Abstract
MicroRNAs (miRNAs or miR) have been integrated into tumorigenic programs as either oncogenes or tumor suppressor genes. The miR-124 was reported to be attenuated in several tumors, such as glioma, medulloblastoma and hepatocellular carcinoma. However, its role in cancer remains greatly elusive. In this study, we show that the miR-124 expression is significantly suppressed in human breast cancer specimens, which is reversely correlated to histological grade of the cancer. More intriguingly, ectopic expression of miR-124 in aggressive breast cancer cell lines MDA-MB-231 and BT-549 strongly inhibits cell motility and invasive capacity, as well as the epithelial–mesenchymal transition process. Also, lentivirus-delivered miR-124 endows MDA-MB-231 cells with the ability to suppress cell colony formation in vitro and pulmonary metastasis in vivo. Further studies have identified the E-cadherin transcription repressor Slug as a direct target gene of miR-124; its downregulation by miR-124 increases the expression of E-cadherin, a hallmark of epithelial cells and a repressor of cell invasion and metastasis. Moreover, knockdown of Slug notably impairs the motility of MDA-MB-231 cells, whereas re-expression of Slug abrogates the reduction of motility and invasion ability induced by miR-124 in MDA-MB-231 cells. These findings highlight an important role for miR-124 in the regulation of invasive and metastatic potential of breast cancer and suggest a potential application of miR-124 in cancer treatment.
Introduction
Breast cancer is the leading cause of cancer death in women with 400 000 deaths annually worldwide (1). It is derived from the sequential alteration of numerous oncogenes and tumor suppressors, such as HER2 (2), c-myc (3), BRCA1 (4) and so on. Like many other solid tumors, metastases are responsible for >90% of breast cancer-related mortality. Metastasis is a multistep process by which primary tumor cells invade adjacent tissue, enter the blood stream, survive in the circulation, extravasate into the surrounding tissue parenchyma and finally form clinically detectable metastases (5). Epithelial–mesenchymal transition (EMT) is thought to be necessary for the progression of benign tumor cells to invasive and metastatic cells or at least an alternative program.
EMT is a process in which adherent epithelial cells shed their epithelial traits and acquire mesenchymal properties, including fibroblastoid morphology, increased potential for motility and in the case of cancer cells, increased invasion, metastasis and resistance to chemotherapy (6,7). Though the signaling pathways are complex, the hallmark of EMT in cancer is the downregulation of E-cadherin, which is also thought to be a repressor of invasion and metastasis. A group of transcription factors have been demonstrated to be capable of orchestrating EMT programmes in cancer progression. These include direct transcriptional repressors of E-cadherin—Snail (SNAI1), Slug (SNAI2), ZEB2 (SIP1)—and others like Twist, ZEB1 and FOXC2 (8).
MicroRNAs (miRNA or miR) represent ~22 nucleotides non-coding RNAs and predominantly perform negative regulation of gene expression at post-transcriptional level through complementarity to the 3′ untranslated regions (3′-UTRs) of messenger RNAs (mRNAs) (9). Attributed to its multiple targets, a specific miRNA may present with diverse functional readouts (10). miRNAs have been proved to involved in almost every biological process, including cell proliferation, apoptosis, differentiation and stress response, exerting a finely tuned regulation of gene expression (11). Meanwhile, accumulated evidence indicates that miRNAs can also function as oncogenes or tumor suppressors and contribute to tumorigenesis by their targets (12). Recently, reports also indicate multiple functions of miRNAs in breast cancer metastasis. For example, miR-9 may initiate EMT and promote breast cancer cell metastasis by directly targeting E-cadherin (13); miR-373 and miR-520c promote breast cancer cell invasion and metastasis by suppression of CD44 (14). Most importantly, the miR-200 family members and miR-205 have been shown to reduce cell migration and invasiveness by targeting ZEB1 and ZEB2 transcription factors, known regulators of EMT (15–17). Therefore, miRNAs and classic genes comprise a complicated network, which plays a critical role in breast cancer progression.
MiR-124 is a brain-enriched miRNA that plays a crucial role in gastrulation and neural development (18,19). Recent reports further demonstrate that deregulation of miR-124 is related to carcinogenesis. The expression level of miR-124 is significantly decreased in glioma, medulloblastoma, oral squamous cell carcinomas and hepatocellular carcinoma (HCC), which suggest a potential tumor suppressive function of miR-124 (20–23). A previous study demonstrated that the upregulation of miR-124 in aggressive breast cancer cell lines suppresses metastasis-relevant traits in vitro (24). However, the function of miR-124 in breast cancer progression, especially its role in human breast cancer patients and in breast cancer mouse model, as well as the molecular mechanisms by which miR-124 exerts its functions and modulates the malignant phenotypes of breast cancer cells, has not been fully understood.
In this study, we demonstrated that miR-124 is pathologically downregulated in breast cancer specimens and cell lines, and that ectopic expression of miR-124 reduced cell motility and invasion capability in human breast cancer cells. In addition, the overexpression of miR-124 determines the epithelial phenotype of breast cancer cells, by targeting the EMT regulator Slug. Furthermore, miR-124 endows MDA-MB-231 cells with the ability to suppress cell colony formation in vitro and metastasis in vivo. Thus, our findings provide valuable clues toward understanding the mechanisms of human breast cancer metastasis and present an opportunity to develop more effective clinical therapies in the future.
Materials and methods
Tissue specimens
Breast cancer and adjacent normal tissue samples were obtained at the time of surgical resection and immediately frozen to −80°C until use. All samples with informed consent were collected from the Comprehensive Breast Health Center, Shanghai Rui-Jin Hospital of Shanghai Jiao Tong University School of Medicine between 2010 and 2011 and were histologically confirmed by staining with hematoxylin and eosin. Use of human tissues was approved by the research ethnics committee of Shanghai Jiao Tong University School of Medicine.
Cell lines and cell culture
Human breast cancer cell line MCF-7 was obtained from ATCC. Human breast cancer cell lines BT-549, BT-474, SK-BR-3, HCC1937 as well as an immortalized breast epithelial cell line MCF-10A were obtained from the cell bank of the Chinese Academy of Sciences (Shanghai, China). Human breast cancer cell lines MDA-MB-231, MDA-MB-468 and MDA-MB-435S were provided by Prof. Ming-Yao Liu (East China Normal University). MDA-MB-231 and MDA-MB-435S cells were cultured in Leibovitz L-15 medium (Gibco) with 10% fetal bovine serum (FBS; Gibco). An immortalized human embryonic kidney cell line HEK293T were cultured in Dulbecco’s modified Eagle’s medium (Hyclone) with 10% FBS. Another breast cancer cell line BT-549 was maintained in RPMI 1640 (Hyclone) with 10% FBS. The cell line MDA-231-D3H2LN (Xenogen, Alameda, CA) stably expressing firefly luciferase used in animal experiment was propagated in Minimum essential medium with Earl’s balanced salts solution (Hyclone) medium supplemented with 10% FBS, 1% non-essential amino acids (Hyclone) and 1% sodium pyruvate (Hyclone). All cells were fostered in a humidified atmosphere of 5% CO2 and 95% air except MDA-MB-231 and MDA-MB-435S cells, which were maintained at 100% air at 37°C.
