Abstract
Harnessing community intelligence in knowledge curation bears significant promise in dealing with communication and education in the flood of scientific knowledge. As knowledge is accumulated at ever-faster rates, scientific nomenclature, a particular kind of knowledge, is concurrently generated in all kinds of fields. Since nomenclature is a system of terms used to name things in a particular discipline, accurate translation of scientific nomenclature in different languages is of critical importance, not only for communications and collaborations with English-speaking people, but also for knowledge dissemination among people in the non-English-speaking world, particularly young students and researchers. However, it lacks of accuracy and standardization when translating scientific nomenclature from English to other languages, especially for those languages that do not belong to the same language family as English. To address this issue, here we propose for the first time the application of community intelligence in scientific nomenclature management, namely, harnessing collective intelligence for translation of scientific nomenclature from English to other languages. As community intelligence applied to knowledge curation is primarily aided by wiki and Chinese is the native language for about one-fifth of the world’s population, we put the proposed application into practice, by developing a wiki-based English-to-Chinese Scientific Nomenclature Dictionary (ESND; http://esnd.big.ac.cn). ESND is a wiki-based, publicly editable and open-content platform, exploiting the whole power of the scientific community in collectively and collaboratively managing scientific nomenclature. Based on community curation, ESND is capable of achieving accurate, standard, and comprehensive scientific nomenclature, demonstrating a valuable application of community intelligence in knowledge curation.
Introduction
With the exponentially increasing volume of knowledge, harnessing community intelligence in knowledge curation has gained significant attention to deal with communication and education in the flood of scientific knowledge [1], [2]. A successful example that engages community intelligence in knowledge aggregation is Wikipedia (http://www.wikipedia.org), an online encyclopedia allowing any user to create/edit any content. Although the openness of editorial capacity to the community may lead to potential vandalism, it is reported that Wikipedia not only achieves more content coverage than BBC (British Broadcasting Corporation) and CNN (Cable News Network) combined [3] but also rivals the traditional Encyclopedia in accuracy [4], [5]. Spirited by the extraordinary success of Wikipedia, it has been advocated, for instance, in life sciences, that biological knowledge databases go wiki [6]. Meanwhile, leading voices in biological knowledge curation (e.g., gene annotation in model organism) published an article in Nature to elaborate the current state and future of knowledge curation; they stated that keeping biological knowledge up-to-date and comprehensive is increasingly lagging behind knowledge creation, inevitably requiring a large number of people getting involved in knowledge curation [7]. In other words, community curation is critical for biological knowledge management due to the burgeoning volume of biological knowledge and insufficient funding for dedicated curators [8]. As wiki features community-based knowledge curation, up-to-date content, and low cost for maintenance [9] – although there are limitations of using open wikis for knowledge management [10], more than a dozen biological knowledge wikis have been constructed to date [11]–[29], attempting to exploit the whole power of the scientific community for collective and collaborative knowledge curation [30], [31].
As knowledge is accumulated at ever-faster rates, a large collection of scientific nomenclature is concurrently developed or evolved in all kinds of fields. Scientific nomenclature, a particular kind of knowledge, is a system of terms used to name things and to describe processes/phenomenon in a particular discipline. Rapid advancements in science and technology further lead to a growing quantity of new scientific terms. Therefore, accurate translation of scientific nomenclature in different languages is of critical significance, not only for effectively facilitating communications and collaborations with English-speaking people, but also for efficiently disseminating knowledge among people in the non-English-speaking world, especially young students and researchers, and thus laying a solid foundation for domestic scientific activities. An accurate translation of a scientific term should be not only expressed equivalently in another language but also widely accepted by the scientific community. However, there are inaccurate and/or unacknowledged translations of scientific nomenclature from English to other languages, particularly for those languages that do not belong to the same language family as English [32]. Taking Chinese as an example, due to differences in language and culture between China and western countries, the English-to-Chinese scientific nomenclature lacks of accuracy and standardization [33]–[36], which can severely affect knowledge exchange and hinder the scientific activities.
To address this issue, here we propose for the first time the application of community intelligence in scientific nomenclature management, namely, harnessing collective intelligence for translation of scientific nomenclature from English to other languages. As community intelligence applied to knowledge curation is primarily aided by wiki and Chinese is the native language for about one-fifth of the world’s population (or over one billion people) [37], we put the proposed application into practice by developing a wiki-based English-to-Chinese Scientific Nomenclature Dictionary (ESND). ESND is a wiki-based, publicly editable and open-content platform, aiming to exploit the whole power of the scientific community in building standard and accurate scientific nomenclature.
