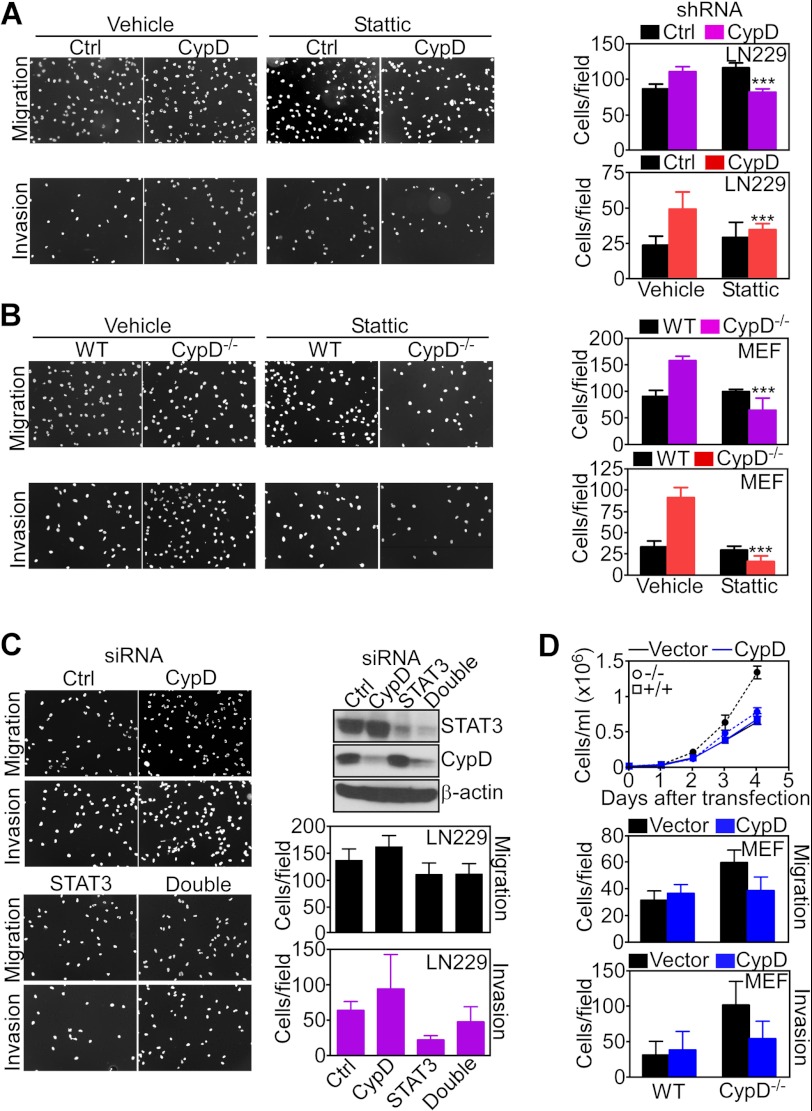
Pharmacologic inhibition of the STAT3 SH2 domain with a small molecule inhibitor, Stattic (21), reversed the increased migration and invasion of CypD-silenced LN229 cells compared with control cultures (Fig. 6A). In contrast, Stattic had no effect on tumor cell migration or invasion in the absence of CypD knockdown (Fig. 6A). Similarly, Stattic reversed the increased migration and invasion of CypD−/− MEFs but was without effect in WT MEFs (Fig. 6B). The more pronounced effect of STAT3 inhibition in CypD−/− MEFs (Fig. 6B) compared with LN229 shRNA transfectants (A) may reflect the incomplete knockdown of CypD in this cell type. As an independent approach, we next silenced STAT3 by siRNA and examined changes in cell motility in the presence or absence of simultaneous CypD knockdown. STAT3 silencing inhibited the migration and invasion of LN229 cells (Fig. 6C), supporting a role of this pathway in cell motility (24). Consistent with the data above, CypD knockdown increased both the migration and invasion of LN229 cells, and these responses were reversed by simultaneous STAT3 knockdown (Fig. 6C). To independently validate a role of CypD in this response, we next carried out additional reconstitution studies by transfecting a control vector or CypD cDNA in CypD−/− MEFs. In these experiments, reconstitution of CypD−/− MEFs with CypD, but not control cDNA, reduced cell proliferation (Fig. 6D, top panel), migration (center panel), and invasion (bottom panel) to the levels of WT CypD MEFs.