RNA extraction and quantitative real-time PCR
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Complementary DNA was synthesized with the Prime-Script RT reagent kit (Promega, Madison, WI). Real-time PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) on an ABI 7900HT fast real-time PCR system (Applied Biosystems). Expression data were uniformly normalized to the internal control U6 and the relative expression levels were evaluated using the ΔΔCt method. Primers for reverse transcription–PCR are listed in Supplementary Table S1, available at Carcinogenesis Online.
Vector construction
In order to prove that miR-124 regulates the expression of human gene Slug (NM_003068) by directly targeting its 3′-UTR, the wild-type full-length 3′-UTR of Slug containing the three putative miR-124 binding sites was amplified from the genomic DNA using primer pairs Slug-UTR-F/R (Supplementary Table S1, available at Carcinogenesis Online) and then cloned into the downstream of the Renilla luciferase gene in the psiCHECK™-2 vector (Promega). Except the wild-type vector, mutant vectors containing four mutated bases on the separately predicted binding sites were constructed using the site-directed mutagenesis kit (Stratagene, La Jolla, CA) with primers Slug-UTR-mut-site1-F/R (Supplementary Table S1, available at Carcinogenesis Online), Slug-mut-UTR-site2-F/R (Supplementary Table S1, available at Carcinogenesis Online) and Slug-mut-UTR-site3-F/R (Supplementary Table S1, available at Carcinogenesis Online). The lentiviral expression vector pLVX-IRES-ZsGreen-miR-124 (Clontech Laboratories) was constructed to stably overexpressing mature sequence of miR-124 in MDA-231-D3H2LN. The 513bp genomic segment including the mature miR-124 sequence and its 289bp 5′ and 204bp 3′-flanking regions were amplified using primers miR-124-pLVX-F/R (Supplementary Table S1, available at Carcinogenesis Online) and subcloned into Xhol and BamHI sites of the pLVX-IRES-ZsGreen vector. The 994bp coding sequences of Slug was amplified from the complementary DNA template of MDA-MB-231 cells and cloned into pcDNA3.1 (-) (Invitrogen) by EcoR V and EcoR I.
Lentivirus production and transduction
Lentivirus was produced and harvested 72h after pLVX-IRES-ZsGreen-miR-124 transfection with the packaging plasmid pCMVΔ8.91 and pMD.G into 293T cells using FuGENE 6 Transfection Reagent (Roche Diagnostics GmbH, Mannheim, Germany ). After filtering through a 0.45 µm low protein binding-polysulfonic filter (Millipore, Bedford, MA) and concentrating with Optima™L-100 XP ultracentrifuge (Beckman Coulter), MDA-231-D3H2LN was infected with virus suspension.
Oligonucleotide transfection
Both miR-124 and negative control (NC) were synthesized by Genepharma (Shanghai, China). The sense sequence of miR-124 is 5ʹ-UAAGG- CACGCGGUGAAUGCC-3ʹ. Small interfering RNA duplex oligonucleotides targeting Slug mRNA including three segments, siSlug-1 (5ʹ-GCAUUUGCAGACAGGUCAAdTdT-3ʹ), siSlug-2 (5ʹ-GGACCACAGUGGCUCAGAAdTdT-3ʹ) and siSlug-3 (5ʹ-CUUCAAGGACACAUU-AGAAdTdT-3ʹ), were synthesized by Ribobio (Guangzhou, China). Oligonucleotide was transfected with Lipofectamine 2000 reagent (Invitrogen) at the concentration of 100nm.
Cell migration and invasion assays
Transwell migration assay and Matrigel invasion assay.
Cell migration and invasion ability were determined by Corning transwell insert chambers (8mm pore size; Corning) and BD BioCoat Matrigel Invasion Chamber (BD Biosciences, Bedford, MA), respectively. The chemoattractant was FBS. Cells were harvested and resuspended in serum-free medium after transfection. About 3 x 104 (migration assay) or 1 x 105 (invasion assay) prepared cells were added into the chamber and incubated for 24h at 37ºC. Cells that had migrated or invaded through the membrane were fixed with 20% methanol and stained with 0.1% crystal violet (Invitrogen), imaged and counted.
xCELLigence system with real-time technology.
Cell migration and invasion assays were also performed using the RTCA DP instrument (Roche Diagnostics GmbH), which was placed in humidified incubator. Cell migration or invasion was assessed using specifically designed 16-well plates (CIM-plate 16; Roche Diagnostics GmbH) with 8 μm pores. These plates are similar to conventional transwells with the microelectrodes located on the underside of the membrane of the upper chamber. The 10% FBS contained medium was added in the lower chamber, and cells were seeded into the upper chamber at 30 000 cells per well in serum-free medium. It is worth mentioning that for invasion assay, the CIM-Plate need to be precoated with 20 μl Matrigel (1:40; BD Biosciences) diluted by L-15 medium. Data analysis was carried out using RTCA software 1.2 supplied with the instrument.
Wound healing assays
Cells were grown to basically 100% confluence in 24-well plates after transfection and serum starvation. Its migration ability was assessed by measuring the movement of cells into a scraped, acellular area created by a sterile tip. The spread of wound closure was observed after 24h and photographed under a microscope. The fraction of cell coverage across the line represents for migration rate.
Colony formation assay
Five hundred MDA-231-D3H2LN cells were placed in complete growth media and allowed to grow until visible colonies formed in a fresh six-well plate (2 weeks). Cell colonies were fixed with cold methanol, stained with 0.1% crystal violet for 30min, washed, air dried, photographed and counted.
Luciferase assays
About 1 x 105 MDA-MB-231 cells or 2 x 105 HEK293T cells per well were seeded in 24-well plates for 24h before transfection. About 100ng of wild-type or mutant Slug 3′-UTR psiCHECK-2 plasmid (Promega) was transiently co-transfected with 60 pmol miR-124 mimic or NC into MDA-MB-231 cells or 293T cells using 1.44 µl Lipofectamine reagent (Invitrogen). Cell lysates were harvested 24h after transfection and then firefly and Renilla luciferase activities were measured by the Dual-Luciferase Reporter Assay System (Promega) on a Berthold AutoLumat LB9507 rack luminometer. Renilla luciferase activities were normalized to firefly luciferase activities to control for transfection efficiency.
