Abstract
This review highlights the recent methods to prepare PNA-based materials through a combination of self-assembly and self-organization processes. The use of these methods allows easy and versatile preparation of structured hybrid materials showing specific recognition properties and unique physicochemical properties at the nano- and micro-scale levels displaying potential applications in several directions, ranging from sensors and microarrays to nanostructured devices for biochips.
Keywords: PNA, monolayers, nanoparticles, self-assembly, self-organisation, materials, surfaces, sensors, microarrays, biochips, DNA-PNA duplexes, hybridization
Introduction
Structurally-ordered nano- and micro-sized architectures based on the controlled assembly/organization of fundamental molecular modules can act as seeds for the growth of novel structured materials, in which the molecular properties are largely modified or completely different from those of the constituting molecular modules. Perceiving the value of structural- and order-dependent properties emerging from such architectures, chemists have focused their attention on the construction of novel materials through the rational control of the assembly process exploiting defined molecular recognition processes. Because of their effective versatility, molecularly organized supramolecular materials have shown to hold great promises for a generational change in different everyday use devices, such as organic semiconductors, solar cells, chemical and biosensors.1-17 Among the different strategies, the use of biomimetic approaches has recently attracted significant attention.18,19 In this context, biomimetic structures based on nucleic acid oligomers are exploited for their recognition properties to prepare supramolecular architectures. The use of DNA for the self-assembly of 2D crystals on surfaces stems from the seminal work of Winfree and Seeman.20 They recognized that the specificity of the H-bonding interactions of DNA’s Watson-Crick complementary base pairs21 would be suitable for the construction of nanostructures via self-assembly.19 In their work, striped lattices were produced on mica surfaces and were imaged by atomic force microscopy (AFM). In order to construct the lattices, either two or four individual DNA units (“tiles”) bearing sticky ends were used in the self-assembly. The tiles were composed of either three or four strands of DNA, each one carefully sequenced in order to form the desired tile upon self-assembly in solution. Furthermore, the DNA sequences allowed for the protrusion of short single-stranded DNA strands from the tile, thus providing the so-called complementary sticky ends, required for the self-assembly of the tiles on the mica surface to form the nanoscale lattices. During the years following their work, the field of structural DNA nanotechnology grew and more complex nanostructures were constructed.19 The level of control which could be achieved was also clearly demonstrated by Rothemund in his article on DNA origami.22 Considering this unprecedented control on the nanostructuration of matter as obtained via the self-assembly of DNA, it is postulated that the use of peptide nucleic acid (PNA) oligonucleotides23 would result in more robust architectures. Indeed, PNA, a DNA analog in which the phosphate backbone is replaced by N-(2-aminoethyl)glycine units, has several differences over DNA as reported by Hyrup and Nielsen:24-26 (1) PNA oligomers can be easily synthesized using established techniques from peptide chemistry i.e. solid phase peptide synthesis; (2) organic moieties can be covalently attached to the C-terminal carboxylic acid group or the N-terminal amino group of the oligomer or on lateral chains; (3) PNA is achiral, so all enantiomeric purity problems are avoided; (4) the PNA peptidic backbone is neutral compared with DNA’s negatively charged sugar-phosphate backbone, therefore, no electrostatic repulsion exists when PNA hybridizes with another PNA or DNA oligomer; (5) partly due to the lack of electrostatic repulsion, PNA-PNA duplexes have a higher thermal stability relative to DNA-PNA and DNA-DNA duplexes; and (6) the possibility to easily control the helix handedness upon introduction of opportunely-modified enantiomerically pure aminoacids. Furthermore, PNAs have a high biostability, since they are not degraded by nucleases nor proteases. They are also chemically resistant to strong acids and weak bases. These properties make them an appealing candidate for a number of applications in material science. However, PNA has mainly been used for biological applications, such as diagnostics, biotechnology and pharmaceuticals.24,27-30 To date the most important materials-driven exploitation of PNA consists of its use as a molecular probe for diagnostics and detection (see below). Indeed, because of the high sensitivity of PNA to a single mismatch, its high duplex stability, and also its ability to cause strand invasions in double-stranded DNA, using PNA in place of DNA as a molecular probe gives more specific, more sensitive and more accurate results for the detection of target nucleic acids sequences. Thus, surface modification utilizing opportunely-modified PNA strands is currently of substantial interest, owing to the possibility of transferring the unique recognition properties to bulk or nanostructured materials by, among others, surface coating through the exploitation of self-assembly monolayers (SAMs). This account paper provides the key works describing the principle approaches toward the preparation of PNA-based nanostructures along with all the potential applications that are currently holding great promises in materials sciences. Three main approaches have successfully been employed in the preparation of such nucleic acids materials: (1) PNA-based self-assembled monolayers on solid surfaces (e.g., on nanoparticles or bulk materials); (2) carbon nanotubes (CNTs) covalently and non-covalently conjugated to PNAs; and (3) self-organized nanostructures.
PNA-Containing Self-Assembled Monolayers on Solid Surfaces
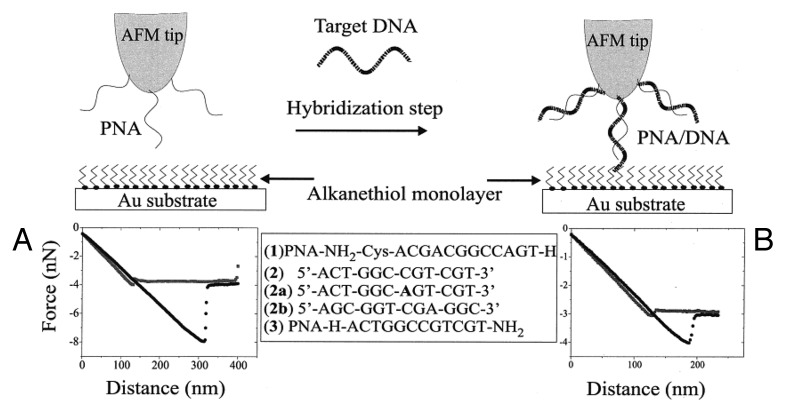
SAMs of PNA have mainly been prepared by covalent attachment of modified PNA oligomers bearing terminal anchoring groups to various surfaces (e.g., gold, pyrite and glass) with the intention of preparing microarray biosensors.30-36 In one of the first reports, thiol-derivatised single-stranded PNAs (ssPNAs) bearing a terminal cysteine group have been self-assembled on Au(111)34,37 using a classical wet technique, by soaking a Au surface in a PNA-containing μM solution. By means of Fourier transform reflection absorption infrared spectroscopy (FT-RAIRS), AFM and X-ray photoemission spectroscopy (XPS) it has been shown that the molecular orientation of the adsorbed ssPNAs (sequence: Cys-O-O-AATCCCCGCAT, where O = –NHCOCH2OCH2CH2OCH2CH2–) strongly depends on the surface coverage. Specifically, at high coverage, the ssPNAs’ molecular axe assumes a normal position and forms SAMs without the need for co-adsorbing adjuvant molecules for filling the undergrowth space. As expected, at low coverage, the ssPNAs lie flat on the surface, thus confirming a concentration-dependent structural transition. Knowing the structural properties for the ssPNAs-based SAMs is essential for engineering efficient biosensors for the characterization of target DNA molecules in solutions.37 Following a similar approach, Willner and coworkers self-assembled a cysteine-modified ssPNA (NH2-Cys-ACGACGGCCAGT-H) on a Au-coated AFM tip for probing and detecting the hybridization interaction with a ssDNA (see Fig. 1).38 Exploiting a chemical force microscopy (CFM), the adhesion force between a ssPNA-modified tip or a DNA/PNA-modified tip with an hydrophobic Au surfaces coated with undecanethiol could be measured in a TRIS-HCl buffer solution (5 mM at pH = 7.4). A substantial decrease in the resulting adhesion force has been observed for the double stranded DNA/PNA-modified tip (F = 1.13 nN) compared with ssPNA-modified tip (F = 4.35 nN) as a consequence of the increased hydrophilic character of the tip upon hybridization with the complementary ssDNA (5′-ACTGGCCGTCGT-3′). Interestingly, soaking the ssPNA-modified tip into a solution of a DNA mutant displaying only one single base mismatch as compared with the fully complementary strand, led to an adhesion force that is identical to the non-hybridized ssPNA-modified tip at a concentration of 5 × 10−10 M. This experiment revealed that such CFM methods are sufficiently sensitive to detect single-base mismatches at very low concentration, thus proving their great potential as nanotools for diagnostic applications.