FIGURE 6.
STAT3 regulation of CypD-directed cell motility. A and B, transfected LN229 cells (A) or WT or CypD−/− MEFs (B) were treated with vehicle or the small molecule STAT3 inhibitor Stattic and analyzed for cell migration (top panel) or cell invasion (bottom panel) by DAPI staining of nuclei. Bar graphs show quantification of cell motility. ***p < 0.001; C, LN229 cells were transfected with control, non-targeting (Ctrl) or CypD- or STAT3-directed siRNA alone or in combination and analyzed by Western blotting or cell invasion by DAPI staining of nuclei of migrated cells. Bar graphs show quantification of cell invasion. Data are mean ± S.D. (n = 10). Magnifications for A–C are ×10. D, WT or CypD−/− MEFs were transfected with vector (black) or WT CypD cDNA (blue) and analyzed for kinetics of cell proliferation by direct cell counting (top panel), cell migration (center panel), or cell invasion (bottom panel). Top panel, data are mean ± S.D. (n = 3). Center and bottom panels, data are mean ± S.D. (n = 5).
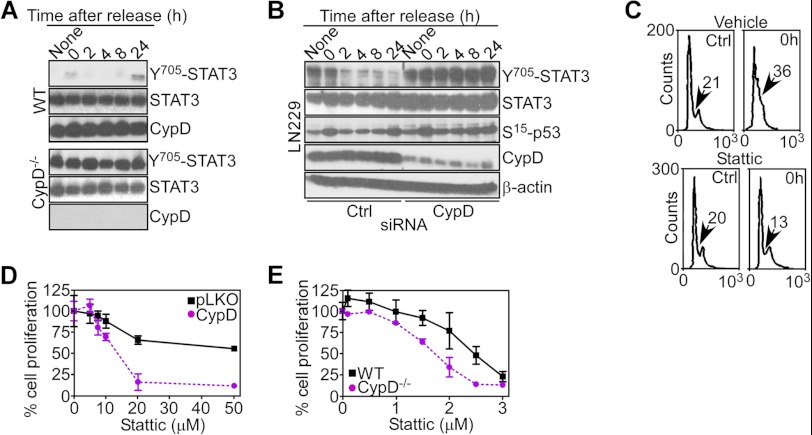
We next asked whether STAT3 signaling was also involved in the increased cell proliferation induced by CypD targeting. Analysis of cell cycle-synchronized cultures revealed that STAT3 was constitutively hyperphosphorylated on Tyr-705 in CypD−/− MEFs throughout cell cycle progression (Fig. 7A). In contrast, synchronized WT MEFs had largely undetectable levels of Tyr-705-phosphorylated STAT3 at all cell cycle phases (Fig. 7A). LN229 cells silenced for CypD also exhibited constitutive hyperphosphorylation of STAT3 on Tyr-705 at all cell cycle transitions (Fig. 7B). In contrast, phosphorylation of p53 on Ser-15 was unchanged in control or CypD-silenced LN229 cells (Fig. 7B). Stattic treatment prevented the accumulation of LN229 cells with S-phase DNA content in response to CypD knockdown, whereas control transfectants treated with Stattic progressed through the cell cycle and transitioned into S-phase (Fig. 7C). Consistent with these data, Stattic inhibited the proliferation of CypD-silenced LN229 cells (Fig. 7D) as well as CypD−/− MEFs (Fig. 7E), compared with control cultures.
FIGURE 7.
STAT3 regulation of CypD-directed cell proliferation. A and B, WT (top panel), CypD−/− MEFs (A), or LN229 cells transfected with control non-targeting (Ctrl) or CypD-directed siRNA (B) were synchronized at the G1/S boundary by double thymidine block and analyzed by Western blotting at the indicated time intervals after release. C, thymidine-synchronized LN229 cells transfected with CypD-directed shRNA were treated with vehicle or STAT3 small molecule inhibitor Stattic and analyzed for DNA content by propidium iodide staining and flow cytometry. The percentage of cells with S-phase DNA content is indicated. D and E, transfected LN229 cells (D) or WT or CypD−/− MEFs (E) were treated with the indicated increasing concentrations of vehicle or small molecule STAT3 inhibitor Stattic and analyzed for cell proliferation by direct cell counting. Data are mean ± S.D. (n = 6).
