Abstract
The common marmoset (Callithrix jacchus) is considered a novel experimental animal model of non-human primates. However, due to antibody unavailability, immunological and pathological studies have not been adequately conducted in various disease models of common marmoset. Quantitative real-time PCR (qPCR) is a powerful tool to examine gene expression levels. Recent reports have shown that selection of internal reference housekeeping genes are required for accurate normalization of gene expression. To develop a reliable qPCR method in common marmoset, we used geNorm applets to evaluate the expression stability of eight candidate reference genes (GAPDH, ACTB, rRNA, B2M, UBC, HPRT, SDHA and TBP) in various tissues from laboratory common marmosets. geNorm analysis showed that GAPDH, ACTB, SDHA and TBP were generally ranked high in stability followed by UBC. In contrast, HPRT, rRNA and B2M exhibited lower expression stability than other genes in most tissues analyzed. Furthermore, by using the improved qPCR with selected reference genes, we analyzed the expression levels of CD antigens (CD3ε, CD4, CD8α and CD20) and cytokines (IL-1β, IL-2, IL-4, IL-5, IL-6, IL-10, IL-12β, IL-13, IFN-γ and TNF-α) in peripheral blood leukocytes and compared them between common marmosets and humans. The expression levels of CD4 and IL-4 were lower in common marmosets than in humans whereas those of IL-10, IL-12β and IFN-γ were higher in the common marmoset. The ratio of Th1-related gene expression level to that of Th2-related genes was inverted in common marmosets. We confirmed the inverted ratio of CD4 to CD8 in common marmosets by flow cytometric analysis. Therefore, the difference in Th1/Th2 balance between common marmosets and humans may affect host defense and/or disease susceptibility, which should be carefully considered when using common marmoset as an experimental model for biomedical research.
Introduction
The common marmoset (Callithrix jacchus) is a New World monkey and is considered potentially useful as an experimental animal model in research fields such as drug toxicology [1], [2], neuroscience [3], [4], autoimmune diseases [5], [6] and infectious diseases [7], [8], because of its size, availability and high genetic similarity with humans [9], [10]. Compared with mice, common marmosets are more useful as an in vivo model to study immune function [11]. However, essential tools and gene information for conducting studies using common marmosets are in short supply or unavailable. For example, monoclonal antibodies specific for common marmosets have been only partially established. Although DNA microarray research for common marmoset brain has been reported [12], sufficient studies have not been performed in other research fields.
Quantitative real-time polymerase chain reaction (qPCR) is the dominant quantitative technique for gene expression analysis due to its broad dynamic range, accuracy, sensitivity, specificity and speed [13]. Thus, qPCR is very useful for investigating physiological and pathological status from a small amount of sample. Normalization to reference genes such as housekeeping genes is usually required for qPCR analysis. However, expression levels of reference genes may vary between tissues, cell types and experimental conditions. Therefore, the validation of suitable reference genes in each experiment is critical for the accurate evaluation of qPCR data. Recently, a set of guidelines for evaluating qPCR experiments was developed [14] and a strict method for the selection of reference genes suitable for normalization was proposed [15]. A freely available program, geNorm applet (http://medgen.ugent.be/~jvdesomp/genorm/), can determine gene stability ranking and the number of reference genes required for normalization in a given panel of samples [15].
To develop an accurate and reliable qPCR method for common marmosets, we examined the expression stabilities of candidate reference genes in various tissues of laboratory common marmosets using geNorm applet. Then, we compared expression levels of immune-related genes in peripheral blood leukocytes between common marmosets and humans. To the best of our knowledge, this is the first such study for the selection of reference genes in common marmosets. The present data will contribute to future studies of gene expression analysis by qPCR for common marmosets.
Materials and Methods
Ethics statement
The study was conducted in accordance with the Act on Welfare and Management of Animals of Japanese government. All animals were housed, cared for, and used according to the principles set forth in the Guide for the Care and Use of Laboratory Animals: Eighth Edition (National Research Council, 2011). All experiments using common marmosets were approved by the committee for animal experiments at the National Institute of Infectious Diseases (Approval Number: 610,007). For humans, whole blood was obtained from eight healthy volunteers (mean age ± sd: 35.7±13.0 years old) after obtaining written informed consent. This study and the consent procedure were approved by the ethics committee of Tokai University School of Medicine (Approval Number: 10I-22).
Animals
Eight common marmosets (1.58±0.29 years old) were obtained from CLEA Japan, Inc. (Tokyo, Japan) and maintained in specific pathogen-free conditions at the National Institute of Infectious Diseases (Tokyo, Japan). Common marmosets were housed solely or in pairs in a single cages 39 cm (W)×55 (D)×70 (H) in size on 12∶12 h light/dark cycles. Room temperature and humidity were maintained at 26–27°C and 40–50%, respectively. Filtered drinking water was delivered by an automatic watering system and total 40–50 g/individual of commercial marmoset chow (CMS-1M, CLEA Japan) were given in a couple of times per day. Dietary supplements (sponge cakes, eggs, banana pudding, honeys, vitamin C and D3) were also given to improve their health status. Machinery noise and dogs' barks were avoided to reduce stress. The cages were equipped with resting perches and a nest box as environmental enrichment. The marmosets were routinely tested to assure the absence of pathogenic bacteria, viruses, and parasite eggs in the animal facilities and did not exhibited abnormal external appearances. Four common marmosets were euthanized by cardiac exsanguinations under anesthesia with Ketamine hydrochroride (50 mg/kg, IM) and Xylazine (3.0 mg/kg, IM). After sacrifice, various tissues removed, and whole blood was obtained from all eight common marmosets.
