Abstract
TolC channel provides a route for the expelled drugs and toxins to cross the outer membrane of Escherichia coli. The puzzling feature of TolC structure is that the periplasmic entrance of the channel is closed by dense packing of twelve α-helices. Efflux pumps exemplified by AcrAB are proposed to drive the opening of TolC channel. How interactions with AcrAB promote the close-to-open transition in TolC remains unclear. In this study, we investigated in vivo the functional and physical interactions of AcrAB with the closed TolC and its conformer opened by mutations in the periplasmic entrance. We found that the two conformers of TolC are readily distinguishable in vivo by characteristic drug susceptibility, thiol modification and proteolytic profiles. However, these profiles of TolC variants respond neither to the in vivo stoichiometry of AcrAB:TolC nor to the presence of vancomycin, which is used often to assess the permeability of TolC channel. We further found that the activity and assembly of AcrAB-TolC tolerates significant changes in amounts of TolC and that only a small fraction of intracellular TolC is likely used to support efflux needs of E. coli. Our findings explain why TolC is not a good target for inhibition of multidrug efflux.
Introduction
E. coli TolC and its homologs in other Gram-negative bacteria enable export of various toxic molecules across the outer membrane (Benz et al., 1993, Fralick, 1996). These proteins function in conjunction with three types of transport systems: ATP-binding cassette (ABC)-type, Resistance Nodulation Division (RND)-type and Major Facilitator superfamily (MF-type) (Paulsen et al., 1997). The association between transporters and TolC is mediated by periplasmic proteins named Membrane Fusion Proteins (MFPs) (Dinh et al., 1994, Zgurskaya et al., 2009). The assembled complexes are proposed to span the entire gram-negative envelope and to expel substrates directly into external medium bypassing the periplasm.
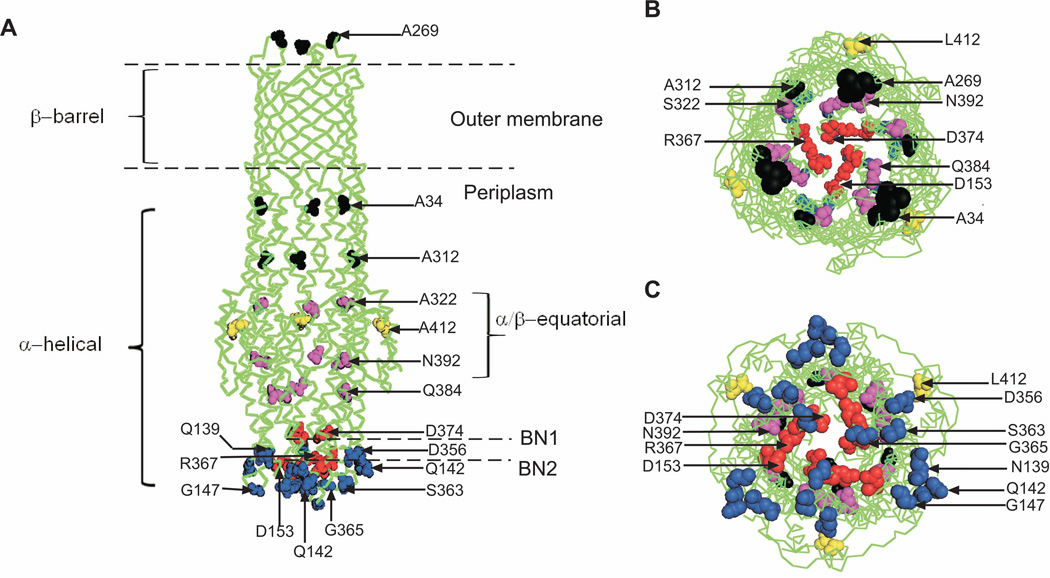
Structural analyses showed that TolC is a trimer folded into a 12-stranded β-barrel embedded into the outer membrane, an α-helical coiled coil domain that spans the nearly 100 Å into the periplasm, and a mixed α/β equatorial domain (Fig. 1) (Koronakis et al., 2000). In crystals, the periplasmic entrance of TolC is tightly closed so that even ions cannot easily diffuse through. In particular, a ring of D371 and D374 residues forms the narrowest pore constriction (BN1). In the proposed allosteric opening mechanism of TolC, the inner coiled coil α-helices (H7/H8) undergo an iris-like movement to realign with the outer coils (H3/H4), thereby enlarging the pore in a “twist-to-open” transition (Fig. 1). This model is supported by finding that disruption of the constraining Y362-R367 bond that tether the inner coiled coil H7/H8 to the outer H3/H4 coiled coils (BN2) increases the conductance of TolC channel by ten fold, whereas locking of the entrance coiled coils by chemical cross-linkers disables export of hemolysin, one of the TolC substrates (Andersen et al., 2002a). Crystal structures of “leaky” TolC mutants are also consistent with the notion that destabilization of ionic bonds in the periplasmic tip could dilate the TolC pore and allow passage of substrates through the channel (Pei et al., 2011, Bavro et al., 2008).
Figure 1. The crystal structure of TolC.
A. The side view of TolC with three domains the β-barrel, α-helical and α/β equatorial indicated. The aminoacid residues targeted in this study are shown as spheres: the extracellular loop and internal walls are in black, the two constrictions BN1 and BN2 are in red, residues of the periplasmic tip are in blue and those of the α/β domain in magenta and yellow.
B. The top extracellular view of the channel.
C. The bottom periplasmic view.
A growing body of evidence indicates that association with the inner membrane complex is needed to trigger opening of TolC channel (Bavro et al., 2008, Pei et al., 2011, Weeks et al., 2010). In the best studied multidrug efflux pump AcrAB-TolC from E. coli, TolC was found to bind both the inner membrane transporter AcrB and the periplasmic MFP AcrA, so that the tri-partite complex is formed between AcrA and TolC engaged by AcrB (Tikhonova et al., 2011). The assembly of the complex presumably leads to a close tip-to-tip AcrB-TolC fit, where D256 and D795 residues of AcrB could initiate the expansion of the periplasmic tip of TolC by interrupting R367-D153 and Y362-R367 links (Bavro et al., 2008, Pei et al., 2011). This expansion however is not sufficient to dilate the inner D371–D374 constriction because in the “leaky” TolC mutant lacking the Y362-R367 bond (YFREHis), the pore remains mostly closed. The full dilation of TolC channel in the assembled complex is believed to be driven by energy-dependent conformational changes in AcrA (Gerken & Misra, 2004, Weeks et al., 2010). Recent kinetic studies showed that the affinity of YFREHis mutant to both AcrA and AcrB is lower than that of the closed wild type TolC (Tikhonova et al., 2011). This finding suggested that opening of TolC could be the final step in the AcrAB-TolC transport cycle resulting in disassembly of the complex and closing of the channel.
In this study, we investigated the function and assembly of AcrAB-TolC complex in vivo by comparing conformations and properties of the wild type closed TolC (TolCHis) and its leaky YFREHis mutant. Our results show that the two conformers can be readily distinguished in vivo by drug susceptibility, proteolytic and thiol labeling profiles. However, these dominant TolC states respond neither to the presence of AcrAB nor other TolC-dependent transporters. We conclude that interactions between TolC and transporters are highly dynamic and only a small fraction of TolC is utilized to support efflux activities of E. coli.
Results
Mutational opening compromises TolC function and structure
Previous studies showed that leaky TolC mutants are able to support AcrAB-dependent efflux of antibiotics, albeit only partially (Bavro et al., 2008). To further characterize how these mutants function with AcrAB, the previously described TolC Y362F/R367E (YFREHis) mutant lacking the two critical salt bridges in the periplasmic tip of TolC and its parental TolCHis protein were either overproduced in ΔtolC cells from a plasmid or integrated back onto the chromosome. The drug efflux activities of two TolC conformers were assessed by measuring minimal inhibitory concentrations (MICs) of various antimicrobial agents, the substrates of AcrAB-TolC. As shown in Table 1, cells producing YFREHis were at least four times more susceptible to erythromycin, oleandomycin and puromycin. However, this mutant retained the full activity (2 fold or less difference in MICs) against novobiocin, lincomycin, SDS, nitrocefin and cloxacillin. The mutant was also fully proficient in secretion of colicin V, a peptide substrate of CvaAB-TolC exporter (data not shown). Thus, mutational opening of TolC does not affect all substrates equally.
Table 1.
Antibiotic susceptibilities of E. coli cells producing TolC or YFRE.
| Drug | MIC (µg/ml) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BW25113(WT) | GD102 (ΔC) | |||||||||
| - | pABHis | - | pUC18 | ::TolC | ::TolCHis | pTolCHis | ::YFRE | ::YFREHis | pYFREHis | |
| CLO | 250–500 | ND | 3.9 | ND | 500 | 250 | ND | 250 | 250 | ND |
| CAR | 7.5 | ND | 3.75 | ND | 3.75–7.5 | 3.75 | ND | 3.75 | 1.875–3.75 | ND |
| NCF | 100–200 | ND | 50 | ND | 100–200 | 100 | ND | 100–200 | 100 | ND |
| PUR | 64 | 128 | 2 | 4 | 64 | 64–128 | 128 | 16 | 16 | 16 |
| CF | 3.2 | 1.6 | 0.4 | 0.8 | 3.2 | 3.2 | 3.2 | 3.2 | 1.6 | 3.2 |
| ERY | 32–64 | 64 | 1–2 | 4 | 32 | 32–64 | 128 | 4 | 4–8 | 16 |
| NOV | 32 | ND | 2 | 1 | 16–32 | 16 | 32 | 16 | 16 | 16 |
| OLE | 625 | ND | 19.5 | 19.5 | 625 | 625 | 625 | 78 | 78 | 312.5 |
| SDS | >10,000 | >10,000 | 10 | 10 | >10,000 | >10,000 | >10,000 | >10,000 | >10,000 | >10,000 |
| VAN | 156 | ND | 312 | 312 | 156 | 156 | 156 | 78 | 39–78 | 39–78 |
CLO, cloxacillin; CAR, carbenicillin; NCF, nitrocefin; PUR, puromycin; CF, chloramphenicol; ERY, erythromycin; NOV, novobiocin; SDS, sodium dodecyl sulfate; OLE, oleandomycin; VAN, vancomycin. ND, not determined because plasmids carry a β-lactam resistance determinant. Data from at least three independent experiments are shown. Plasmid pABHis is a pUC18 derivative carrying acrAB under the native promoter (Tikhonova & Zgurskaya, 2004).
