Abstract
Archaeal and eukaryotic cytosols contain group II chaperonins, which have a double-barrel structure and fold proteins inside a cavity in an ATP-dependent manner. The most complex of the chaperonins, the eukaryotic TCP-1 ring complex (TRiC), has eight different subunits, chaperone containing TCP-1 (CCT1–8), that are arranged so that there is one of each subunit per ring. Aspects of the structure and function of the bovine and yeast TRiC have been characterized, but studies of human TRiC have been limited. We have isolated and purified endogenous human TRiC from HeLa suspension cells. This purified human TRiC contained all eight CCT subunits organized into double-barrel rings, consistent with what has been found for bovine and yeast TRiC. The purified human TRiC is active as demonstrated by the luciferase refolding assay. As a more stringent test, the ability of human TRiC to suppress the aggregation of human γD-crystallin was examined. In addition to suppressing off-pathway aggregation, TRiC was able to assist the refolding of the crystallin molecules, an activity not found with the lens chaperone, α-crystallin. Additionally, we show that human TRiC from HeLa cell lysate is associated with the heat shock protein 70 and heat shock protein 90 chaperones. Purification of human endogenous TRiC from HeLa cells will enable further characterization of this key chaperonin, required for the reproduction of all human cells.
Keywords: TRiC, CCT, Chaperonin, Crystallin, Protein folding
Introduction
Molecular chaperones in the cell guide nascent and misfolded proteins to their native states and protect misfolded proteins from aggregation. The three main ATP-dependent chaperone classes that assist in folding proteins are heat shock protein 70 and 90 (Hsp70 and Hsp90) and chaperonin (Hartl et al. 2011). These broad chaperone classes may function in pathways by transferring proteins that cannot be sufficiently folded from one chaperone to another (Frydman et al. 1994; Hartl et al. 2011). Hsp70 chaperones bind to nascent proteins immediately after exiting the ribosome, while Hsp90 chaperones assist their client proteins at the end of the folding process (Hartl et al. 2011). The most complex of the ATP-dependent chaperones, the chaperonins, fold substrates in a cavity within the chaperonin where the substrate is sequestered, in whole or in part, away from the environment of the cell (Thulasiraman et al. 1999; Tang et al. 2006).
Chaperonin ATPases are composed of two back-to-back rings with seven to nine subunits each (Horwich et al. 2007). Each subunit has three domains: the apical domain that recognizes substrates, the equatorial domain that contains the ATP-binding site and facilitates subunit–subunit contacts, and the intermediate domain that acts as a hinge region between the two other domains (Braig et al. 1994; Spiess et al. 2004). Chaperonins are divided into two groups: group I (found in prokaryotes and in the chloroplasts and mitochondria of eukaryotes) and group II (found in the Archaeal and eukaryotic cytosol) (Hartl et al. 2011). The Archaeal group II chaperonins consist of two seven to nine subunit rings that have one to three different subunits (Bigotti and Clarke 2008), while the eukaryotic group II chaperonin TCP-1 ring complex (TRiC) consists of two identical rings, each with eight different chaperone containing TCP-1 (CCT) subunits (Cong et al. 2010).
TRiC was first identified in rabbit reticulocytes when it was found that newly made tubulin subunits entered a 900-kDa complex before becoming competent to assemble into microtubules (Yaffe et al. 1992). This complex consisted of a set of polypeptides between 55 and 60 kDa; one of which was recognized by the TCP-1 antibody (Yaffe et al. 1992). Concurrently, preliminary characterization of human and mouse TCP-1 found that in both species TCP-1 made a complex of approximately 900 kDa and that TCP-1 associated with four to six unidentified polypeptides and two Hsp70 homologs (Lewis et al. 1992). Another simultaneous study showed that TRiC was capable of binding and folding proteins to their native states (Frydman et al. 1992). Structures of TRiC with tubulin and actin have since been resolved and showed that TRiC recognizes and binds these essential protein substrates that first led to its discovery (Hynes and Willison 2000; Llorca et al. 2001; Muñoz et al. 2011; Neirynck et al. 2006).
TRiC does not only bind actin and tubulin; Thulasiraman et al. demonstrated that TRiC binds 9–15 % of newly synthesized proteins in [35 S]-methionine pulse labeled baby hamster kidney cells (Thulasiraman et al. 1999). The mechanism and high-resolution structure of group II chaperonins has been elucidated using the archaeal chaperonin of Methanococcus maripauldis, Mm-Cpn (Douglas et al. 2011; Pereira et al. 2012, 2010; Zhang et al. 2010). Due to the increased complexity of TRiC, much of the structural and biochemical research on group II chaperonins has been carried out with less complex archaeal chaperonins, such as Mm-Cpn.
The most common preparation of TRiC for biomedical research is endogenous TRiC from bovine testes tissue (Feldman et al. 2003; Ferreyra and Frydman 2000; Frydman et al. 1992). Purification of endogenous TRiC from rabbit reticulocytes has also been effective (Frydman et al. 1994; Gao et al. 1992; Nimmesgern and Hartl 1993; Norcum 1996). More recently, purification of exogenously tagged yeast TRiC in yeast has been developed (Dekker et al. 2011; Pappenberger et al. 2006), along with purification of endogenous yeast TRiC by exogenously tagging an interacting protein (Leitner et al. 2012). Co-expression of all eight human CCT subunits in baby hamster kidney cells has been attempted but resulted in very low yields (Machida et al. 2012). While these purifications have increased the opportunities to study TRiC, characterization of human TRiC has been lacking.