Methods
ESND is built on wiki, which was first proposed by Ward Cunningham [38]. Briefly speaking, a wiki is an open website that allows users to add, modify, or delete its content via a web browser using a simplified markup language or a rich-text editor. As illustrated by Wikipedia and other wiki-based projects [39]–[41], wiki has several key features. First, it allows any user to create new pages or to edit any page (with customized permission control for editing), using the web browser without any extra add-ones. Second, it enables web contents to be written collectively and collaboratively by multiple users. Third, it builds a quite simple website, facilitating users to be involved in an ongoing process of creation and collaboration that constantly changes the website layout. Thus, wiki can significantly ease the process of knowledge creation, curation, and sharing. The wide adoption of community intelligence in knowledge management is to some extent attributable to free wiki software in aid of its implementation, such as MediaWiki (http://www.mediawiki.org), a free, open-source, and widely used wiki engine (e.g., adopted by Wikipedia). To develop ESND, we used MediaWiki (Version 1.19.1), MySQL (http://www.mysql.org; a free and popular relational database management system, Version 5.1.58), and PHP (http://www.php.net; a widely-used general-purpose scripting language, Version 5.2.17) on a Red Hat Enterprise Linux Server.
Results and Discussion
Community Curation of Scientific Nomenclature
ESND is a wiki-based, publicly editable, open-content platform for collaborative management of English-to-Chinese scientific nomenclature. Scientific nomenclature is a system of terms in different fields. In ESND, terms are extracted from the scientific literature and classified according to their corresponding fields. Currently, ESND primarily focuses on biological terms; it contains 2679 terms in life sciences, 770 terms in computer science and 135 terms in chemistry.
In ESND, each wiki page represents a specific term, which generally contains a collection of relevant information, including English abbreviation, English interpretation, Chinese interpretation, Chinese translation(s) and the corresponding detailed note(s) (Figure 1). Specifically, English and Chinese interpretations are definitions expressed in the two languages. Each term may contain more than one Chinese translation (detailed below) and the note provides a detailed explanation for the corresponding translation.
Figure 1. Screenshot of the term “Exon” in ESND (http://esnd.big.ac.cn/index.php/Exon).
It is likely that one English term may have multiple Chinese translations. In life sciences, for instance, there are two terms “exon” and “intron” that are often translated as “ ” (Pinyin: wài xiǎn zǐ and “
” (Pinyin: wài xiǎn zǐ and “ ” (Pinyin: nèi hán zǐ). However, recent studies have demonstrated that not all exons are encoded into proteins and not all introns are not transcribed, advocating that “
” (Pinyin: nèi hán zǐ). However, recent studies have demonstrated that not all exons are encoded into proteins and not all introns are not transcribed, advocating that “ ” (Pinyin: wài yuán zǐ) and “
” (Pinyin: wài yuán zǐ) and “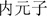 ” (Pinyin: nèi yuán zǐ) are more appropriate translations, respectively [42], [43]. Another example is “evolution”, which also has multiple translations: “
” (Pinyin: nèi yuán zǐ) are more appropriate translations, respectively [42], [43]. Another example is “evolution”, which also has multiple translations: “ ” (Pinyin: jìn huà) and “
” (Pinyin: jìn huà) and “ ” (yǎn huà). Nowadays, some people believe that it might be better translated as “
” (yǎn huà). Nowadays, some people believe that it might be better translated as “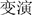 ” (Pinyin: biàn yǎn), since “
” (Pinyin: biàn yǎn), since “ ” (Pinyin: biàn) indicates mutation and “
” (Pinyin: biàn) indicates mutation and “ ” (Pinyin: yǎn) represents the process of natural selection.
” (Pinyin: yǎn) represents the process of natural selection.
To tackle this issue, ESND incorporates a “vote” function for each wiki page to engage community intelligence in dealing with multiple translations for a given English term. The number of votes indicates the recognition of the corresponding translation accepted by the scientific community. For the term “exon” as mentioned above, higher vote for a Chinese translation indicates that it is more acknowledged by the scientific community (Figure 1). Therefore, community-based votes effectively solve this issue by harnessing community intelligence in standardizing English-to-Chinese nomenclature. In addition, every term in ESND incorporates an open comment function as well as a corresponding discussion page, which can help users make comment and raise discussion for any debatable issue.
To facilitate adding new terms, ESND provides an open template for creating new wiki pages in correspondence to new terms. Users can adopt the template (http://esnd.big.ac.cn/index.php/template:AddNor) to easily add a new term. For example, when adding the term “Gene” (Figure 2), “Abbr” is the abbreviation of the term if available, “En” is its English interpretation, “Cn” is its Chinese interpretation, and “Category” is the field that the term belongs to.