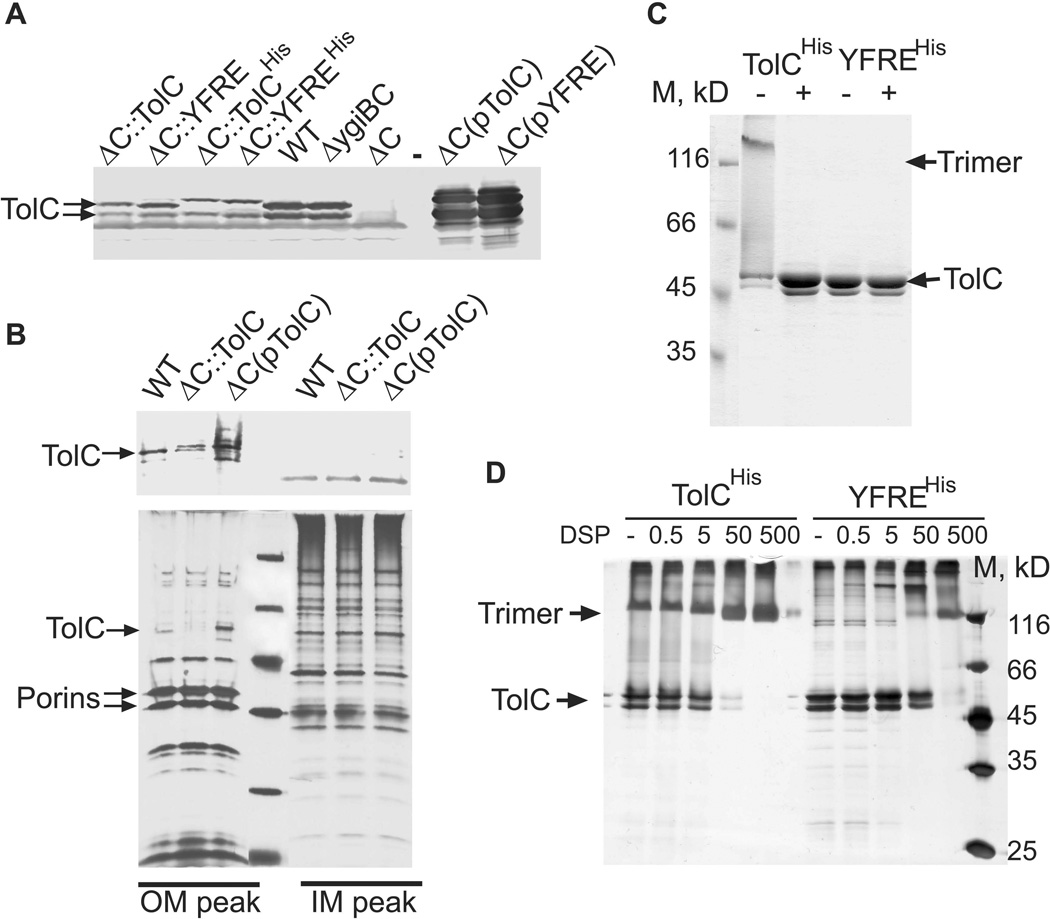
Growth of cells overproducing either TolCHis or YFREHis from a plasmid and cells carrying respective genes on a chromosome was inhibited by the same concentrations of antimicrobials (Table 1). However, immunoblotting analysis showed that amounts of TolC in membranes of the wild type BW25113 (WT) cells were at least 5–10 fold higher than the amounts of both YFREHis and TolCHis proteins produced from the genes reintegrated onto the E. coli chromosome (Fig. 2A, ΔC::TolCHis and ΔC::YFREHis). This reduced expression was not due to the presence of the C-terminal 6His-tags because the re-integrated tagless TolC variants were produced at the same low levels and were functionally indistinguishable from the 6His-tagged proteins (Fig. 2A and Table 1). On the other hand, amounts of the plasmid-borne TolC variants exceeded the native TolC levels in WT membranes by more than 100 fold. Furthermore, separation of inner and outer membranes in sucrose density gradients showed that amounts of TolC specifically localized to the outer membrane paralleled the amounts of protein in the total membrane fractions (Fig. 2B). These results show that fluctuations in TolC amounts by several orders of the magnitude do not affect its export activity and that in this range of concentrations TolC does not limit AcrAB mediated drug efflux.
Figure 2. Expression and oligomerization of the plasmid-encoded and chromosomal TolCHis and YFREHis.
A. Immunoblotting analysis of membrane fractions isolated from E. coli WT and GD102 cells carrying the chromosomal or the plasmid-encoded TolC variants. Membrane fractions (10 µg of total protein) were separated by 12% SDS-PAGE and probed with polyclonal anti-TolC antibodies. In this Figure and everywhere else pTolC=pTolCHis and pYFRE=pYFREHis.
B. Immunoblotting and silver staining analyses of the inner (IM) and outer (OM) membranes of E. coli cells producing the chromosomal and the plasmid-encoded TolC. Membrane fractions were separated by ultracentrifugation on a 30–60% (w/w) sucrose gradient. The IM and OM fractions were pooled together analyzed by the SDS-PAGE followed by silver staining (bottom panel) and anti-TolC immunoblotting (top panel).
C. Purified TolCHis and YFREHis (0.5 µg) each were mixed with SDS-sample buffer and either boiled for 5 min (+) or incubated at room temperature (−). Proteins were separated by 10% SDS-PAGE and stained with Coomassie Brilliant Blue (CBB).
D. Purified TolCHis and YFREHis (0.5 µg) were mixed with increasing concentrations of DSP. After incubation for 30 min at room temperature, reactions were terminated by addition of Tris-HCl (pH 8.0) to final concentration 100 mM, proteins were separated by 10% SDS-PAGE and stained with silver nitrate.
To compare structural stability of TolCHis and YFREHis, we used the instrinsic resistance of TolC trimers to denaturation by SDS. Purified TolCHis appears on SDS-polyacrylamide gels as a double band because its C-terminal His-tag is cleaved at approx. R459 by E. coli proteases (see Methods and (Koronakis et al., 1997)). Without heat denaturation, a large fraction of TolCHis migrates during SDS-PAGE as a trimer with the apparent molecular mass of 130 kD (Fig. 2C). In contrast, YFREHis trimers were unstable in SDS-sample buffer and during electrophoresis, the protein completely dissociated into its monomeric state (Fig. 2C). The instability of YFREHis trimers was further confirmed using dithiobis succinimidyl propionate (DSP) cross-linking approach. More than 90% of TolCHis have been cross-linked into trimers at 0.05 mM DSP, whereas a ten-fold higher concentration of DSP was needed to cross-link trimers of YFREHis (Fig. 2D). Thus, the disruption of salt bridges destabilizes TolC trimers.
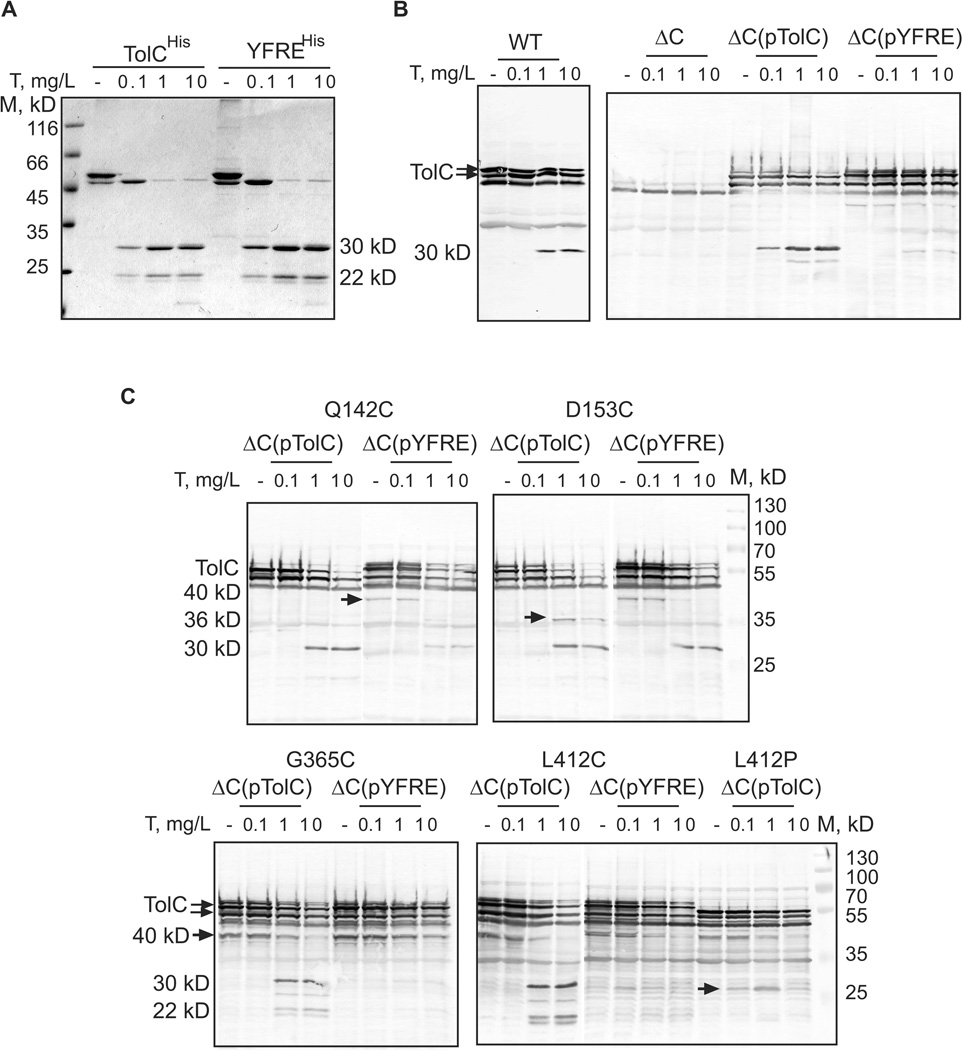
We next used limited proteolysis to compare conformations of the two TolC variants in vivo and in vitro. The proteolytic profile of the purified YFREHis was similar to that of TolCHis (Fig. 3A). Both proteins were digested by trypsin into stable 30 and 22 kD fragments that correspond to the N- and C-terminal halves of the TolC cleaved immediately following R267 located in the extracellular loop (Koronakis et al., 1997). In whole WT cells permeabilized to trypsin by osmotic shock, the chromosomally produced TolC is also digested by trypsin into a stable ~30 kDa fragment but without visible accumulation of the C-terminal 22 kD fragment (Fig. 3B). We could not detect the ~30 kDa fragment in ΔC::TolCHis and ΔC::YFREHis cells because of the low amounts of TolCHis and YFREHis proteins produced in cells (Fig. 2A). However, the plasmid-borne TolCHis produced in ΔtolC cells was also readily cleaved by trypsin with the formation of this fragment. In contrast, under the same conditions, the overproduced YFREHis was significantly more resistant to trypsin digest. Even at highest concentration of trypsin, most of YFREHis remained undigested with only trace amounts of a 30 kDa fragment showing on western blots (Fig. 3B). Taken together, these results suggest that a significant fraction of overproduced YFREHis could be misfolded in vivo, whereas YFREHis solubilized from membranes with non-ionic detergents is structurally similar to TolCHis.