DISCUSSION
In this study, we have shown that deletion or decreased expression of mitochondrial CypD activates interorganelle signaling in normal or tumor cell types, leading to transcriptional changes in gene expression, modulation of a chemokine/chemokine receptor signature, and activation of the pleiotropic inflammatory mediator, STAT3. In turn, this resulted in accelerated cell cycle progression through S-phase and enhanced chemokine-dependent autocrine/paracrine cell migration and invasion.
CypD is the only component of a mitochondrial permeability transition pore (2) required for certain forms of cell death, for instance, oxidative stress (1, 3, 27). Although the role of CypD in this process has not been completely elucidated (11), there is evidence that inhibition of CypD protects from apoptosis and may have therapeutic potential in vivo (28), limiting the extent of tissue damage during cerebral (1), vascular (27), or pancreatic β cell (29) injury. Although additional functions of CypD in autophagy (12) and regulation of glycolysis (11) have been proposed, a role of this molecule in nuclear control of gene expression, chemokine responses, and STAT3-dependent signaling (this study) has not been proposed previously.
In this context, active STAT3 is a recognized effector of multiple inflammatory and homeostatic responses (30). Activated by cytokines, IL-6, IL-10, and IL-23, growth factor receptors, and non-receptor tyrosine kinases (Abl, Src, Syk), STAT3-dependent transcription of target genes maintains Th17 cells (31), modulates B and T regulatory populations (32), and contributes to antigen presentation by dendritic cells (33). STAT3 signaling is also exploited in cancer (34), influencing disparate mechanisms of cell differentiation, proliferation, apoptosis, angiogenesis, and cell motility/invasion (24, 35). Here, loss of mitochondrial CypD resulted in increased production of IL-6 (35), and, to a much greater extent, Cxcl12 (36), two well known inducers of STAT3. In turn, this was associated with canonical markers of STAT3 activation, including phosphorylation on Tyr-705, enhanced promoter activity, increased mRNA expression of STAT3 target genes, and STAT3-dependent increased cell proliferation and cell motility. Pharmacologic blockade of JAK2, but not Src (35), reversed STAT3 responses, consistent with a role of cytokine signaling in STAT3 induction after CypD loss.
Intriguingly, an oncogenic pool of STAT3 has been identified in mitochondria (37) and implicated in regulation of oxidative phosphorylation (38), potentially modulated by the SIRT1 deacetylase (25). Although the biochemical requirements of this pathway are still controversial (39), there is evidence that mitochondrial STAT3 may improve organelle performance and antagonize apoptosis during acute vascular injury (40), consistent with a non-transcriptional function of STAT3 in cellular responses (41). At variance with this paradigm, STAT3 induced by CypD depletion did not significantly accumulate in mitochondria, and its phosphorylation on Ser-727, a marker of the mitochondrial pool of the molecule (25, 26), was not affected in CypD-targeted cells.
The mechanistic links that connect changes in mitochondrial CypD levels to STAT3 activation remain to be fully elucidated. There is prior evidence that changes in mitochondrial integrity or function can induce retrograde modulation of nuclear gene expression (18). Potential mediators of these interorganelle responses may include modulation of Ca2+ homeostasis (42), production of ROS (43), proteotoxic stress (44), or, more recently, metabolic unbalance with secondary activation of an endoplasmic reticulum-dependent unfolded protein response (9). At variance with previous models of retrograde signaling, CypD−/− cells did not produce excess ROS, which could influence gene expression, and their enhanced proliferative kinetics were unaffected by a ROS scavenger. Conversely, other soluble mediators of mitochondria retrograde signaling can be postulated, in particular changes in Ca2+ homeostasis, which, reminiscent of CypD depletion (3, 27), may influence nuclear gene expression (45). Consistent with this model, CypD−/− cells exhibited increased NFκB-dependent transcription3. This is a common hallmark of mitochondria → nuclei retrograde signaling (18), including in response to Ca2+ perturbations (45), and pivotal for the activation of many of the cytokines found up-regulated by CypD deficiency (24).