Investigations of the arrangement of the CCT subunits in TRiC purified from different species have given conflicting results (Cong et al. 2010; Dekker et al. 2011). However, a recent novel mass spectrometry method established that bovine TRiC purified from testes tissue and yeast TRiC purified via an interacting protein had the same arrangement with CCT2 and CCT6 forming the homotypic contacts (Leitner et al. 2012). This does not rule out that other CCT subunit arrangements of TRiC may exist, especially in different tissues and at different developmental stages. Previous research has shown that TRiC complexes containing specific subunits may have different roles (Roobol et al. 1995) and even that the CCT subunits may have functions in the cell independent of the TRiC complex (Roobol and Carden 1999).
The largest divergence of sequence among the CCT subunits is in the apical substrate–recognition domain, suggesting that the different subunits of TRiC may have evolved distinctive subunit specificities (Frydman 2001; Kim et al. 1994; Spiess et al. 2006). Although only a few substrates have been studied, it is clear that not all CCT subunits are involved in binding of non-native-state substrates to TRiC (Feldman et al. 2003; Hynes and Willison 2000; Llorca et al. 2001; Spiess et al. 2006). The apical domains of CCT1 and CCT4 have been implicated in TRiC binding to exon one of the huntingtin protein (Tam et al. 2006). Spiess et al. demonstrated that TRiC subunits CCT1 and CCT7 bind the von Hippel–Lindau tumor suppressor protein (pVHL) (Spiess et al. 2006). While these studies show that not all CCT subunits are required to bind a substrate to TRiC, we cannot firmly conclude whether only specific CCT subunits can bind particular substrates.
While there is emerging understanding of the CCT subunit arrangement and the recognition of substrates by specific CCT subunits, there has been little study on the assembly of TRiC from the CCT subunits. With the eight CCT subunits expressed from seven different genes, the assembly of TRiC must be very finely regulated (Kubota et al. 1999). This regulation may mean that the TRiC rings can contain a different arrangement and ratio of CCT subunits at some point in their lifetime. To investigate whether human TRiC found in epithelial cells is the same as bovine TRiC from testes tissue, we purified endogenous TRiC from an epithelial cell derivative line. HeLa cells have previously been shown to express all eight CCT proteins at high levels (Fountoulakis et al. 2004), making this an appropriate cell line for endogenous TRiC purification. For consistency, the new nomenclature of Hsp70, α-crystallin, Hsp90, and the chaperonins GroE/TRiC is HSPA, HSPB4, HSPC, and HSPD/CCT, respectively, but the older names have been used herein (Kampinga et al. 2009).
Materials and methods
TRiC purification from HeLa cells
A starter culture of HeLa suspension cells (HeLa-S3; ATCC) was grown in S-MEM (Sigma) supplemented with 10 % fetal bovine serum, 1 % l-Glu, and 1 % penicillin and streptomycin. From this starter culture, Cell Essentials, Inc. (Boston, MA) grew a 20-L suspension culture of HeLa cells, resulting in a cell pellet of approximately 100 g.
All of the following steps were performed at 4 °C. The HeLa cells were lysed following the HeLa nuclear extraction protocol (Tran et al. 2001). Briefly, the pellet was washed with iced phosphate buffer (137 mM NaCl, 2.68 mM KCl, 4.29 mM Na2HPO4, and 1.47 mM KH2PO4). The packed cell volume (PCV) of the pellet was determined and the pellet was resuspended in two PCVs of hypotonic buffer A (10 mM Tris, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, and 0.5 mM dithiothreitol (DTT)) and mixed thoroughly. The HeLa cells were dounced 35 times with pestle B. The dounced cells were centrifuged at 2,500 × g for 15 min, resulting in three layers. The top cytoplasmic layer contained TRiC and was therefore supplemented with 1 mM ATP and used in the subsequent purification.
The human TRiC purification hereafter loosely follows the bovine TRiC purification described by Ferreyra and Frydman (2000). Two ammonium sulfate precipitations (25 then 55 %) were performed on the cytosolic fraction isolated above. Human TRiC was found in the supernatant of the 25 % ammonium sulfate cut and the pellet of the 55 % ammonium sulfate cut. This pellet was dissolved in a minimal volume of MQ-A (20 mM HEPES/KOH, pH 7.4, 50 mM NaCl, 5 mM MgCl2, 10 % glycerol, 1 mM DTT, 0.1 mM PMSF, 0.1 mM EDTA, and 1 mM ATP) and placed in 50-kDa MWCO dialysis tubing (SpectraPor) and dialyzed twice (2 h to overnight) against MQ-A at 4 °C.
The dialyzed sample was centrifuged at 15,000 × g to remove aggregates and passed over a HiLoad 26/10 Q sepharose column (GE Healthcare). Human TRiC was eluted off of this column by 40 % MQ-B (MQ-A with 1 M NaCl). The fractions containing TRiC were pooled, diluted in half by MQ-A, and applied to a Heparin HiTrap HP 5 × 5 mL column (GE Healthcare). Human TRiC eluted during a 14-column volume gradient from 20 to 65 % MQ-B. The fractions containing TRiC were pooled and concentrated down to 1 mL using Vivaspin ultraconcentrators (Satorius Stedim). This sample was loaded on a Superose 6 10/300 GL size exclusion column (GE Healthcare). Human TRiC was eluted by MQ-A around 12–14.5 mL of the size exclusion column, consistent with that of a 1-MDa complex. These fractions were pooled, concentrated, and the protein concentration was measured using the BCA assay (Pierce) with BSA as the standard.
SDS-PAGE and immunoblots
Proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (14 %) at 165 V for 1 h after boiling in reducing buffer (60 mM Tris, pH 6.8, 2 % SDS, 5 % β-mercaptoethanol, 10 % glycerol, and bromophenol blue for color) for 5 min. The gels were stained with Coomassie blue or Krypton (Pierce). Transfer was conducted for 1.5 h at 300 mA in transfer buffer (10 % methanol, 25 mM Tris, and 192 mM glycine) onto 0.45 μm polyvinylidene difluoride membranes (Millipore).