Figure 3. In vivo and in vitro proteolytic profiles of TolCHis and YFREHis.
A. Tryptic digest of purified TolCHis and YFREHis. For each reaction, 0.5 µg of protein in buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl and 0.05% DDM were mixed with indicated concentrations of trypsin (T) and incubated for 15 min at 37°C. Reactions were terminated by boiling in SDS-sample buffer and analyzed by 12% SDS-PAGE followed by CBB staining.
B. Tryptic digest of WT and ΔC cells carrying the plasmid-encoded TolC variants. For each reaction, 1.8×108 cells were washed and resuspended in buffer containing 20 mM Tris-HCl (pH 7.5), 5 mM EDTA and 20% sucrose. Trypsin (T) was added to reactions in indicated concentrations. After incubation for one hour at 37°C, proteolysis was terminated by boiling in SDS-sample buffer, total proteins were separated by 12% SDS-PAGE and probed with polyclonal anti-TolC antibodies.
C. Tryptic digest of ΔC cells carrying the plasmid-encoded TolCHis and YFREHis mutants with Q142C, D153C, G365C, L412C and L412P substitutions. Reactions were set-up as described in B and analyzed by SDS-PAGE followed by anti-TolC immunostaining. Stable tryptic fragments are indicated.
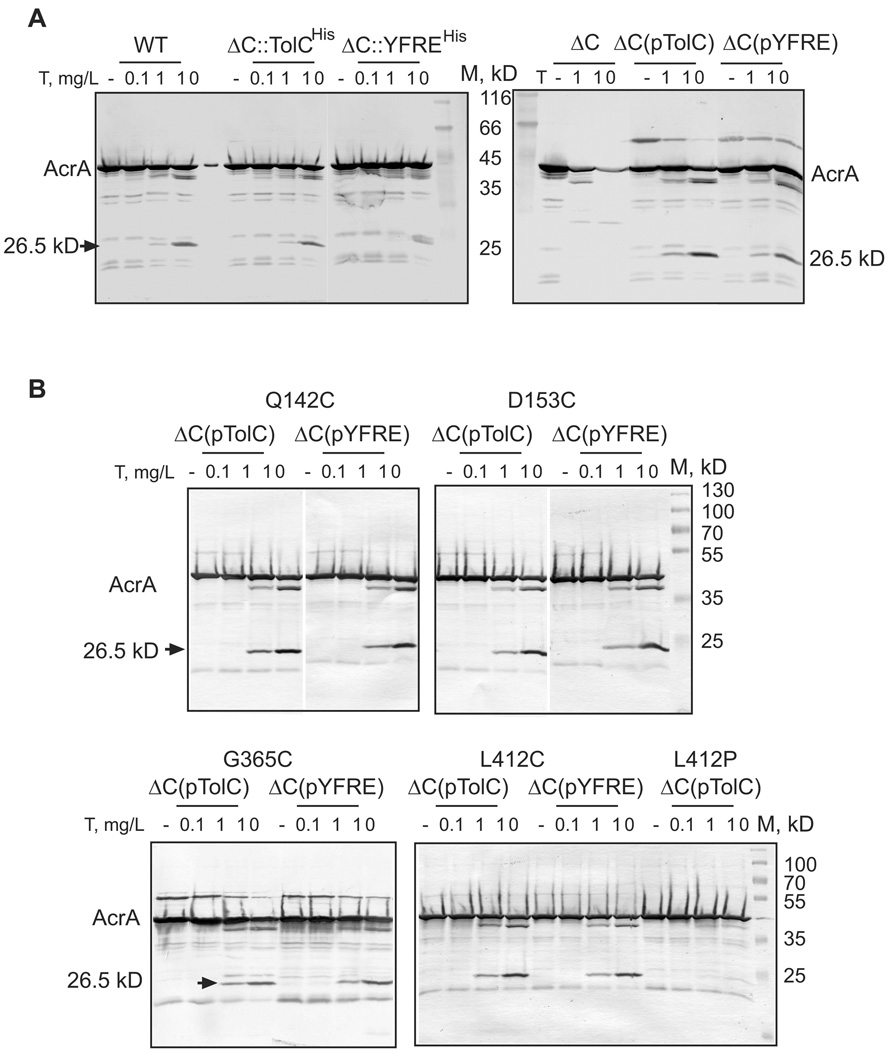
Our previous studies suggested that TolC-induced conformational changes in the membrane proximal domain of MFPs could be a limiting factor in activities of efflux pumps (Ge et al., 2009, Modali & Zgurskaya, 2011). We next determined whether YFREHis is proficient in activation of AcrAB. For this purpose, we analyzed changes in the proleolytic profile of AcrA, which is highly sensitive to assembly of the tri-partite AcrAB-TolC complex but not its bi-partite intermediates (Ge et al., 2009). Consistent with previous results, in the absence of TolC (ΔC), AcrA is rapidly digested by trypsin, as seen from the disappearance of the full length protein, without accumulation of stable products (Fig. 4A). In WT cells containing all three AcrA, AcrB and TolC components and in ΔC cells with TolCHis produced either from chromosome or from plasmid, AcrA is cleaved by trypsin into a stable 26.5 kDa fragment. This result shows that TolCHis variant interacts with AcrA and induces conformational changes in its membrane proximal domain. Surprisingly, as with MIC measurements, changes in the proteolytic profile of AcrA did not depend on concentration of TolCHis in cells. The overproduction from plasmid and the very low expression of TolCHis after re-integration of the respective gene onto chromosome resulted in the same efficiency of AcrA conversion into its proteolytically stable form (Fig. 4A). This result suggests that TolC interactions with transporters are highly dynamic. Similar to drug susceptibility phenotypes the protection of AcrA does not change with changes in amounts of TolC protein.
Figure 4. In vivo proteolytic profiles of AcrA in cells with different genetic backgrounds.
A. WT and ΔC cells carrying the plasmid-encoded and chromosomal TolCHis and YFREHis were treated with indicated concentrations of trypsin (T) as described in Fig. 3B. Total proteins were separated by 12%SDS-PAGE and probed with polyclonal anti-AcrA antibodies. The 26.5 kD tryptic fragment of AcrA protected in the presence of TolC is indicated.
B. Tryptic digest of ΔC cells carrying the plasmid-encoded TolCHis and YFREHis mutants with Q142C, D153C, G365C, L412C and L412P substitutions. Reactions were set-up and analyzed as described in A. Stable tryptic fragments are indicated.
The plasmid-borne YFREHis was effective in protection of the 26.5 kDa AcrA fragment (Fig. 4A). In contrast, only small amounts of this AcrA fragment could be detected in the trypsin-treated ΔC:: YFREHis cells with low levels of YFREHis (Fig. 4A). Thus, the reduced activity of YFREHis in drug susceptibility assays (Table 1) correlates with the low amounts of the properly folded YFREHis in cells (Fig. 2A) and the decreased amounts of the protected AcrA protein (Fig. 4A). We conclude that the defects in assembly of TolC-AcrAB complex contribute to the increased susceptibility of YFREHis producing cells to antibiotics.
Cysteine-scanning mutagenesis of TolC
To further characterize the permeability and conformations of TolC in vivo, we introduced 16 unique cysteine residues into the four major regions of TolC: (i) an external loop, (ii) internal walls of the channel, (iii) both inside and outside of the periplasmic tip and (iv) the equatorial domain (Fig. 1 and Table 2). The same set of cysteine residues was introduced into TolCHis and YFREHis. As seen in Table 3, most of the mutations either in TolCHis or YFREHis had no effect on their activity in multidrug efflux. The exception is D153, which forms a critical salt bridge with R367 and contributes to constriction of TolC pore from the periplasmic side (Fig. 1, BN2). The D153C mutation increased susceptibility of TolCHis to erythromycin (16 fold) and novobiocin (2 fold) but not to puromycin and SDS. On the other hand, the same D153C substitution did not further compromise the activity of YFREHis and in fact partially (2 fold increase) rescued the erythromycin and puromycin susceptibilities of this TolC variant. Interestingly, the reciprocal R367C substitution did not affect drug susceptibility of TolCHis, which contrasts with the previous finding that either D153A or R367S substitutions increase conductance of the purified of TolC channel and their simultaneous disruption has an additive effect on TolC conductance (Andersen et al., 2002a). This result suggests that D153 of TolC has an additional role in drug efflux, which is different from the constriction of TolC pore.
Table 2.
