Abstract
Mammalian gene regulation is often mediated by distal enhancer elements, in particular, for tissue specific and developmental genes. Computational identification of enhancers is difficult because they do not exhibit clear location preference relative to their target gene and also because they lack clearly distinguishing genomic features. This represents a major challenge in deciphering transcriptional regulation. Recent ChIP-seq based genome-wide investigation of epigenomic modifications have revealed that enhancers are often enriched for certain epigenomic marks. Here we utilize the epigenomic data in human heart tissue along with validated human heart enhancers to develop a Support Vector Machine (SVM) model of cardiac enhancers. Cross-validation classification accuracy of our model was 84% and 92% on positive and negative sets respectively with ROC AUC = 0.92. More importantly, while P300 binding has been used as gold standard for enhancers, our model can distinguish P300-bound validated enhancers from other P300-bound regions that failed to exhibit enhancer activity in transgenic mouse. While GWAS studies reveal polymorphic regions associated with certain phenotypes, they do not immediately provide causality. Next, we hypothesized that genomic regions containing a GWAS SNP associated with a cardiac phenotype might contain another SNP in a cardiac enhancer, which presumably mediates the phenotype. Starting with a comprehensive set of SNPs associated with cardiac phenotypes in GWAS studies, we scored other SNPs in LD with the GWAS SNP according to its probability of being an enhancer and choose one with best score in the LD as enhancer. We found that our predicted enhancers are enriched for known cardiac transcriptional regulator motifs and are likely to regulate the nearby gene. Importantly, these tendencies are more favorable for the predicted enhancers compared with an approach that uses P300 binding as a marker of enhancer activity.
Keywords: Enhancer, Epigenomics, SVM, heart disease
1. Introduction
Eukaryotic transcription is intricately regulated at multiple levels including chromatin reorganization through epigenomic modifications and sequence specific binding of transcription factors (TF) to either proximal promoter or to distal enhancer/repressor regions of the gene.1,2 Enhancers can regulate their target genes from long distances, up to a megabase away and are especially important in regulating developmental and tissue-specific genes.3,4 Numerous genome wide association studies (GWAS) have revealed genomic loci associated with various human traits.5 Going from association to causality is however a major challenge, because a vast majority of GWAS signals lie in non-coding regions, often far from any gene, and our understanding of functional consequences of non-coding mutations is incomplete. It is possible that many of these associations are mediated via regulatory regions.6 By investigating putative polymorphic enhancers near GWAS signals, we might be able to identify the causal links between genetic variability and disease, at least in some cases. Thus, both for our fundamental understanding of transcriptional regulation as well as for interpretation of genotype-phenotype relationships, a comprehensive knowledge of context-specific enhancers is critical.
Large scale identification of enhancers is challenging because they do not have sufficiently discriminating sequence properties (except for their tendency to harbor homotypic binding motifs7) and their location is not restricted relative to the location of the target gene. Moreover, enhancers are often tissue and cell-type specific and are detectable only under the appropriate conditions. Recent revolution in sequencing technologies have triggered several large scale profiling of epigenomic marks and analysis of these marks have revealed strong associations between enhancers and specific epigenomic marks (either positive or negative8–10). Using genome-wide profiling of several epigenomic marks, Ernst et al. segmented the genome into 51 segment classes, where each segment class is defined by a specific combination of epigenomic marks.8,11 They designated two of these segment classes as strong and weak enhancers. Apart from epigenomic marks, histone acetylase P300 is known to bind to tissue-specific enhancers, with high rate of experimental validation using mouse transgenic.10,12 However, it is argued that while P300 may mark tissue-specific enhancers, those enhancers are not necessarily active in a specific context.13 This assertion is consistent with less than perfect validation rate of P300 bound regions as enhancers. Despite this, previous approaches to predict enhancers have used P300 bound regions as the gold standard to assess the methods prediction accuracy.14,15