RNA isolation
Heparinized venous blood samples from common marmosets were obtained before sacrifice and incubated in erythrocyte lysis buffer (155 mM NH4Cl, 10 mM KHCO3, and 0.1 mM EDTA). Following incubation on ice for 5 min, cells were centrifuged at 300×g for 10 min at 4°C and washed with lysis buffer and then PBS. Leukocytes were lysed with QIAzol® Lysis Reagent (Qiagen, Hilden, Germany) and total RNA was extracted using an RNeasy® Plus Universal Mini Kit (Qiagen) according to the manufacturer's instructions. Tissue samples (spleen, mesenteric lymph node, jejunum, ileum, descending colon, cerebrum, cerebellum, brainstem, heart, lung, liver and kidney) were excised from each animal and immediately submerged in RNAlater® RNA Stabilization Reagent (Qiagen). Then total RNA was extracted using RNeasy® Plus Universal Mini Kit (Qiagen). RNA concentration and integrity were assessed using the Agilent RNA 6,000 Nano Kit (Agilent Technologies, Inc., CA, USA) in an Agilent 2100 Bioanalyzer. All RNA samples were confirmed to have no degradation and were of optimal quality for downstream qPCR applications.
Candidate reference genes
Based on a literature search, eight commonly used candidate internal control genes were selected for analysis: GAPDH (glyceraldehyde-3-phosphate dehydrogenase), ACTB (actin, beta), rRNA (18S ribosomal RNA), B2M (beta-2-microglobulin), UBC (ubiquitin C), HPRT (hypoxanthine phosphoribosyltransferase 1), SDHA (succinate dehydrogenase complex, subunit A) and TBP (TATA-box binding protein). All genes chosen have independent cellular functions and are not thought to be co-regulated. The sequences of primers specific for each reference gene are shown in Table 1.
Table 1. Sequences of qPCR primers for housekeeping genes.
| Target gene | Species | 5′-primer sequence -3′ a) , b) | Product size (bp) | PCR efficiency | Reference | |
| Forward | Reverse | |||||
| GAPDH | Cj | TCGGAGTCAACGGATTTGGTC | TTCCCGTTCTCAGCCTTGAC | 181 | 0.920 | DD279474 |
| Hs | --------------------- | -------------------- | 181 | 0.921 | AF261085 | |
| ACTB | Cj | GATGGTGGGCATGGGTCAGAA | AGCCACACGCAGCTCGTTGT | 163 | 0.901 | DD279463 |
| Hs | --------------------- | ---------------A---- | 163 | 0.883 | NM_001101 | |
| HPRT | Cj | ATCCAAAGATGGTCAAGGTCG | GTATTCATTATAGTCAAGGGCATA | 134 | 0.842 | DD289567 |
| Hs | --------------------- | ------------------------ | 134 | 0.880 | M31642 | |
| B2M | Cj | CTATTCAGCATGCTCCAAAGA | AAGACAAGTCTGAATGCTCCAC | 168 | 0.928 | AF084623 |
| Hs | ----C----G-A--------- | ---------------------- | 168 | 0.950 | AB021288 | |
| UBC | Cj | TCCCTTCTCGGCGGTTCTG | . TGCATTGTCAAGCGGCGAT | 158 | 0.922 | AB571242 |
| Hs | -------------A----- | TC----------T-A---- | 160 | 0.936 | NM_021009 | |
| rRNA | Cj | CGACCATAAACGATGCCGAC | GGTGGTGCCCTTCCGTCAAT | 145 | 0.918 | AB571241 |
| Hs | -------------------- | -------------------- | 145 | 0.940 | M10098 | |
| SDHA | Cj | TGGGAACAAGAGGGCATCTG | CCACCACGGCATCAAATTCATG | 86 | 0.934 | XM_002745154 |
| Hs | -------------------- | -------T-------------- | 86 | 0.948 | BC001380 | |
| TBP | Cj | CCATGACTCCCGGAATCCCTAT | ATAGGCTGTGGGGTCAGTCCA | 70 | 0.920 | EU796973 |
| Hs | ---------------------- | --------------------- | 70 | 0.954 | M55654 | |
Hyphen indicates a nucleotide identical to human sequences.
Dot indicates a shift nucleotide to marmoset sequences.
Quantitative real-time PCR
First-strand cDNA was synthesized using PrimeScript® RT reagent Kit (Takara Bio, Otsu, Japan) with attached random hexamers and oligo(dT) primers. Reactions were incubated at 37°C for 15 min followed by 85°C for 5 sec according to the manufacturer's instructions. Then each cDNA sample was diluted with RNase/DNase-free water to 25 ng/µL. The expression level of each gene was analyzed by qPCR using the Bio-Rad CFX96 system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). PCR reactions consisted of 5 µL of SsoFast™ EvaGreen® Supermix (Bio-Rad), 3.5 µL of RNase/DNase-free water, 0.5 µL of 5 µM primer mix, 1 µL of cDNA in a total volume of 10 µL. The primer sequences are shown in Tables 1 and 2. Cycling conditions were as follows: 30 sec at 95°C followed by 45 rounds of 95°C for 1 sec and 60°C for 5 sec. Melting curve analysis to determine the dissociation of PCR products was performed between 65°C and 95°C. Data were expressed as mean values of experiments performed in triplicate. Seven points of a 10-fold serial dilution of standard DNA was used for absolute quantification. Standard DNA was generated by cloning PCR products into pGEM-T Easy Vector (Promega, WI, USA). Sequences of the cloned plasmid were confirmed by DNA sequencing using the CEQ8000 Genetic Analysis System (Beckman Coulter). Quality and concentration of the plasmid DNA were validated using Agilent DNA 7,500 Kit in an Agilent 2100 Bioanalyzer.