The primary antibodies used for CCT1–8 were from Santa Cruz Biotechnology: CCT1, sc-53454; CCT2, sc-28556; CCT3, sc-33145; CCT4, sc-48865; CCT5, sc-13886; CCT6, sc-100958; CCT7, sc-130441; and CCT8, sc-13891. The Hsp70 (HSPA) and Hsp90 (HSPC) antibodies were from Enzo Life Sciences: Hsc70/Hsp70 (HSPA1A/HSPA8), SPA-820; and Hsp90α (HSPC2), SPA-840. The secondary antibodies were alkaline phosphatase (AP)-conjugated (Millipore) and the membranes were visualized using the AP-conjugate substrate kit (BioRad).
Electron microscopy
Copper grids with Formvar carbon coating (400 mesh, Ted Pella) were glow discharged for 20 s, and 5 μL of purified human TRiC was placed on the grids for 5 min. Excess sample on the grids was blotted off using filter paper, and the grids were floated onto a drop of filtered 1.5 % uranyl acetate (Sigma-Aldrich) for 45 s. Grids were visualized under a JEOL 1200 SX transmission electron microscope (TEM), and digital micrographs were taken using an AMT 16000 S camera system.
Luciferase refolding assay
The luciferase refolding assay was performed as described in Thulasiraman et al. (2000). Briefly, 8.2 μM of luciferase (Promega) was unfolded in unfolding buffer (6 M guanidine hydrochloride, 25 mM HEPES/KOH pH 7.4, 50 mM KOAc, and 5 mM DTT) at room temperature for 1 h with mixing. The unfolded luciferase was diluted 1:40 (205 nM) in unfolding buffer and then further diluted 1:25 (8.2 nM) into refolding buffer (25 mM HEPES/KOH, pH 7.4, 100 mM KOAc, 10 mM Mg(OAc)2, 2 mM DTT, 1 mM ATP, 10 mM creatine phosphate, 40 U/mL creatine kinase, and 2 % DMSO) with or without 400 nM of purified human TRiC. At various time points, an aliquot of the refolding reaction was diluted 1:25 into Steady-Glo Assay Reagent buffer (Promega) and luminescence was measured on a FLUOstar Optima plate reader (BMG Labtech) with FLUOstar Optima software.
Human γD-crystallin aggregation suppression assay
The aggregation suppression assay is described in detail in Knee et al. (2011). Briefly, 23 μM human γD-crystallin was unfolded overnight at 37 °C in unfolding buffer (5.5 M guanidine hydrochloride, 50 mM Tris-HCl, pH 7.5, and 5 mM DTT). To initiate aggregation, the unfolded protein was diluted 1:10 (2.3 μM) into refolding buffer (50 mM Tris-HCl, pH 7.5, 1 mM DTT, 50 mM KCl, 5 mM MgCl2, and 1 mM ATP) with or without 2.3 μM purified human TRiC. Aggregation kinetics were measured at 350 nm on a Cary UV/Vis spectrophotometer (Varian) using the Varian Kinetics program.
Results
The first step in purifying endogenous human TRiC from HeLa cells was separating the cytoplasmic fraction of the cells from the nuclei because TRiC is a cytoplasmic chaperonin. This was accomplished using a HeLa nuclear extraction protocol (Tran et al. 2001) and verified by immunoblots probed with the CCT1 primary antibody (Fig. 1). Next, a series of ammonium sulfate cuts further purified TRiC from other HeLa proteins. The resuspended pellet was passed over three chromatography columns: anion exchange (Q sepharose)-, heparin-affinity-, and Superose-6 size exclusion chromatography. The elution peak from the size exclusion column was between 12 and 14.5 mL (Fig. 2). This was consistent with other TRiC purifications and the purification of Mm-Cpn (Frydman et al. 1994; Reissmann et al. 2007). The average yield of this purification from a 100 g HeLa cell pellet was 5 mg of purified human TRiC with ~90 % purity (Fig. 3a).
Fig. 1.
Human TRiC recovered from the cytoplasmic fraction of HeLa cells. Three layers recovered from low-speed centrifugation after cell lysis: cytoplasmic layer (C), middle layer (M), and nuclei layer (N). The samples were electrophoresed through SDS-PAGE, and immunoblots were prepared and probed with anti-CCT1. Most of the CCT1, and therefore TRiC, was found in the cytoplasm of the lysed cells
Fig. 2.

Human TRiC purified by size exclusion chromatography. The input (inp) and various elution volumes (7.5–14.5 mL) were electrophoresed through 14 % SDS-PAGE and stained with Coomassie blue. TRiC appears as a series of bands ~60 kDa in size eluted in volumes of 12-14.5 mL consistent with a 1-MDa complex
Fig. 3.