Western blot analysis
For the protein expression analyses, standard western blotting was carried out. Whole cell protein lysates were electrophoresed on 10% sodium dodecyl sulfate–polyacrylamide gels and transferred onto polyvinylidene difluoride membranes (Millipore). The membranes were incubated with primary antibodies overnight at 4°C and then with the appropriate horseradish peroxidase-conjugated secondary antibody. The following antibodies were used: β-catenin antibody (9587; Cell Signaling, Beverly, MA), phosphor-β-catenin antibody (9561s; Cell Signaling), Slug (9585; Cell Signaling), Snail (3879s; Cell Signaling), Twist (ab 50887; Abcam, Cambridge, UK), ZEB1 (3396s; Cell Signaling), SIP1 (ab25837; Abcam), occludin (5446s; Cell Signaling), claudin-1 (BS 1063; Biworld, St Louis Park, MN), vimentin (5741; Cell Signaling), E-cadherin (4065; Cell Signaling), β-actin (CP01; Calbiochem, San Diego, CA), purified mouse-anti-fibronectin (610078; BD Biosciences, San Jose, CA)
Immunofluorescence analysis
MDA-MB-231 and BT-549 cells were transfected with miRNAs oligonucleotide, as described in the section of Oligonucleotide transfection, plated onto glass coverslips in 24-well plates and stained after 48h. For β-catenin, Slug and vimentin staining, cells were fixed in 4% paraformaldehyde, permeabilized in 0.1% Triton X-100 and probed with a rabbit-anti-β-catenin antibody (1:200; Cell Signaling), rabbit-anti-Slug antibody (1:400; Cell Signaling) or rabbit-anti-vimentin antibody (1:100; Cell Signaling) overnight. The primary antibody was detected using anti-rabbit-Alexa 594-conjugated antibodies (1:200; Invitrogen). To detect nuclei, cells were co-stained with 4′,6-diamidino-2-phenylindole (H-1200; Vector Laboratories). Fluorescence was observed and imaged using a Nikon Eclipse TE300 confocal laser microscope (Nikon Instruments, Melville, NY).
In vivo metastasis assays
MDA-MB-231 cells that had been engineered to stably express firefly luciferase (MDA-MB-231-D3H2LN) (Xenogen) were infected with lentiviruses carrying empty vector or miR-124 expression construct. For in vivo pulmonary metastasis assays, 5 x 105 MDA-MB-231-D3H2LN cells were suspended in 200 µl phosphate-buffered saline and injected into the lateral tail vein of 6-week-old female severe combined immunodeficiency mice. Two weeks after the injection of MDA-MB-231-D3H2LN cells, the lung metastasis burden was monitored weekly by bioluminescence imaging (BLI). Mice were anesthetized each time and given intraperitoneal injection of d-luciferin (150 μg/g body wt prepared in phosphate-buffered saline), and 10–15min after the injection, bioluminescence images were captured with a charge-coupled device camera (IVIS; Xenogen). Four weeks later, the mice were killed. Mice were manipulated and housed according to protocols approved by Shanghai Medical Experimental Animal Care Commission.
Immunohistochemistry
To visualize lung metastases, the mice were killed. The lungs were dissected and fixed in 4% paraformaldehyde before paraffin embedding. The tissue was then sliced as 4 µm sections and stained in hematoxylin and eosin. Immunohistochemical staining for human vimentin (5741S, Cell Signaling) was also performed on the lung sections. Images were captured at ×40 magnification, and then the number of metastases in the lung were counted and analyzed.
Statistical analysis
All data are presented as the mean ± SD; groups were compared using two-tailed Student’s t-test.
P values < 0.05 were considered significant.
Results
Downregulation of miR-124 in breast cancer
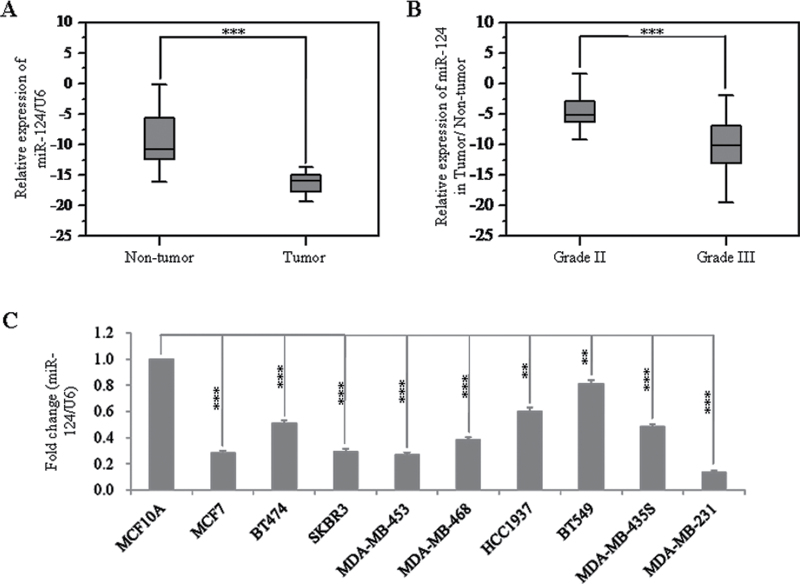
To investigate the role of miR-124 in initiation and progression of human breast cancer, we first compared the expression levels between clinical breast carcinomas and paired adjacent non-neoplastic tissues from 38 cases of breast cancer patients. By stem-loop quantitative reverse transcription–PCR, we showed that the expression levels of miR-124 were reduced in 37 of 38 cases of breast tumor specimens (P < 0.001), compared with those of adjacent non-neoplastic tissues (Figure 1A and Supplementary Table S2, available at Carcinogenesis Online). Furthermore, correlation analysis showed that the miR-124 expression level was reversely correlated to histological grade (P < 0.001), with lower expression in grade III (higher degree of malignancy) (Figure 1B and Supplementary Table S3, available at Carcinogenesis Online). Besides, we also found that the age of breast cancer patients was significantly associated with the expression level of miR-124 (Supplementary Table S3, available at Carcinogenesis Online).
Fig. 1.
Expression of miR-124 in breast cancer specimens and cell lines. (A) Comparison of the miR-124 abundance in 38 paired tumor and adjacent non-tumor tissues. Alteration of expression is shown as box plot presentations, with vertical axis indicating the relative expression level of miR-124 normalized to the internal control U6. The mean level of miR-124 expression in breast cancer tissues was significantly lower than that in non-tumor tissues (P < 0.001, independent t-test). (B) Expression levels of mature miR-124 between grade II and III in breast cancer tissues. The vertical axis represents the relative expression level of miR-124 in tumor tissue normalized to the adjacent non-tumor part. The mean level of miR-124 expression in breast cancer tissues with grade III was significantly lower than that in tissues with grade II (P < 0.001, independent t-test, grade II, n = 15; grade III, n = 15). (C) Expression level of mature miR-124 in MCF-10A and nine breast cancer cell lines. For all detected cell lines, fold changes of relative expression of miR-124 versus that of MCF-10A are represented in the vertical axis. Experiments were performed three times. Data are presented as means ± SE. The symbol ** and *** represents great significant difference (P < 0.01 and P < 0.001) by a two-tailed Student’s t-test.
We further evaluated the expression levels of miR-124 in breast cancer cell lines. The result showed that the expression levels of miR-124 evaluated by real-time PCR in all nine breast cancer cell lines were significantly reduced at different degrees compared with MCF-10A, an immortalized breast epithelial cell line (Figure 1C). In addition, the highly metastatic MDA-MB-231 cells showed the lowest expression level of miR-124. These results indicate that the reduced miR-124 expression was a frequent event in human breast cancer cells and tissues, which may be involved in breast carcinoma progression.