Figure 2. Example for adding a term to ESND.

ESND is freely available at http://esnd.big.ac.cn, allowing any user to view and search for any content. Most important, ESND is a collaborative platform for collective management of English-to-Chinese scientific nomenclature, allowing people to participate in sharing their knowledge on specific terms of their own interest. If one term is not available, or incomplete, or you think of a better translation, you can share your knowledge by adding information to ESND. In addition, any user can make vote when there are multiple translations available for a given English term and thus the most voted translation indicates the wide acceptance by the scientific community. “With enough eyeballs, all bugs are shallow” [44]. Aided by wiki technology, ESND is a community-based dynamic resource of English-to-Chinese scientific nomenclature.
Future Developments and Perspective
Future developments include the addition of more scientific nomenclature from different fields and the improvement of user interface for term management. To our knowledge, it is for the first time that collective intelligence is applied for community management of scientific nomenclature and ESND is the first implementation to harnessing community intelligence for scientific nomenclature management. Based on community curation, ESND is capable of achieving accurate, standard, and comprehensive scientific nomenclature, demonstrating a valuable application of community intelligence in knowledge curation.
Funding Statement
This work was supported by grants from National Natural Science Foundation of China (60803050, 61132009), the “100-Talent Program” of Chinese Academy of Sciences (Y1SLXb1365), and National Programs for High Technology Research and Development (863 Program; Grant No. 2012AA020409), the Ministry of Science and chnology of the People’s Republic of China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Salzberg SL (2007) Genome re-annotation: a wiki solution? Genome Biol 8: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hu JC, Aramayo R, Bolser D, Conway T, Elsik CG, et al. (2008) The emerging world of wikis. Science 320: 1289–1290. [DOI] [PubMed] [Google Scholar]
- 3. McLean R, Richards BH, Wardman JI (2007) The effect of Web 2.0 on the future of medical practice and education: Darwikinian evolution or folksonomic revolution? Med J Aust 187: 174–177. [DOI] [PubMed] [Google Scholar]
- 4. Giles J (2005) Internet encyclopaedias go head to head. Nature 438: 900–901. [DOI] [PubMed] [Google Scholar]
- 5.Livemint website. Available: http://www.livemint.com/Politics/vgnLWa30gH8ng9GvfaW1MN/Jimmy-Wales-Wikipedia-rivals-traditional-encyclopedias-in.html. Accessed 2013 January 6.
- 6. Giles J (2007) Key biology databases go wiki. Nature 445: 691. [DOI] [PubMed] [Google Scholar]
- 7. Howe D, Costanzo M, Fey P, Gojobori T, Hannick L, et al. (2008) Big data: The future of biocuration. Nature 455: 47–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baker M (2012) Databases fight funding cuts. Nature 489: 19. [DOI] [PubMed] [Google Scholar]
- 9. Zhang Z, Cheung KH, Townsend JP (2009) Bringing Web 2.0 to bioinformatics. Brief Bioinform 10: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Finn RD, Gardner PP, Bateman A (2012) Making your database available through Wikipedia: the pros and cons. Nucleic Acids Res 40: D9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kumar S, Schiffer PH, Blaxter M (2012) 959 Nematode Genomes: a semantic wiki for coordinating sequencing projects. Nucleic Acids Res 40: D1295–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stokes TH, Torrance JT, Li H, Wang MD (2008) ArrayWiki: an enabling technology for sharing public microarray data repositories and meta-analyses. BMC Bioinformatics 9 Suppl 6S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoehndorf R, Bacher J, Backhaus M, Gregorio SE Jr, Loebe F, et al. (2009) BOWiki: an ontology-based wiki for annotation of data and integration of knowledge in biology. BMC Bioinformatics 10 Suppl 5S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McIntosh BK, Renfro DP, Knapp GS, Lairikyengbam CR, Liles NM, et al. (2012) EcoliWiki: a wiki-based community resource for Escherichia coli. Nucleic Acids Res 40: D1270–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Good BM, Clarke EL, de Alfaro L, Su AI (2012) The Gene Wiki in 2011: community intelligence applied to human gene annotation. Nucleic Acids Res 40: D1255–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Renfro DP, McIntosh BK, Venkatraman A, Siegele DA, Hu JC (2012) GONUTS: the Gene Ontology Normal Usage Tracking System. Nucleic Acids Res 40: D1262–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bolser DM, Chibon PY, Palopoli N, Gong S, Jacob D, et al. (2012) MetaBase–the wiki-database of