Figure 1. Schematic representation of the CFM-based diagnostic nanotool for detecting DNA sequences through a measurement of the force. Adapted with permission from reference 38.
Similar ssPNAs containing SAMs for DNA detection have also been reported by Burgener et al. following a pre-modification of a Au surface with a dextran layer, which is then functionalized by an organic monolayer bearing terminal thiol functionalities that undergo a covalent bond-forming reaction with a ssPNA bearing a terminal reactive maleimide group.39 Au-coated surfaces with PNA have also been prepared for ultrasensitive nanoparticle-enhanced surface plasmon resonance imaging (SPRI) detection of DNA sequences down to 1 fM (SPRI has recently emerged as an extremely versatile method for detecting interactions of biomolecules in a micro-array format through the spatial monitoring of local differences in the reflectivity of incident light from an array of biomolecules linked to a chemically-modified Au surface, usually detected as bright and dark spots in the SPR image). In this device configuration, gold nanoparticles (Au-NPS) were functionalized with DNA, and dithiobis(N-succinimidylpropionate, DTPS) was used for linking PNA to the gold surface of sensor microchannels. Following a sandwich-like strategy, a solution of a full matching DNA sequence was then injected into a microchannel containing immobilized PNA together with Au-NPs bearing a 12-mer DNA both complementary for the terminal tracts of the targeting DNA strands. The specificity of the full matched DNA strands has been checked by comparing the NPS-enhanced SPRI signals with those obtained when singly mismatched DNA sequences were allowed to interact with the surface-immobilized ssPNA. From the Δ %Rb/Δ %R ratio between the NPs-enhanced responses, an average value of 7, 4 was calculated for 1 fM concentration. This increase is attributed to the high dissociation constants of the mismatched sequences, the response of which dropped to the background level below 1 pM, thus dramatically increasing the mismatch recognition in connection with the decreased detection limits.40
Exploiting surface plasmon fluorescence spectroscopy (SPFS), Knoll and coworkers have developed new sensors for the detection of DNA sequences using PNA-based microchips.41-43 SPFS technique permits to increase, compared with bulk signals, the fluorophores emission confined onto metallic surfaces covering a 90° glass prism. This effect is generated when the UV-laser beam is reflected at the glass-metal interface at the surface plasmon resonance angle, generating an enhanced electromagnetic field at several tens of nanometers around the surface. This can excite neighboring fluorophores, resulting in an enhanced fluorescence signal. The SPFS allows a sensitive and high-performance quantitative detection of a fluorophore placed at the metal interface. In their studies, Knoll and coworkers used a Au-coated glass prism functionalized with a biotinylated SAMs monolayer that undergoes self-assembly with a streptavidin monolayer formed by specific binding to the biotin-sites exposed on the SAM and finally a biotinylated oligonucleotide of 15 bases monolayer linked to the streptavidin layer. Exposing the modified surface toward different concentrations of complementary oligonucleotide sequences terminally bearing a fluorophore, the system records the fluorescence signal as a function of time under UV irradiation. In this way, it can be possible to follow the hybridization between two oligonucleotides and estimate the kinetic and the thermodynamic equilibrium parameters. In these devices, the uncharged ssPNA disregard inter-strand columbic interactions between two adjacent oligonucleotides and the hybridization follows the Langmuir model, thus allowing for a quantitative interpretation of the hybridization kinetics. Among the many uses, this technique hold great potentials for applications in the detection of polymerase chain reaction (PCR) products or identification of point mutations associated with cancer and other diseases (Fig. 2).
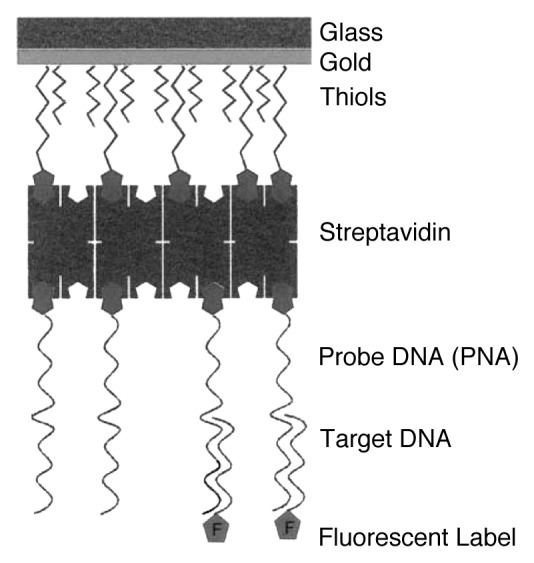
Figure 2. Schematic representation of the sensor structure employed for the SPFS studies of DNA-DNA hybridization. Adapted with permission from reference 41.
In a very recent example, Calabretta et al.44 reported the patterning of PNA on glass surfaces using reactive microcontact printing. Glass surfaces were modified with aldehyde functional groups, and the PNA oligomers were functionalized with an amino group attached via a short aminoethyl ethoxy linker. To pattern the surfaces, a freshly oxidized polydimethylsiloxane (PDMS) stamp was “inked” with a 20 µM solution of PNA in acetonitrile for 5 min, dried with N2 and then brought into contact with the aldehyde modified glass surface for 15 min. This allowed the amino groups to react with the aldehyde functional groups forming an imine linkage between the PNA and the glass substrate. The imine bonds were then reduced to the corresponding amines in the presence of NaBH4. This process is summarized in Figure 3A. Fluorescence microscopy of a glass surface printed with a fluorescein-bearing PNA oligomer confirmed the micropattern of covalently-bounded PNA on the glass surface (Fig. 3B). The patterned surfaces were then hybridized with complementary and single-base mismatched dye-labeled DNA oligomers to test their ability to recognize cDNA sequences. It was shown that both sequences successfully hybridized to PNA, however, the mismatched sequence’s PNA/DNA duplex displayed a lower melting temperature of approximately 5°C compared with the fully complementary sequence, therefore confirming that the microarray biosensor could be used to selectively detect sequence mismatches and point mutations in DNA.
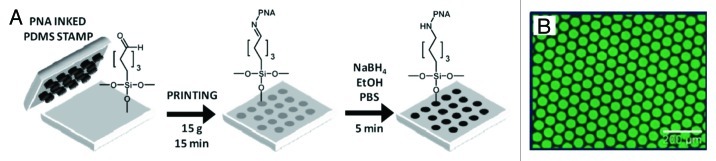
Figure 3. (A) Schematic representation of the patterning of amino-capped PNA oligomers onto aldehyde-functionalized glass slides via reactive contact printing. (B) Fluorescence microscope image (scale bar: 200 µm) of a glass surface following microcontact printing with fluorescein-labeled PNA. Adapted with permission from reference 44.
In a parallel work, Messere and coworkers reported on the functionalization of PNA-based magnetic conjugates using an alternative strategy, in which dextran-magnetic nanoparticles have opportunely been modified introducing reversible bonds for the immobilization of unmodified oligonucleotides.45 When complementary ssPNA and ssDNA nanohybrids were mixed, the base pairing led to an extended nanoparticle-based assembly (Fig. 4). When a magnetic field is applied, the ssPNA/ssDNA particle assembly shows higher responses rate to the external field, thus supporting the hypothesis of the formation of the assembly and its enhanced activities in the presence of the magnetic field because of the aggregate nature of the material. These materials are on the way to be tested for the local and thermal-induced release of DNA strands for in vivo applications.45
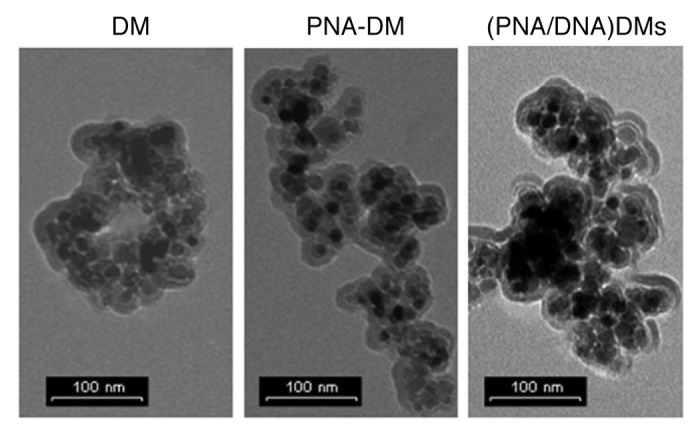
Figure 4. TEM images of the ssPNA/ssDNA hybridization inducing nanoparticle aggregation. Adapted with permission from reference 45.