Exogenous overexpression of miR-124 suppresses the migration and invasion of breast cancer cell lines in vitro
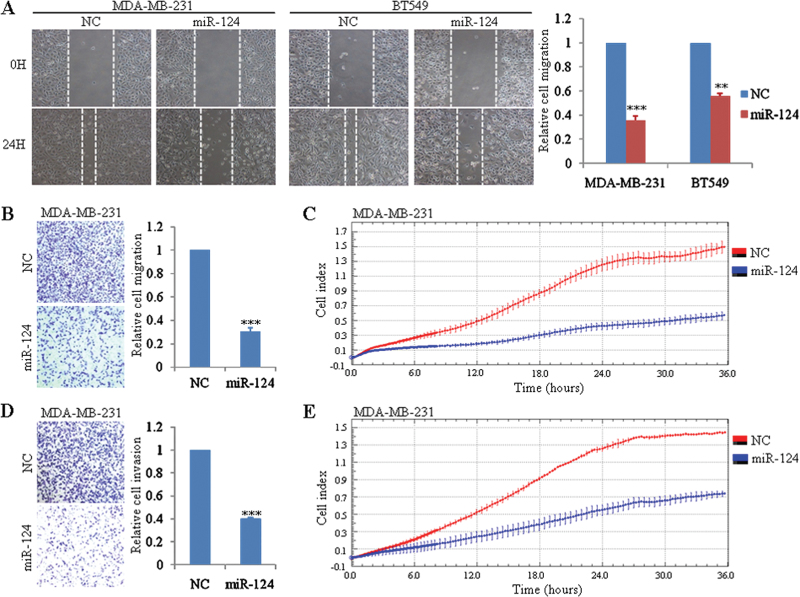
To define the function of clinically decreased miR-124 in the progression of breast cancer, we transiently transfected MDA-MB-231 cells, which shows the lowest expression of miR-124 in nine tested breast cancer cell lines, with miR-124 mimics or NC. The proliferation rate of cells with ectopic expression of miR-124 was not changed (Supplementary Figure S1A, available at Carcinogenesis Online). Considering MDA-MB-231 is a highly metastatic cell, we then investigated the effect of miR-124 on its migration and invasion capacity. The wound healing assay as well as migration chamber assay indicated that miR-124 overexpression can significantly inhibit cell migration compared with control group (Figure 2A and B). The same results were observed in other two tested breast cancer cell lines—BT-549 (Figure 2A, Supplementary Figure S1C, available at Carcinogenesis Online) and SK-BR-3 (Supplementary Figure S1B and C, available at Carcinogenesis Online). In addition, xCELLigence system with real-time technology allows us to observe the suppression of migration ability of miR-124 in MDA-MB-231 cells dynamically. Migration curves indicated that disparity between miR-124-transfected cells and control MDA-MB-231 cells was expanding with the extension of time (Figure 2C). Furthermore, invasion capability of MDA-MB-231 cells transfected with NC or miR-124 was evaluated by Matrigel invasion chamber assay. Indeed as expected, ectopic expression of miR-124 in MDA-MB-231 can strikingly decrease its invasion ability (Figure 2D). This was also confirmed by xCELLigence system using CIM-Plate precoated with Matrigel (Figure 2E).
Fig. 2.
Overexpression of miR-124 inhibits the migration and invasion of breast cancer cell lines in vitro. (A) Wound healing assay of MDA-MB-231 and BT-549 cells transfected with NC or mature miR-124 mimics. A scratch wound was made and images from five different fields were taken. Representative pictures of one field at the beginning (t = 0) and at the end of the recording (t = 24h) in each condition are shown. (B) Transwell migration assay of MDA-MB-231 cells transfected with NC or mature miR-124 mimics. Cell migration was analyzed 24h after seeding in Transwells. (C) Dynamic monitoring of cell migration using the xCELLigence system. MDA-MB-231 cells transfected with NC or mature miR-124 mimics were seeded to a CIM-Plate and subjected to a dynamic migration assay lasting for 36h. (D) Matrigel invasion assay of MDA-MB-231 cells transfected with NC and mature miR-124 mimics. Cell invasion was analyzed 24h after seeding in Transwells. (E) Dynamic monitoring of cell invasion using the xCELLigence system. MDA-MB-231 cells transfected with NC or mature miR-124 mimics were seeded to a CIM-Plate precoated with Matrigel and subjected to a dynamic invasion assay lasting for 36h. Error bars indicate ±SEM. The symbols ** and *** represent great significant difference (P < 0.01 and P < 0.001) by a two-tailed Student’s t-test.
Exogenous overexpression of miR-124 reverses EMT in breast cancer cell lines
When we transfected MDA-MB-231 cells with miR-124 mimics, an obvious morphological change from the spindle-shaped, mesenchymal form to a rounded or cobblestone-shaped, epithelial-like form with many cells aggregating together in groups was observed (Figure 3A). In addition, the same situation of morphological change was also observed in another tested breast cancer cell line BT-549 (Figure 3A). Accompanied with the apparent morphological changes, a set of protein markers related with EMT were also changed. Regardless of the MDA-MB-231 or the BT-549 cells, the epithelial marker E-cadherin showed robust upregulation and protein level of β-catenin was also increased to a certain extent, which was reversely correlated with the decrease of its phosphorylated form (Figure 3B). On the contrary, the expression levels of two mesenchymal markers (fibronectin and vimentin) decreased (Figure 3B). Above results were also supported by immunofluorescence staining. The expression of β-catenin was induced and localized to the plasma membrane in miR-124-transfected MDA-MB-231 cells, typical of the pattern observed in epithelial cells, whereas in miR-124-transfected BT-549 cells, we did not observe the cellular relocalization as well as the increase in β-catenin expression (Figure 3C). On the opposite, the mesenchymal marker vimentin expression was notably decreased and its intracellular distribution was changed (Figure 3C). The tight junction molecules such as claudin-1 and occludin were also studied and our results showed that the transient overexpression of miR-124 had no significant effect on the protein level of claudin-1 and occludin (Supplementary Figure S2C, available at Carcinogenesis Online). In summary, the ectopic expression of miR-124 in above mesenchymal cancer cells partially represses the EMT phenotype in breast cancer.
Fig. 3.
Exogenous overexpression of miR-124 reverses EMT in MDA-MB-231 and BT-549 cells. (A) Morphological changes of MDA-MB-231 and BT-549 cells transiently transfected with NC or mature miR-124 mimics. (B) Immunoblotting of epithelial and mesenchymal markers in MDA-MB-231 and BT-549 cells transiently transfected with NC or mature miR-124 mimics for 24 or 48h. β-Actin was used as a loading control. (C) Immunofluorescence analysis of β-catenin and vimentin in MDA-MB-231 and BT-549 cells transfected with NC or mature miR-124 mimics for 48h. 4′,6-Diamidino-2-phenylindole staining was used to detect nuclei and is merged with β-catenin and vimentin in their respective panels.