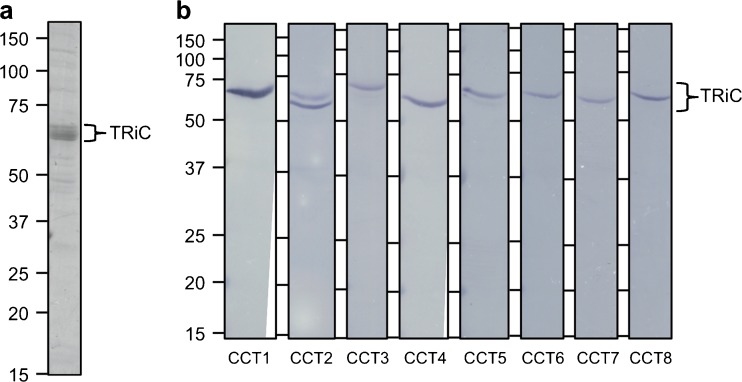
All eight subunits present in purified human TRiC. a A series of bands consistent with TRiC were present in the final purified human TRiC sample as shown on Coomassie-stained 14 % SDS-PAGE. b Immunoblots probed with all eight anti-CCT primary antibodies show that the purified human TRiC sample contains all eight CCT subunits in approximately equal ratios. There were no degradation products of the subunits in the purified human TRiC sample
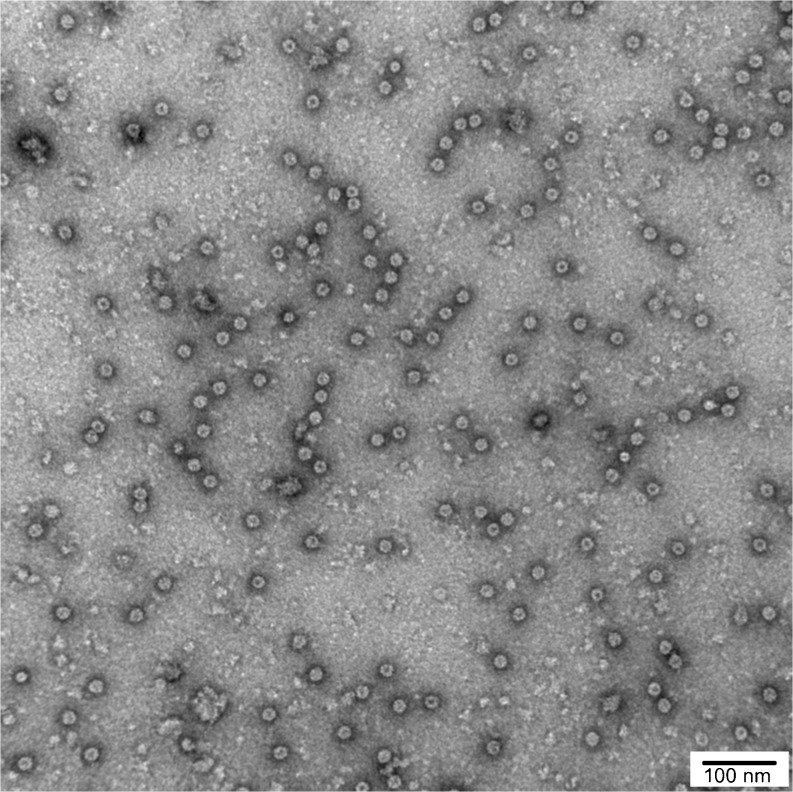
All eight CCT subunits were present in the purified human TRiC sample as seen by immunoblots probed with antibodies against each of the eight subunits (Fig. 3b). They were all present in roughly equal stoichiometry. By negative stain TEM, purified human TRiC appeared as two back-to-back rings approximately 185 Å in height and 165 Å in diameter (Fig. 4). The morphology of purified human TRiC was consistent with that of purified TRiC reported in the literature (Cong et al. 2010; Dekker et al. 2011).
Fig. 4.
Negative stain TEM of purified human TRiC reveal double rings. The morphology of human TRiC is consistent with that of TRiC purified from other species. The complexes were ~165 Å in diameter and ~185 Å in height and shown here at × 150,000 magnification
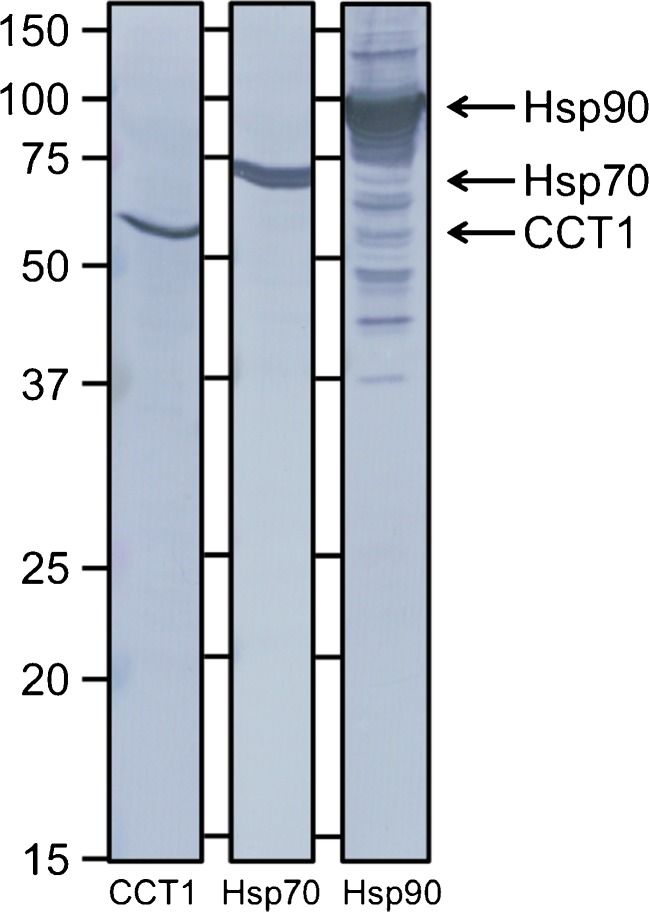
When heparin-affinity chromatography was omitted from the purification, Hsp70/Hsc70 and Hsp90 co-purified with human TRiC (Fig. 5). This interaction may be due to human TRiC binding to Hsp70/Hsc70 and Hsp90 in HeLa cells, while exchanging substrates between the chaperones. When heparin-affinity chromatography was utilized, Hsp70/Hsc70 and Hsp90 were not seen in the purified human TRiC sample, demonstrating that while this interaction was quite robust, it could be eliminated.
Fig. 5.