Table 2. Sequences of qPCR primers for CD markers and cytokines.
| Target gene | Species | 5′-primer sequence -3′ a) , b) | Product size (bp) | PCR efficiency | Reference | |
| Forward | Reverse | |||||
| CD3ε | Cj | GGCTTGCTGCTGCTGGTTTAC | CCGGATGGGCTCATAGTCTG | 150 | 0.865 | DQ189218 |
| Hs | --------------------- | -------------------- | 150 | 0.848 | NM_000733 | |
| CD4 | Cj | GGAAAACGGGAAAGTTGCATCA | GCCTTCTCCCGCTTAGAGAC | 163 | 0.926 | AF452616 |
| Hs | C------A-------------- | --------------C----- | 162 | 0.907 | M35160 | |
| CD8α | Cj | TCTCCCAAACCAAGTCCAAGG | AGTTTCTCAGGGCCGAGCAG | 144 | 0.940 | DQ189217 |
| Hs | ---------A----C------ | . ---G--------------- | 143 | 0.912 | NM_001768 | |
| CD20 | Cj | GGGCTGTCCAGATTATGAATG | GAGTTTTTCTCCGTTGCTGC | 166 | 0.942 | DQ189220 |
| Hs | --------------------- | -------------------- | 166 | 1.002 | X07203 | |
| IL-1β | Cj | TGCACCTGTACGATCCCTGAAC | TTGCACAAAGGACATGGAGAACAC | 145 | 0.806 | AB539804 |
| Hs | ---------------A------ | ---T-------------------- | 145 | 0.780 | NM_000576 | |
| IL-2 | Cj | CCCAAGAAGGCCAAAGAATTG | CTTAAGTGAAAGTTTTTGCTTTGAG | 104 | 0.773 | DQ826674 |
| Hs | -------------C----C-- | ------------------------- | 103 | 0.797 | BC070338 | |
| IL-4 | Cj | CATTGTCACAGAGCAAAAGACTC | CTCAGTTGTGTTCTTGGAGGCA | 79 | 0.910 | XM_002744606 |
| Hs | . GCC----------G------- | ---------------------- | 77 | 0.878 | NM_000589 | |
| IL-5 | Cj | AATCACCAACTGTGCACTGAAGAA | . TTTGGCGGTCAATGTGTTCCTT | 130 | 0.871 | DQ658152 |
| Hs | ------------------------ | TT------C--------A--T--- | 132 | 0.860 | NM_000879 | |
| IL-6 | Cj | GATTCAATGAGGAGACTTGCC | TGTTCTGGAGGTACTCTAGGTA | 81 | 0.920 | DQ658153 |
| Hs | --------------------- | ---------------------- | 81 | 0.990 | NM_00600 | |
| IL-10 | Cj | CTGCCTCACATGCTTCGAGA | TGGCAACCCAGGTAACCCTTA | 134 | 0.970 | DQ658154 |
| Hs | ------A------------- | --------------------- | 134 | 0.920 | M57627 | |
| IL-12β | Cj | . GGACGGCAAGGAGTATGAGTA | TTGAGCTTGTGAACGGCATC | 111 | 0.935 | AB539805 |
| Hs | G----AA--------------- | -------------------- | 112 | 0.900 | M65272 | |
| IL-13 | Cj | TCCAGCTTGCTTGTCCGAG | CTGCAAATAATGATGCGTT-GATGT | 127 | 0.916 | AB571243 |
| Hs | ----------A-------- | . ---------------T--C--A-- | 127 | 0.964 | NM_002188 | |
| IFN-γ | Cj | GGGTTCTCTTGGCTGTTACTG | TGTCTAAGAAAAGAGTTCCATTATC | 116 | 0.838 | FJ598593 |
| Hs | --------------------- | . -C---------------------- | 115 | 0.856 | NM_000619 | |
| TNF-α | Cj | AGCCTGTAGCCCATGTTGTAG | CTCTCAGCTCCACGCCATTG | 102 | 0.887 | DQ520835 |
| Hs | --------------------- | -------------------- | 102 | 0.817 | NM_000594 | |
Hyphen indicates a nucleotide identical to human sequences.
Dot indicates a shift nucleotide to marmoset sequences.
Analysis of gene expression stability
The expression stability of selected reference genes was evaluated using a publicly available program, geNorm applet [15]. geNorm calculates the stability of tested reference genes according to the similarity of their expression profiles by pairwise comparison and M value, where the gene with the highest value is the least stable one. It is possible to perform sequential elimination of the least stable gene in any given experimental group, thus resulting in the exclusion of all but the two most stable genes in each case.
Flow cytometry
Heparinized peripheral blood was collected from common marmosets and centrifuged in Lymphocepal (IBL Co. Takasaki, Japan) at 2,000 rpm for 30 min. Mononuclear cells were collected and re-suspended in RPMI1640 medium containing 10% fetal calf serum. Cells were stained with anti-common marmoset CD8 antibody (Mar8–10) [16] for 15 min at 4°C and washed with 1% (w/v) bovine serum albumin-containing PBS. Subsequently, cells were stained with phycoerythrin-labeled secondary antibody, peridinin chlorophyll protein cyanin5.5 (PerCPCy5.5)-conjugated anti-human CD3 (SP34-2) and Alexa488-conjugated anti-common marmoset CD4 (Mar4-33) antibodies [16]. Peripheral blood from healthy human volunteers was collected and mononuclear cells isolated by Ficoll-Paque (GE Healthcare Biosciences, Uppsala, Sweden) gradient centrifugation. The monoclonal antibodies used for cell staining were as follows: PerCPCy5.5-conjugated anti-human CD3 (SP34-2), allophycocyanin-conjugated anti-human CD4 (SK3), fluorescein isothiocyanate-conjugated anti-human CD8 (HIT8a) (BD PharMingen). Cells were analyzed by FACSCalibur (Becton Dickinson, Franklin Lakes, NJ, USA).