Furthermore, considering a group of transcription factors including direct transcriptional repressors of E-cadherin-Snail, Slug, SIP1 and others like Twist, ZEB1 are capable of orchestrating EMT programmes in cancer progression, therefore we also investigated the expression of the above transcription factors in the presence of ectopic miR-124 expression. Intriguingly, our results showed that with the transient miR-124 expression, the protein level of Slug decreased significantly, whereas the protein levels of other transcription factors had no significant changes (Supplementary Figure S2A and B, available at Carcinogenesis Online), which suggest a potential role of Slug in the miR-124-induced cell effects.
MiR-124 directly regulates the EMT mediator Slug
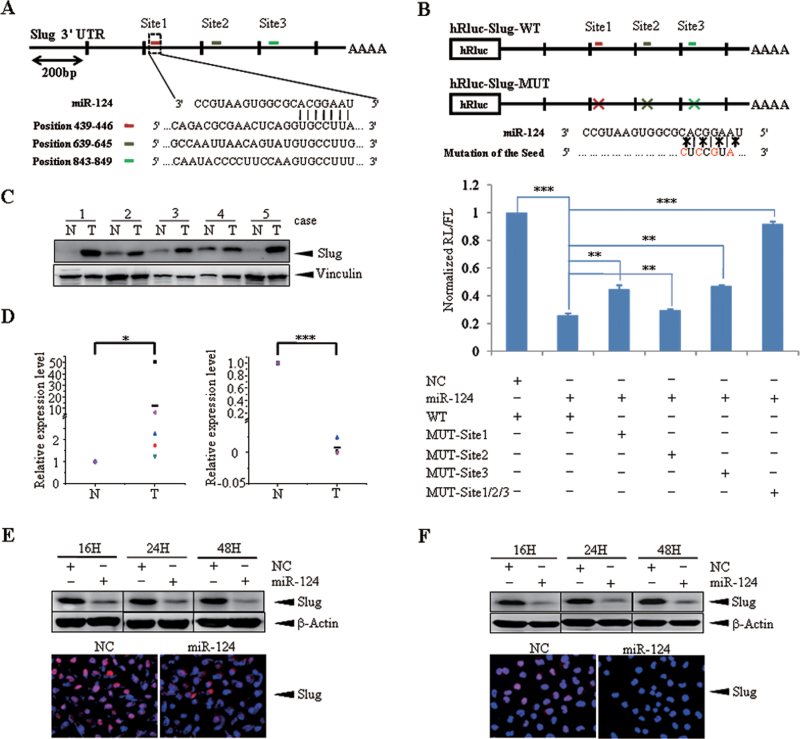
As well known, miRNAs execute post-transcriptional regulation by binding to the 3′-UTR of mRNAs. To investigate the target involved in the EMT regulation triggered by miR-124, we searched putative target genes using bioinformatics prediction software Targetscan (version 6.0, November 2011, Whitehead Institute for Biomedical Research) and 1654 conserved targets were predicted. Among these candidates, we focused on transcription factor Slug because of its high score (ranked ninth) and function related with EMT, most importantly, its expression is decreased with the ectopic expression of miR-124 (Supplementary Figure S2A, available at Carcinogenesis Online). In turn, there are three binding sites of miR-124 that is broadly conserved in vertebrates on Slug 3′-UTR (Figure 4A). Another prediction program, miRanda (August 2010 Release, Memorial Sloan-Kettering Cancer Center) verified this information. In order to prove that miR-124 directly targets Slug 3′-UTR, we performed luciferase reporter assay. The wild-type full-length 3′-UTR of Slug was cloned into the downstream of the Renilla luciferase gene in the psiCHECK™-2 vector with a firefly luciferase coding gene as internal control. Besides, mutant vectors containing four mutated bases on the predicted binding sites were constructed (Figure 4B, upper panel). MDA-MB-231 cells were transiently transfected with these constructs and miR-124 mimics or NC. MiR-124 mimics rather than NC significantly suppressed the luciferase activity of reporter genes containing 3′-UTR of Slug (Figure 4B, lower panel). Moreover, the inhibition was partially rescued when one of the binding sites was mutated or almost fully relieved with all sites mutated (Figure 4B, lower panel). The result was also confirmed in 293T cells (Supplementary Figure S3, available at Carcinogenesis Online). These results indicate that the effect of miR-124 is due to specific and direct interaction with the putative binding sites of the 3′-UTR of Slug.
Fig. 4.
Slug is a direct target of miR-124. (A) Schematic representation of 3′-UTR of Slug gene and three predicted binding sites of miR-124. (B) Upper panel: Schematic representation of the luciferase reporter constructs. The T7 RNA polymerase promoter drives constitutive transcription of a chimeric mRNA containing the Renilla luciferase coding sequence fused to the wide-type full-length Slug 3′-UTR (Slug-3′-UTR-WT) or to the same UTR mutated with four base pairs of the miR-124 seed sequences in three sites, respectively, or combined (Slug-3′-UTR-MUT). Lower panel: Relative activity of the luciferase gene fused with the wild-type or mutant 3′-UTR of Slug genes. The data are mean ± SEM of separate transfections (n = 3) and are shown as the ratio of Renilla luciferase activity to firefly. (C) Expression of Slug in five paired clinical breast cancer specimens. N and T are mean adjacent normal tissue and paired breast carcinoma, respectively. Vinculin serves as an internal control. Expression and correlation of miR-124 and Slug in paired clinical breast cancer samples. (D) The expression level of Slug was designated as the ratio of optical intensity between Slug and vinculin in each lane in Figure 4C, whereas the relative abundance of miR-124 of the same breast cancer samples were determined by stem-loop reverse transcription–PCR. For miR-124 and Slug, their expression level was assumed as 1 in adjacent normal tissues and corresponding calculated in paired tumor tissues. ‘−’ indicates the mean of miR-124 or Slug expression level. (E) Decreased expression of endogenous Slug due to miR-124 overexpression in MDA-MB-231 cells. Upper panel: Immunoblotting of endogenous Slug protein in MDA-MB-231 cells transfected with NC or miR-124 mimics for 16, 24 and 48h; β-Actin serves as loading control. Lower panel: Immunofluorescence assay was carried out when MDA-MB-231 cells was transfected with NC or miR-124 mimics for 48h. (F) Decreased expression of endogenous Slug due to miR-124 overexpression in BT-549 cells. Upper panel: Immunoblotting of endogenous Slug protein in BT-549 cells transfected with NC or miR-124 mimics for 16, 24 and 48h; β-Actin serves as loading control. Lower panel: Immunofluorescence assay was carried out when BT-549 cells was transfected with NC or miR-124 mimics for 48h. The symbol * denotes statistical difference (P < 0.05), whereas ** and *** represent great significant difference (P < 0.01 and P < 0.001) by a two-tailed Student’s t-test.