Here we report an SVM trained specifically on 83 validated cardiac enhancers using four epigenomic profiles marks (H3K4me1, H3K27me3, P300 and DNase hypersensitivity) in human heart tissue. Our model achieves a cross-validation classification accuracy of 84% and 92% on positive and negative sets respectively. It was encouraging that our model can distinguish validated enhancers from those that were bound by P300 but failed to exhibit enhancer activity in transgenic mouse. Next, starting with a comprehensive set of 229 SNPs associated with cardiac phenotypes in 36 GWAS studies, we identified putative enhancers harboring SNPs in linkage disequilibrium (LD) with the GWAS SNP. We found that our predicted enhancers are enriched for binding sites for all known core cardiac transcriptional regulators GATA, MEF2, STAT, NF-AT, Nkx, and FOX. Using a novel approach we show that the predicted enhancers are likely to regulate the nearby gene. Our predicted enhancers uniquely point to a few genes highly relevant to the heart disease. Moreover, these tendencies of having enriched cardiac transcriptional motifs and likelihood of regulating nearby genes are more favorable for the predicted enhancers compared with an approach that uses P300 binding as a marker of enhancer activity. Overall, we show that a SVM model trained exclusively on validated enhancers performs better than those that use P300 binding as gold standard and that GWAS studies can be better interpreted in light of predicted polymorphic enhancers.
2. Results
2.1. SVM model for cardiac enhancers
2.1.1. Data
Heart tissue was chosen for our analysis because of the availability of both relevant epigenetic data (H3K4me1, H3K27me3, P300 and DNase hypersensitivity) and validated human enhancers. We collected 83 experimentally heart enhancers validated in mouse transgenic from VISTA browse and split them into 1kb regions (step size 500 bps) to be used as positive training set. Negative set was constructed by mixing random samples of 1 Kb long regions from the genome and randomly selected promoters. H3K4me1, H3K4me3, H3K27me3, P300 and DNase-I epigenetic markers, which have previously been shown to be associated with tissue-specific enhancers, were collected for the heart tissue from the GEO database. For each epigenetic mark we calculated its average signal strength across every 1 Kb genomic region as feature vector of the region. In order to normalize the feature vectors of the positive and negative set to zero mean and unit variance, we randomly sampled 40,000 1 Kb regions across the genome to estimate mean and variance of feature vector.
2.1.2. Training
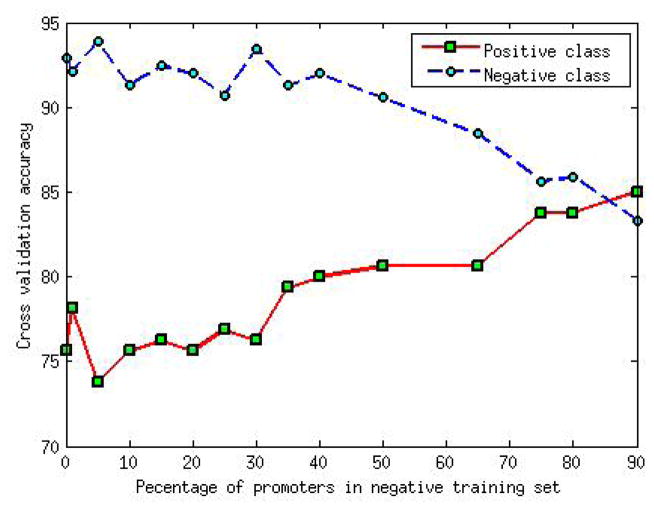
Epigenetic marks relevant to enhancers are relatively sparse in the genome. If the negative example in the training set only included random regions then SVM would choose subset of these inactive regions as its support vectors and would create a classifier hyperplane separating inactive regions from any epigenetically active region, resulting in high false positive rate. Therefore, in our negative set, in addition to random genomic regions, we added gene promoters as examples of epigenetically active non-enhancer regions. Figure 1 shows the effect of varying the proportion of promoters region in negative training set. In general, we found that a greater proportion of promoters in negative set improves positive set accuracy with relatively smaller decline in negative set accuracy, at least initially. This suggests that including a small fraction of promoters in the negative training set results in a better classification. Therefore, we constructed the negative training set by mixing 1000 random genomic regions and 250 randomly selected gene promoters.