Statistical analysis
Student's t-test was used for statistical analysis to assess significant differences in qPCR assays. A P value<0.05 was considered to be statistically significant.
Results
The expression levels of candidate reference genes in tissues
Eight housekeeping genes were chosen as reference genes: GAPDH, ACTB, rRNA, B2M, UBC, HPRT, SDHA and TBP. We determined the transcription levels of these eight genes in 13 tissues (leukocyte, spleen, lymph node, jejunum, ileum, colon, cerebrum, cerebellum, brainstem, heart, lung, liver and kidney) from four individual common marmosets by qPCR. The sequences of primers specific for each reference gene are shown in Table 1. The expression level of each gene in each tissue is shown as the copy number per µg of purified total RNA (Figure 1). The most abundant gene was rRNA while the rarest gene was UBC and the difference in expression level between the two genes was more than 100,000-fold. For several genes, the expression levels were highly different among tissues. For example, B2M expression in heart and brain segments (cerebrum, cerebellum and brainstem) was markedly lower than in other tissues. HPRT expression also showed a large variability among tissues. In addition, the expression levels of rRNA, B2M and HPRT varied among individuals; the mean values of standard deviation were 0.224, 0.235 and 0.303, respectively, while those of the other genes were below 0.2.
Figure 1. Absolute copy numbers of candidate reference genes.
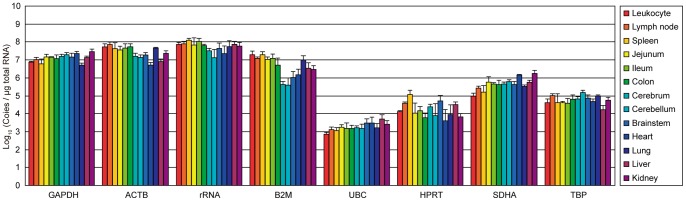
The expression level of each gene in 13 tissues is shown as a logarithmic histogram of absolute copy numbers per µg of total RNA. Means and standard deviations of four individuals are indicated. GAPDH: glyceraldehyde-3-phosphate dehydrogenase; ACTB: actin, beta; rRNA: 18S ribosomal RNA; B2M: beta-2-microglobulin; UBC: ubiquitin C; HPRT: hypoxanthine phosphoribosyltransferase 1; SDHA: succinate dehydrogenase complex, subunit A; TBP: TATA-box binding protein.
A variety of gene expression stabilities among tissues
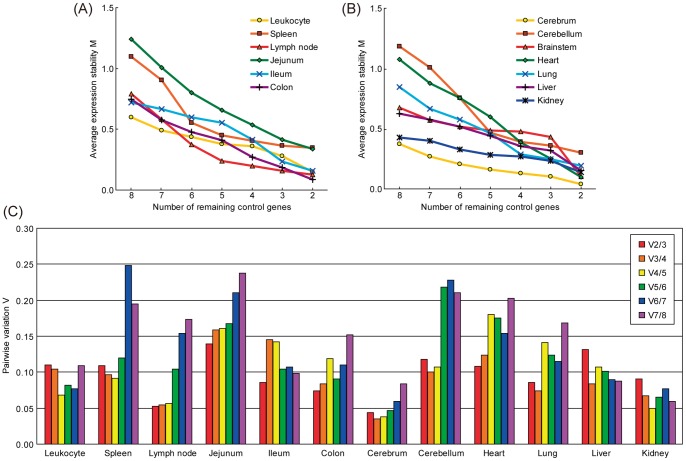
To evaluate the expression stability of selected reference genes, we used a publicly available program, geNorm applets. geNorm provides a ranking of tested genes based on the reference gene stability measure M, which is defined as the average pairwise variation of a particular gene compared with all other control genes. Thus, genes with higher M values have greater variations of expression. In addition, assessment of the pairwise variations (Vn/n+1) between each combination of sequential normalization factors allows identification of the optimal number of reference genes. In the original publication describing geNorm [15], a threshold of 0.15 for pairwise variation was established, below which the inclusion of additional reference genes was not necessary.
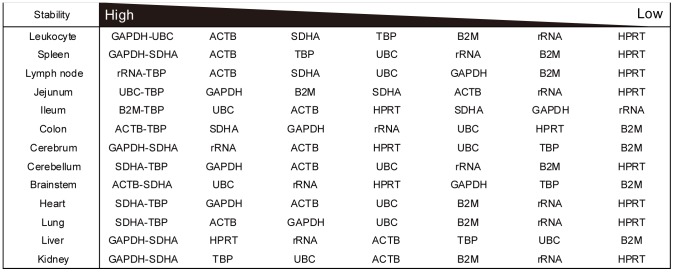
geNorm analysis produced line plots indicating the mean expression stability M of the remaining candidate reference genes in each round of the analysis (Figure 2A and 2B), the pairwise variation V (Figure 2C) and ranking of the candidate reference genes from the least stable to the two most stable genes (Figure 3). The stability score M indicated that gene expression in spleen, jejunum and cerebellum were relatively less stable than other tissues (Figure 2A and B). However, all tissues tested exhibited high stabilities, as M values were less than 1.5, which was the default limit even when all eight genes were analyzed. According to pairwise variation V (Figure 2C), the two most stable genes were sufficient for a stable and valid reference for each tissue analyzed by qPCR because V2/3 values were less than 0.15 in all tissues. Jejunum was the most variable tissue with a V2/3 value of 0.139. Figure 3 shows ranking of gene expression stability based on M values. GAPDH, ACTB, SDHA and TBP had higher stability, while HPRT, rRNA and B2M were variable in most tissues. TBP in intestinal segments (jejunum, ileum and colon) and SDHA in brain segments (cerebrum, cerebellum and brain stem) were particularly stable. HPRT ranked as the worst of the eight genes in the 13 tissues tested.