The pathophysiological context of these observations is highlighted by the critical role of STAT3 and chemokine signaling in tumors and their microenvironment. In particular, the ligand-receptor pair Cxcl12-Cxcr4, which in our studies mediated autocrine/paracrine cell motility after depletion of CypD, has been implicated in the interplay between tumor cells and stroma, promoting stem cell mobilization (46), cell invasion, recruitment of bone marrow-derived myeloid cells, angiogenesis (47), and further activation of STAT3 (36). In this scenario, constitutively decreased levels of CypD in malignancies of the cervix and pancreas, as predicted by bioinformatics analysis3, may contribute to tumor progression via inhibition of oxidative cell death (1, 27) and activation of chemokine- and STAT3-dependent inflammatory signaling (this study). As an alternative scenario, therapeutic inhibition of CypD, either as a strategy to enhance cardioprotection during ischemia-reperfusion injury (28) or as a consequence of cyclosporine A-mediated immunosuppression (48), may enhance local inflammation and cooperate with loss of immune surveillance to favor malignant transformation in organ transplant patients (49).
In summary, we have identified CypD (1) as a pivotal effector of a novel form of mitochondrial retrograde signaling (18) that promotes STAT3 activation and chemokine/chemokine receptor-dependent cell motility. These studies reinforce the concept of interorganelle signaling as a regulatory network poised to affect broad cellular homeostatic responses (9) and identify an unexpected role of CypD (1) in extramitochondrial control of inflammatory gene expression.
Supplementary Material
Acknowledgments
We thank Dr. Nika N. Danial (Dana Farber Cancer Institute, Harvard Medical School) for CypD−/− MEFs.
This work was supported, in whole or in part, by National Institutes of Health Grants CA140043, CA78810, and CA118005. This work was also supported by Cancer Center Support Grant (CCSG) CA010815 (to The Wistar Institute).
Microarray data were submitted to the GEO database and are available by using accession number GSE41280.

This article contains supplemental Fig. S1.
M. Tavecchio, S. Lisanti, A. Lam, J. C. Ghosh, N. M. Martin, M. O'Connell, A. T. Weeraratna, A. V. Kossenkov, L. C. Showe, and D. C. Altieri, unpublished observations.
- CypD
- cyclophilin D
- MEF
- mouse embryonic fibroblast
- ROS
- reactive oxygen species
- FDR
- false discovery rate.
REFERENCES
- 1. Schinzel A. C., Takeuchi O., Huang Z., Fisher J. K., Zhou Z., Rubens J., Hetz C., Danial N. N., Moskowitz M. A., Korsmeyer S. J. (2005) Cyclophilin D is a component of mitochondrial permeability transition and mediates neuronal cell death after focal cerebral ischemia. Proc. Natl. Acad. Sci. U.S.A. 102, 12005–12010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Green D. R., Kroemer G. (2004) The pathophysiology of mitochondrial cell death. Science 305, 626–629 [DOI] [PubMed] [Google Scholar]
- 3. Baines C. P., Kaiser R. A., Purcell N. H., Blair N. S., Osinska H., Hambleton M. A., Brunskill E. W., Sayen M. R., Gottlieb R. A., Dorn G. W., Robbins J., Molkentin J. D. (2005) Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature 434, 658–662 [DOI] [PubMed] [Google Scholar]
- 4. Kinnally K. W., Peixoto P. M., Ryu S. Y., Dejean L. M. (2011) Is mPTP the gatekeeper for necrosis, apoptosis, or both? Biochim. Biophys. Acta 1813, 616–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hausenloy D. J., Boston-Griffiths E. A., Yellon D. M. (2012) Cyclosporin A and cardioprotection. From investigative tool to therapeutic agent. Br. J. Pharmacol. 165, 1235–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fulda S., Galluzzi L., Kroemer G. (2010) Targeting mitochondria for cancer therapy. Nat. Rev. Drug Discov. 9, 447–464 [DOI] [PubMed] [Google Scholar]