Fig. 1.
Effect of variation of proportion of promoter region on accuracy of model. Two fold cross validation is used for positive set. Negative set accuracy is calculated by running the trained model on large number of random 1 kb genomic regions not including those used for training.
2.1.3. Testing
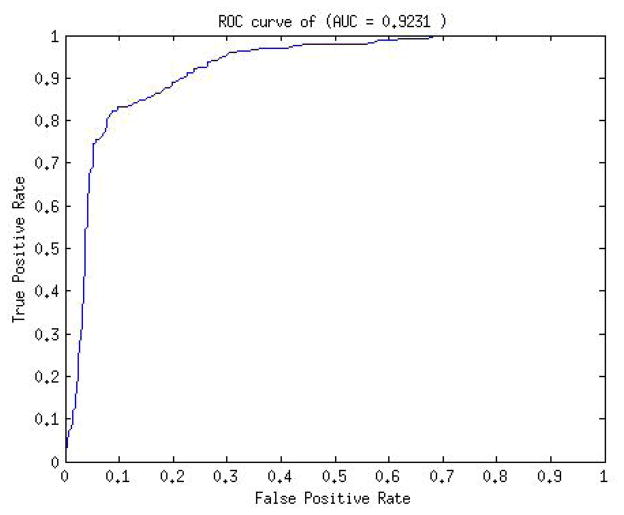
We used 5-fold cross validation for positive set accuracy estimate. For negative test set we randomly sampled 1000 1kb genomic regions. On performing grid search (see Methods) to train the SVM model the average testing classification accuracy on positive set was 84.1% and on negative set was 92%. The roc curve for the model prediction is shown in Figure 2. The AUC of the model was 0.9231.
Fig. 2.
ROC curve of SVM model
Despite some evidence to the contrary, a number of previous works have assumed P300-bound regions to be active enhancers and used them as gold standards to train and evaluate enhancer prediction tools. Next, we tested whether our model trained on validated enhancer and oblivious of P300 binding can nevertheless distinguish active and inactive P300-bound regions. We tested our model with 12 P300 peaks in human heart which were found not to have enhancer activity.16 Interestingly, the model classified 10(83%) of these cases as non-enhancers. Although based on a small set of examples, this suggests that our model can distinguish inactive P300-bound regions from active enhancers.
Narlikar et al.17 proposed a model based on specific motifs as features for cardiac enhancer identification. To compare performance of our model with their’s, 83 validated enhancers were separated into 60 training and 23 testing instances. SVM was trained on the 60 instances. We extracted the 1Mb regions flanking each of the 23 test enhancers and predicted enhancer in those genomic regions using the trained SVM. We first checked how well P300 can retrieve the validated enhancers. We found that there are only 69 P300 peaks in adult human heart in the 23 genomic regions, out of which only one overlapped with a validated enhancer. In other words, P300 peaks are poor predictor of enhancer activity in this context.