Figure 2. Gene expression stability and pairwise variation of candidate reference genes using geNorm analysis.
(A) and (B): Average gene expression stability values M of the remaining reference genes during stepwise exclusion of the least stable gene in the different tissue panels are shown. Data are divided into two figures to avoid closely-packed lines. See also figure 3 for the ranking of genes according to their expression stability. (C) Pairwise variation analysis was used to determine the optimal number of reference genes for use in qPCR data normalization. The recommended limit for V value is 0.15, the point at which it is unnecessary to include additional genes in a normalization strategy.
Figure 3. Ranking of gene expression stability of candidate reference genes using geNorm analysis.
Candidate reference genes are ranked in order of stability for each tissue with the two most stable genes at the left and the least stable at the right.
Comparison of gene expression levels between human and common marmoset leukocytes
Subsequently, we analyzed gene expression levels of four CD antigens (CD3ε, CD4, CD8α, and CD20) and ten cytokines, interleukin (IL)-1β, IL-2, IL-4, IL-5, IL-6, IL-10, IL-12β, IL-13, interferon (IFN)-γ and tumor necrosis factor (TNF)-α, in peripheral blood leukocytes from humans and common marmosets (Figure 4). The sequences of primers specific for these immune-related genes are shown in Table 2. The normalization factor for common marmoset leukocytes was calculated using GAPDH and UBC based on the geNorm analysis as described above. For human leukocytes, we found that the expression of all eight genes were stable (M value = 0.363), of which ACTB and HPRT had the best score (M value = 0.163, V2/3 = 0.062) and were selected for use. The expression levels of CD4 and IL-4 were significantly lower in common marmosets than in humans while those of IL-10, IL-12β and IFN-γ were significantly higher in common marmosets compared with humans. Of interest, the expression level of IL-4 was notably lower in common marmosets than humans, and was close to the detection limit. There was no statistical difference in the expression levels of the other genes tested between common marmosets and humans.
Figure 4. The expression levels of CD antigens and cytokine genes in common marmoset and human leukocytes.
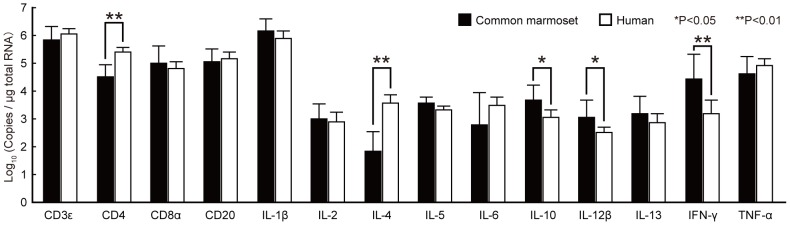
The expression level of each gene is shown as a logarithmic histogram of absolute copy numbers per µg of total RNA. Means and standard deviations of eight individuals are indicated. Asterisk indicates statistically significant differences between marmosets and humans by Student's t-test (*P value<0.05, **P value<0.01).
Difference of CD4/CD8 ratio between humans and common marmosets
We calculated ratios of the expression levels of CD4 to CD8 (CD4/CD8 ratio) in human and common marmoset leukocytes (Figure 5, left panel). CD4/CD8 ratios were significantly higher in human leukocytes compared with common marmoset leukocytes (mean ± sd, 0.59±0.22 vs. −0.49±0.41, P<0.01). To confirm the difference in CD4/CD8 ratios, we examined the proportion of CD4+ and CD8+ in CD3+ T cells by flow cytometric analysis. As shown in Figure 6, the rates of CD3+ cells in the lymphocyte gate were similar between common marmosets (30%) and humans (38%). However, the rates of CD4+/CD3+ cells and CD8+/CD3+ cells was 36% and 61% in common marmosets, respectively, and 75% and 21% in humans, respectively. Similarly, the CD4/CD8 ratio was markedly different between common marmosets and humans (mean ± sd, 0.56±0.08 vs. 3.22±0.35, P<0.01) by qPCR. This indicated a good correlation between the results from FACS analysis and that of qPCR analysis. To examine whether the CD4/CD8 ratio is affected by age, we further performed FACS analyses with PBMCs from young and old marmosets (Table 3). The result showed that the inverted CD4/CD8 ratio was fairly constant among individuals and over ages.
Figure 5. The expression ratios of CD8 to CD4 (CD8∶CD4) and Th1-related genes to Th2-related genes.
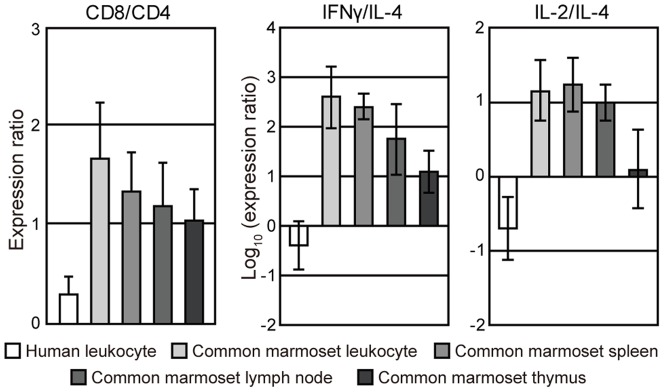
The ratio of CD8∶CD4 (left panel), IFN-γ∶IL-4 (middle panel) and IL-2∶IL-4 (right panel) in human and common marmoset leukocytes, spleen, lymph node and thymus are shown. Significant differences in the CD8∶CD4, IFN-γ∶IL-4 and IL-2∶IL-4 ratios were found between human leukocytes and common marmoset tissues (*P<0.05).