To further prove that Slug is a target gene of miR-124, MDA-MB-231 cells were transiently transfected with miR-124 mimics or NC. Notably, the expression of Slug substantially decreased after miR-124 transfection as early as 16h and arrived at its minimum level at 48h (Figure 4E, upper panel). Immunofluorescence also validated this point (Figure 4E, lower panel). Similarly, we also observed the same phenomenon in the BT-549 cells (Figure 4F). Moreover, there is a similar inverse correlation between the expression level of miR-124 and Slug in five tested clinical samples (Figure 4C and D). Collectively, these data support that Slug, an EMT-related transcription factor, was a direct target of miR-124.
Slug participates in miR-124 imposing EMT-related migration and invasion
To explore whether miR-124 exerts its function through its target gene Slug, we performed an RNA interference directed against Slug gene in MDA-MB-231 cells and evaluated the expression of Slug as well as EMT markers by western blot. The silencing of Slug was confirmed by western blot (Supplementary Figure S4B, available at Carcinogenesis Online). Importantly, consistent with the effect of miR-124, the suppression of slug changed the cell morphology from a spindle-shaped, mesenchymal form to a cobblestone-shaped, epithelial-like form (Supplementary Figure S4A, available at Carcinogenesis Online). Meanwhile with the highly efficient knockdown, the protein level of epithelial marker E-cadherin was increased, whereas the protein level of mesenchymal marker fibronectin was decreased to a certain extent (Supplementary Figure S4B, available at Carcinogenesis Online). Furthermore, we assayed migration and invasion ability of MDA-MB-231 cells transfected with small interfering RNA fragments using CIM-Plate precoated with Matrigel or not on xCELLigence system. In varying degrees, knockdown of Slug notably impaired the motility of cells (Supplementary Figure S4C and D, available at Carcinogenesis Online).
Next, to investigate contribution of Slug to EMT-related migration and invasion, we ectopically expressed Slug together with miR-124 in MDA-MB-231 cells to evaluate whether it may overcome the suppression effect of miR-124 on cell migration and invasion. MDA-MB-231 cells were co-transfected with NC or miR-124 together with pcDNA3.1 (-)-vector or pcDNA3.1 (-)-Slug for 48h. Overexpression of Slug rescued the morphological change caused by ectopic expression of miR-124 in MDA-MB-231 partially (Figure 5A). Western blot analysis revealed that mesenchymal markers (vimentin and fibronectin) positively regulated, whereas epithelial marker (E-cadherin and β-catenin) negatively regulated accompanied with restoration of Slug (Figure 5B). Migration and invasion assay were also carried out on xCELLigence system. Overexpression of Slug abrogated the reduction of migration and invasion ability caused by ectopic expression of miR-124 in MDA-MB-231 cells (Figure 5C and D). Altogether, the above results suggest that Slug is a functional target of miR-124 contributing to its role in EMT-related migration and invasion in MDA-MB-231 cells.
Fig. 5.
Overexpression of Slug impairs miR-124-induced inhibition of migration and invasion in MDA-MB-231 cells. (A) Morphology of MDA-MB-231 cells co-transfected with NC or miR-124 mimics together with either pcDNA3.1 (-)-vector or pcDNA3.1 (-)-Slug for 48h. Overexpression of Slug partially rescued the morphological change caused by ectopic expression of miR-124 in MDA-MB-231 cells. (B) Immunoblotting of Slug and EMT-related markers in MDA-MB-231 cells with the co-transfection of NC or miR-124 mimics together with either pcDNA3.1 (-)-vector or pcDNA3.1 (-)-Slug. (C) and (D) Dynamic monitoring of cell migration and invasion using the xCELLigence system. MDA-MB-231 cells co-transfected with NC or miR-124 mimics together with either pcDNA3.1 (-)-vector or pcDNA3.1 (-)-Slug. Overexpression of Slug impaired the reduction of migration (C) and invasion ability (D) caused by ectopic expression of miR-124 in MDA-MB-231 cells. Error bars indicate ±SEM. The symbol * denotes statistical difference (P < 0.05), whereas ** and *** represent great significant difference (P < 0.01 and P < 0.001) by a two-tailed Student’s t-test.
Ectopic overexpression of miR-124 reduces metastatic potential of breast cancer cells in vivo
All in vitro results prompted us to explore whether miR-124 overexpression can inhibit tumor metastasis in vivo. To answer this question, we chose an engineered cell line MDA-231-D3H2LN derived from breast cancer cell line MDA-MB-231 stably expressing luciferase as our cell model of in vivo studies. After tail vein injection, the cells localize at lung with blood circulation and the whole dynamic metastasis procedure can be observed by capturing fluorescence emitted from cells with bioluminescence imager. First, we constructed two cell lines with or without miR-124 overexpression by infecting MDA-231-D3H2LN cells with MDA-231-D3H2LN-miR-ZsGreen and MDA-231-D3H2LN-miR-124 lentivirus, respectively, and carried out a series of in vitro validation. By comparing the cell morphology of the two cell lines, we can easily find that with the overexpression of miR-124 (Supplementary Figure S5A, available at Carcinogenesis Online), MDA-231-D3H2LN cells changed from the spindle-shaped, mesenchymal form to a cobblestone-shaped, epithelial-like form (Supplementary Figure S5B, available at Carcinogenesis Online). In addition, epithelial markers (E-cadherin and β-catenin) were enhanced, whereas mesenchymal markers (vimentin and fibronectin) were decreased accompanied with the reduction of Slug protein caused by miR-124 overexpression, (Supplementary Figure S5C, available at Carcinogenesis Online). Also, the low expression of Slug induced the downregulation of vimentin and forced significant localization of β-catenin to cell membrane (Supplementary Figure S5D, available at Carcinogenesis Online). Furthermore, we tested the cell migration and invasion ability using CIM-Plate precoated with Matrigel or not on xCELLigence system. There was no doubt that MDA-231-D3H2LN cells stably expressing miR-124 had reduced migration and invasion capacity (Figure 6A). Tumor metastasis is a multistep process influenced with multifactors. Apart from migration and invasion, metastatic colonization is also a very important aspect, so we examined whether exogenous expression miR-124 can inhibit the colony formation of MDA-231-D3H2LN cells. Compared with the control group, the size and number of colony were reduced due to the overexpression of miR-124 (P < 0.05) (Figure 6B).
Fig. 6.
MiR-124 overexpression inhibits metastasis in vivo. (A) Dynamic monitoring of cell migration and invasion of MDA-231-D3H2LN cells infected with pLVX-ZsGreen vector or pLVX-ZsGreen-124 on xCELLigence system. Exogenous expression of miR-124 significantly impaired the migration and invasion ability of MDA-231-D3H2LN cells. (B) Influence of miR-124 on colony formation of MDA-231-D3H2LN cells. Representative dishes are presented (left panel). The number of clones was counted for each well of six-well plates and shown in the y-axis of the right panel. The results were reproducible in three independent experiments. (C) In vivo metastasis assays of MDA-231-D3H2LN cells with or without miR-124 overexpression. Pulmonary metastasis burden of xenografted animals was monitored weekly using BLI (middle, total flux). The BLI images of representative mice at the fourth week after injection are shown. The color scale depicts the photon flux emitted from the metastasis cells (left). Wet lung weights in tumor-bearing mice are measured. Bars correspond to mean ± SD. (D) Representative images of immunohistochemical staining for vimentin (left) and hematoxylin and eosin staining (right) of lungs isolated from miR-ZsGreen mice or miR-124-mice at ×40 and ×100 magnification. Visible micrometastases were counted and analyzed (five fields per group). The symbol * denotes statistical difference (P < 0.05), whereas ** represents great significant difference (P < 0.01) by a two-tailed Student’s t-test.