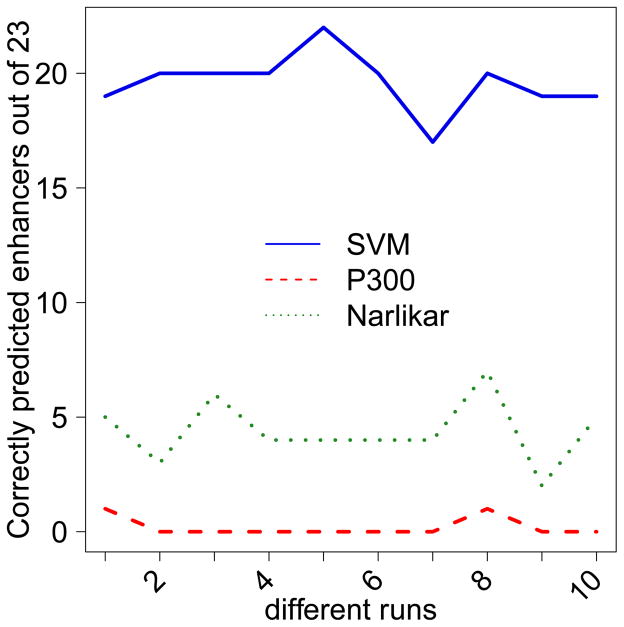
Using our trained SVM model we scored each 1 Kb region in the test set. Cardiac enhancer predicted in Narlikar et al. 17 are typically much shorter. For fair comparison with Narlikar et al. 17 (1) we extended each of their enhancer to 1 Kb region flanking the reported location, and (2) used a threshold on the enhancer score such that the predictions made by our SVM and the Motif based model cover almost the same number of enhancers (same basepair coverage as well due to extension) in the genomic test set. Among the 8522 enhancer regions predicted by the SVM, 21 of the 23 validated enhancers were included, while among 8551 enhancer regions predicted by Narlikar et al.17 only 13 were covered. we repeated the above comparison between our method, P300 peaks and Narlikar et. al. 10 times with different sets of 60 training and 23 testing instances out of total 83 enhancers. Figure 3 shows the number of enhancer predicted by each method across different iterations.
Fig. 3.
Number of enhancers (out of 23) predicted by SVM, P300 peaks and Narlikar et. al.
Taken together, these results suggest that the SVM model trained on epigenomic data is more suitable for identifying cardiac enhancers than are P300 binding or motif based models.
2.2. Identification of cardiac enhancers near SNPs associated with cardiac phenotypes
Next, we hypothesized that the causal variants underlying GWAS signals might lie within an enhancer element and affect gene regulation. We tested this hypothesis on SNPs associated with a variety of cardiomyopathies. Starting with NHGRI’s GWAS catalog,5 which includes 1332 studies revealing 6852 SNPs, we manually selected studies for cardiovascular disease traits. This yielded 229 SNPs from 36 studies. We then extended this seed SNPs set to include all other SNPs in Linkage Disequilibrium (LD) with a seed SNP using Broad Institutes SNAP server.18 We included all SNPs within 500kb from a seed SNP with r2 ≥ 0.3. The extended SNP were merged from the 1000 Genome Project and multiple HapMap releases (Consortium 2003; Consortium 2010). For each of the resulting 14233 SNPs, we scored 1kb flanking region using our SVM model to prioritize them as potential cardiac enhancers. Of all SNPs, the SVM scored 1054 as having enhancer probability ≥ 0.8. We found that distance of these enhancers from the corresponding GWAS SNP was significantly shorter than expected (Wilcoxon p-value = 3.9E-05).