Figure 6. The ratio of CD4+ to CD8+ cells in common marmoset and human peripheral blood mononuclear cells (PBMCs) by flow cytometry.
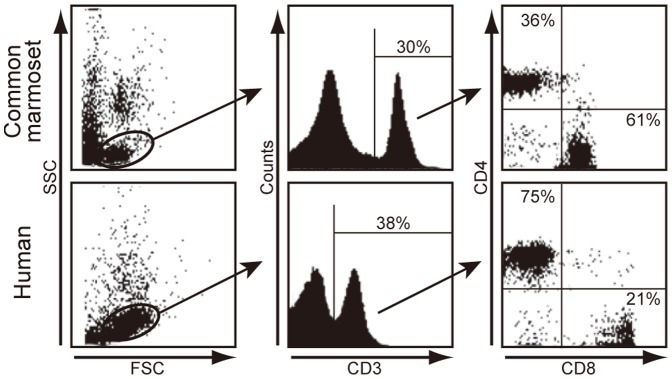
Representative scattered plots of FSC and SSC are shown in the left panels. Middle panels represent a histogram of CD3 analyzed in the lymphocyte gate. Gated CD3+ cells were analyzed for CD4 and CD8 expression (right panels).
Table 3. CD8/CD4 ratio in PBMCs from young and old marmosets.
| Age | Sex | % positive | CD8/CD4 ratio | |
| CD8 | CD4 | |||
| 3 month* | male | 58.3 | 38.4 | 1.52 |
| 1.5 year | female | 60.7 | 36.1 | 1.68 |
| 1.5 year* | male | 55.1 | 41.5 | 1.33 |
| 2.0 year | male | 52.7 | 44.6 | 1.18 |
| 10 year* | female | 58.6 | 37.8 | 1.55 |
| Mean ± sd | 57.1±3.2 | 39.7±3.4 | 1.45±0.20 | |
Only FACS analysis, but not qPCR, was done with PBMCs from these three marmosets.
Difference in T helper 1 (Th1)/T helper 2 (Th2) balance between humans and common marmosets
We compared the ratios of expression levels of Th1-related genes (IFN-γ or IL-2) and Th2-related genes (IL-4) (IFN-γ∶IL-4 or IL-2∶IL-4 ratio) (Figure 5, middle and right panels). Both logarithmic values of the IFN-γ∶IL-4 and IL-2∶IL-4 ratios were negative in human leukocytes whereas those of common marmoset leukocytes, spleen, lymph node and thymus indicated positive values, showing a clear difference in the Th1/Th2 balance between humans and common marmosets.
Discussion
In the present study, we evaluated the expression stability of common marmoset housekeeping genes in various tissues. To the best of our knowledge, this is the first report of a systematic evaluation of potential reference genes in common marmosets. We chose eight commonly used classical housekeeping genes. Of all genes tested, rRNA showed the most abundant expression and UBC showed the lowest expression. The UBC gene contains multiple directly repeated ubiquitin coding sequences (i.e., polyubiquitin precursor protein) [17]. However, the primer set we used enabled amplification of the unrepeated sequence at the 5′ region of the UBC gene only. Thus, low UBC expression in our data does not reflect the amount of ubiquitin C protein. B2M expression levels were markedly lower in brains and hearts than in other tissues. Resident brain cells normally express few or no MHC class I and B2M molecules [18]–[20]. In addition, B2M expression is upregulated by infection or autoimmune disease [21]–[23]. Therefore, in disorders with cellular infiltration such as inflammation (especially encephalitis) or cancer cell invasion, B2M expression levels may be significantly varied compared with normal tissue. Thus, we predict that B2M may be unsuitable as a reference gene in many cases.
We assessed gene expression stability using the geNorm applet. As shown in Figure 2, geNorm analysis indicated that all tested genes were stable in each tissue. However, there were some trends in the stability ranking (Figure 3). For example, TBP in intestine segments and SDHA in brain segments represented prominently high stabilities. GAPDH, ACTB, SDHA and TBP were generally ranked high followed by UBC. In contrast, the stability of rRNA was generally low. This suggests the amount of mRNA is not always proportional to that of total RNA as reported by other studies [24], [25]. In addition, HPRT, rRNA and B2M varied widely among tissues and rarely ranked high.
We analyzed the expression levels of CD antigens and cytokines by qPCR to compare the characteristics of peripheral blood leukocytes between common marmosets and humans (Figure 4). We observed that the expression levels of CD4 and IL-4 were lower in common marmosets than in humans. In contrast, the expression levels of IL-10, IL-12β and IFN-γ were higher in common marmosets. We calculated PCR efficiency of each primer set and found there was no great difference between primers for common marmosets and those for humans (Tables 1 and 2). Thus, the differences in the gene expression levels between common marmosets and humans are not attributable to the differences in PCR efficiency.
We also observed that the CD4∶CD8 ratio and Th1/Th2 balance were inverted in common marmosets by qPCR analysis (Figure 5). In particular, we confirmed the inverted CD4∶CD8 ratio by flow cytometric analysis (Figure 6 and Table 3). The inverted CD4∶CD8 ratio was stable over age. Of interest, we noted that the Th1/Th2 balance is different between common marmosets and humans, although we can only speculate on the cause of the difference. First, intestinal parasite infections may affect the Th1/Th2 balance by regulating expression of genes encoding cytokines [26]–[28]. In particular, protozoan parasites are potent stimulators of IFN-γ expression and Th1 responses [29]. Moreover, humans living in poor hygienic conditions in developing countries had higher Th1 cytokine levels compared with people in developed countries [30]. Although the common marmosets used in this study were maintained in specific pathogen-free conditions, we cannot rule out that such infectious agents may be one of a number of factors responsible for the difference in Th1/Th2 balance.