TolC mutants used in this study.
| Mutations in TolC |
Location in structure (color in Fig. 1) |
Known phenotypes | References |
|---|---|---|---|
| Y362F R367E |
Outer bottleneck BN2 (red) | Increases conductance, dilates the constriction | (Bavro et al., 2008, Pei et al., 2011) |
| R367C | Outer bottleneck (red) | R367S, R367E, R367H disrupt a salt bridge with D153 | (Andersen et al., 2002a, Bavro et al., 2008) |
| D153C | Outer bottleneck (red) | D153A disrupts a salt bridge with R367, increases conductance | (Andersen et al., 2002a) |
| D374C | Inner bottleneck BN1 (red) | D374A increases conductance | (Andersen et al., 2002b, Higgins et al., 2004, Eswaran et al., 2003) |
| Q139C | α-helix 3 (H3) of the outer coiled coil (blue) | Cross-links to AcrA in vivo | (Lobedanz et al., 2007) |
| Q142C | α-helix 3 (H3) of the outer coiled coil (blue) | Cross-links to AcrA and AcrB in vivo | (Lobedanz et al., 2007, Tamura et al., 2005) |
| G147C | H3/H4 Turn 1 (blue) | G147A affects folding/stability | (Weeks et al., 2010) |
| S363C | α-helix 7(H7) of the inner coiled coil (blue) | Cross-links to AcrA | (Lobedanz et al., 2007) |
| G365C | H7/H8 Turn 2 (blue) | Cross-links to AcrB | (Bavro et al., 2008, Tamura et al., 2005) |
| D356C | α-helix 7(H7) of the inner coiled coil (blue) | NA | NA |
| Q384C | H8 at the bottom of the α/β equatorial domain (magenta) | NA | NA |
| N392C | H8 at the bottom of the α/β equatorial domain (magenta) | NA | NA |
| S322C | H7, on the top of α/β domain (magenta) | NA | NA |
| L412C and L412P |
H9 of α/β domain (yellow) | Mutations in L412 affect drug efflux | (Yamanaka et al., 2002) |
| A312C | H7, inside the channel (black) | NA | NA |
| A34C | H2, inside the channel (black) | NA | NA |
| A269C | Extracellular loop (black) | NA | NA |
Table 3.
Impact of cysteine substitutions on activities of TolCHis and YFREHis in drug susceptibility complementation assays.
| Cys position | Relative MICsa | |||||||
|---|---|---|---|---|---|---|---|---|
| PUR | NOV | ERY | SDS | |||||
| TolCHis | YFREHis | TolCHis | YFREHis | TolCHis | YFREHis | TolCHis | YFREHis | |
| A269C | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 1 |
| A34C | 1 | 1 | 1 | 1 | 0.5 | 1 | 1 | 1 |
| A312C | 1 | 1 | 1 | 1 | 0.5 | 2 | 1 | 1 |
| S322C | 0.5 | 2 | 0.5 | 1 | 1 | 2 | 1 | 1 |
| N392C(ch)b | 1(1) | 2 | 1(1) | 1 | 1(1) | 2 | 1(1) | 1 |
| Q384C | 1 | 2 | 1 | 2 | 2 | 2 | 1 | 1 |
| D374C(ch)b | 1(1) | 1 | 1(1) | 0.5 | 1(1) | 1 | 1(1) | 1 |
| R367C | 1 | - | 1 | - | 0.5 | - | 1 | - |
| D153C(ch)b | 1(1) | 2 | 0.5(0.5) | 1 | 0.06(0.25) | 2 | 1(1) | 1 |
| G365C | 1 | 0.5 | 1 | 0.25 | 1 | 1 | 1 | 0.03 |
| S363C(ch)b | 1(1) | 1 | 1(1) | 1 | 1(1) | 2 | 1(1) | 1 |
| G147C(ch)b | 1(1) | 1 | 0.5(1) | 1 | 1(1) | 2 | 1(1) | 1 |
| Q142C | 1 | 2 | 1 | 0.5 | 1 | 2 | 1 | 1 |
| Q139C | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 |
| D356C(ch)b | 1(1) | 1 | 1(1) | 1 | 0.5(1) | 2 | 1(1) | 1 |
| L412C(ch)b | 2(1) | 1 | 1(1) | 0.5 | 1(1) | 2 | 1(1) | 1 |
| L412P | 0.125 | - | 0.25 | - | 0.25 | - | 0.008 | - |
Relative MICs were calculates as ratios between MICs for GD100 cells containing the plasmid -encoded mutant and wild-type TolCHis or YFREHis, respectively.
Relative MICs for cells carrying chromosomal (ch) TolCHis and its variants are shown in parentheses. See footnote of Table 1 for abbreviations. Data from at least two independent experiments are shown.
The only substitution that further compromised YFREHis function was G365C located in the turn region between H7 and H8 (Turn 2) of TolC. This mutant is more susceptible to all tested antimicrobials including the detergent SDS and at the same time, is less susceptible than the parent YFREHis to vancomycin (Table 4 and see below). In the crystal structure of the open TolCYFRS, G365 moves by ~7 Å from its position in the closed protein, due to realignment of H7 and H8 helices upon disruption Y362 and R367 bonds. The effect of this substitution on TolC function appears to be additive to and different from those of Y362F and R367E.
Table 4.
Vancomycin susceptibility of E.coli GD100 (acrAB+tolC−) and KG101 (acrAB−tolC−) containing the plasmid-encoded mutant and wild-type TolCHis or YFREHis.
|
Cys position |
MICs (µg/ml) | |||
|---|---|---|---|---|
| acrAB+tolC− | acrAB−tolC− | |||
| TolCHis | YFREHis | TolCHis | YFREHis | |
| - | 78–156 | 39–78 | 312 | 78 |
| A269C | 156 | 39 | 156 | 156 |
| A34C | 39 | 39 | 156 | 78 |
| A312C | 78 | 39 | 156 | 78 |
| S322C | 156 | 78 | 312 | 78 |
| N392C | 156 | 39 | 156 | 78 |
| Q384C | 78 | 19.5 | 156 | 78 |
| D374C | 156 | 39 | 312 | 78 |
| R367C | 39 | nd | 78 | nd |
| D153C(ch) | 19.5 (78) | 39 | 19.5 (78) | 78 |
| G365C | 156 | 78 | 156 | 156 |
| S363C | 156 | 19.5 | 312 | 78 |
| G147C | 78 | 78 | 312 | 156 |
| Q142C | 156 | 19.5 | 312 | 78 |
| Q139C | 78 | 39 | 156 | 39 |
| D356C | 156 | 78 | 312 | 78 |
| L412C | 78 | 156 | 156 | 78 |
| L412P | 156 | nd | nd | nd |
MICs of vancomycin for cells carrying chromosomal (ch) D153C variant are shown in parentheses. Data from at least three independent experiments are shown.
The role of the equatorial α/β domain in TolC function is still obscure but genetic studies suggested that it could play a role in opening of TolC (Bavro et al., 2008, Weeks et al., 2010, Augustus et al., 2004). The L412 residue in the helix H9 of the α/β domain was previously identified as important for TolC function (Yamanaka et al., 2002). Although the L412C substitution did not significantly change MICs of antimicrobials, a more drastic L412P substitution significantly diminished TolC activity as seen from decreased MICs of all tested AcrAB-TolC substrates (Table 3).
To determine whether changes in MICs correlate with changes in TolC structure, we compared proteolytic profiles of D153C, G365C and L412 TolC mutants. As an additional control we used Q142C mutants, which showed minor changes in drug susceptibilities assay (Table 3). Both TolCHis and YFREHis containing Q142C were indistinguishable from the parent variants (Fig. 3C). The proteolytic pattern of D153C contained a unique 35 kD fragment, which is not present in patterns of any other mutants, indicating that the conformation of this mutant is different from that of TolCHis. In addition, D153C appears to partially rescue YFREHis conformation as seen from the D153CYFRE proteolytic pattern, which contains a 30 kD fragment characteristic of TolCHis but not YFREHis patterns. In agreement, D153CYFRE was less susceptible to erythromycin and puromycin than YFREHis.
The proteolytic patterns of G365C and L412C were similar to each other and different from other mutants (Fig. 3C). In addition to a characteristic 30 kD tryptic product, profiles of these mutants contained a ~40 kD fragment apparently generated by internal proteases, and ~21/22 kD fragments. The 40 kD band is also prominent in G365CYFRE and L412CYFRE, but as in other YFREHis derivatives the 30 kD fragment is not detectable. The most damaging L412P substitution destabilized the 30 kD fragment even in TolCHis (Fig. 3C, indicated by an arrow). Thus, despite distant locations, substitutions in G365 (turn 2) and L412 (α/β domain) induce similar changes in the TolC proteolytic profiles. It appears that the loss of the 30 kD fragment in TolC patterns correlates with increased drug susceptibility of the corresponding cells, whereas this fragment is present in functionally proficient TolC variants.
In agreement with their partial activities in drug efflux (Table 3), TolC variants containing D153C, G365C and L412C interacted with AcrA as seen from the accumulation of a 26.5 kD fragment of AcrA in cells producing these proteins (Fig. 4B). The L412P mutant with the most severe phenotype also failed to protect the 26.5 kD fragment of AcrA. Thus, the loss of activity of this mutant is likely due to the loss of its functional association with AcrAB.