Shiraishi and coworkers have associated the high sensitivity of fluorescence detection46,47 with the specificity of the simultaneous self-assembly of two beads, templated by the hybridization with an external nucleic acid48 for sensing natural nucleic acids.49 Specifically, because of a significant demand for the detection of Trypanosoma brucei parasite, and because of its high rRNA content (ca. 1 million copies per parasite), they targeted two unique sequences within its 18S rRNA. To do so, they prepared two biotinylated 18-mer PNA oligomers coupled to (strept)avidin coated fluorescent polystyrene beads complementary to the two targeting 18S rRNA (Fig. 5). The beads were chosen to allow an easy and simultaneous location and distinction from each other by conventional fluorescence microscopy. PNA3534 [Bio-(eg2)3-GAA ACA CCG ACC CAA GGC-Lys-NH2] has been bound to a red bead of 5.9 μm, while PNA3533 [Bio-(eg2)3-CCG CTC CCG TGT TTC TTG-Lys-NH2] to a green bead of 1 μm. When the two beads will hybridize to the same nucleic acid molecule, they will co-locate signaling the presence of the target sequence. Exploiting this approach, Shiraishi and coworkers were able to detect co-localization of beads of 1.6 ng of RNA per sample, corresponding to ca 0.5 fmol of 18S rRNA (assuming that 18S rRNA constitutes 20% of the total RNA).
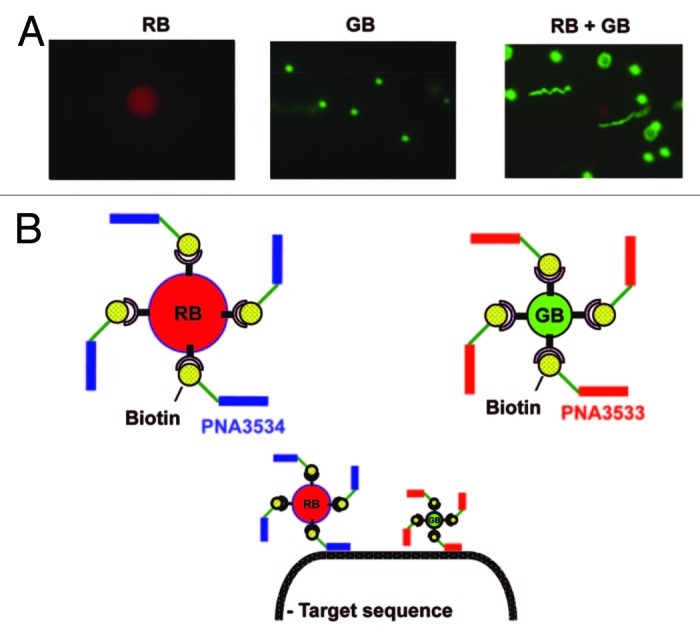
Figure 5. (A) Fluorescent microscopic images of the green bead (GB), the red bead (RB) and a mixture of the two. (B) Representation of PNA3534 which has been coupled to RB, of PNA3533 which has been coupled to GB, and of the target sequence detection by two bead/PNA complexes. Adapted with permission from reference 49.
Wang et al. were the first to use PNA immobilized onto carbon paste electrode (CPE) for the recognition of specific DNA sequences using Co(phen)33+ as electroactive hybridization indicator.50 They demonstrated that PNA-based devices present higher thermal stability (30°C) and are able to recognize ssDNAs at low ionic strengths (1–5 mM buffer concentrations) compared with their analogous DNA-based devices (Fig. 6). Upon hybridization at low buffer concentrations, the DNA biosensors are not able to bind complementary strands, while the PNA-based sensors gradually increase their response as a function of the buffer concentration and it decreases slightly at high ionic strength (10–20 mM buffer concentrations). The effect of the temperature shows that at rt, both devices give a hybridization response, but the signal of PNA-based sensor revealed to be more than 3-fold higher: in fact, temperature increasing does not affect the response of the PNA-based device, whereas that only based on DNA vanishes at 40°C.
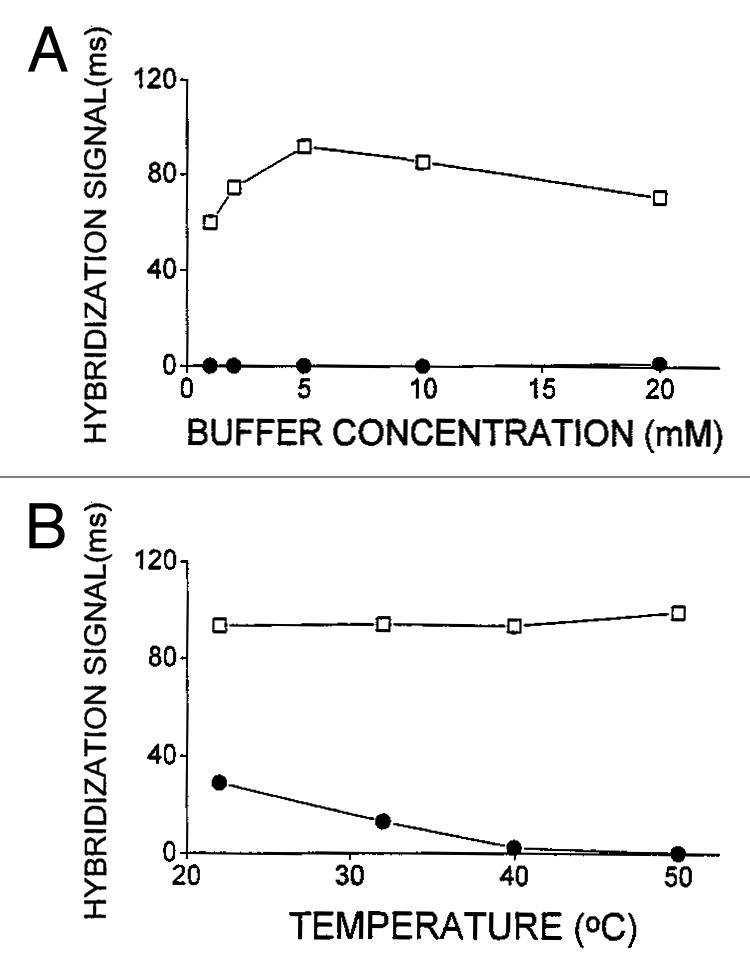
Figure 6. (A) Effect of the buffer concentration on the hybridization of PNA-DNA (squares) and DNA-DNA (dots) duplexes. (B) Effect of the temperature on the PNA-DNA (squares) and DNA-DNA (dots) hybridization. Adapted with permission from reference 50.
Similarly, Wang and coworkers have also developed PNA-based probes using thiol-derivatized PNA strands immobilized on quartz crystal microbalance (QCM) transducers. Specifically, they have demonstrated that PNA-functionalized QCM biosensors are able to bind single-stranded DNA oligomers displaying high selectivity (5-fold higher) with respect to those displaying one-base mismatch.51
Winssinger and coworkers have also exploited PNA/DNA’s high thermal stability to develop sensing devices based on a PNA-encoding method.30,52 The latter allow a convenient and fast determination of protease activity and substrate specificity in a microarray format.30,52 Their approach consists of (1) incubating a solution of fluorogenic protease substrates libraries encoded with PNA tags, with the protease (or lysate) of interest and (2) detecting the cleavage products by spatial deconvolution of the library via hybridization of the PNA tags to a DNA microarray (as it is shown on Fig. 7). The proteolysis is detected by the change in the electronic properties of the fluorophore when the substrate is cleaved from the fluorophore-PNA adduct, resulting in a large increase in fluorescence that is resolved during the hybridization with the DNA chip. The location of hybridization is dictated by the sequence of the PNA tag, which is bound to the sequence of the substrate. PNA-encoded libraries are prepared via a split and mix combinatorial method, while microarrays are prepared by photocrosslinking of unmodified oligonucleotides on aminosilane-coated glass slides.53
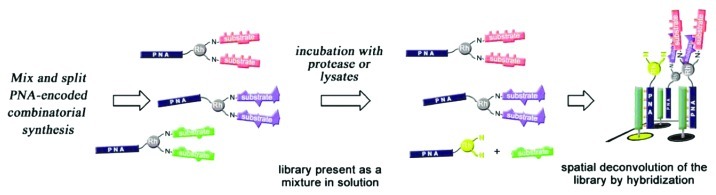
Figure 7. Representation of the PNA-encoded fluorogenic substrate libraries as developed by Winssinger and coworkers. Adapted with permission from reference 52.