2.3. Cardiac enhancers near cardiac GWAS SNPs are enriched for cardiac regulator motifs
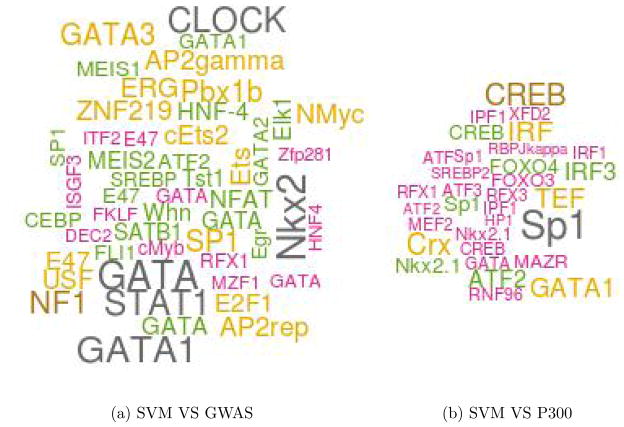
Cardiac transcription is primarily regulated by members of GATA, MEF2, STAT, NF-AT, Nkx, and FOX families of TFs.19–22 Next, we tested whether predicted enhancers near GWAS SNPs are enriched for known cardiac TF binding motifs. We first constructed three SNP sets: (1) eSNPs: comprised of the top 500 SNPs in LD with a GWAS SNP ranked by the SVM score, (2) pSNPs: the top 500 SNPs in the LD with a GWAS SNP ranked by mean P300 tag density (using bigwig summary tool from UCSC) in human heart, (3) gSNPs: The GWAS SNPs themselves. For each SNP we extracted the 1kb genomic flanking region resulting in three sets of sequences. For each sequence we determined the binding sites corresponding to 981 vertebrate motifs in TRANSFAC23 whose motif match score (using our own tool24) was in the top 95th percentile of scores achievable by that motif. We then determined the enriched motifs in one set of sequences relative to the other using Fisher Exact Test. Because enhancers have distinctive compositions which can bias motif enrichment, we normalized the two sequence sets for their GC composition via random sampling prior to motif enrichment analysis. When comparing SVM SNPs to the GWAS SNPs, 50 motifs were enriched with p-value ≤ 0.05, 11 of which corresponded to multiple representatives of GATA, STAT, NF-AT, Nkx families. When we compared the P300 SNPs with GWAS SNPs, among the 34 enriched motifs with GATA, Nkx and STAT families were represented by 4 motifs. Importantly, when we compare SVM SNPs directly to the P300 SNPs, GATA, FOX, MEF2 families of TF motifs were found to be enriched among the 32 enriched motifs. Figure 4 shows the top 50 motifs significantly enriched in SVM SNPs compared to GWAS SNPs or P300 SNPs. When we restrict the motif search to 20 bps flanking the SNP using same parameters, we still observe enrichment of NF-AT and STAT motifs in SVM SNPs relative to GWAS SNPs. However similar enrichment is also observed in P300 SNPs. It is possible that the SNP affect the formation of cis regulatory modules indirectly. Further investigation is required. In summary, all core cardiac TF families are enriched near eSNP loci, relative either to GWAS SNPs or to P300-bound regions. The overall conclusion was comparable when we used top 200 SVM scores and top 200 P300 score to be construct eSNP and pSNP sets. We note that because of small numbers, the p-values were modest and did not qualify a strict FDR threshold.
Fig. 4.
Significantly enriched motifs in SVM SNPs. The size of each TF label is proportionsal to its significance. For instance, the p-value for GATA1 in (a) is 0.001 and in (b) is 0.004. The largest p-value is 0.05.
2.4. Cardiac enhancers near cardiac GWAS SNPs are likely to regulate the nearby genes
Next we tested whether the predicted enhancers are likely to regulate genes. While enhancers can in principle regulate non-neighboring genes, a majority of them do regulate nearby genes,25 therefore, we focused only on the gene promoter closest to the SNP. For a SNP locus and a gene promoter, we estimated the likelihood of SNP locus to regulate the gene as the correlation between the DNase-I hypersensitivity (DHS) at the locus and the expression of the genes across 15 cell types in which DHS and RNA-seq was performed in parallel (see Methods); this approach to link a putative enhancer to a target genes is similar to Ref. 11. We constructed three comparison SNP sets. gSNP comprised of 229 GWAS SNPs. To construct eSNP set, we selected the SNP with highest SVM score in LD with each GWAS SNP as long as the SVM score was ≥ 0.8, resulting in 115 eSNP, all of which were intronic or intergenic. Similarly, to construct pSNP set, we selected the SNP with highest P300 mean tag density in LD with each GWAS SNP as long as the P300 tag density was ≥ 1, resulting in 58 pSNP. For each SNP we obtained the closest gene promoter. We then performed three pair-wise comparisons. For instance, when comparing eSNPs with gSNPs, we focused on genes that were closest to both an eSNP and a gSNP. Then we computed two DHS-expression correlations - between eSNP locus and the gene and between gSNP and the same gene. Given all such pairs of correlations we tested whether eSNP-gene correlation was greater than the gSNP gene correlation using paired one-side Wilcoxon test. We found that eSNP loci were more likely than gSNP loci to regulate the closest gene (based on 124 genes, p-value = 0.03), eSNP loci were more likely than pSNP loci to regulate the closest gene (based on 50 genes, p-value = 0.01), and pSNP loci were not more likely than eSNP loci to regulate the closest gene (based on 23 genes, p-value = 0.87). We also checked whether the distance of eSNPs from the closest gene promoter was shorter than that for gSNP or pSNP and we did not observe a statistical difference. The results suggest that SVM predicted enhancers are more likely to regulate the nearby genes relative to both the original GWAS SNPs and P300 predicted enhancers.