Substrate path in TolC channel
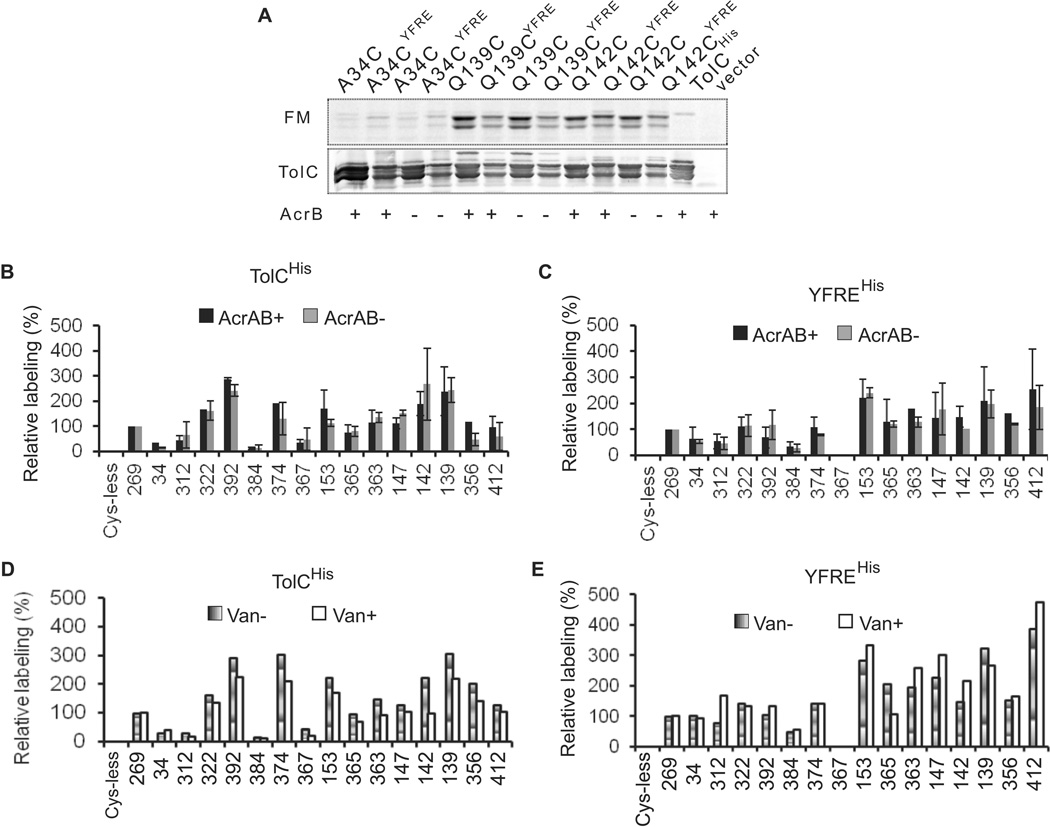
Combined mutagenesis-conductance experiments and structural studies indicated the existence of two periplasmic bottlenecks that constrict the TolC channel (Andersen et al., 2002a, Bavro et al., 2008, Pei et al., 2011). During transport these constrictions are thought to be released by interactions with AcrAB transporter. To identify residues of TolC that are exposed to its substrates, acrAB plus cells carrying various Cys-containing TolC constructs were incubated with a thiol-reacting probe fluorescein-5-maleimide (FM), the substrate of AcrAB-TolC transporter (Kim & Nikaido, 2012). The fluorescence intensity of protein bands was normalized to the amounts of respective proteins. The efficiency of labeling was expressed as a percentage of the fluorescence intensity of TolC with A269C in the extracellular loop, which is expected to be readily accessible to FM in external medium. Fig. 5 shows that accessibility of Cys residues differs notably in TolCHis and YFREHis backgrounds. The largest differences between the two variants are seen in their periplasmic tips. Cys residues in positions 153 (BN2), 365 and 356 (turn 2) are more accessible to FM in YFREHis than in TolCHis protein, whereas Cys in position 142 (turn 1) was better labeled in TolCHis than in YFREHis (Fig. 5B and 5C). This result is consistent with structural analyses that demonstrated a significant rearrangement in the periplasmic tip of TolC brought about by disruption of salt bridges holding together the inner and outer coiled coils (Bavro et al., 2008, Pei et al., 2011, Weeks et al., 2010). This dilation of the periplasmic tip of TolC apparently affects the equatorial domain as well. The L412C located in the external H9 of the α/β domain was better labeled in YFREHis, whereas S322C and N392C in the inner H7 and H8, respectively, were more accessible to FM in TolCHis. In both variants, Q384C, also located in the proximity to the α/β domain, was labeled only slightly above the background, suggesting that this residue has very limited contact with FM.
Figure 5. Labeling of the plasmid-borne TolC-Cys mutants with fluorescein-5-maleimide (FM).
A. Representative fluorescence labeling profiles of TolCHis and YFREHis mutants. E. coli ΔtolC or ΔacrAB ΔtolC cells carrying the indicated plasmids-encoded TolC variants were incubated with 250 µM FM for 30 min and reactions terminated by addition of 10 mM DTT. Membrane fractions were isolated by ultracentrifugation and 5 µg of total protein separated by 12% SDS-PAGE. Fluorescent proteins in gels were visualized using Storm Imager (top panel, FM). The same gels were next electroblotted onto PVDF membrane and probed with polyclonal anti-TolC antibody (bottom panel, TolC).
B. Relative intensities of fluorescein-labeled TolCHis and YFREHis mutants. E. coli ΔtolC or ΔacrAB ΔtolC cells carrying the indicated plasmids-encoded TolCHis variants were labeled with FM and analyzed as described in A. ImageQuant software was used to quantify fluorescence intensities and immunostaining of bands corresponding to TolC. Fluorescence intensities of each band normalized to amounts of each mutant protein were expressed as a percent of the normalized fluorescence intensity of A269C mutant.
C. The same as in B but cells carried plasmid-encoded YFREHis variants.
D. E. coli ΔtolC cells carrying the indicated plasmids-encoded TolCHis variants were pre-incubated with 2.5 mM vancomycin for 30 min, then labeled with FM and analyzed as described in A.
E. The same as in C but cells carried plasmid-encoded YFREHis variants.
Surprisingly, D374C (BN1) was labeled with FM in both TolCHis and YFREHis variants but this residue was consistently better labeled in TolCHis. Side chains of D374 form a ring of negative charges that constricts the channel to a diameter of ~0.44 nm with the distance of only 0.61–0.62 nm between adjunct D374 side chains (Koronakis et al., 2000). With the average diameter of 0.69+/−0.2 nm for an FM molecule, this constriction is not passable for this probe. However, Cys residue in position 374 is well exposed to FM, indicating that with D374C substitution, the pore of TolC is less constricted than in the native protein. In addition, previous conductance studies suggested that substitutions in position 374 do not affect the second constriction BN2. We found that D374C remains accessible to FM whether BN2 is intact (TolCHis) or disrupted by removal of critical salt bridges (YFREHis). This result suggested that D374C is accessible from both the extracellular and periplasmic sides. The same is true for the residues located in BN2. In the crystal structure of YFREHis, the D374 pore remains closed (Bavro et al., 2008, Pei et al., 2011). Yet, D153C was fully accessible to FM in both TolCHis and YFREHis, indicating that FM likely gains its access to this Cys from both sides of the outer membrane.
Taken together these results demonstrate that TolC is largely accessible to small molecules from both the external medium and from the periplasm. The labeling profiles of TolCHis and YFREHis differ from each other, providing further evidence that these two variants adopt different conformations in vivo. The increased labeling of the periplasmic tip of YFREHis is consistent with the large movement of coiled-coils seen in crystal structures of this mutant.
TolC channel is not occluded in the periplasm
The FM labeling described above outlined the dominant states of TolCHis and YFREHis in vivo in the presence of AcrAB. In the proposed models of AcrAB-TolC complex (Bavro et al., 2008, Symmons et al., 2009), AcrA and AcrB form multiple contacts with the periplasmic tip and α-helical domain of TolC and presumably enable the close-to-open conformational transition of the channel. To determine whether these AcrAB-TolC interactions can be visualized in vivo, we next determined the accessibility of Cys-containing TolCHis and YFREHis mutants to FM in cells lacking AcrAB pump. We found that in the absence of AcrB the labeling profiles of TolCHis and YFREHis remain different and that these profiles do not change significantly when compared to those of acrAB plus cells (Figs. 5B and 5C). Thus, the dominant conformations of TolCHis and YFREHis are not sensitive to the presence of AcrAB.
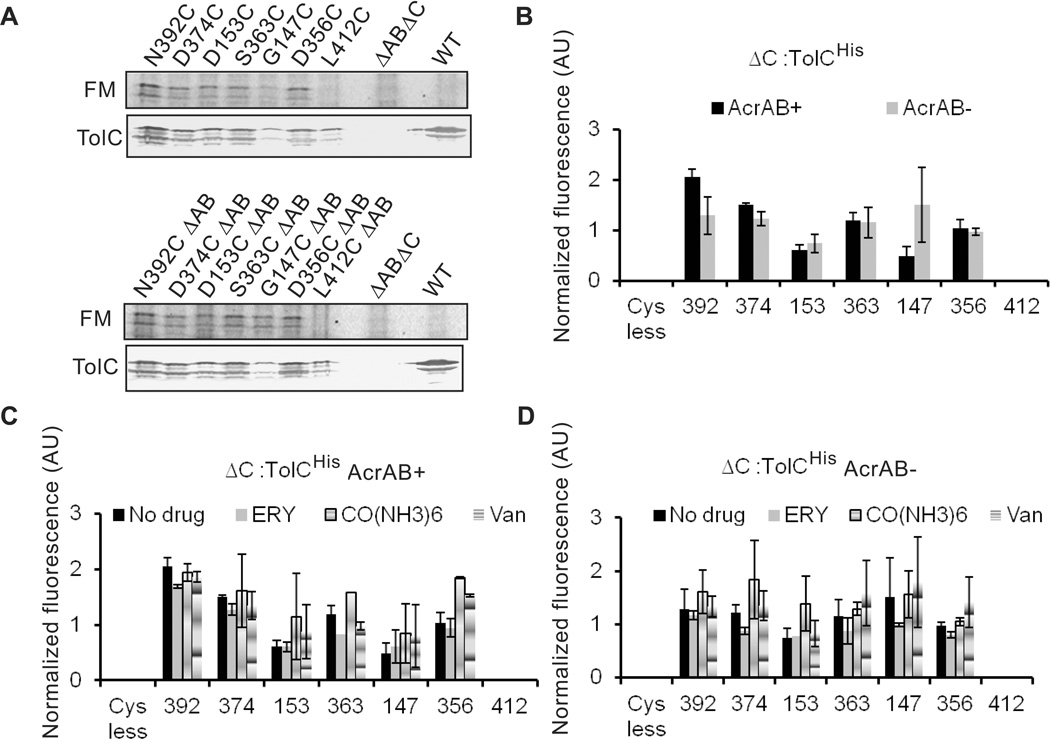
Overproduction of TolC from plasmids could potentially bias the FM labeling experiments toward free TolC, which is not engaged into transport complexes. Therefore, we next re-integrated genes encoding TolCHis with G147C, D153C, D356C, S363C, D374C, N392C and L412C substitutions back into the native tolC locus of acrAB plus and minus strains. We selected these TolC variants because their cysteine residues were labeled differently in the plasmid-borne TolCHis and YFREHis proteins (Fig. 5B and 5C); in addition, D153C substitution significantly reduced TolCHis function in drug efflux (Table 3). As with TolCHis, when compared to the basal level of TolC in the WT cells, integration onto a chromosome led to about 5–10 fold reduction in amounts of all Cys-containing TolC variants (Fig. 6A). However, only traces of G147C could be detected in membranes. This result is consistent with previous reports that TolC with substitutions in G147 position is expressed at lower levels, possibly because of misfolding and loss of stability (Weeks et al., 2010). AcrAB had no effect on the expression levels of TolC variants. However, even at such reduced amounts, all TolC-Cys variants conferred the levels of antibiotic resistance identical to those of the plasmid-borne proteins (Table 3). ΔC::D153C cells were a surprising exception being less susceptible to erythromycin than cells carrying D153C variant on a plasmid. This result suggests that the high copy number of the plasmid-borne D153C variant contributes to the increased susceptibility to erythromycin.