- 7. Nguyen T. T., Stevens M. V., Kohr M., Steenbergen C., Sack M. N., Murphy E. (2011) Cysteine 203 of cyclophilin D is critical for cyclophilin D activation of the mitochondrial permeability transition pore. J. Biol. Chem. 286, 40184–40192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shulga N., Wilson-Smith R., Pastorino J. G. (2010) Sirtuin-3 deacetylation of cyclophilin D induces dissociation of hexokinase II from the mitochondria. J. Cell Sci. 123, 894–902 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9. Chae Y. C., Caino M. C., Lisanti S., Ghosh J. C., Dohi T., Danial N. N., Villanueva J., Ferrero S., Vaira V., Santambrogio L., Bosari S., Languino L. R., Herlyn M., Altieri D. C. (2012) Control of tumor bioenergetics and survival stress signaling by mitochondrial HSP90s. Cancer Cell 22, 331–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wolf A., Agnihotri S., Micallef J., Mukherjee J., Sabha N., Cairns R., Hawkins C., Guha A. (2011) Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J. Exp. Med. 208, 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Machida K., Ohta Y., Osada H. (2006) Suppression of apoptosis by cyclophilin D via stabilization of hexokinase II mitochondrial binding in cancer cells. J. Biol. Chem. 281, 14314–14320 [DOI] [PubMed] [Google Scholar]
- 12. Carreira R. S., Lee Y., Ghochani M., Gustafsson A. B., Gottlieb R. A. (2010) Cyclophilin D is required for mitochondrial removal by autophagy in cardiac cells. Autophagy 6, 462–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hetz C., Glimcher L. H. (2009) Fine-tuning of the unfolded protein response. Assembling the IRE1α interactome. Mol. Cell 35, 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Walter P., Ron D. (2011) The unfolded protein response. From stress pathway to homeostatic regulation. Science 334, 1081–1086 [DOI] [PubMed] [Google Scholar]
- 15. Tabas I., Ron D. (2011) Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 13, 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haynes C. M., Ron D. (2010) The mitochondrial UPR. Protecting organelle protein homeostasis. J. Cell Sci. 123, 3849–3855 [DOI] [PubMed] [Google Scholar]
- 17. Siegelin M. D., Dohi T., Raskett C. M., Orlowski G. M., Powers C. M., Gilbert C. A., Ross A. H., Plescia J., Altieri D. C. (2011) Exploiting the mitochondrial unfolded protein response for cancer therapy in mice and human cells. J. Clin. Invest. 121, 1349–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Butow R. A., Avadhani N. G. (2004) Mitochondrial signaling. The retrograde response. Mol. Cell 14, 1–15 [DOI] [PubMed] [Google Scholar]
- 19. Srinivasan V., Kriete A., Sacan A., Jazwinski S. M. (2010) Comparing the yeast retrograde response and NF-κB stress responses. Implications for aging. Aging Cell 9, 933–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ghosh J. C., Siegelin M. D., Dohi T., Altieri D. C. (2010) Heat shock protein 60 regulation of the mitochondrial permeability transition pore in tumor cells. Cancer Res. 70, 8988–8993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schust J., Sperl B., Hollis A., Mayer T. U., Berg T. (2006) Stattic. A small-molecule inhibitor of STAT3 activation and dimerization. Chem. Biol. 13, 1235–1242 [DOI] [PubMed] [Google Scholar]
- 22. Storey J. D., Tibshirani R. (2003) Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods Mol. Biol. 224, 149–157 [DOI] [PubMed] [Google Scholar]
- 23. Huang D. W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 24. Sansone P., Bromberg J. (2012) Targeting the interleukin-6/Jak/Stat pathway in human malignancies. J. Clin. Oncol. 30, 1005–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bernier M., Paul R. K., Martin-Montalvo A., Scheibye-Knudsen M., Song S., He H. J., Armour S. M., Hubbard B. P., Bohr V. A., Wang L., Zong Y., Sinclair D. A., de Cabo R. (2011) Negative regulation of STAT3 protein-mediated cellular respiration by SIRT1 protein. J. Biol. Chem. 286, 19270–19279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou L., Too H. P. (2011) Mitochondrial localized STAT3 is involved in NGF induced neurite outgrowth. PLoS ONE 6, e21680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nakagawa T., Shimizu S., Watanabe T., Yamaguchi O., Otsu K., Yamagata H., Inohara H., Kubo T., Tsujimoto Y. (2005) Cyclophilin D-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature 434, 652–658 [DOI] [PubMed] [Google Scholar]
- 28. Malouitre S., Dube H., Selwood D., Crompton M. (2010) Mitochondrial targeting of cyclosporin A enables selective inhibition of cyclophilin-D and enhanced cytoprotection after glucose and oxygen deprivation. Biochem. J. 425, 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fujimoto K., Chen Y., Polonsky K. S., Dorn G. W., 2nd (2010) Targeting cyclophilin D and the mitochondrial permeability transition enhances β-cell survival and prevents diabetes in Pdx1 deficiency. Proc. Natl. Acad. Sci. U.S.A. 107, 10214–10219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mertens C., Darnell J. E., Jr. (2007) SnapShot. JAK-STAT signaling. Cell 131, 612. [DOI] [PubMed] [Google Scholar]