2.5. Genes near cardiac enhancers are enriched for cardiac function
Next we tested whether the genes uniquely closest to the eSNPs provide greater insight into the cardiovascular disease phenotype, relative to genes uniquely closest either to gSNPs or the pSNPs. We used the same criteria as above to obtain the closest gene lists, but unlike the expression analysis above we retained only the unique genes in each list. Unfortunately, the uniqueness requirement greatly reduced the number of genes with 94 for gSNP, 17 for eSNPs and only 2 for pSNPs. We then used ToppGene26 to compare enrichment of disease categories in the three gene lists. ToppGene uses three sources for disease ontology terms -GWAS, Comparative Toxicogenomics Database, and OMIM. We excluded GWAS to avoid circularity. As expected, the pSNP gene list did not show any enrichment. At FDR ≤ 0.05 the genes near gSNP also did not show enrichment for any disease term. The 17 genes in the eSNP list include NOS3 and MYH7. NOS3 alone showed enrichment for 2 terms - “Hypertension, Pregnancy-Induced” and “Coronary Vasospasm”. MYH7 alone was enriched for 5 distinct terms from OMIM database, all immediately related to myopathy or cardiomayopathy. The results are based on very limited dataset and one cannot draw general conclusion but they suggest that SVM can uniquely lead to genes directly relevant to the phenotype.
3. Conclusion
Here we present a SVM model for human cardiac enhancers based on four epigenomic marks H3K4me1, H3K27me3, DHS and P300, each of which have previously shown to be associated with enhancers in various cell types. While P300 is known to bind to tissue specific enhancers,12 and have been used as the gold standard for estimating accuracy of previous enhancer prediction approaches,14,15,17 many P300 bound regions fail to exhibit enhancer activity.12,13 Our SVM trained specifically on experimentally human cardiac enhancers validated in trangenic mouse, can not only predict other validated enhancers with high accuracy, it can also distinguish validated enhancers from the regions that were bound by P300 but failed to exhibit enhancer activity in transgenic mouse.
There are three prior approaches to predict enhancers. Narlikar et al. use clusters of known cardiac TF motifs as predictor of cardiac enhancers.17 Lee at al. train a SVM model based on genomic features based on cardiac P300 bound regions.14 Another SVM model for CD4+ T-cell enhancers based on epigenomic features, again, using P300-bound regions as the gold standard was proposed in.15 We have demonstrated the ability of our SVM model to distinguish between active and inactive P300 bound sites. Additionally, direct comparison of prediction accuracy on novel validated cardiac enhancers of our SVM model with that of P30014 and Narlikar et al.,17 explicitly shows that active enhancers have specific epigenomic properties not captured just by P300 binding or by clusters of putative binding sites. Genomic regions bound by P300 may not be active. Therefore, use of additional features add the tissue specific context to the model. Furthermore, kernel transformation of feature space used by SVM builds a non-linear classifiers. Thus it captures a greater variety of enhancers by recognizing a wider combination of epigenetic factors.