A second possible reason may be a difference in the number of cells producing the respective cytokines. As shown in Figure 6, the ratio of CD4+ to CD8+ cells were markedly different in total leukocytes from common marmosets and humans. Since IL-4 is mainly produced by CD4+ T cells [31], [32], its expression level may be influenced by the CD4∶CD8 ratio. However, this is not true for all the cytokines tested. For example, the expression levels of IL-2, IL-5 and IL-13, largely produced by T cells, were not significantly different between common marmosets and humans. Therefore, we suggest that the CD4∶CD8 ratio has little effect on Th1/Th2 balance. IL-10 is produced by T cells and monocytes [33] and IL-12β is naturally produced by dendritic cells and macrophages [34], [35]. However, we could not verify these cell numbers in the common marmoset. Further studies are required to determine whether the numbers of cytokine-producing cells influence the expression levels of IL-10 and IL-12β.
Another possibility is genetic variation. Bostik et al., reported distinct sequence differences in the promoter region or the proximal region of cytokine genes including IL-4, IL-10, IL-12β and TNF-γ among humans, macaque and mangabey monkeys, which affected regulation of cytokine synthesis [36]. Jeong et al., reported that the expression level of IL-4 was lower in monkeys (baboon and macaque) than in hominoids (human and chimpanzee) while the expression levels of IL-12β and the IFN-γ were higher in monkeys [37]. It is likely that Th1 dominant expression is common to primates other than hominoids and the difference in Th1/Th2 balance may be caused by genetic differences between common marmosets and humans.
The use of common marmoset is growing in popularity as a non-human primate model in many fields including autoimmune disease and infectious disease. In this study, we presented data regarding gene expression stabilities of common marmoset housekeeping genes and differences in the Th1/Th2 balance between common marmosets and humans. This difference may affect host defense and/or disease susceptibility, which should be carefully considered in biomedical research using common marmoset as an experimental model. We believe our data will contribute to future investigations using common marmoset models of various diseases.
Acknowledgments
We would like to acknowledge the efforts of Yasushi Ami in animal experiments. We also thank Ms. Hiro Yamada for technical assistance.
Funding Statement
This work was supported in part by Grants-in-Aid for Research on Emerging and Re-emerging Infectious Diseases from the Ministry of Health, Labor, and Welfare, Japan (Grants H20-shinkou-ippan-013 and H23-shinkou-ippan-010) as well as by Grant-in-Aid for Challenging Exploratory Research 23659237 from the Japan Society for the Promotion of Science. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Klug S, Neubert R, Stahlmann R, Thiel R, Ryffel B, et al. (1994) Effects of recombinant human interleukin 6 (rhIL-6) in marmosets (Callithrix jacchus). 1. General toxicity and hematological changes. Arch Toxicol 68: 619–631. [DOI] [PubMed] [Google Scholar]
- 2. Zühlke U, Weinbauer G (2003) The common marmoset (Callithrix jacchus) as a model in toxicology. Toxicol Pathol 31 Suppl: 123–127. [DOI] [PubMed] [Google Scholar]
- 3. Yaguchi M, Tabuse M, Ohta S, Ohkusu-Tsukada K, Takeuchi T, et al. (2009) Transplantation of dendritic cells promotes functional recovery from spinal cord injury in common marmoset. Neurosci Res 65: 384–92. [DOI] [PubMed] [Google Scholar]
- 4. Ando K, Maeda J, Inaji M, Okauchi T, Obayashi S, et al. (2008) Neurobehavioral protection by single dose l-deprenyl against MPTP-induced parkinsonism in common marmosets. Psychopharmacology (Berl) 195: 509–516. [DOI] [PubMed] [Google Scholar]
- 5. Genain CP, Lee-Parritz D, Nguyen MH, Massacesi L, Joshi N, et al. (1994) In healthy primates, circulating autoreactive T cells mediate autoimmune disease. J Clin Invest 94: 1339–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Genain CP, Hauser SL (1997) Creation of a model for multiple sclerosis in Callithrix jacchus marmosets. J Mol Med 75: 187–197. [DOI] [PubMed] [Google Scholar]
- 7. Bright H, Carroll AR, Watts PA, Fenton RJ (2004) Development of a GB virus B marmoset model and its validation with a novel series of hepatitis C virus NS3 protease inhibitors. J Virol 78: 2062–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Adams AP, Aronson JF, Tardif SD, Patterson JL, Brasky KM, et al. (2008) Common marmosets (Callithrix jacchus) as a nonhuman primate model to assess the virulence of eastern equine encephalitis virus strains. J Virol 82: 9035–9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mansfield K (2003) Marmoset models commonly used in biomedical research. Comp Med 53: 383–392. [PubMed] [Google Scholar]
- 10. Abbott DH, Barnett DK, Colman RJ, Yamamoto ME, Schultz-Darken NJ (2003) Aspects of common marmoset basic biology and life history important for biomedical research. Comp Med 53: 339–350. [PubMed] [Google Scholar]
- 11. Quint DJ, Buckham SP, Bolton EJ, Solari R, Champion BR, et al. (1990) Immunoregulation in the common marmoset, Calithrix jaccus: functional properties of T and B lymphocytes and their response to human interleukins 2 and 4. Immunology 69: 616–621. [PMC free article] [PubMed] [Google Scholar]