Figure 6. Labeling of chromosomally produced TolC-Cys mutants with fluoresceine-5-maleimide (FM).
A. Representative fluorescence labeling profiles of TolCHis variants containing indicated Cys substitutions. TolCHis variants were re-integrated into the native locus of either GD102 (acrAB+) or KG101 (acrAB−) chromosomes. Cells were labeled, membrane fractions isolated and analyzed as described in Fig. 5A.
B. Relative intensities of fluorescein-labeled TolCHis mutants. E. coli acrAB plus and minus cells carrying the indicated TolCHis variants on chromosomes were labeled with FM and analyzed as described in Fig. 5A. ImageQuant software was used to quantify fluorescence intensities and immunostaining of bands corresponding to TolC. Fluorescence intensities of each band were normalized to amounts of each mutant protein.
C. E. coli cells carrying the indicated chromosome-encoded TolCHis variants were pre-incubated for 30 min with one of 2.5 mM erythromycin, 2.5 mM cobalt hexamine trichloride or 2.5 mM vancomycin, then labeled with FM and analyzed as described in B.
D. The same as C but cells do not produce AcrAB pump.
Intriguingly, even when produced at very low levels, Cys residues in the two constriction bottlenecks of TolC channel (D153C, D374C and S363C) were still labeled with FM and their accessibilities were similar to the plasmid-borne proteins (Fig. 6). Furthermore, D374C and N392C of TolC were somewhat better labeled with FM in cells carrying acrAB. At the same time, G147C was consistently better labeled in acrAB minus cells. These labeling profiles remained the same in cells overproducing MacAB macrolide efflux pump or in cells producing colicin exporter CvaAB (data not shown). This result strongly suggests that even when present at the level significantly below that of transporters, the periplasmic tip of TolC is not occluded and the D374 pore is accessible for small molecules. The addition of erythromycin, a substrate of AcrAB-TolC, or cobalt hexamine, a TolC inhibitor which binds to D374 (Andersen et al., 2002b), did not affect labeling profiles of chromosomally produced TolC-Cys variants (Fig. 6C and 6D).
Effect of vancomycin on drug susceptibility and FM labeling of TolC mutants
Vancomycin is a large 3.2 nm×2.2 nm molecule with multiple functionalities for potential interactions and hydrogen bonding. The efficacy of this antibiotic in Gram-negative bacteria is thought to be low because of the slow diffusion across the outer membrane. Previously, the expression of YFREHis in a tolC null strain of E. coli was found to increase susceptibility to vancomycin, independent of AcrA or AcrB (Bavro et al., 2008). This result suggested that these mutations facilitate the passive diffusion of vancomycin across the outer membrane. If YFREHis compromises the permeability of the outer membrane, one would expect that susceptibility to vancomycin will vary with amounts of the protein. We found however that ΔC:: YFREHis cells producing very low amounts of this protein and cells carrying the plasmid-encoded YFREHis reduce MIC of vancomycin by the same 2–4 fold, whereas the deletion of tolC gene increases the vancomycin MIC by two fold (Table 1). Thus, similar to other antibiotics susceptibility to vancomycin is not affected by large changes in TolC concentrations. This lack of concentration dependence suggests that by itself YFREHis does not increase diffusion of vancomycin across the outer membrane.
Most of the Cys substitutions did not affect vancomycin susceptibility of cells carrying the plasmid-encoded TolC variants (Table 4). The notable exception is D153C, which sensitized cells to vancomycin when present in TolCHis to a higher degree than Y362F and R367E mutations combined, but had no effect in YFREHis mutant. Interestingly, the effect of D153C on vancomycin susceptibility was dependent on the amounts of this protein in cells. Cells producing the chromosomally-encoded D153C and the WT were killed by 78–156 µg/ml of vancomycin, whereas cells carrying the plasmid-borne D153C were 4–8 fold more susceptible to this antibiotic. This dependency of MICs on the amounts of D153C in cells was also seen for erythromycin (Table 3), indicating that increased influx of antibiotics through D153C could be the major reason for the drop in MICs. In addition to D153C, cells carrying A34C, R367C, Q142CYFRE, S363CYFRE and Q384CYFRE variants were two-fold more susceptible to vancomycin than cells producing the respective TolCHis or YFREHis variants. In contrast, cells producing L412CYFRE were two-fold more resistant to this antibiotic than YFREHis.
For the majority of the TolC constructs, the increased susceptibility of cells to vancomycin was also dependent on the presence of AcrAB. On average, cells lacking acrAB genes were less susceptible to vancomycin than their acrAB plus counterparts (Table 4). But AcrAB dependence was more pronounced for cells with TolCHis than for the cells with YFREHis constructs. The exceptions are the acrAB null cells carrying D153C and R367C mutants that were as susceptible to vancomycin as those producing AcrAB. Since both these mutants disrupt the periplasmic constriction of TolC, this finding is in the line with the previous observations that open TolC mutants sensitize E. coli cells to vancomycin independently whether they are bound by AcrAB or not (Bavro et al., 2008).
Given its large size, vancomycin entering TolC channel from the external medium might be expected to reduce or even block accessibility of Cys located on the internal walls of the channel and in the constriction pore. We found however, that in cells preincubated with a ten-fold excess of vancomycin over FM, Cys-containing TolCHis and YFREHis variants were labeled to the same extent in cells without vancomycin (Fig. 5D, 5E). The same result was obtained in cells carrying TolC-Cys variants on the chromosome (Fig. 6C and 6D). In the reverse assay, the presence of 250 µM of FM had no effect on MICs of vancomycin in AcrAB+ cells with Cys-containing TolC constructs (data not shown). Thus, similar to other substrates, vancomycin and FM do not compete for the same sites neither in labeling nor in drug susceptibilities assays.
Discussion
In this study, we compared in vivo properties and activities of two the best characterized TolC variants. Cells carrying YFREHis mutant were consistently more susceptible to macrolides and puromycin than those with the parental TolCHis variant. Surprisingly, drug susceptibilities of cells did not match large changes in concentrations of either of these TolC variants (Table 1). When the concentration of TolC was lower or higher than that in WT cells by at least ten fold in both directions, the MICs of the broad range of antibiotics in Table 1 remained unchanged. This result suggested that even when present at very low concentrations TolC does not limit multidrug efflux capacity.
If we assume that antibiotics expelled through TolC are delivered only by AcrAB pump, then the rate of transport through TolC channel is limited by AcrAB capacity (Vmax). By reducing the number of TolC available for efflux, one could expect a decrease in MICs when TolC becomes limiting. With TolCHis this did not happen even when we reduced the number of TolC significantly (Table 1, Fig. 2A and 6A). Thus, under tested conditions AcrAB-TolC operates below its capacity and TolC does not limit drug efflux. We envision three possible explanations for this result: (i) only a small fraction of AcrAB-TolC operates in cells and this low number is sufficient to maintain resistance (for example, a single AcrAB-TolC complex satisfies all efflux needs); (ii) drug efflux does not need to be continuous to provide resistance (efflux is active only at certain threshold concentrations of drugs); (iii) one TolC trimer keeps several AcrAB pumps operational (TolC has a catalytic/regulatory role and is not part of the active complex). A large body of evidence supports the notion that the active efflux complex contains TolC. Hence, a small number of AcrAB-TolC complexes and non-continuous efflux are the most likely explanations for why antibiotic susceptibilities do not change in a broad range of TolC concentrations.
Chemical cross-linking and co-purification approaches do not provide accuracy needed to quantify TolC assembled into AcrAB-TolC complexes. To determine whether amounts of AcrAB-TolC complexes vary with TolC, we used the in vivo proteolytic patterns of AcrA (Fig. 4). We previously showed that the appearance of 26.5 kD fragment of AcrA reports on conformational changes in the membrane proximal domain of AcrA that are specific for the assembly of the tri-partite AcrAB-TolC but not the bi-partite AcrA-AcrB or AcrA-TolC complexes (Ge et al., 2009). The 26.5 kD fragment in the proteolytic patterns of AcrA also correlates with the functionality of TolC in drug efflux. Cells producing L412P mutant with the most severe reduction in drug susceptibility phenotype lacked this fragment (Table 3 and Fig. 4B). Surprisingly, despite variations in TolC amounts by several orders of magnitude, we found only small differences in the amounts of 26.5 kD fragment of AcrA in ΔC::TolCHis cells, WT and ΔC(pTolC) cells carrying the plasmid-encoded TolCHis (Fig. 4A). This result is consistent with the lack of changes in MICs and demonstrates that amounts of AcrAB-TolC complexes remain mostly constant in the range of TolCHis concentrations tested in this study. We conclude that the assembly and function of AcrAB-TolC complex are not driven by amounts of TolC protein. Since E. coli cells produce AcrAB in excess over TolC (Tikhonova & Zgurskaya, 2004), AcrAB is also unlikely to limit the number of AcrAB-TolC complexes. In agreement, overproduction of AcrAB in the WT E. coli cells results in a two fold increase in MICs of some (for example, puromycin) but not other (chloramphenicol) antibiotics (Table 1). Therefore, only a certain fraction of TolC is accessible to AcrAB to form a complex. This result further suggests that the assembly of AcrAB-TolC complex could be regulated allosterically. In the case of TolC-dependent type I secretion systems such as CvaAB and HlyBD, the regulation of the complex assembly by substrate has been demonstrated before using the in vivo cross-linking and proteolysis experiments (Balakrishnan et al., 2001, Hwang et al., 1997). However, in similar experiments as well as in studies of protein-protein interactions in vitro, no effect of small molecule substrates and inhibitors was found for AcrAB-TolC (Tikhonova et al., 2011, Tikhonova & Zgurskaya, 2004, Touze et al., 2004). It appears that yet unknown factors could be involved in regulation of AcrAB-TolC assembly in vivo.