It has been previously suggested that a better knowledge of context-specific enhancers can help interpret GWAS signals.8 However, this reasonable assertion has not been tested explicitly on a specific disease area. Here we use our enhancer prediction tool to interpret GWAS studies related to cardiovascular phenotypes. We found an enrichment of high scoring cardiac enhancers near cardiac GWAS SNPs. Analysis of these putative enhancers suggest that (1) they are enriched for known core cardiac transcription factor binding sites, (2) they are likely to regulate nearby genes, and (3) they can uniquely point to certain genes involved with cardiac function and heart disease.
4. Methods
4.1. Correlating DNase Hypersensitivity and Gene Expression
To assess correlation of chromatin accessibility at a putative enhancer to expression level of a putative target gene, we extracted genome wide DHS as well as RNA-seq data from 15 cell types from a single study (GSE29692, GSE23316) representing a breadth of cell types HepG2, GM12878, A549, HeLa-S3, AG04450, BJ, NHLF, NHEK, HUVEC, h1-Hesc, HMEC, HSMM, K562, MCF-7, SK-N-SH RA. For the enhancer region we extracted the DHS tag density in each of the 15 cell types using bigWigSummary tool. Correspondingly, for the putative target genes we obtained the gene expression (RPKM) in the same set of cell types. We then estimated the pearson correlation between DHS and gene expression as an indicator of interaction between the enhancer and the gene.
4.2. More on SVM and grid search criteria
There are several references available for SVM.27–30 Here we give a brief review to appreciate our criteria for cross validation and grid search on the parameter space. In SVM, vector in original feature space is projected onto a higher dimensional feature space using kernel function (usually non-linear). Because of this the data which in original space is not linearly separable, becomes separable in transformed space, where the SVM tries to find a maximum margin hyperplane that separates the positive and negative set in the kernel space. SVM, employs a structural risk minimization (SRM) method31,32 to obtain the hyperplane, which tries to balance complexity of the model while minimizing the empirical risk. Therefore, relative to traditional methods based on empirical risk minimization, SVM is better suited to handle the problem of overfitting. SVM chooses a maximum margin hyperplane by identifying subset of training data (called support vectors), which would be closer to the optimal separating plane. Support vectors are cases which are most difficult to classify as positive or negative. Therefore to ensure good performance of SVM classifier, it is necessary to have a set of extreme examples (in both positive and negative example in the training set) that would qualify as support vectors.
Our positive training set included 330 (80% of 415) regions while the negative training set included 1000 regions. We weighted the positive and negative examples to accommodate for the difference in sizes. An exhaustive search over the weight space was conducted to obtain best possible cross-validation result. The weight used for negative and positive set respectively was 1 and 1.2. Furthermore, we defined our criteria for grid search based on the observation that randomly sampled negative set may contain enhancer regions and therefore, it is not desirable to minimize false positive rate to extreme. In addition, we required that difference between two rates is below a fixed threshold. This is equivalent to maximizing the F-score, while keeping difference of true positive (TP) and true negative (TN) rate below a fixed threshold.
Acknowledgments
This work is support by NIH grant R01GM085226 to S.H. and UMD-UMB seed grant to S.H. and C.C.