biological databases. Nucleic Acids Res 40: D1250–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stehr H, Duarte JM, Lappe M, Bhak J, Bolser DM (2010) PDBWiki: added value through community annotation of the Protein Data Bank. Database (Oxford) 2010: baq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hodis E, Prilusky J, Martz E, Silman I, Moult J, et al. (2008) Proteopedia - a scientific ‘wiki’ bridging the rift between three-dimensional structure and function of biomacromolecules. Genome Biol 9: R121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, et al. (2011) Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res 39: D141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li JW, Robison K, Martin M, Sjodin A, Usadel B, et al. (2012) The SEQanswers wiki: a wiki database of tools for high-throughput sequencing analysis. Nucleic Acids Res 40: D1313–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cariaso M, Lennon G (2012) SNPedia: a wiki supporting personal genome annotation, interpretation and analysis. Nucleic Acids Res 40: D1308–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mader U, Schmeisky AG, Florez LA, Stulke J (2012) SubtiWiki–a comprehensive community resource for the model organism Bacillus subtilis. Nucleic Acids Res 40: D1278–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Csosz E, Mesko B, Fesus L (2009) Transdab wiki: the interactive transglutaminase substrate database on web 2.0 surface. Amino Acids 36: 615–617. [DOI] [PubMed] [Google Scholar]
- 25. Zhao D, Wu J, Zhou Y, Gong W, Xiao J, et al. (2012) WikiCell: a unified resource platform for human transcriptomics research. OMICS 16: 357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kelder T, van Iersel MP, Hanspers K, Kutmon M, Conklin BR, et al. (2012) WikiPathways: building research communities on biological pathways. Nucleic Acids Res 40: D1301–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mons B, Ashburner M, Chichester C, van Mulligen E, Weeber M, et al. (2008) Calling on a million minds for community annotation in WikiProteins. Genome Biol 9: R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huss JW, 3rd, Orozco C, Goodale J, Wu C, Batalov S, et al (2008) A gene wiki for community annotation of gene function. PLoS Biol 6: e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Orii N, Ganapathiraju MK (2012) Wiki-pi: a web-server of annotated human protein-protein interactions to aid in discovery of protein function. PLoS One 7: e49029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, Bajic VB, Yu J, Cheung K-H, Townsend JP (2011) Data Integration in Bioinformatics: Current Efforts and Challenges. In: Mahdavi MA, editor. Bioinformatics - Trends and Methodologies. Rijeka, Croatia: InTech - Open Access Publisher.
- 31.Groza T, Tudorache T, Dumontier M (2012) State of the art and open challenges in community-driven knowledge curation. J Biomed Inform doi: 10.1016/j.jbi.2012.11.007. [DOI] [PubMed]
- 32.Wikipedia website. Available: http://en.wikipedia.org/wiki/List_of_language_families. Accessed 2012 October 16.
- 33. Lu Y (2006) The Task of the Standardization and Unification for Scientific and Technological Terms in China is Still Heavy and the Goal Is Still Faraway (in Chinese). Acta Editologica 4: 241–242. [Google Scholar]
- 34. Li J (2000) Serveral questions about Scientific Nomenclature Designation (in Chinese). Chinese Science and Technology Terms Journal 2: 6–8. [Google Scholar]
- 35. Feng Z (2005) Science and Technology Terms-Its past and present (in Chinese). Terminology Standardization & Information Technology 2: 4–8. [Google Scholar]
- 36. Meng L (2011) Studies on Term Translation in the Terminological Perspective (in Chinese). Chinese Science & Technology Translators Journal 24: 28–30. [Google Scholar]
- 37.Wikipedia website. http://en.wikipedia.org/wiki/Chinese_language. Accessed 2012 October 16.
- 38.Leuf B, Cunningham W (2001) The Wiki way: Collaboration and sharing on the Internet. Addison-Wesley Professional 3–15.
- 39. Waldrop M (2008) Big data: Wikiomics. Nature 455: 22–25. [DOI] [PubMed] [Google Scholar]
- 40. Callaway E (2010) No rest for the bio-wikis. Nature 468: 359–360. [DOI] [PubMed] [Google Scholar]
- 41. Daub J, Gardner PP, Tate J, Ramskold D, Manske M, et al. (2008) The RNA WikiProject: community annotation of RNA families. RNA 14: 2462–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun N, Sun D, Zhu D (1990) Molecular Genetics (in Chinese). Nanjing: Nanjing University Press. 490 p.
- 43. Zhang H, Yi H (2007) A Discussion on Study for the Comparison of the Biological Terms between Chinese and Foreign Language (in Chinese). CHINA TERMINOLOGY 9: 49–50. [Google Scholar]
- 44.O’Reilly T (2005) What is Web 2.0: design patterns and business models for the next generation of software. O’Reilly Media.