Figure 8A shows the results obtained on the microarray after proteolysis and deconvolution of test substrates. An equimolar mixture of two different substrates (Caspase-3 and Thrombin probes) was hybridized to chips at concentrations ranging from 10 pM to 800 pM. This was performed prior to and after incubation with the enzyme Caspase-3. Only the feature on the chip corresponding to caspase-3 showed appreciable fluorescence with a linear relationship to the concentration of the substrate (Fig. 8B).52
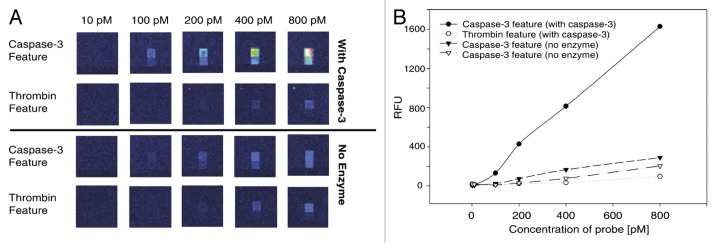
Figure 8. (A) Affymetrix microarray after proteolysis of two different substrates (Caspase-3 and Thrombine feature) prior to and after incubation with the enzyme Caspase-3, at different concentrations in substrates. (B) Plots of relative fluorescence intensities as a function of the concentration in substrates. Adapted with permission from reference 52.
ssPNA-CNTs Hybrids
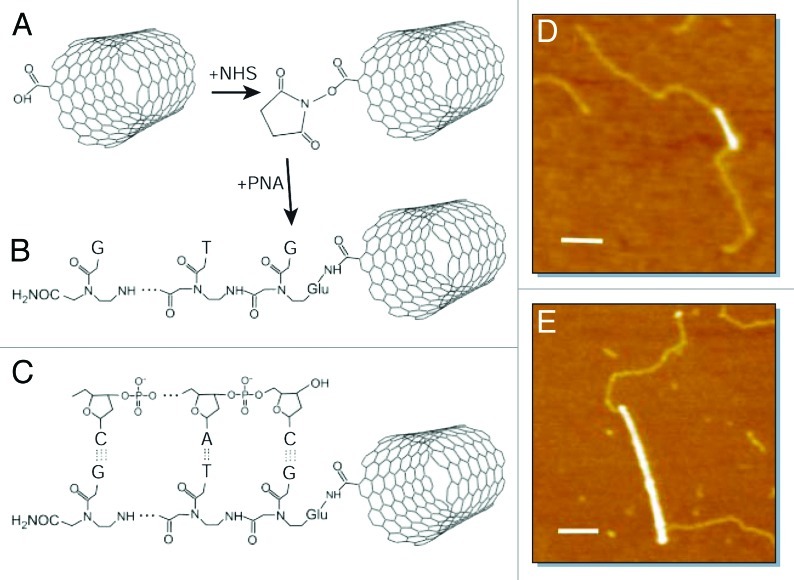
Exploiting the unique conductive and mechanical properties of CNTs with the specific recognition properties of ssPNA, Dekker and coworkers reported the engineering of carbon-based nucleic acid hybrids as new versatile functional modules for the bottom-up fabrication of field-effect transistors, logic circuits and single-electron transistors displaying potential uses in electronic and biosensing devices.54 In fact, the recognition properties imparted to SWNTs by oligonucleotide adducts could be used for the attachment of SWNTs to each other and/or to other substrate features, such as electrodes, on which monolayers of complementary sequences can be self-assembled. The hybridization properties of ssPNA-SWCNTs conjugates might also be exploited in a biological context, for example in biosensors for the detection of DNA sequences. Exploiting amide-forming reactions in the presence of N-hydroxysuccinimide (NHS), COOH-functionalized SWCNTs (obtained through oxidation reaction in the presence of a 3:1 mixture of concentrated H2SO4 and HNO3) have been coupled to ssPNAs (NH2-Glu-GTGCTCATGGTG-CONH2), as shown in Figure 9A and B, respectively. To investigate the recognition properties of the PNA-SWCNTs conjugates toward DNA strands, 12-base-pair fragments of double-stranded exposing single-stranded “sticky” ends complementary to the PNA sequence have been synthesized (Fig. 9C). Hybridization of the PNA-SWCNTs conjugates with the DNA oligomers in water and deposition on a mica surface with 5 mM MgCl2 allowed the material to be studied via AFM microscopy (Fig. 9D and E). Specifically, AFM studies at rt showed that DNA attachment occurs predominantly at or near the nanotube ends. Because the oxidation reaction mainly takes place at the extremities of the nanotubes bundles, individual SWCNTs are rare. The unusual attachment of DNA to the sidewall of SWNTs also indicates that this results from the sequence-specific PNA/DNA base-pairing, rather than from nonspecific interactions.
Figure 9. (A and B) Covalent functionalization of SWCNTs with ssPNA via an amide linkage; (C) a DNA fragment with a single-stranded, “sticky” end hybridizes to the PNA-SWCNTs hybrids; (D and E) tapping-mode AFM images of the PNA-SWCNTs and DNA-PNA-SWNTs. Scale bars: 100 nm. Adapted with permission from reference 54.
Following a similar approach, Matsumoto and coworkers reported on the engineering of a CNT-based field-effect transistor (FET) in which the reverse side of the CNTs-based electrode has been coupled to ssPNA-bearing SAMs as detecting probes conjugated to the tumor necrosis factor-α-gene (TNF-α).55 Upon exposure to wild-type DNA samples through microfluidic channels, time-dependent conductance measurements enabled rapid and simple discrimination against single-nucleotide polymorphism or non-cDNA strands. For 11-mer PNA oligonucleotides, full cDNA with concentration as low as 6.8 fM could be effectively detected, showing that this kind of CNT-based biochip FET are promising candidates for the development of integrated, high-throughput DNA sensors for medical, forensic and environmental diagnostics.56 Beside the covalent approach, nucleic acids have been also non-covalently conjugated to CNTs. The effectiveness of the binding and recognition properties of ssPNAs non-covalently adsorbed on SWCNTs, have been reported in an independent work by Rodger and Rajendra, who described the use of flow linear dichroism (LD) to study the interaction of nucleic acids and CNTs.57 Specifically, the LD signal of the PNA-SWCNTs, when subtracted to that of SWCNTs alone, gives the values of the non-covalently adsorbed nucleic acid. Unsurprisingly, the binding of the ssPNA is quite different from that of ssDNA and double stranded DNAs (dsDNA). It is likely that the nucleobases lie almost perpendicularly (the bases are flat in DNA) on the CNTs surface with the peptidic backbone wrapping around the nanotube framework at a tilted angle of 45°. With respect to PNA, DNA wraps with an inverted base orientation as shown by oppositely signed LD signals.