Current models supported by structural and genetic studies suggest that in the assembled complex TolC undergoes a close-to-open conformational transition (Misra & Bavro, 2009, Symmons et al., 2009). FM labeling of Cys residues positioned inside of TolC channel, its two constriction bottlenecks and the periplasmic tip clearly showed that the structural differences between TolCHis and YFREHis persist in vivo (Fig. 5). In particular, TolC residues in the periplasmic tip that underwent significant changes upon mutational opening were more accessible to FM in the open YFREHis than in the closed TolCHis variant. These conformational differences however were not limited to the periplasmic tip of TolC and also involved the α/β equatorial domain (N392 and L412) and the D374 constriction pore. The impact of Y362F/R367E substitutions on the overall structure of TolC is also obvious in the SDS stability and proteolytic profiles (Figs 2 and 3). Purified YFREHis trimers were unstable during SDS-PAGE and required higher concentrations of DSP to be cross-linked (Fig. 2C, 2D). In vivo, YFREHis resisted the tryptic digest with only small amounts of the protein cleaved into a 30 kD fragment characteristic for TolCHis. A similar defect in tryptic profile was seen for an even more functionally deficient L412P mutant. Also, at low concentrations, such as in ΔC::YFREHis cells, YFREHis did not protect the 26.5 kD fragment of AcrA as effectively as TolCHis. These results show that YFREHis is structurally defective and fails to establish the same interactions with AcrA as TolCHis. This conclusion is further supported by recent kinetic studies that showed that the affinity of YFREHis mutant to both AcrA and AcrB is lower than that of the closed TolCHis (Tikhonova et al., 2011).
Yet, both TolCHis and YFREHis variants are incorporated into AcrAB complexes and confer similar levels of antibiotic resistance to most of AcrAB substrates except macrolides and puromycin (Table 3). The interactions with AcrAB however did not bring detectable changes to conformation of either one of TolC variants. The accessibility of TolC Cys residues to FM was largely the same in acrAB plus and minus backgrounds (Fig. 5 and 6). The only exception is the chromosomally-produced G147C mutant, which was consistently better labeled with FM in acrAB minus than in acrAB plus cells (Fig. 6). It appears that the amount of G147C in the membrane is so low that a significant fraction of this TolC variant is assembled into the complex with AcrAB and can be detected through differences in accessibility to FM.
For other TolC mutants, overproduction of various TolC-dependent transporters did not block the FM access to the periplasmic entrance of TolC. Even when expressed at levels below those of WT cells, the Cys in position 363, which was previously found to be cross-linked to AcrA (Lobedanz et al., 2007), was still labeled with FM (Fig. 6B). Although sensitive enough to detect the differences in conformations of TolCHis and YFREHis, the FM labeling did not provide evidence that AcrAB or other transporters induce conformational changes in TolC channel. This result however implies that if such changes occur, they are limited to a minor fraction of intracellular TolC.
If the periplasmic entrance of TolC is not occluded, one would expect that increasing amounts of TolC variants with disrupted constriction pores will increase the rate of diffusion of drugs across the outer membrane and therefore, increase E. coli susceptibility to drugs. The phenotype of D153C mutant meets this expectation. MICs of both erythromycin and vancomycin decreased with increasing amounts of D153C in cells, suggesting that the elevated levels of this TolC variant compromise the permeability of the outer membrane (Tables 3 and 4). The overproduction of D153C seems to significantly increase influx of these anibiotics into the cell and in the case of erythromycin the rate of influx exceeds its active efflux by AcrAB-G153C pump. In contrast, the susceptibility to antibiotics of cells carrying YFREHis was mostly unaffected by changes in the intracellular concentrations of this protein (Table 1). Perhaps, only a small fraction of the overproduced YFREHis is inserted into the outer membrane, limiting influx of vancomycin through this channel. In agreement, MICs of erythromycin and oleandomycin increased with increasing amounts of YFREHis, further suggesting that assembly of this protein into a complex with AcrAB is not efficient and limits efflux of these antibiotics.
As seen in Table 4, with the exception of D153C, vancomycin susceptibility of cells that produce either TolCHis or YFREHis is higher in the presence of AcrAB. This result suggests that changes in vancomycin susceptibility are linked to the activity of AcrAB-TolC pump. One possible interpretation of this AcrAB dependence is that vancomycin leaks through TolC, which is engaged by AcrAB transporter (Weeks et al., 2010). We found no competition between vancomycin and FM in drug susceptibility and labeling assays. It is possible that vancomycin diffusion through TolC is very fast without interactions with cysteine residues. Alternatively, cysteines in selected positions are accessible to FM but not to vancomycin because of steric constrains.
The localization in the outer membrane, its structure and the proposed mechanism indicate that TolC could be a good target for drug efflux inhibitors. However, various whole cell screens so far did not identify a single compound that could block the activity of TolC channel in vivo. In the conductance experiments trivalent hexamine cobalt bound the D374 pore of TolC with low nanomolar affinity and blocked the channel but the compound is toxic to E.coli and its efficacy in inhibition of efflux could not be tested (Higgins et al., 2004). In the in vivo experiments we found that the ten-fold molar excess of cobalt hexamine did not inhibit FM labeling of chromosomal TolC-Cys variants (Fig. 6C and 6D). Furthermore, D374C residues were accessible to FM modification in both closed and open conformers of TolC, better so in TolCHis than in YFREHis (Fig. 5), suggesting that D374 pore is flexible and passable to FM from both extracellular and periplasmic sides of the outer membrane. The disruption of R367-D153 and Y362-R367 links in the periplasmic tip of TolC has a long range effects on the structure of the entire α-helical and equatorial domains of TolC that negatively impact the interactions with AcrAB transporter. It is conceivable therefore that these salt bridges remain intact in the active AcrAB-TolC complex and drug efflux does not require large conformational rearrangements in the periplasmic entrance of the channel.
An additional problem with targeting TolC for development of drug efflux inhibitors is the finding that the activity and assembly of AcrAB-TolC tolerates significant depletion of TolC protein through changes in its expression levels or mutational modifications. We estimate that ΔC::YFREHis cells contain about 100 copies of YFREHis, of those even smaller number is functional. Yet, with the exception of macrolides, drug susceptibility profiles of these cells are identical to those of WT cells.
Experimental Procedures
Construction of strains and mutants
TolC mutants were constructed using Quick change site-directed mutagenesis lightning kit (Agilent technologies) using pTolC as template (Tikhonova et al., 2009). All mutations were verified by sequencing at OMRF sequencing facility. E. coli strains GD100 (ΔtolC) and GD102 (ΔtolC–ygiBC) described previously (Dhamdhere & Zgurskaya, 2010), and KG101 (ΔtolC ΔacrAB) constructed in this study, are derivatives of BW25113 lacIqrrnBT14ΔlacZWJ16hsdR514ΔaraBADAH33ΔΔrhaBADLD78) (Datsenko & Wanner, 2000). TolCHis and its Cys-containing variants were re-integrated into the native locus of GD102 chromosome using primers homologous to 48 bp upstream of the first codon of tolC genes and 38 bp downstream from the stop codon of ygiC gene. We previously showed that the lack of ygiBC, the two genes downstream of tolC, does not affect localization, folding and activity of TolC (Dhamdhere & Zgurskaya, 2010). All mutants were constructed using λ red recombinase system and antibiotic resistance genes were removed as described in Datsenko and Wanner, 2000.
Growth conditions and MIC determination
All strains grown in Luria-Bertani (LB) broth with ampicillin (100 µg/ml) at 37°C with shaking at 200 rpm. MICs of various antimicrobial agents were measured using the two-fold dilution technique in 96-well microtiter plates. For this purpose, exponentially growing cultures (OD600 ~1.0) were inoculated at a density of 104 cells per ml into LB medium containing ampicillin (100 µg/ml) in the presence of two-fold increasing concentrations of drugs under investigation. Cell growth was determined visually after incubation of the microtiter plates at 37°C for 16 h.
Protein purification and analyses
TolCHis and YFREHis were purified as described before (Tikhonova et al., 2009). Purified TolC (as well as TolC in membrane fractions) appears on gels as two bands with only the top band reacting with a monoclonal anti-6His antibody. This result and previous studies (Koronakis et al., 1997) suggested that the lower band is the product of the proteolytic cleavage at the C-terminal R459 of TolC. Inner and outer membranes of E. coli cells were fractionated on sucrose gradient as described previously (Tikhonova & Zgurskaya, 2004, Osborn et al., 1972). Protein concentrations were measured by Bradford assay using bovine serum albumin as a standard. Immunoblotting analyses were carried out by incubation with primary polyclonal anti-TolC and anti-AcrA antibodies, followed by incubation with secondary alkaline-phosphatase-conjugated anti-rabbit antibody. The 5-bromo-4-chloro-3-indoyl phosphate (BCIP) and nitroblue tetrazolium (NBT) substrates were used to visualize the bands.