Hsp70 and Hsp90 co-purified with TRiC when heparin-affinity chromatography was omitted. Immunoblots probed with antibodies against CCT1, Hsp70, and Hsp90 clearly show Hsp70 and Hsp90 present in the purified human TRiC sample when heparin-affinity chromatography was not used
Human TRiC purified from HeLa cells was active in the luciferase refolding assay, which has been previously used to test activity of TRiC purified from bovine testes (Frydman et al. 1992) and rabbit reticulocytes (Frydman et al. 1994; Nimmesgern and Hartl 1993). In the assay, luciferase was unfolded and then diluted into buffer with purified human TRiC (Thulasiraman et al. 2000). The presence of refolded luciferase in the mixture was assayed by addition of luciferin and subsequent luminescence production monitoring. Purified human TRiC refolded luciferase for over 2 h at room temperature (Fig. 6).
Fig. 6.
Purified human TRiC active in refolding luciferase. Human TRiC (blue) refolds luciferase more efficiently than the BSA (green) or water (red) controls. Human TRiC was active over 2 h at room temperature
Though luciferase has frequently been used to assay the refolding activity of a variety of chaperonins, it is not an authentic substrate for human TRiC. A human protein whose folding and competing aggregation has been systematically studied is human γD-crystallin (HγD-Crys) (Flaugh et al. 2005; Kosinski-Collins and King 2003). TRiC is almost certainly present in lens epithelium cells and primary lens fibers (Hoehenwarter et al. 2008). In characterizing the activity of the archaeal chaperonin Mm-Cpn, Knee et al. found that Mm-Cpn not only suppressed the aggregation of HγD-Crys but also enhanced its refolding in vitro (2011). The major lens chaperone, α-crystallin, suppresses HγD-Crys aggregation in vitro but does not refold the molecules (Acosta-Sampson and King 2010; Moreau and King 2012a).
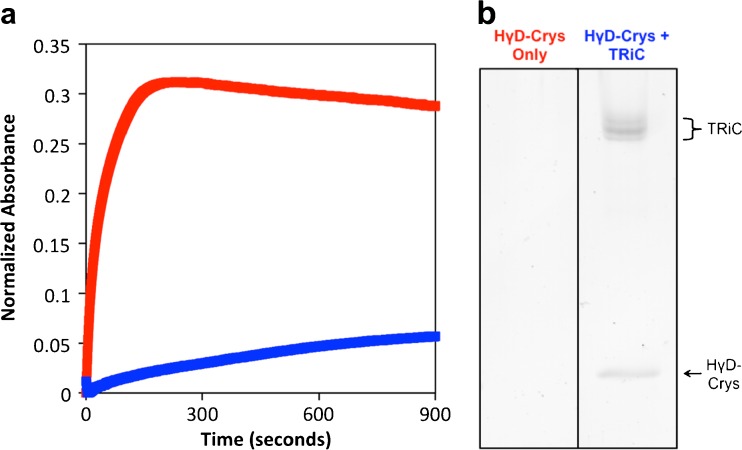
Therefore, we decided to assess whether human TRiC was active with respect to the HγD-Crys substrate. In this assay, when unfolded HγD-Crys was diluted from denaturant into buffer at concentrations of 50 μg/mL, partially folded intermediates partitioned between productive refolding and off-pathway aggregation. This aggregation was monitored by sample turbidity. When purified human TRiC was added to the buffer, aggregation was significantly suppressed (Fig. 7a). Furthermore, native-like HγD-Crys could be detected when the filtered sample was assessed on SDS-PAGE (Fig. 7b), suggesting that HγD-Crys was refolded by human TRiC.
Fig. 7.
Purified human TRiC suppression of HγD-Crys aggregation and HγD-Crys native-like state refolding. a Aggregation of HγD-Crys (red) can be suppressed by the addition of human TRiC (blue) by approximately 80 % after 15 min at 37 °C. b When filtered, the aggregation suppression samples were observed on Krypton-stained 14 % SDS-PAGE. The sample with purified human TRiC showed a band corresponding to HγD-Crys (a control lane with only HγD-Crys was run in parallel for comparison but not shown here) indicating that human TRiC can refold HγD-Crys to a native-like state
In summary, human TRiC was purified from HeLa cells by first extracting the cytoplasmic layer of the cells and then performing three chromatography steps. The purified material contained all eight CCT subunits in approximately equal ratios. TRiC could be effectively separated from other chaperones in the cells by heparin-affinity chromatography. Purified human TRiC possessed the back-to-back ring morphology that defines the structure of chaperonins. Our purified human TRiC was not only active in refolding the model substrate luciferase but also suppressed the aggregation and refolded the authentic substrate HγD-Crys.
Discussion
While TRiC has been readily purified endogenously from bovine testes (Frydman et al. 1992; Leitner et al. 2012) and pseudo-exogenously from yeast (Leitner et al. 2012; Pappenberger et al. 2006), purification of human TRiC has been limited. Expression of all eight subunits exogenously from the cloned genes is difficult. The direct approach of growing a large amount of human epithelial cells and purifying endogenous TRiC has been successful in producing the authentic human chaperonin.
Molecular chaperones have been proposed to be therapeutic targets (Almeida et al. 2011). TRiC may play a role in suppressing Huntington's disease by decreasing huntingtin aggregate formation (Kitamura et al. 2006; Tam et al. 2009). If TRiC is to be used therapeutically in the clinic, it will be necessary to study human TRiC to further understand chaperonin functions inside human cells. While the TRiC isolated from bovine testes may be similar overall to the human version, there are differences in all eight subunits between bovine and human TRiC, let alone any type of arrangement or assembly differences.