- 31. Korn T., Bettelli E., Oukka M., Kuchroo V. K. (2009) IL-17 and Th17 Cells. Annu. Rev. Immunol. 27, 485–517 [DOI] [PubMed] [Google Scholar]
- 32. Littman D. R., Rudensky A. Y. (2010) Th17 and regulatory T cells in mediating and restraining inflammation. Cell 140, 845–858 [DOI] [PubMed] [Google Scholar]
- 33. Nefedova Y., Gabrilovich D. I. (2007) Targeting of Jak/STAT pathway in antigen presenting cells in cancer. Curr. Cancer Drug. Targets 7, 71–77 [DOI] [PubMed] [Google Scholar]
- 34. Yu H., Pardoll D., Jove R. (2009) STATs in cancer inflammation and immunity. A leading role for STAT3. Nat. Rev. Cancer 9, 798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bollrath J., Greten F. R. (2009) IKK/NF-κB and STAT3 pathways. Central signalling hubs in inflammation-mediated tumour promotion and metastasis. EMBO Rep. 10, 1314–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Soriano S. F., Hernanz-Falcón P., Rodríguez-Frade J. M., De Ana A. M., Garzón R., Carvalho-Pinto C., Vila-Coro A. J., Zaballos A., Balomenos D., Martínez-A C., Mellado M. (2002) Functional inactivation of CXC chemokine receptor 4-mediated responses through SOCS3 up-regulation. J. Exp. Med. 196, 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gough D. J., Corlett A., Schlessinger K., Wegrzyn J., Larner A. C., Levy D. E. (2009) Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science 324, 1713–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wegrzyn J., Potla R., Chwae Y. J., Sepuri N. B., Zhang Q., Koeck T., Derecka M., Szczepanek K., Szelag M., Gornicka A., Moh A., Moghaddas S., Chen Q., Bobbili S., Cichy J., Dulak J., Baker D. P., Wolfman A., Stuehr D., Hassan M. O., Fu X. Y., Avadhani N., Drake J. I., Fawcett P., Lesnefsky E. J., Larner A. C. (2009) Function of mitochondrial Stat3 in cellular respiration. Science 323, 793–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Phillips D., Reilley M. J., Aponte A. M., Wang G., Boja E., Gucek M., Balaban R. S. (2010) Stoichiometry of STAT3 and mitochondrial proteins: Implications for the regulation of oxidative phosphorylation by protein-protein interactions. J. Biol. Chem. 285, 23532–23536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heusch G., Musiolik J., Gedik N., Skyschally A. (2011) Mitochondrial STAT3 activation and cardioprotection by ischemic postconditioning in pigs with regional myocardial ischemia/reperfusion. Circ. Res. 109, 1302–1308 [DOI] [PubMed] [Google Scholar]
- 41. Szczepanek K., Lesnefsky E. J., Larner A. C. (2012) Multi-tasking. Nuclear transcription factors with novel roles in the mitochondria. Trends Cell Biol. 22, 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hayashi T., Su T. P. (2007) Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell 131, 596–610 [DOI] [PubMed] [Google Scholar]
- 43. Wallace D. C. (2005) A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer. A dawn for evolutionary medicine. Annu. Rev. Genet. 39, 359–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Haynes C. M., Yang Y., Blais S. P., Neubert T. A., Ron D. (2010) The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol. Cell 37, 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Biswas G., Anandatheerthavarada H. K., Zaidi M., Avadhani N. G. (2003) Mitochondria to nucleus stress signaling. A distinctive mechanism of NFκB/Rel activation through calcineurin-mediated inactivation of IκBβ. J. Cell Biol. 161, 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. To L. B., Levesque J. P., Herbert K. E. (2011) How I treat patients who mobilize hematopoietic stem cells poorly. Blood 118, 4530–4540 [DOI] [PubMed] [Google Scholar]
- 47. Duda D. G., Kozin S. V., Kirkpatrick N. D., Xu L., Fukumura D., Jain R. K. (2011) CXCL12 (SDF1α)-CXCR4/CXCR7 pathway inhibition. An emerging sensitizer for anticancer therapies? Clin. Cancer Res. 17, 2074–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kahan B. D. (2011) Fifty years in the vineyard of transplantation. Looking back. Transplant. Proc. 43, 2853–2859 [DOI] [PubMed] [Google Scholar]
- 49. Rama I., Grinyó J. M. (2010) Malignancy after renal transplantation. The role of immunosuppression. Nat. Rev. Nephrol. 6, 511–519 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.