Self-Organized PNA-Based Nanoarchitectures
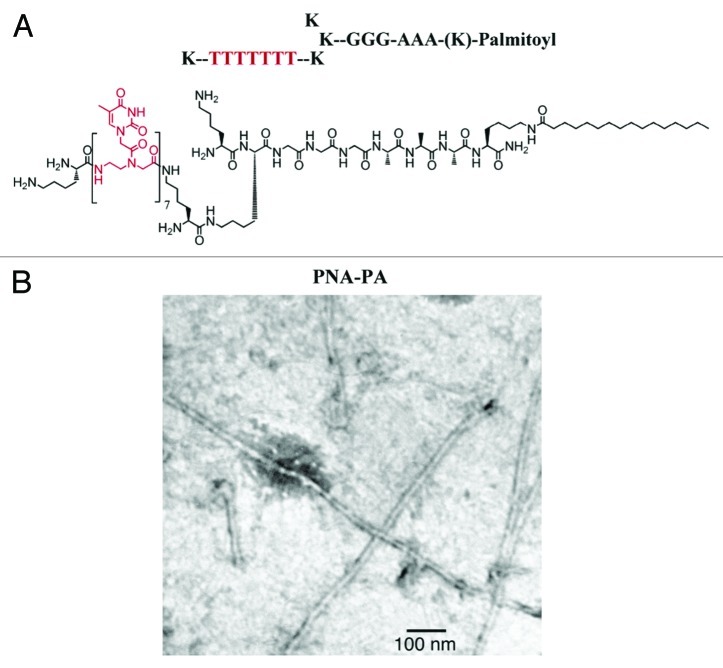
Nanostructured hydrogel-type materials prepared through self-organization are important as they can specifically bind oligonucleotides that, through a change of gelification temperature or fluorescence, can be exploited as sensing materials for their analytical determination. In these systems, the organic modules usually self-assemble into nanofibers networks. In this respect, Stupp and coworkers58 reported a novel amphiphilic nucleic acid hybrid in which a polythymine heptameric ssPNA is linked to a peptide fragment (KGGGGAAAK sequence, Fig. 10A), the latter governing the fiber formation promoted by the folding of β-sheets. The thermal stability of duplexes formed by the peptide/PNA nanofibers with nucleotide oligomers, as evaluated with variable temperature UV-spectroscopy, showed that the DNA and RNA form stronger complexes compared with previous work with other ssPNAs. This resulted in about ~25°C increase of the Tm (melting temperature) over the corresponding polythymine PNA heptamer. Furthermore, hybridization with 5′-AAATAAA-3′ ssDNA resulted in a 15°-lower Tm as compared with that of the fully-matching 5′-AAAAAAA-3′ ssDNA, thus showing the high sequence specificity. In addition, fluorescence polarization measurements with the fluorescein-tagged polyadenylic ssDNA heptamer revealed to be dramatically increased (~33 mP) as observed in the presence of self-assembled PNA-PA nanofibers (Fig. 10B). This confirms the binding to the nanofiber as a consequence of the reduced rotational freedom. Since such architectures can easily be endocytosed by cells, this type of hybrid materials are thought to be attractive scaffolds for the carriage and in situ release of siRNA sequences.
Figure 10. (A) Structure of the self-organizing PNA-peptide amphiphilic hybrid; (B) TEM images of the nanostructured fibers. Adapted with permission from reference 58.
In a very recent work, Janssen et al. utilized a polythymine dodecamer ssPNA to template the assembly of a chiral oligo (p-phenylenevinylene) derivative bearing a diaminotriazine H-bonding recognition motif in organic solution (methylcyclohexane), as shown on Figure 11.59 Nanostructured chiral helices, as studied in solution by variable temperature UV-vis steady-state absorption and circular dichroism (CD) spectroscopic measurements, were obtained. Their size was controlled by the length of the PNA template. AFM analysis of the structures drop-casted onto an HOPG surface confirmed uniform particles with a height of 3–4 nm.
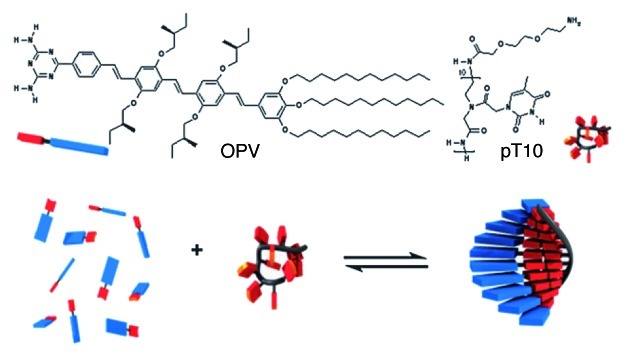
Figure 11. (Top) Structure of the ssPNA and OPV-based chromophore and (bottom) cartoon representation of the ssPNA-templated self-assembly/self-organization of the OPV molecules. Adapted with permission from reference 59.
Structural DNA nanotechnology exploits reciprocal exchange between DNA double helices or hairpins to produce branched DNA motifs, which are combined using sticky-ended cohesion to generate predictable and definite structures or arrays. Incorporating PNA along DNA into these motifs would lead to thermally more stable scaffolds thanks to PNA’s functional advantages for hybrid duplexes. Yet, one should consider that this might be achieved if the structural and chemical differences of PNA can be taken into account in the design and assembly of new constructs. Lukeman et al. used PNA fragments into a DNA double crossover (DX) molecules.60 Specifically, an “AB*” system consisting of two tiles (Fig. 12), both terminated with sticky ends: a DX tile (A tile) and a DX + J tile (B* tile, appended to a hairpin between the two crossovers), forms two-dimensional arrays when the tiles assemble correctly. Two different A tiles were synthesized: Ad, which is entirely constituted of DNA and Ap in which the two short crossover strands (in green and yellow on Fig. 12) are terminated with PNA strands; while eight versions of B* tiles were prepared: B0–B7, in which the number of nucleotides is altered (the numeral designates how many nucleotide pairs have been added). AFM studies showed that the Ad tile forms clean arrays with tiles B0, B1 and to some extent with B2. However, as the number of added bases increases, random aggregates are observed. The self-assembly between the Ap tile with B0, B1, B2 or B3 is also inconclusive while well-patterned arrays with the expected ~34 nm periodicity are obtained when the Ap tile is combined with B4, B5 and to some extent with B6. Moreover, the helical pitch of the DNA/PNA fragment of Ap can be calculated assuming that the average helical repeat always consists of 10.5 base pairs per turn and that the helices between crossovers on neighboring tiles contain 2.5 turns. It thus means that AB* systems can used for analyzing the helical pitch of polynucleotides, if they can form the outer arms of a DX molecule.
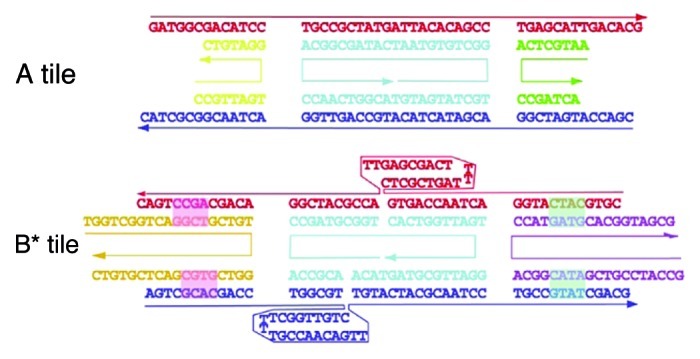
Figure 12. Representation of the DX tiles. The upper (A) tile contains yellow and green strands which consist of DNA in Ad and PNA in Ap. In the lower B0 tile, nucleotide base pairs are added alternately to the left and right pairs of outer arms to form tiles B1–B7 as indicated by the green and pink highlights. Adapted with permission from reference 60.
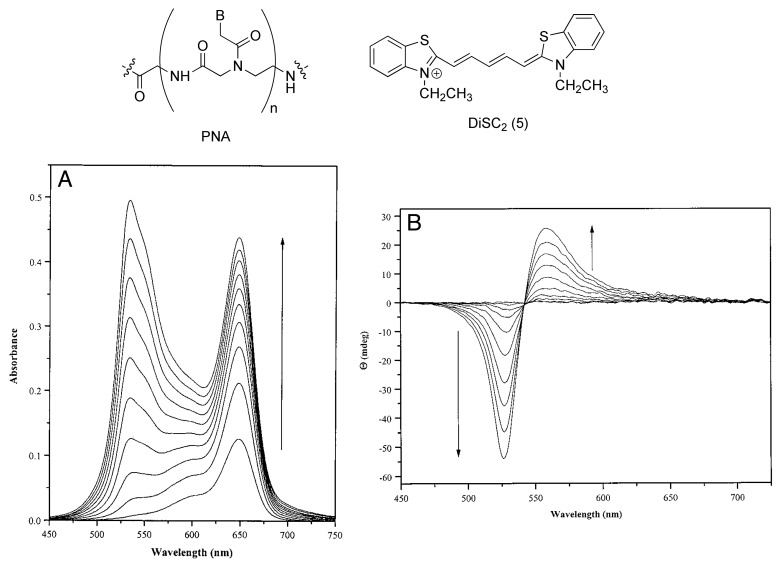
Armitage and coworkers first reported in 1999 the possibility to organize dye molecules using PNA templates in PNA-DNA hybrid sequences.61 They have demonstrated that symmetrical cyanine dyes (e.g., DiSC2-5) are able to bind a variety of PNA-DNA duplexes at the minor groove as shown by UV-vis and CD measurements (Fig. 13). In particular, UV-vis titration spectra of DiSC2-5 with PNA-DNA duplex (PD1) show the appearance of a new band corresponding to the electronic transition at 534 nm, hypsochromically shifted with respect to the absorption spectrum of the dye. This new absorption is due to the supramolecular binding between the dye and PD1, since this 534 nm absorption band is not observed for those spectra taken in the presence of ssPNA or ssDNA. The bisignate bands observed in the induced CD spectra (Fig. 13B) are related to exciton coupling between multiple cyanine dyes, indicating that multiple DiSC2-5 molecules are simultaneously bound to the PNA-DNA hybrid.