Human TRiC activity in assisting the refolding of firefly luciferase corresponds to activities reported for other mammalian TRiC complexes (Frydman et al. 1992, 1994; Nimmesgern and Hartl 1993). The lens crystallins represent a more rigorous test for the activity of human TRiC. The lens crystallins must remain stable and folded throughout life for the aggregated state results in the lens disease cataract (Moreau and King 2012b). The α-crystallin chaperone present at high concentrations in the lens suppresses the aggregation of HγD-Crys but cannot refold it (Acosta-Sampson and King 2010; Evans et al. 2008; Horwitz 1992). The results showed that human TRiC both suppressed the aggregation of partially folded HγD-Crys and, in the presence of ATP, was able to refold the chains. Human TRiC may in fact play a role in protecting the lens fibers from cortical cataract (Mitchell et al. 1997).
We found that Hsp70 and Hsp90 remained bound to purified human TRiC if one of the chromatography steps is omitted. This is not surprising since TRiC co-purifies with Hsp70 and Hsp90 in rabbit reticulocytes (Frydman et al. 1994; Nimmesgern and Hartl 1993). However, while the shuttling of substrates between Hsp70 and TRiC has been widely studied (Kabir et al. 2011), substrate exchange from TRiC to Hsp90 is much less understood. The substantial amounts of Hsp90 that co-purified with TRiC from HeLa cells may make this a good system for further studying the TRiC-Hsp90 interaction.
Other directions with human TRiC will be to attempt high-resolution structural studies for comparison to yeast and bovine TRiC. The arrangement of human TRiC purified from HeLa cells may be different than that of bovine TRiC purified from testes, not only because of the differences in species as mentioned above but also due to differences between the tissue and the cells in culture. Furthermore, it will be interesting to see whether purified human TRiC can refold actin and tubulin, the two largest substrates of TRiC, as efficiently as bovine TRiC. We plan to study whether each CCT subunit is needed to recognize and refold particular substrates, such as actin and tubulin.
It has been postulated that the eight different CCT subunits of TRiC are needed to recognize a variety of substrates (Feldman et al. 2003; Llorca et al. 2001). The CCT subunits may recognize different types of proteins, e.g., CCT2 may recognize beta-propeller proteins, while CCT8 may recognize hydrophobic beta sheets. Even more specifically, the CCT subunits may recognize different proteins, e.g., CCT1 may bind huntingtin (Tam et al. 2006) while CCT7 recognizes pVHL (Spiess et al. 2006). However, with our limited knowledge on the substrate recognition of TRiC, it is unknown if different CCT subunits specifically or redundantly recognize these various substrates. It may be that while CCT1 recognizes huntingtin with the highest efficiency, CCT4 or CCT7 can bind it when CCT1 is not present or defective.
The arrangement of TRiC has been determined for TRiC purified from bovine testes and from yeast, but it is unknown how this arrangement varies among tissues or at different developmental stages. Also unknown is how TRiC can assemble into this arrangement. As with other large complexes, it is likely that a highly regulated sequence of events is needed for the final arrangement. Chaperonin-like Biedel–Bradet Syndrome subunits assemble into the final BBSome complex by sequential addition of each subunit (Zhang et al. 2012). Such sequential assembly is possible for TRiC as well. Our purified TRiC material is an initial step in furthering the knowledge on this crucial human chaperonin.
Acknowledgments
The authors thank Dr. Kate Moreau and Daniel Goulet for their helpful discussions. This work was funded by NIH Roadmap grant EY016525 and NEI grant EY015834.
References
- Acosta-Sampson L, King J. Partially folded aggregation intermediates of human gammaD-, gammaC-, and gammaS-crystallin are recognized and bound by human alphaB-crystallin chaperone. J Mol Biol. 2010;401(1):134–152. doi: 10.1016/j.jmb.2010.05.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida MB, Nascimento JL, Herculano AM, Crespo-López ME. Molecular chaperones: toward new therapeutic tools. Biomed Pharmacother. 2011;65(4):239–243. doi: 10.1016/j.biopha.2011.04.025. [DOI] [PubMed] [Google Scholar]
- Bigotti MG, Clarke AR. Chaperonins: the hunt for the group II mechanism. Arch Biochem Biophys. 2008;474(2):331–339. doi: 10.1016/j.abb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Braig K, Otwinowski Z, Hegde R, Boisvert DC, Joachimiak A, Horwich AL, Sigler PB. The crystal structure of the bacterial chaperonin GroEL at 2.8 A. Nature. 1994;371(6498):578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- Cong Y, Baker ML, Jakana J, Woolford D, Miller EJ, Reissmann S, Kumar RN, Redding-Johanson AM, Batth TS, Mukhopadhyay A, Ludtke SJ, Frydman J, Chiu W. 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proc Natl Acad Sci USA. 2010;107(11):4967–4972. doi: 10.1073/pnas.0913774107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker C, Roe SM, McCormack EA, Beuron F, Pearl LH, Willison KR (2011) The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins. EMBO J 30(15):3078–3090 [DOI] [PMC free article] [PubMed]
- Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, Frydman J. Dual action of ATP hydrolysis couples lid closure to substrate release into the group II chaperonin chamber. Cell. 2011;144(2):240–252. doi: 10.1016/j.cell.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P, Slingsby C, Wallace BA. Association of partially folded lens betaB2-crystallins with the alpha-crystallin molecular chaperone. Biochem J. 2008;409(3):691–699. doi: 10.1042/BJ20070993. [DOI] [PubMed] [Google Scholar]