References
- 1.Maston GA, Evans SK, Green MR. Annu Rev Genomics Hum Genet. 2006;7:29. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- 2.White RJ. Nat Rev Genet. 2011;12:459. doi: 10.1038/nrg3001. [DOI] [PubMed] [Google Scholar]
- 3.Naranjo S, Voesenek K, de la Calle-Mustienes E, Robert-Moreno A, Kokotas H, Grigoriadou M, Economides J, Van Camp G, Hilgert N, Moreno F, Alsina B, Petersen MB, Kremer H, Gomez-Skarmeta JL. Human genetics. 2010;128:411. doi: 10.1007/s00439-010-0864-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E. Hum Mol Genet. 2003;12:1725. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 5.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Proc Natl Acad Sci U S A. 2009;106:9362. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaffney DJ, Veyrieras JB, Degner JF, Pique-Regi R, Pai AA, Crawford GE, Stephens M, Gilad Y, Pritchard JK. Genome Biol. 2012;13:R7. doi: 10.1186/gb-2012-13-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gotea V, Visel A, Westlund JM, Nobrega MA, Pennacchio LA, Ovcharenko I. Genome Res. 2010;20:565. doi: 10.1101/gr.104471.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ernst J, Kellis M. Nat Biotechnol. 2010;28:817. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zentner GE, Tesar PJ, Scacheri PC. Genome Res. 2011;21:1273. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birnbaum R, Clowney E, Agamy O, Kim M, Zhao J, Yamanaka T, Pappalardo Z, Clarke S, Wenger A, Nguyen L, et al. Genome Research. 2012;22:1059. doi: 10.1101/gr.133546.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Nature. 2011;473:43. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. Nature. 2009;457:854. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Proc Natl Acad Sci U S A. 2010;107:21932. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee D, Karchin R, Beer MA. Genome Res. 2011;21:2167. doi: 10.1101/gr.121905.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez M, Miranda-Saavedra D. Nucleic Acids Res. 2012;40:e77. doi: 10.1093/nar/gks149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.May D, Blow M, Kaplan T, McCulley D, Jensen B, Akiyama J, Holt A, Plajzer-Frick I, Shoukry M, Wright C, et al. Nature genetics. 2011 doi: 10.1038/ng.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narlikar L, Sakabe N, Blanski A, Arimura F, Westlund J, Nobrega M, Ovcharenko I. Genome research. 2010;20:381. doi: 10.1101/gr.098657.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. Bioinformatics. 2008;24:2938. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frey N, Olson EN. Annu Rev Physiol. 2003;65:45. doi: 10.1146/annurev.physiol.65.092101.142243. [DOI] [PubMed] [Google Scholar]
- 20.Hannenhalli S, Putt ME, Gilmore JM, Wang J, Parmacek MS, Epstein JA, Morrisey EE, Margulies KB, Cappola TP. Circulation. 2006;114:1269. doi: 10.1161/CIRCULATIONAHA.106.632430. [DOI] [PubMed] [Google Scholar]
- 21.Manukyan I, Galatioto J, Mascareno E, Bhaduri S, Siddiqui MA. J Cell Mol Med. 2010;14:1707. doi: 10.1111/j.1582-4934.2009.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlesinger J, Schueler M, Grunert M, Fischer JJ, Zhang Q, Krueger T, Lange M, Tonjes M, Dunkel I, Sperling SR. PLoS Genet. 2011;7:e1001313. doi: 10.1371/journal.pgen.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E. Nucleic Acids Res. 2006;34:D108. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levy S, Hannenhalli S. Mamm Genome. 2002;13:510. doi: 10.1007/s00335-002-2175-6. [DOI] [PubMed] [Google Scholar]
- 25.West AG, Fraser P. Hum Mol Genet. 2005;14(Spec No 1):R101. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- 26.Chen J, Bardes EE, Aronow BJ, Jegga AG. Nucleic Acids Res. 2009;37:W305. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burges C. Data mining and knowledge discovery. 1998;2:121. [Google Scholar]
- 28.Suykens J, Vandewalle J. Neural processing letters. 1999;9:293. [Google Scholar]
- 29.Boser B, Guyon I, Vapnik V. 1992;144 [Google Scholar]
- 30.Cristianini N, Shawe-Taylor J. An introduction to support Vector Machines: and other kernel-based learning methods. Cambridge Univ Pr; 2000. [Google Scholar]
- 31.Vapnik V. The nature of statistical learning theory. Springer-Verlag New York Inc; 2000. [Google Scholar]
- 32.Cortes C, Vapnik V. Machine learning. 1995;20:273. [Google Scholar]