Figure 13. (A) UV-vis titration spectra of PD1 with increasing concentrations of DiSC2-5. (B) CD titration spectra of PD1 with increasing concentration of DiSC2-5. Adapted with permission from reference 61.
Armitage and coworkers62 also reported on the synthesis and characterization of a novel three-component biopolymer microgel composed by a protein (avidin), DNA and PNA strands, in which the cross-linking gelification is directed by Watson-Crick H-bonded base pairing (Fig. 14). They assembled a trifunctional DNA-based three-way junction (TWJ), peripherally exposing sticky ends composed of free unmatched nucleobases, with complementary ssPNA oligomers bearing terminal biotin groups able to bind a tetrafunctional protein, avidin. Using variable temperature UV-Vis, fluorescence and static light scattering techniques, they have shown that the self-assembly of these structures lead to a controlled and thermoreversible gel, the gelation temperature of which can be fine-tuned by a selected choice of the nucleotide sequence of the recognition PNA/DNA sites. Specifically, variable temperature dynamic and static light scattering measurements have shown the micro-size nature of the self-assembled particles displaying a fractal nature of 1.85. The microgel dissociates completely at ca. 43°C (corresponding to the Tm for a fully-matching TTGAGAT-biotin ssPNA conjugate) due to disruption of DNA-PNA double strand, while cooling restores the H-bonded PNA/DNA base pairing yielding the reassembled microgel. Both biotin-avidin interaction and DNA-TWJ assembly are stable under these temperature conditions, and thus the bioinylated PNA remains associated with the avidin. Upon introduction of a sequence mistmacth in the DNA-TWJ sticky ends, a decrease of the gelation temperature from 43°C to 40°C and then to 23°C could be progressively achieved.
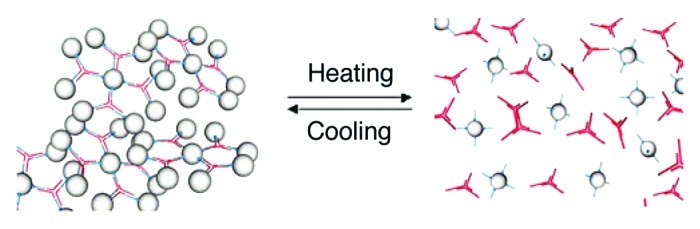
Figure 14. Schematic representation of the thermoreversible gelation process of the avidin-PNA/TWJ-DNA system (spheres, avidin-PNA conjugates; red crosses, DNA-TWJ junctions). Adapted with permission from reference 62.
Sharma et al. studied the formation of C-C+ PNA tetraplexes.63 To do so, four PNA oligomers of different sequences and length were used. The formation of the semiprotonated C-C+ tetraplexes has been monitored via UV-thermal transitions at 295 nm. Indeed, since C and C+ show a large absorption difference at this wavelength, a reverse sigmoidal pattern is observed when C-C+ tetraplex are formed.64 Tetraplex formation was not observed for PNA1 (H2N-T-C-C-βala-COOH) and PNA2 (H2N-T-C-C-C-βala-COOH) at any pH, while PNA3 (H2N-T-C-C-C-C-βala-COOH) and PNA4 (AcHN-Lvs-T-C-C-C-C-C-C-C-C-CONH2) showed formation of strong C-C+ tetraplexes more stabilized (by 10–20°C) than those formed with isosequential DNA strands, up to pH 5.5. The tetraplexes are no longer observed above pH 6. This example represents the first C-C+ tetraplexes consisting of unmodified PNA sequences TC4 and TC8.
Conclusions
In this Special Focus Review, an overview on the different supramolecular routes toward the preparation of nanostructured PNA-based materials has been given through a selection of the most important examples in the field. By acknowledging a detailed description of templated (i.e., by surface or CNTs) and non-templated (i.e., depending on the ssPNA itself) self-assembly and self-organization protocols undertaken to supramolecularly-organized ssPNA modules, this article discusses the main strategies toward the engineering of nanostructured PNA-based materials. Therefore, the reader has been guided through a systematic exploration of the most common avenues to prepare such organized materials accompanied by a discussion of their main potential applications. Although prototypes of various functional PNA-based materials (e.g., microarrays of various nature) and preparative approaches have been proposed, the examples reported in this article represent the initial steps of an increasing effort aimed at the design and preparation of further soft nucleotide-based materials which possess all the necessary characteristics (such as structural stability under thermal and electrical stresses) for technological applications in devices, others that those used in biotechnology. From the above described examples, it clearly appears that PNA can be a reliable alternative to DNA in those assemblies lacking of the intrinsic properties for device applications, for instance sizable conductivity, redox or photoactive properties, soft and very fragile properties, high thermal lability at relatively low temperatures, low synthetic versatility for side-chain functionalization. Together with these advantages, overcoming PNA’s physical (e.g., tendency to aggregate and limited solubility, to name a few) and synthetic protocol (e.g., it needs large excess of non-recyclable monomers for each SPPS step) limitations is the challenge that chemists working in this area have to tackle to ensure a dramatic impact of PNA in material science. Although the research is still in its beginning, more types of new PNA derivatives can be manufactured carrying out innovative and imaginative research to develop new complex biomimetic architectures creeping ever closer to the ultimate goal of producing real-world technologies, showing its limits only in the human imagination.