- 12. Fukuoka T, Sumida K, Yamada T, Higuchi C, Nakagaki K, et al. (2010) Gene expression profiles in the common marmoset brain determined using a newly developed common marmoset-specific DNA microarray. Neurosci Res 66: 62–85. [DOI] [PubMed] [Google Scholar]
- 13. Bustin SA, Nolan T (2004) Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech 15: 155–166. [PMC free article] [PubMed] [Google Scholar]
- 14. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, et al. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611–622. [DOI] [PubMed] [Google Scholar]
- 15. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kametani Y, Suzuki D, Kohu K, Satake M, Suemizu H, et al. (2009) Development of monoclonal antibodies for analyzing immune and hematopoietic systems of common marmoset. Exp Hematol 37: 1318–1329. [DOI] [PubMed] [Google Scholar]
- 17. Wiborg O, Pedersen MS, Wind A, Berglund LE, Marcker KA, et al. (1985) The human ubiquitin multigene family: some genes contain multiple directly repeated ubiquitin coding sequences. EMBO J 4: 755–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Drezen JM, Babinet C, Morello D (1993) Transcriptional control of MHC class I and beta 2-microglobulin genes in vivo. J Immunol 150: 2805–2813. [PubMed] [Google Scholar]
- 19. Lampson LA (1995) Interpreting MHC class I expression and class I/class II reciprocity in the CNS: reconciling divergent findings. Microsc Res Tech 32: 267–285. [DOI] [PubMed] [Google Scholar]
- 20. Daar AS, Fuggle SV, Fabre JW, Ting A, Morris PJ (1984) The detailed distribution of HLA-A, B, C antigens in normal human organs. Transplantation 38: 287–292. [DOI] [PubMed] [Google Scholar]
- 21. Kimura T, Griffin DE (2000) The role of CD8(+) T cells and major histocompatibility complex class I expression in the central nervous system of mice infected with neurovirulent Sindbis virus. J Virol 74: 6117–6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keskinen P, Ronni T, Matikainen S, Lehtonen A, Julkunen I (1997) Regulation of HLA class I and II expression by interferons and influenza A virus in human peripheral blood mononuclear cells. Immunology 91: 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mátrai Z, Németh J, Miklós K, Szabó Z, Masszi T (2009) Serum beta2-microglobulin measured by immunonephelometry: expression patterns and reference intervals in healthy adults. Clin Chem Lab Med 47: 585–589. [DOI] [PubMed] [Google Scholar]
- 24. Solanas M, Moral R, Escrich E (2001) Unsuitability of using ribosomal RNA as loading control for Northern blot analyses related to the imbalance between messenger and ribosomal RNA content in rat mammary tumors. Anal Biochem 288: 99–102. [DOI] [PubMed] [Google Scholar]
- 25. Valente V, Teixeira SA, Neder L, Okamoto OK, Oba-Shinjo SM, et al. (2009) Selection of suitable housekeeping genes for expression analysis in glioblastoma using quantitative RT-PCR. BMC Mol Biol 10: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bentwich Z, Weisman Z, Moroz C, Bar-Yehuda S, Kalinkovich A (1996) Immune dysregulation in Ethiopian immigrants in Israel: relevance to helminth infections? Clin Exp Immunol 103: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sacks D, Sher A (2002) Evasion of innate immunity by parasitic protozoa. Nat Immunol 3: 1041–1047. [DOI] [PubMed] [Google Scholar]
- 28. Sher A, Coffman RL (1992) Regulation of immunity to parasites by T cells and T cell-derived cytokines. Annu Rev Immunol 10: 385–409. [DOI] [PubMed] [Google Scholar]
- 29. Denkers EY, Gazzinelli RT (1998) Regulation and function of T-cell-mediated immunity during Toxoplasma gondii infection. Clin Microbiol Rev 11: 569–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Malhotra I, Ouma J, Wamachi A, Kioko J, Mungai P, et al. (1997) In utero exposure to helminth and mycobacterial antigens generates cytokine responses similar to that observed in adults. J Clin Invest 99: 1759–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Abbas AK, Murphy KM, Sher A (1996) Functional diversity of helper T lymphocytes. Nature 383: 787–793. [DOI] [PubMed] [Google Scholar]
- 32. Mosmann TR, Sad S (1996) The expanding universe of T-cell subsets: Th1, Th2 and more. Immunol Today 17: 138–146. [DOI] [PubMed] [Google Scholar]
- 33. de Waal Malefyt R, Abrams J, Bennett B, Figdor CG, de Vries JE (1991) Interleukin 10(IL-10) inhibits cytokine synthesis by human monocytes: an autoregulatory role of IL-10 produced by monocytes. J Exp Med 174: 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hsieh CS, Macatonia SE, Tripp CS, Wolf SF, O'Garra A, et al. (1993) Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science 260: 547–549. [DOI] [PubMed] [Google Scholar]
- 35. Macatonia SE, Hosken NA, Litton M, Vieira P, Hsieh CS, et al. (1995) Dendritic cells produce IL-12 and direct the development of Th1 cells from naive CD4+ T cells. J Immunol 154: 5071–5079. [PubMed] [Google Scholar]
- 36. Bostik P, Watkins M, Villinger F, Ansari AA (2004) Genetic analysis of cytokine promoters in nonhuman primates: implications for Th1/Th2 profile characteristics and SIV disease pathogenesis. Clin Dev Immunol 11: 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jeong AR, Nakamura S, Mitsunaga F (2008) Gene expression profile of Th1 and Th2 cytokines and their receptors in human and nonhuman primates. J Med Primatol 37: 290–296. [DOI] [PubMed] [Google Scholar]