Fluorescein-5-maleimide labeling assay
Freshly transformed GD100 or KG101 cells carrying the plasmid-encoded TolCHis and YFREHis variants were grown in 5 ml of LB medium containing ampicillin (100 µg/ml) to an OD600 of ~1.0. Cells were harvested by centrifugation at 4,000 rpm at 4°C. The cell pellet was resuspended in 1 ml phosphate buffered saline (PBS), fluorescein-5-maleimide (FM) was added to a final concentration of 250 µM (Invitrogen) and cells were incubated on a rotator for 30 min at room temperature. In experiments with vancomycin, the cells were resuspended in 1 ml PBS and divided into two equal aliquots. To one aliquot vancomycin was added to a final concentration of 2.5 mM and both the aliquots were incubated for 30 min at room temperature on a rotator. Further, FM was added to these cells to a final concentration of 250 µM and incubated for 30 min at room temperature. In all cases the reaction was stopped by the addition of dithioerythritol (DTT) to a final concentration of 10 mM. Cells were pelleted and resuspended in 1 ml of buffer containing 50 mM Tris-HCl (pH 8.0), 1 mM EDTA, 1mM PMSF and 0.1 mg/ml lysozyme. Cells were broken by sonication and the unbroken cells were removed by centrifugation. The resulting supernatant was centrifuged at 50,000 rpm for 40 min to collect membrane fraction. The membrane pellet was resuspended in 50 µl of buffer containing 50 mM Tris-HCl (pH 8.0), 100 mM NaCl and 1 mM PMSF. Total protein concentration was measured by Bradford method (Bio-Rad) and 5 µg of each sample were boiled in non-reducing SDS-sample buffer and separated by 12% SDS-PAGE. FM labeling of TolC cysteine mutants was monitored by scanning SDS-PAGE gels using a STORM 840 fluorescent scanner (Molecular Dynamics). After scanning the gel, the proteins were transferred to a polyvinylidene fluoride (PVDF) membrane at 70 V for 1h. The membrane was probed with polyclonal anti-tolC antibodies and visualized using chromogenic substrates NBT/BCIP. Intensity of TolC bands after FM labeling and after immunoblotting was calculated using the ImageQuant TL program. Total fluorescence intensity of the TolC bands obtained after fluorescence scanning were quantified and normalized to the intensity of TolC bands obtained after immunoblotting. Further, the labeling intensity for each mutant was expressed as a percentage of the TolC A269C mutant.
Acknowledgement
This work was supported by National Institutes of Health Grant R21 AI092486 to HIZ. We thank Jon Weeks for helpful discussions.
Footnotes
Authors declare no conflict of interest.
References
- Andersen C, Koronakis E, Bokma E, Eswaran J, Humphreys D, Hughes C, Koronakis V. Transition to the open state of the TolC periplasmic tunnel entrance. Proc Natl Acad Sci U S A. 2002a;99:11103–11108. doi: 10.1073/pnas.162039399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen C, Koronakis E, Hughes C, Koronakis V. An aspartate ring at the TolC tunnel entrance determines ion selectivity and presents a target for blocking by large cations. Mol Microbiol. 2002b;44:1131–1139. doi: 10.1046/j.1365-2958.2002.02898.x. [DOI] [PubMed] [Google Scholar]
- Augustus AM, Celaya T, Husain F, Humbard M, Misra R. Antibiotic-sensitive TolC mutants and their suppressors. J Bacteriol. 2004;186:1851–1860. doi: 10.1128/JB.186.6.1851-1860.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan L, Hughes C, Koronakis V. Substrate-triggered recruitment of the TolC channel-tunnel during type I export of hemolysin by Escherichia coli. J Mol Biol. 2001;313:501–510. doi: 10.1006/jmbi.2001.5038. [DOI] [PubMed] [Google Scholar]
- Bavro VN, Pietras Z, Furnham N, Perez-Cano L, Fernandez-Recio J, Pei XY, Misra R, Luisi B. Assembly and channel opening in a bacterial drug efflux machine. Mol Cell. 2008;30:114–121. doi: 10.1016/j.molcel.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz R, Maier E, Gentschev I. TolC of Escherichia coli functions as an outer membrane channel. Zentralbl Bakteriol. 1993;278:187–196. doi: 10.1016/s0934-8840(11)80836-4. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhamdhere G, Zgurskaya HI. Metabolic shutdown in Escherichia coli cells lacking the outer membrane channel TolC. Mol Microbiol. 2010;77:743–754. doi: 10.1111/j.1365-2958.2010.07245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh T, Paulsen IT, Saier IT., Jr A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J Bacteriol. 1994;176:3825–3831. doi: 10.1128/jb.176.13.3825-3831.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaran J, Hughes C, Koronakis V. Locking TolC entrance helices to prevent protein translocation by the bacterial type I export apparatus. J Mol Biol. 2003;327:309–315. doi: 10.1016/s0022-2836(03)00116-5. [DOI] [PubMed] [Google Scholar]
- Fralick JA. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Q, Yamada Y, Zgurskaya H. The C-terminal domain of AcrA is essential for the assembly and function of the multidrug efflux pump AcrAB-TolC. J Bacteriol. 2009;191:4365–4371. doi: 10.1128/JB.00204-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken H, Misra R. Genetic evidence for functional interactions between TolC and AcrA proteins of a major antibiotic efflux pump of Escherichia coli. Mol Microbiol. 2004;54:620–631. doi: 10.1111/j.1365-2958.2004.04301.x. [DOI] [PubMed] [Google Scholar]
- Higgins MK, Eswaran J, Edwards P, Schertler GF, Hughes C, Koronakis V. Structure of the ligand-blocked periplasmic entrance of the bacterial multidrug efflux protein TolC. J Mol Biol. 2004;342:697–702. doi: 10.1016/j.jmb.2004.07.088. [DOI] [PubMed] [Google Scholar]
- Hwang J, Zhong X, Tai PC. Interactions of dedicated export membrane proteins of the colicin V secretion system: CvaA, a member of the membrane fusion protein family, interacts with CvaB and TolC. J Bacteriol. 1997;179:6264–6270. doi: 10.1128/jb.179.20.6264-6270.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Nikaido H. Different functions of MdtB and MdtC subunits in the heterotrimeric efflux transporter MdtB(2)C complex of Escherichia coli. Biochemistry. 2012;51:4188–4197. doi: 10.1021/bi300379y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronakis V, Li J, Koronakis E, Stauffer K. Structure of TolC, the outer membrane component of the bacterial type I efflux system, derived from two-dimensional crystals. Mol Microbiol. 1997;23:617–626. doi: 10.1046/j.1365-2958.1997.d01-1880.x. [DOI] [PubMed] [Google Scholar]
- Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- Lobedanz S, Bokma E, Symmons MF, Koronakis E, Hughes C, Koronakis V. A periplasmic coiled-coil interface underlying TolC recruitment and the assembly of bacterial drug efflux pumps. Proc Natl Acad Sci U S A. 2007;104:4612–4617. doi: 10.1073/pnas.0610160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra R, Bavro VN. Assembly and transport mechanism of tripartite drug efflux systems. Biochim Biophys Acta. 2009;1794:817–825. doi: 10.1016/j.bbapap.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modali SD, Zgurskaya HI. The periplasmic membrane proximal domain of MacA acts as a switch in stimulation of ATP hydrolysis by MacB transporter. Mol Microbiol. 2011;81:937–951. doi: 10.1111/j.1365-2958.2011.07744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn MJ, Gander JE, Parisi E, Carson J. Mechanism of Assembly of the outer membrane of Salmonella typhimurium. J. Biol. Chem. 1972;247:3962–3972. [PubMed] [Google Scholar]
- Paulsen IT, Park JH, Choi PS, Saier MH., Jr A family of gram-negative bacterial outer membrane factors that function in the export of proteins, carbohydrates, drugs and heavy metals from gram-negative bacteria. FEMS Microbiol Lett. 1997;156:1–8. doi: 10.1111/j.1574-6968.1997.tb12697.x. [DOI] [PubMed] [Google Scholar]
- Pei XY, Hinchliffe P, Symmons MF, Koronakis E, Benz R, Hughes C, Koronakis V. Structures of sequential open states in a symmetrical opening transition of the TolC exit duct. Proc Natl Acad Sci U S A. 2011;108:2112–2117. doi: 10.1073/pnas.1012588108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons MF, Bokma E, Koronakis E, Hughes C, Koronakis V. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc Natl Acad Sci U S A. 2009;106:7173–7178. doi: 10.1073/pnas.0900693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura N, Murakami S, Oyama Y, Ishiguro M, Yamaguchi A. Direct Interaction of Multidrug Efflux Transporter AcrB and Outer Membrane Channel TolC Detected via Site-Directed Disulfide Cross-Linking. Biochemistry. 2005;44:11115–11121. doi: 10.1021/bi050452u. [DOI] [PubMed] [Google Scholar]
- Tikhonova EB, Dastidar V, Rybenkov VV, Zgurskaya HI. Kinetic control of TolC recruitment by multidrug efflux complexes. Proc Natl Acad Sci U S A. 2009;106:16416–16421. doi: 10.1073/pnas.0906601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonova EB, Yamada Y, Zgurskaya HI. Sequential Mechanism of Assembly of Multidrug Efflux Pump AcrAB-TolC. Chem Biol. 2011;18:454–463. doi: 10.1016/j.chembiol.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonova EB, Zgurskaya HI. AcrA, AcrB, and TolC of Escherichia coli Form a Stable Intermembrane Multidrug Efflux Complex. J Biol Chem. 2004;279:32116–32124. doi: 10.1074/jbc.M402230200. [DOI] [PubMed] [Google Scholar]
- Touze T, Eswaran J, Bokma E, Koronakis E, Hughes C, Koronakis V. Interactions underlying assembly of the Escherichia coli AcrAB-TolC multidrug efflux system. Mol Microbiol. 2004;53:697–706. doi: 10.1111/j.1365-2958.2004.04158.x. [DOI] [PubMed] [Google Scholar]
- Weeks JW, Celaya-Kolb T, Pecora S, Misra R. AcrA suppressor alterations reverse the drug hypersensitivity phenotype of a TolC mutant by inducing TolC aperture opening. Mol Microbiol. 2010;75:1468–1483. doi: 10.1111/j.1365-2958.2010.07068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka H, Nomura T, Morisada N, Shinoda S, Okamoto K. Site-directed mutagenesis studies of the amino acid residue at position 412 of Escherichia coli TolC which is required for the activity. Microb Pathog. 2002;33:81–89. doi: 10.1006/mpat.2002.0519. [DOI] [PubMed] [Google Scholar]
- Zgurskaya HI, Yamada Y, Tikhonova EB, Ge Q, Krishnamoorthy G. Structural and functional diversity of bacterial membrane fusion proteins. Biochim Biophys Acta. 2009;1794:794–807. doi: 10.1016/j.bbapap.2008.10.010. [DOI] [PubMed] [Google Scholar]