- Feldman DE, Spiess C, Howard DE, Frydman J. Tumorigenic mutations in VHL disrupt folding in vivo by interfering with chaperonin binding. Mol Cell. 2003;12(5):1213–1224. doi: 10.1016/S1097-2765(03)00423-4. [DOI] [PubMed] [Google Scholar]
- Ferreyra RG, Frydman J. Purification of the cytosolic chaperonin TRiC from bovine testis. Methods Mol Biol. 2000;140:153–160. doi: 10.1385/1-59259-061-6:153. [DOI] [PubMed] [Google Scholar]
- Flaugh SL, Kosinski-Collins MS, King J. Contributions of hydrophobic domain interface interactions to the folding and stability of human gammaD-crystallin. Protein Sci. 2005;14(3):569–581. doi: 10.1110/ps.041111405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountoulakis M, Tsangaris G, J-e O, Maris A, Lubec G. Protein profile of the HeLa cell line. J Chromatogr A. 2004;1038(1–2):247–265. doi: 10.1016/j.chroma.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall J, Tempst P, Hartl F. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO J. 1992;11(13):4767–4778. doi: 10.1002/j.1460-2075.1992.tb05582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994;370(6485):111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- Gao Y, Thomas J, Chow R, Lee G, Cowan N. A cytoplasmic chaperonin that catalyzes beta-actin folding. Cell. 1992;69(6):1043–1050. doi: 10.1016/0092-8674(92)90622-J. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hoehenwarter W, Tang Y, Ackermann R, Pleissner K-P, Schmid M, Stein R, Zimny-Arndt U, Kumar NM, Jungblut PR. Identification of proteins that modify cataract of mouse eye lens. Proteomics. 2008;8(23–24):5011–5024. doi: 10.1002/pmic.200800380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich AL, Fenton WA, Chapman E, Farr GW. Two families of chaperonin: physiology and mechanism. Annu Rev Cell Dev Biol. 2007;23:115–145. doi: 10.1146/annurev.cellbio.23.090506.123555. [DOI] [PubMed] [Google Scholar]
- Horwitz J. Alpha-crystallin can function as a molecular chaperone. Proc Natl Acad Sci USA. 1992;89(21):10449–10453. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes GM, Willison KR. Individual subunits of the eukaryotic cytosolic chaperonin mediate interactions with binding sites located on subdomains of beta-actin. J Biol Chem. 2000;275(25):18985–18994. doi: 10.1074/jbc.M910297199. [DOI] [PubMed] [Google Scholar]
- Kabir MA, Uddin W, Narayanan A, Reddy PK, Jairajpuri MA, Sherman F, Ahmad Z. Functional subunits of eukaryotic chaperonin CCT/TRiC in protein folding. J Amino Acids. 2011;2011:843206. doi: 10.4061/2011/843206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, Bruford EA, Cheetham ME, Chen B, Hightower LE (2009) Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones 14:105–111 [DOI] [PMC free article] [PubMed]
- Kim S, Willison K, Horwich A. Cytosolic chaperonin subunits have a conserved ATPase domain but diverged polypeptide-binding domains. Trends Biochem Sci. 1994;19(12):543–548. doi: 10.1016/0968-0004(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Kitamura A, Kubota H, Pack C-G, Matsumoto G, Hirayama S, Takahashi Y, Kimura H, Kinjo M, Morimoto RI, Nagata K. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat Cell Biol. 2006;8(10):1163–1170. doi: 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- Knee KM, Goulet DR, Zhang J, Chen B, Chiu W, King JA. The group II chaperonin Mm-Cpn binds and refolds human γD crystallin. Protein Sci. 2011;20(1):30–41. doi: 10.1002/pro.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski-Collins MS, King J. In vitro unfolding, refolding, and polymerization of human gammaD crystallin, a protein involved in cataract formation. Protein Sci. 2003;12(3):480–490. doi: 10.1110/ps.0225503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H, Yokota S, Yanagi H, Yura T. Structures and co-regulated expression of the genes encoding mouse cytosolic chaperonin CCT subunits. Eur J Biochem. 1999;262(2):492–500. doi: 10.1046/j.1432-1327.1999.00405.x. [DOI] [PubMed] [Google Scholar]
- Leitner A, Joachimiak LA, Bracher A, Mönkemeyer L, Walzthoeni T, Chen B, Pechmann S, Holmes S, Cong Y, Ma B, Ludtke S, Chiu W, Hartl FU, Aebersold R, Frydman J. The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure. 2012;20(5):814–825. doi: 10.1016/j.str.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis V, Hynes G, Dong Z, Saibil H, Willison K. T-complex polypeptide-1 is a subunit of a heteromeric particle in the eukaryotic cytosol. Nature. 1992;358(6383):249–252. doi: 10.1038/358249a0. [DOI] [PubMed] [Google Scholar]
- Llorca O, Martín-Benito J, Gómez-Puertas P, Ritco-Vonsovici M, Willison KR, Carrascosa JL, Valpuesta JM. Analysis of the interaction between the eukaryotic chaperonin CCT and its substrates actin and tubulin. J Struct Biol. 2001;135(2):205–218. doi: 10.1006/jsbi.2001.4359. [DOI] [PubMed] [Google Scholar]
- Machida K, Masutani M, Kobayashi T, Mikami S, Nishino Y, Miyazawa A, Imataka H. Reconstitution of the human chaperonin CCT by co-expression of the eight distinct subunits in mammalian cells. Protein Expr Purif. 2012;82(1):61–69. doi: 10.1016/j.pep.2011.11.010. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Cumming RG, Attebo K, Panchapakesan J. Prevalence of cataract in Australia: the Blue Mountains eye study. Ophthalmology. 1997;104(4):581–588. doi: 10.1016/s0161-6420(97)30266-8. [DOI] [PubMed] [Google Scholar]