Acknowledgments
Dedicated to Prof R. Marchelli. D.B. gratefully acknowledges the EU through the ERC Starting Grant “COLORLANDS” project, the FRS-FNRS (FRFC contracts n° 2.4.550.09 and 2.4.617.07.F and MIS n° F.4.505.10.F), the “Loterie Nationale,” the “TINTIN” ARC project (09/14–023) and the University of Namur and University of Trieste (internal funding). V.C. thanks the University of Trieste for her doctoral fellowship; L.-E.C. and A.D. thank FRIA for their doctoral fellowships. Dr S. Mohnani is acknowledged for his efforts to have started our research endeavors in the field of PNA science.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/artificialdna/article/21941
References
- 1.Ajayaghosh A, Praveen VK. Pi-organogels of self-assembled p-phenylenevinylenes: soft materials with distinct size, shape, and functions. Acc Chem Res. 2007;40:644–56. doi: 10.1021/ar7000364. [DOI] [PubMed] [Google Scholar]
- 2.Ajayaghosh A, Varghese R, Mahesh S, Praveen VK. From vesicles to helical nanotubes: a sergeant-and-soldiers effect in the self-assembly of oligo(p-phenyleneethynylene)s. Angew Chem Int Ed Engl. 2006;45:7729–32. doi: 10.1002/anie.200603238. [DOI] [PubMed] [Google Scholar]
- 3.Antonietti M, Förster S. Vesicles and Liposomes: A Self-Assembly Principle Beyond Lipids. Adv Mater. 2003;15:1323–33. doi: 10.1002/adma.200300010. [DOI] [Google Scholar]
- 4.Gao Y, Zhang X, Ma C, Li X, Jiang J. Morphology-controlled self-assembled nanostructures of 5,15-di[4-(5-acetylsulfanylpentyloxy)phenyl]porphyrin derivatives. Effect of metal-ligand coordination bonding on tuning the intermolecular interaction. J Am Chem Soc. 2008;130:17044–52. doi: 10.1021/ja8067337. [DOI] [PubMed] [Google Scholar]
- 5.Muñoz-Bonilla A, Ibarboure E, Papon E, Rodriguez-Hernandez J. Self-organized hierarchical structures in polymer surfaces: self-assembled nanostructures within breath figures. Langmuir. 2009;25:6493–9. doi: 10.1021/la9003214. [DOI] [PubMed] [Google Scholar]
- 6.Cui S, Liu H, Gan L, Li Y, Zhu D. Fabrication of Low-Dimension Nanostructures Based on Organic Conjugated Molecules. Adv Mater. 2008;20:2918–25. doi: 10.1002/adma.200800619. [DOI] [Google Scholar]
- 7.Gomar-Nadal E, Puigmartí-Luis J, Amabilino DB. Assembly of functional molecular nanostructures on surfaces. Chem Soc Rev. 2008;37:490–504. doi: 10.1039/b703825a. [DOI] [PubMed] [Google Scholar]
- 8.Zhao YS, Fu HB, Hu FQ, Peng AD, Yang WS, Yao JN. Tunable Emission from Binary Organic One-Dimensional Nanomaterials: An Alternative Approach to White-Light Emission. Adv Mater. 2008;20:79–83. doi: 10.1002/adma.200700542. [DOI] [Google Scholar]
- 9.An B-K, Kwon S-K, Jung S-D, Park SY. Enhanced emission and its switching in fluorescent organic nanoparticles. J Am Chem Soc. 2002;124:14410–5. doi: 10.1021/ja0269082. [DOI] [PubMed] [Google Scholar]
- 10.Chen L, McBranch DW, Wang HL, Helgeson R, Wudl F, Whitten DG. Highly sensitive biological and chemical sensors based on reversible fluorescence quenching in a conjugated polymer. Proc Natl Acad Sci U S A. 1999;96:12287–92. doi: 10.1073/pnas.96.22.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones RM, Lu L, Helgeson R, Bergstedt TS, McBranch DW, Whitten DG. Building highly sensitive dye assemblies for biosensing from molecular building blocks. Proc Natl Acad Sci U S A. 2001;98:14769–72. doi: 10.1073/pnas.251555298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim Y, Swager TM. Sensory Polymers for Electron-Rich Analytes of Biological Interest. Macromolecules. 2006;39:5177–9. doi: 10.1021/ma060692x. [DOI] [Google Scholar]
- 13.Mohnani S, Llanes-Pallas A, Bonifazi D. Mastering nanostructured materials through H-bonding recognitions at interfaces. Pure Appl Chem. 2010;82:917–29. doi: 10.1351/PAC-CON-10-01-06. [DOI] [Google Scholar]
- 14.Yoosaf K, Llanes-Pallas A, Marangoni T, Belbakra A, Marega R, Botek E, et al. From Molecular to Macroscopic Engineering: Shaping Hydrogen-Bonded Organic Nanomaterials. Chemistry. 2011;17:3262–73. doi: 10.1002/chem.201002103. [DOI] [PubMed] [Google Scholar]
- 15.Marangoni T, Mezzasalma SA, Llanes-Pallas A, Yoosaf K, Armaroli N, Bonifazi D. Thermosolutal self-organization of supramolecular polymers into nanocraters. Langmuir. 2011;27:1513–23. doi: 10.1021/la104276y. [DOI] [PubMed] [Google Scholar]
- 16.Yoosaf K, Belbakra A, Llanes-Pallas A, Bonifazi D, Armaroli N. Engineering supramolecular photoactive nanomaterials by hydrogen-bonding interactions. Pure Appl Chem. 2011;83:899–912. doi: 10.1351/PAC-CON-10-10-22. [DOI] [Google Scholar]
- 17.Bonifazi D, Mohnani S, Llanes-Pallas A. Supramolecular chemistry at interfaces: molecular recognition on nanopatterned porous surfaces. Chemistry. 2009;15:7004–25. doi: 10.1002/chem.200900900. [DOI] [PubMed] [Google Scholar]
- 18.Gothelf KV, LaBean TH. DNA-programmed assembly of nanostructures. Org Biomol Chem. 2005;3:4023–37. doi: 10.1039/b510551j. [DOI] [PubMed] [Google Scholar]
- 19.Pitchiaya S, Krishnan Y. First blueprint, now bricks: DNA as construction material on the nanoscale. Chem Soc Rev. 2006;35:1111–21. doi: 10.1039/b602886c. [DOI] [PubMed] [Google Scholar]
- 20.Winfree E, Liu F, Wenzler LA, Seeman NC. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–44. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 21.Otero R, Xu W, Lukas M, Kelly REA, Laegsgaard E, Stensgaard I, et al. Specificity of watson-crick base pairing on a solid surface studied at the atomic scale. Angew Chem Int Ed Engl. 2008;47:9673–6. doi: 10.1002/anie.200803333. [DOI] [PubMed] [Google Scholar]
- 22.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 24.Hyrup B, Nielsen PE. Peptide nucleic acids (PNA): synthesis, properties and potential applications. Bioorg Med Chem. 1996;4:5–23. doi: 10.1016/0968-0896(95)00171-9. [DOI] [PubMed] [Google Scholar]
- 25.Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009;459:73–6. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 26.Han D, Pal S, Nangreave J, Deng Z, Liu Y, Yan H. DNA origami with complex curvatures in three-dimensional space. Science. 2011;332:342–6. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 27.Demidov VV, Potaman VN, Frank-Kamenetskii MD, Egholm M, Buchard O, Sönnichsen SH, et al. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem Pharmacol. 1994;48:1310–3. doi: 10.1016/0006-2952(94)90171-6. [DOI] [PubMed] [Google Scholar]
- 28.Uhlmann E, Peyman A, Breipohl G, Will DW. PNA: Synthetic Polyamide Nucleic Acids with Unusual Binding Properties. Angew Chem Int Ed. 1998;37:2796–823. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2796::AID-ANIE2796>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen PE. PNA Technology. Mol Biotechnol. 2004;26:233–48. doi: 10.1385/MB:26:3:233. [DOI] [PubMed] [Google Scholar]
- 30.Harris JL, Winssinger N. PNA encoding (PNA=peptide nucleic acid): from solution-based libraries to organized microarrays. Chemistry. 2005;11:6792–801. doi: 10.1002/chem.200500305. [DOI] [PubMed] [Google Scholar]
- 31.Briones C, Mateo-Martí E, Gómez-Navarro C, Parro V, Román E, Martín-Gago JA. Structural and functional characterization of self-assembled monolayers of peptide nucleic acids and its interaction with complementary DNA. J Mol Catal Chem. 2005;228:131–6. doi: 10.1016/j.molcata.2004.09.076. [DOI] [Google Scholar]