To further investigate the relationship between miR-124 and metastasis in vivo, MDA-231-D3H2LN-miR-ZsGreen or MDA-231-D3H2LN-miR-124 cells were transplanted into severe combined immunodeficiency mice via tail vein injection and lung metastasis is observed since the second week by bioluminescence imaging. Strikingly, at the fourth week after injection, the group of miR-ZsGreen was going through higher lung metastasis burden than the group of miR-124 (Figure 6C, left). Moreover, statistics show that the metastasis suppression caused by miR-124 overexpression was becoming more and more apparent (Figure 6C, middle). Parallel with the BLI results, lung weights of the miR-ZsGreen group showed obvious increase compared with the miR-124 group because of the pulmonary metastasis burden (Figure 6C, right). Immunostaining for human vimentin and hematoxylin and eosin staining of lungs showed that the number and size of micrometastases were significantly lower in miR-124 mice than miR-ZsGreen mice (P < 0.01) (Figure 6D). Taken together, these results demonstrated that miR-124 is capable of suppressing tumor metastasis in vivo.
Discussion
Global deregulation of miRNAs has been observed in various cancer tissues from numerous profile data (25,26). The characteristic pattern of miRNAs expression is well correlated with different stages of human tumors (27). Moreover, these small non-coding RNAs can function as oncogenes or tumor suppressor genes and contribute to tumorigenesis and progression (12). In this study, we showed that the expression level of miR-124 was significantly lower in human breast cancer tissues than that of adjacent normal tissues. We further analyzed whether the expression level of miR-124 is correlated with clinical characteristics of 38 breast cancer patients, such as tumor size, histological grade, TNM (T describes the size of the tumor and whether it has invaded nearby tissue, N describes regional lymph nodes that are involved, M describes distant metastasis) stage and so on. Our analytic results showed that the expression level of miR-124 was reversely correlated to histological grade as well as the age of this cohort of patients (Supplementary Table S3, available at Carcinogenesis Online). In functional studies, reintroduction of miR-124 dramatically repressed the migration and invasion of breast cancer cells in vitro and tumor metastasis in vivo. Moreover, this study has identified Slug as the direct target of miR-124 in breast cancer cells; its downregulation by miR-124 increases the expression of E-cadherin, which is a hallmark of epithelial cells and also a repressor of cell invasion and metastasis. These findings suggest that miR-124 plays an important role in the invasive and/or metastatic potential of breast cancer.
It is well known that invasion and metastasis, two of the most important hallmarks of malignant tumors, are the prominent fatal factors for human cancers. EMT is thought to be a key step in the progression of tumors toward invasion and metastasis. EMT is implicated in physiological processes, such as embryonic development and wound healing, and in pathological processes, such as the progression of early-stage non-invasive tumors to invasive malignancies and fibrosis. EMT can be induced in vitro by exposing normal and neoplastic epithelial cells to various growth factors, including transforming growth factor-β, platelet-derived growth factor and hepatocyte growth factor (8,12,28). Downstream of these growth factors and their receptors lie a panel of transcriptional factors. These transcriptional factors include the zinc-finger proteins (Snail and Slug), the forkhead box proteins (FOXC1 and FOXC2), the basic helix-loop-helix protein (Twist) and the zinc-finger, E-box-binding proteins (ZEB1 and ZEB2).
Our findings emphasize the importance of miRNA-triggered downregulation of transcriptional factors related to EMT during cancer cell invasion and metastasis and are in concordance with those recently reported by Park et al. (16) and Gregory et al. (17), which underline the role of miR-200 family and miR-205 targeting ZEB1 and ZEB2. Our study suggests that miR-124, through downregulation of its target Slug, may contribute to the establishment and maintenance of the epithelial phenotype, thus significantly modifying the migration and invasion behavior of aggressive mesenchymal breast cancer cells like MDA-MB-231 cells.
Notably, although miR-124 robustly increases the expression of E-cadherin, which is a hallmark of epithelial cells, it downregulates the expression levels of two mesenchymal markers (fibronectin and vimentin) in MDA-MB-231 and BT-549 cells. However, the regulation of β-catenin and vimentin in BT-549 cells are small compared with MDA-MB-231 cells. We extrapolated that besides Slug, which have been proved to be downregulated by miR-124, other transcriptional factors such as Twist and Snail could also have some influence on the expression of β-catenin and vimentin. The expression levels of these transcription factors in these two cell lines are different. For example, the expression of Twist can hardly be detected in MDA-MB-231 cells, whereas its expression level is quite high in BT-549 cells (Supplementary Figure S2A, available at Carcinogenesis Online). Our results also showed that the transient overexpression of miR-124 had no significant effect on the protein level of two tight junction molecules claudin-1 and occludin (Supplementary Figure S2C, available at Carcinogenesis Online), suggesting that miR-124 partially reverses EMT in breast cancer cell lines. Other transcription factors such as Snail could also be necessary for the complete reverse of EMT in these breast cancer cells.
Slug, as well as another member of Snail family, Snail, has been associated with EMT during both embryonic development and cancer metastasis (29,30). It can directly repress the transcription of E-cadherin, which is thought to be a repressor of invasion and metastasis, as well as a subset of genes that encode cadherins, claudins and cytokeratins to induce EMT. Slug has also been reported to be more relevant for generating breast cancer cells with cancer stem cell phenotype than Snail (31). The Slug gene is located on human chromosome 8q11.21. Comparative genomic hybridization has shown an amplification of this region in many types of cancer (32). Slug is a labile protein with a short half-life. It can be regulated by mouse double minute 2 (MDM2)-mediated ubiquitination and degradation in a p53-dependent manner (33). Although mutant p53 represses MDM2 expression and stabilizes Slug protein, which induces EMT, this study has provided another mechanism to regulate the protein level of Slug in breast cancer cells. We confirmed that miR-124 directly targets Slug by binding its 3′-UTR. Therefore, downregulation of miR-124 may contribute to the increased expression of Slug in post-transcription level and in turn facilitate breast carcinogenesis and progression.