- Moreau KL, King JA. Cataract-causing defect of a mutant γ-crystallin proceeds through an aggregation pathway which bypasses recognition by the α-crystallin chaperone. PLoS One. 2012;7(5):e37256. doi: 10.1371/journal.pone.0037256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau KL, King JA. Protein misfolding and aggregation in cataract disease and prospects for prevention. Trends Mol Med. 2012;18(5):273–282. doi: 10.1016/j.molmed.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz IG, Yébenes H, Zhou M, Mesa P, Serna M, Park AY, Bragado-Nilsson E, Beloso A, de Cárcer G, Malumbres M, Robinson CV, Valpuesta JM, Montoya G. Crystal structure of the open conformation of the mammalian chaperonin CCT in complex with tubulin. Nat Struct Mol Biol. 2011;18(1):14–19. doi: 10.1038/nsmb.1971. [DOI] [PubMed] [Google Scholar]
- Neirynck K, Waterschoot D, Vandekerckhove J, Ampe C, Rommelaere H. Actin interacts with CCT via discrete binding sites: a binding transition-release model for CCT-mediated actin folding. J Mol Biol. 2006;355(1):124–138. doi: 10.1016/j.jmb.2005.10.051. [DOI] [PubMed] [Google Scholar]
- Nimmesgern E, Hartl FU. ATP-dependent protein refolding activity in reticulocyte lysate. Evidence for the participation of different chaperone components. FEBS Lett. 1993;331(1–2):25–30. doi: 10.1016/0014-5793(93)80290-B. [DOI] [PubMed] [Google Scholar]
- Norcum MT. Novel isolation method and structural stability of a eukaryotic chaperonin: the TCP-1 ring complex from rabbit reticulocytes. Protein Sci. 1996;5(7):1366–1375. doi: 10.1002/pro.5560050715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappenberger G, McCormack EA, Willison KR. Quantitative actin folding reactions using yeast CCT purified via an internal tag in the CCT3/gamma subunit. J Mol Biol. 2006;360(2):484–496. doi: 10.1016/j.jmb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Pereira JH, Ralston CY, Douglas NR, Kumar R, Lopez T, McAndrew RP, Knee KM, King JA, Frydman J, Adams PD. Mechanism of nucleotide sensing in group II chaperonins. EMBO J. 2012;31(3):731–740. doi: 10.1038/emboj.2011.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JH, Ralston CY, Douglas NR, Meyer D, Knee KM, Goulet DR, King JA, Frydman J, Adams PD. Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle. J Biol Chem. 2010;285(36):27958–27966. doi: 10.1074/jbc.M110.125344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissmann S, Parnot C, Booth CR, Chiu W, Frydman J. Essential function of the built-in lid in the allosteric regulation of eukaryotic and archaeal chaperonins. Nat Struct Mol Biol. 2007;14(5):432–440. doi: 10.1038/nsmb1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roobol A, Carden MJ. Subunits of the eukaryotic cytosolic chaperonin CCT do not always behave as components of a uniform hetero-oligomeric particle. Eur J Cell Biol. 1999;78(1):21–32. doi: 10.1016/S0171-9335(99)80004-1. [DOI] [PubMed] [Google Scholar]
- Roobol A, Holmes FE, Hayes NV, Baines AJ, Carden MJ. Cytoplasmic chaperonin complexes enter neurites developing in vitro and differ in subunit composition within single cells. J Cell Sci. 1995;108(Pt 4):1477–1488. doi: 10.1242/jcs.108.4.1477. [DOI] [PubMed] [Google Scholar]
- Spiess C, Meyer AS, Reissmann S, Frydman J. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004;14(11):598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess C, Miller EJ, McClellan AJ, Frydman J. Identification of the TRiC/CCT substrate binding sites uncovers the function of subunit diversity in eukaryotic chaperonins. Mol Cell. 2006;24(1):25–37. doi: 10.1016/j.molcel.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam S, Geller R, Spiess C, Frydman J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat Cell Biol. 2006;8(10):1155–1162. doi: 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam S, Spiess C, Auyeung W, Joachimiak L, Chen B, Poirier MA, Frydman J. The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat Struct Mol Biol. 2009;16(12):1279–1285. doi: 10.1038/nsmb.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y-C, Chang H-C, Roeben A, Wischnewski D, Wischnewski N, Kerner MJ, Hartl FU, Hayer-Hartl M. Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell. 2006;125(5):903–914. doi: 10.1016/j.cell.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Thulasiraman V, Ferreyra RG, Frydman J. Folding assays. Assessing the native conformation of proteins. Methods Mol Biol. 2000;140:169–177. doi: 10.1385/1-59259-061-6:169. [DOI] [PubMed] [Google Scholar]
- Thulasiraman V, Yang CF, Frydman J. In vivo newly translated polypeptides are sequestered in a protected folding environment. EMBO J. 1999;18(1):85–95. doi: 10.1093/emboj/18.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran DP, Kim SJ, Park NJ, Jew TM, Martinson HG. Mechanism of poly(A) signal transduction to RNA polymerase II in vitro. Mol Cell Biol. 2001;21(21):7495–7508. doi: 10.1128/MCB.21.21.7495-7508.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Farr GW, Miklos D, Horwich AL, Sternlicht ML, Sternlicht H. TCP1 complex is a molecular chaperone in tubulin biogenesis. Nature. 1992;358(6383):245–248. doi: 10.1038/358245a0. [DOI] [PubMed] [Google Scholar]
- Zhang J, Baker ML, Schröder GF, Douglas NR, Reissmann S, Jakana J, Dougherty M, Fu CJ, Levitt M, Ludtke SJ, Frydman J, Chiu W. Mechanism of folding chamber closure in a group II chaperonin. Nature. 2010;463(7279):379–383. doi: 10.1038/nature08701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Yu D, Seo S, Stone EM, Sheffield VC (2012) Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndrome protein complex, the BBSome. J Biol Chem 287(24):20625–20635 [DOI] [PMC free article] [PubMed]