- 32.Liu Z-C, Shin D-S, Shokouhimehr M, Lee K-N, Yoo B-W, Kim Y-K, et al. Light-directed synthesis of peptide nucleic acids (PNAs) chips. Biosens Bioelectron. 2007;22:2891–7. doi: 10.1016/j.bios.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Mateo-Martí E, Briones C, Pradier CM, Martín-Gago JA. A DNA biosensor based on peptide nucleic acids on gold surfaces. Biosens Bioelectron. 2007;22:1926–32. doi: 10.1016/j.bios.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 34.Mateo-Martí E, Briones C, Román E, Briand E, Pradier CM, Martín-Gago JA. Self-assembled monolayers of peptide nucleic acids on gold surfaces: a spectroscopic study. Langmuir. 2005;21:9510–7. doi: 10.1021/la050366v. [DOI] [PubMed] [Google Scholar]
- 35.Mateo-Martí E, Rogero C, Briones C, Martín-Gago JA. Do peptide nucleic acids form self-assembled monolayers on pyrite surfaces? Surf Sci. 2007;601:4195–9. doi: 10.1016/j.susc.2007.04.081. [DOI] [Google Scholar]
- 36.Singh RP, Oh B-K, Choi J-W. Application of peptide nucleic acid towards development of nanobiosensor arrays. Bioelectrochemistry. 2010;79:153–61. doi: 10.1016/j.bioelechem.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Briones C, Mateo-Marti E, Gómez-Navarro C, Parro V, Román E, Martín-Gago JA. Ordered self-assembled monolayers of Peptide nucleic acids with DNA recognition capability. Phys Rev Lett. 2004;93:208103. doi: 10.1103/PhysRevLett.93.208103. [DOI] [PubMed] [Google Scholar]
- 38.Lioubashevski O, Patolsky F, Willner I. Probing of DNA and Single-Base Mismatches by Chemical Force Microscopy Using Peptide Nucleic Acid-Modified Sensing Tips and Functionalized Surfaces. Langmuir. 2001;17:5134–6. doi: 10.1021/la0105174. [DOI] [Google Scholar]
- 39.Burgener M, Sänger M, Candrian U. Synthesis of a stable and specific surface plasmon resonance biosensor surface employing covalently immobilized peptide nucleic acids. Bioconjug Chem. 2000;11:749–54. doi: 10.1021/bc0000029. [DOI] [PubMed] [Google Scholar]
- 40.D’Agata R, Corradini R, Grasso G, Marchelli R, Spoto G. Ultrasensitive detection of DNA by PNA and nanoparticle-enhanced surface plasmon resonance imaging. Chembiochem. 2008;9:2067–70. doi: 10.1002/cbic.200800310. [DOI] [PubMed] [Google Scholar]
- 41.Kambhampati D, Nielsen PE, Knoll W. Investigating the kinetics of DNA-DNA and PNA-DNA interactions using surface plasmon resonance-enhanced fluorescence spectroscopy. Biosens Bioelectron. 2001;16:1109–18. doi: 10.1016/S0956-5663(01)00239-1. [DOI] [PubMed] [Google Scholar]
- 42.Tawa K, Knoll W. Mismatching base-pair dependence of the kinetics of DNA-DNA hybridization studied by surface plasmon fluorescence spectroscopy. Nucleic Acids Res. 2004;32:2372–7. doi: 10.1093/nar/gkh572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao D, Kim J, Yu F, Nielsen PE, Sinner E-K, Knoll W. Surface density dependence of PCR amplicon hybridization on PNA/DNA probe layers. Biophys J. 2005;88:2745–51. doi: 10.1529/biophysj.104.051656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calabretta A, Wasserberg D, Posthuma-Trumpie GA, Subramaniam V, van Amerongen A, Corradini R, et al. Patterning of peptide nucleic acids using reactive microcontact printing. Langmuir. 2011;27:1536–42. doi: 10.1021/la102756k. [DOI] [PubMed] [Google Scholar]
- 45.Milano G, Musumeci D, Gaglione M, Messere A. An alternative strategy to synthesize PNA and DNA magnetic conjugates forming nanoparticle assembly based on PNA/DNA duplexes. Mol Biosyst. 2010;6:553–61. doi: 10.1039/b915680a. [DOI] [PubMed] [Google Scholar]
- 46.Truskey GA, Burmeister JS, Grapa E, Reichert WM. Total internal reflection fluorescence microscopy (TIRFM). II. Topographical mapping of relative cell/substratum separation distances. J Cell Sci. 1992;103:491–9. doi: 10.1242/jcs.103.2.491. [DOI] [PubMed] [Google Scholar]
- 47.Mertz J, Xu C, Webb WW. Single-molecule detection by two-photon-excited fluorescence. Opt Lett. 1995;20:2532. doi: 10.1364/OL.20.002532. [DOI] [PubMed] [Google Scholar]
- 48.Stadler AL, Sun D, Maye MM, van der Lelie D, Gang O. Site-selective binding of nanoparticles to double-stranded DNA via peptide nucleic acid “invasion”. ACS Nano. 2011;5:2467–74. doi: 10.1021/nn101355n. [DOI] [PubMed] [Google Scholar]
- 49.Shiraishi T, Deborggraeve S, Büscher P, Nielsen PE. Sensitive detection of nucleic acids by PNA hybridization directed co-localization of fluorescent beads. Artif DNA PNA XNA. 2011;2:60–6. doi: 10.4161/adna.2.2.16562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Palecek E, Nielsen PE, Rivas G, Cai X, Shiraishi H, et al. Peptide Nucleic Acid Probes for Sequence-Specific DNA Biosensors. J Am Chem Soc. 1996;118:7667–70. doi: 10.1021/ja9608050. [DOI] [Google Scholar]
- 51.Wang J, Rivas G, Cai X, Chicharro M, Parrado C, Dontha N, et al. Detection of point mutation in the p. 53 gene using a peptide nucleic acid biosensor. Anal Chim Acta. 1997;344:111–8. doi: 10.1016/S0003-2670(97)00039-1. [DOI] [Google Scholar]
- 52.Winssinger N, Damoiseaux R, Tully DC, Geierstanger BH, Burdick K, Harris JL. PNA-encoded protease substrate microarrays. Chem Biol. 2004;11:1351–60. doi: 10.1016/j.chembiol.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 53.Urbina HD, Debaene F, Jost B, Bole-Feysot C, Mason DE, Kuzmic P, et al. Self-assembled small-molecule microarrays for protease screening and profiling. Chembiochem. 2006;7:1790–7. doi: 10.1002/cbic.200600242. [DOI] [PubMed] [Google Scholar]
- 54.Williams KA, Veenhuizen PTM, de la Torre BG, Eritja R, Dekker C. Nanotechnology: carbon nanotubes with DNA recognition. Nature. 2002;420:761. doi: 10.1038/420761a. [DOI] [PubMed] [Google Scholar]
- 55.Kerman K, Morita Y, Takamura Y, Tamiya E, Maehashi K, Matsumoto K. Peptide Nucleic Acid–Modified Carbon Nanotube Field-Effect Transistor for Ultra-Sensitive Real-Time Detection of DNA Hybridization. Nanobiotechnology. 2005;1:065–070. doi: 10.1385/NBT:1:1:065. [DOI] [Google Scholar]
- 56.Maehashi K, Matsumoto K, Kerman K, Takamura Y, Tamiya E. Ultrasensitive Detection of DNA Hybridization Using Carbon Nanotube Field-Effect Transistors. JPN J Appl Phys 2. 2004;43:L1558–L1560. doi: 10.1143/JJAP.43.L1558. [DOI] [Google Scholar]
- 57.Rajendra J, Rodger A. The binding of single-stranded DNA and PNA to single-walled carbon nanotubes probed by flow linear dichroism. Chemistry. 2005;11:4841–7. doi: 10.1002/chem.200500093. [DOI] [PubMed] [Google Scholar]
- 58.Guler MO, Pokorski JK, Appella DH, Stupp SI. Enhanced oligonucleotide binding to self-assembled nanofibers. Bioconjug Chem. 2005;16:501–3. doi: 10.1021/bc050053b. [DOI] [PubMed] [Google Scholar]
- 59.Janssen PGA, Meeuwenoord N, van der Marel G, Jabbari-Farouji S, van der Schoot P, Surin M, et al. ssPNA templated assembly of oligo(p-phenylenevinylene)s. Chem Commun (Camb) 2010;46:109–11. doi: 10.1039/b913307k. [DOI] [PubMed] [Google Scholar]
- 60.Lukeman PS, Mittal AC, Seeman NC. Two dimensional PNA/DNA arrays: estimating the helicity of unusual nucleic acid polymers. Chem Commun (Camb) 2004;1694-5:1694–5. doi: 10.1039/b401103a. [DOI] [PubMed] [Google Scholar]
- 61.Smith JO, Olson DA, Armitage BA. Molecular Recognition of PNA-Containing Hybrids: Spontaneous Assembly of Helical Cyanine Dye Aggregates on PNA Templates. J Am Chem Soc. 1999;121:2686–95. doi: 10.1021/ja9837553. [DOI] [Google Scholar]
- 62.Cao R, Gu Z, Hsu L, Patterson GD, Armitage BA. Synthesis and characterization of thermoreversible biopolymer microgels based on hydrogen bonded nucleobase pairing. J Am Chem Soc. 2003;125:10250–6. doi: 10.1021/ja035211t. [DOI] [PubMed] [Google Scholar]
- 63.Sharma NK, Ganesh KN. PNA C-C+ i-motif: superior stability of PNA TC8 tetraplexes compared to DNA TC8 tetraplexes at low pH. Chem Commun (Camb) 2005;4330-2:4330–2. doi: 10.1039/b506870c. [DOI] [PubMed] [Google Scholar]
- 64.Phan AT, Mergny JL. Human telomeric DNA: G-quadruplex, i-motif and Watson-Crick double helix. Nucleic Acids Res. 2002;30:4618–25. doi: 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]