Recent studies indicated the abnormal expression and pathological significance of miR-124 in various human cancers like HCC, anaplastic astrocytomas and glioblastoma multiforme. The decreased expression of miR-124 in carcinomas may result from multiple regulatory events, including the methylation of CpG islands. DNA methylation of miR-124 was first reported by Lujambio et al. (34) in colon, breast and lung cancer, as well as in leukemia and lymphoma (35). Subsequent studies confirmed frequent miR-124 methylation in leukemia affecting clinical outcome and additionally showed frequent miR-124 methylation in gastric cancer, cervical cancer and HCC (21,36,37). Most recently, Lv et al. (24) reported that miR-124 could be downregulated by hypermethylation in its promoters in MDA-MB-231 cells; however, they did not measure the expression level as well as the methylation situation of miR-124 genes in human breast cancer patients, therefore, future studies should be conducted to understand the mechanism of its downregulation in breast cancer.
In conclusion, our observations imply that the loss of miR-124 may result in gained expression of Slug, which endows breast cancer cells with the ability of improved migration and invasion capacity in vitro and metastasis in vivo and in turn favors tumor progression. We may reasonably speculate that the restoration of miR-124 activity may represent an attractive strategy for breast cancer therapy.
Supplementary material
Supplementary Table S1–S3 and Figures S1–S5 can be found at http://carcin.oxfordjournals.org/
Funding
National Key Program (973) for Basic Research of China (2009CB918404); National Science Foundation of China (90813034, 81172521 and 81170505); ‘Shu Guang’ project by Shanghai Municipal Education Commission and Shanghai Education Development Foundation (09SG18) and SMC Program of Shanghai Jiao Tong University.
Supplementary Material
Acknowledgements
The authors thank Prof. Ming-Yao Liu (East China Normal University) for providing the human breast cancer cell lines MDA-MB-231, MDA-MB-435S and MDA-MB-468 cells.
Conflict of Interest Statement: The authors declare no conflict of interests.
Glossary
Abbreviations:
- BLI
bioluminescence imaging
- EMT
epithelial–mesenchymal transition
- FBS
fetal bovine serum;
- HCC
hepatocellular carcinoma
- miRNA
microRNAs
- miR-124
miRNA-124
- mRNA
messenger RNAs
- NC
negative control
- UTR
untranslated region.
References
- 1. Gonzalez-Angulo A.M., et al. (2007). Overview of resistance to systemic therapy in patients with breast cancer. Adv. Exp. Med. Biol., 608, 1–22 [DOI] [PubMed] [Google Scholar]
- 2. Slamon D.J., et al. (1987). Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science., 235, 177–182 [DOI] [PubMed] [Google Scholar]
- 3. Varley J.M., et al. (1987). Alterations to either c-erbB-2(neu) or c-myc proto-oncogenes in breast carcinomas correlate with poor short-term prognosis. Oncogene., 1, 423–430 [PubMed] [Google Scholar]
- 4. Ford D., et al. (1995). Estimates of the gene frequency of BRCA1 and its contribution to breast and ovarian cancer incidence. Am. J. Hum. Genet., 57, 1457–1462 [PMC free article] [PubMed] [Google Scholar]
- 5. Valastyan S., et al. (2011). Tumor metastasis: molecular insights and evolving paradigms. Cell., 147, 275–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Taube J.H., et al. (2010). Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl. Acad. Sci. U.S.A., 107, 15449–15454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Singh A., et al. (2010). EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene., 29, 4741–4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Polyak K., et al. (2009). Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat. Rev. Cancer., 9, 265–273 [DOI] [PubMed] [Google Scholar]
- 9. Bartel D.P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell., 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 10. Valastyan S., et al. (2009). A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell., 137, 1032–1046 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11. Marilena V, et al. (2012). microRNA involvement in human cancer. Carcinogenesis, 33, 1126–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen C.Z. (2005). MicroRNAs as oncogenes and tumor suppressors. N. Engl. J. Med., 353, 1768–1771 [DOI] [PubMed] [Google Scholar]
- 13. Ma L., et al. (2010). miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol., 12, 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang Q., et al. (2008). The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat. Cell Biol., 10, 202–210 [DOI] [PubMed] [Google Scholar]
- 15. Burk U., et al. (2008). A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep., 9, 582–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park S.M., et al. (2008). The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev., 22, 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gregory P.A., et al. (2008). The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol., 10, 593–601 [DOI] [PubMed] [Google Scholar]
- 18. Lee M.R., et al. (2010). miR-124a is important for migratory cell fate transition during gastrulation of human embryonic stem cells. Stem Cells., 28, 1550–1559 [DOI] [PubMed] [Google Scholar]
- 19. Cheng L.C., et al. (2009). miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci., 12, 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xia H., et al. (2012). Loss of brain-enriched miR-124 microRNA enhances stem-like traits and invasiveness of glioma cells. J. Biol. Chem., 287, 9962–9971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hunt S., et al. (2011). MicroRNA-124 suppresses oral squamous cell carcinoma motility by targeting ITGB1. FEBS Lett., 585, 187–192 [DOI] [PubMed] [Google Scholar]
- 22. Furuta M., et al. (2010). miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis., 31, 766–776 [DOI] [PubMed] [Google Scholar]
- 23. Zheng F., et al. (2012). The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut., 61, 278–289 [DOI] [PubMed] [Google Scholar]
- 24. Lv X.B., et al. (2011). miR-124 suppresses multiple steps of breast cancer metastasis by targeting a cohort of pro-metastatic genes in vitro . Chin. J. Cancer., 30, 821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Calin G.A., et al. (2006). MicroRNA signatures in human cancers. Nat. Rev. Cancer., 6, 857–866 [DOI] [PubMed] [Google Scholar]
- 26. Iorio M.V., et al. (2012). MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med., 4, 143–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lu J., et al. (2005). MicroRNA expression profiles classify human cancers. Nature., 435, 834–838 [DOI] [PubMed] [Google Scholar]
- 28. Kalluri R., et al. (2009). The basics of epithelial-mesenchymal transition. J. Clin. Invest., 119, 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tanno B., et al. (2010). Expression of Slug is regulated by c-Myb and is required for invasion and bone marrow homing of cancer cells of different origin. J. Biol. Chem., 285, 29434–29445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mittal M.K., et al. (2011). SLUG-induced elevation of D1 cyclin in breast cancer cells through the inhibition of its ubiquitination. J. Biol. Chem., 286, 469–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bhat-Nakshatri P., et al. (2010). SLUG/SNAI2 and tumor necrosis factor generate breast cells with CD44+/CD24- phenotype. BMC Cancer., 10, 411– 426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cobaleda C., et al. (2007). Function of the zinc-finger transcription factor SNAI2 in cancer and development. Annu. Rev. Genet., 41, 41–61 [DOI] [PubMed] [Google Scholar]
- 33. Wang S.P., et al. (2009). p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat. Cell Biol., 11, 694–704 [DOI] [PubMed] [Google Scholar]
- 34. Lujambio A., et al. (2007). Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res., 67, 1424–1429 [DOI] [PubMed] [Google Scholar]
- 35. Wong K.Y., et al. (2011). Epigenetic inactivation of the miR-124-1 in haematological malignancies. PLoS ONE., 6, e19027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Silber J., et al. (2008). miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med., 6, 14–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wilting S.M., et al. (2010). Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol. Cancer., 9, 167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.