Abstract
The basement membrane of the corneal epithelium presents biophysical cues in the form of topography and compliance that can modulate cytoskeletal dynamics, which, in turn, can result in altering cellular and nuclear morphology and alignment. In this study the effect of topographic patterns of alternating ridges and grooves on nuclear and cellular shape and alignment was determined. Primary corneal epithelial cells were cultured on either planar or topographically patterned (400–4000nm pitch) substrates. Alignment of individual cell-body was correlated with respective nucleus for analysis of orientation and elongation. A biphasic response in alignment was observed. Cell bodies preferentially aligned perpendicular to the 800nm pitch; and with increasing pitch, cells increasingly aligned parallel to the substratum. Nuclear orientation largely followed this trend with the exception of those on 400nm. On this biomimetic size-scale some nuclei oriented perpendicular to the topography while their cytoskeleton elements aligned parallel. Both, nuclei and cell bodies were elongated on topography compared with those on flat surfaces. Our data demonstrate that nuclear orientation and shape are differentially altered by topographic features that is not mandated by alignment of the cell-body. This novel finding suggests that nuanced differences in alignment of the nucleus vs the cell body exist and that these differences could have consequences on gene and protein regulation that ultimately regulate cell behaviors. A full understanding of these mechanisms could disclose novel pathways that would better inform evolving strategies in cell, stem cell and tissue engineering as well as the design and fabrication of improved prosthetic devices.
Keywords: Cornea, epithelial cells, nucleus, contact guidance, biophysical cues, alignment, topography, mechano-transduction
Introduction
Corneal opacities, the third major cause of blindness in the world1, may be a result of endothelial dystrophies, epithelial dystrophies, stromal scarring or a haze in the stroma associated with the persistence of myofibroblasts. A healthy corneal epithelium is essential for a positive wound healing response and prevention of opacities2. Transplantation with corneal donor tissue and keratoprosthesis are two currently available treatment options for corneal blindness. However, the numbers of donor tissues available do not meet the demand and the alternative is the use of a bioengineered synthetic cornea. Synthetic materials have inherent physical properties such as stiffness3–6, surface chemistry7–11 and topography12–15, and these attributes can influence corneal cellular behavior. It is therefore imperative that we understand the biological response of corneal cells to biophysical cues in order to aid in the development of improved keratoprosthetics as well as to advance evolving strategies for bioengineering corneal constructs and the engineering of corneal wound beds.
Corneal wound healing is influenced by biophysical cues and soluble signals within the extracellular environment16–18. Corneal epithelial cells are anchored to the underlying stroma through the basement membrane (BM), a specialization of the extracellular matrix (ECM). The structural elements of the BM (collagen, laminin, proteoglycans and other fibrils) create a rich 3-dimensional nanoscale topographical environment (20–200 nm) comprised of pores, fibers and raised islands19,20. The constituents of the BM provide both mechanical and biochemical stimuli to the anchored cells and are known to modulate a wide menu of fundamental cell behaviors21–24.
In order to better understand the mechanisms that influence cellular responses to topographic cues, synthetic substrates have been employed which mimic defined features of the biophysical environment of BM. Many studies have utililized topographically patterned substrates of anisotropically ordered alternating ridges and grooves25–28. A common cellular response to anisotropic substratum topographies is contact guidance resulting in the cells aligning parallel to the long axis of the underlying topographic features. We have previously demonstrated that the alignment of the actin cytoskeleton of human corneal epithelial cells (HCECs) is strongly dependent on the direction, pitch (pitch=ridge width + groove width).. The depth of the groove also plays a critical role in the cellular recognition of the underlying topography28–31.
In a recent study from our group, it was shown that human umbilical vein endothelial cells (HUVEC), cultured on a topographically patterned substrates possessing biomimetic-scale and anisotropic surface order, had significantly altered expression of more than 3000 genes. Many of the genes identified were segregated into families associated with ECM and cell adhesion, cytoskeletal reorganization, DNA replication and repair and spindle organization32. In other cells, changes in cell shape by remodeling of cytoskeleton have been reported to control cyclins and regulate the entry of cells from G1 to S phase33.
While multiple studies have reported alignment of cell bodies to underlying micron-scale topography, few have reported changes in nuclear morphology as a result of contact guidance. Previous studies describing cell orientation have used data from either the actin cytoskeleton or the nucleus and assumed a direct correlation between the two34,35. The nucleus, the largest organelle in the cell, is mechanically stabilized by structural proteins in the nuclear membrane which link the cytoskeleton to the nucleus. While the cytoskeleton is capable of rapid and dynamic remodeling during cell alignment and migration, the nucleus, which is approximately 5–10 times stiffer than the cytoskeleton36, appears to undergo only slight variations. The mechanical link between the cytoskeleton and nucleus suggests that mechanotransduction, from the extracellular environment, may affect chromatin binding proteins in the nuclear membrane thereby influencing spatial positioning of chromatin and chromosomes that will influence the regulation of gene expression. Though it is well known that changes in nuclear shape are capable of influencing gene expression37 there is a paucity of information available regarding nuclear response to substratum topographic cues.
Therefore, to ascertain the effect of substratum topography on nuclear morphology compared with overall shape and morphology of the cell body, primary HCECs were cultured for 12 h on polymeric (Norland Optical adhesive- NOA81) substrates that were either planar or topographically patterned with alternating parallel ridges and grooves of various pitches. Because soluble constituents of the culture environment have been demonstrated to interact with topographic cues in modulating cell behaviors29, we further studied how two routinely used culture media influence the topographic responses of these cells. One of the media was a well defined serum-free epithelial growth medium with a defined growth supplement (EpiLife medium, Invitrogen, NY) while the other media (EP medium) was Dulbecco’s modified Eagle’s medium (Life Technologies, Grand Island, NY) containing fetal bovine serum in addition to a number of supplemented factors. Morphology and alignment of the nuclei and cytoskeleton were quantified with fluorescence microscopy.
Materials and Methods
Fabrication of NOA81 chips with ridge/groove topographic features
Silicon chips containing 6 regions of ridge and groove features and flat control areas were fabricated using X-ray lithography as described previously (referred to as “six packs”)38. The dimensions of the various topographies are listed in table 1. Briefly, a composite stamp of the silicon chip master was made by curing a “hard” layer of poly(dimethylsiloxane) (PDMS) to retain the topographic features and then a pliable PDMS layer for easy removal and handling of the stamp. The pattern could then be reproduced into a thin layer of NOA81 optical adhesive (Norland Products, NJ) 39 deposited onto 35-mm tissue culture plates using a spin coater (4000 rpm for 40 sec) and cured in a XL-1500 UV cross-linker under 365 nm light for 100 minutes. NOA81, a proprietary mercapto-ester compound of Norland Products, is supplied as a single component liquid adhesive that readily cures as a rigid polymer with exposure to UV light. Research from our laboratory has previously demonstrated NOA81 as a suitable material for cell culture40–42.
Table 1.
Dimensions of topographic features on substrates
| Pitch | Ridge width (nm) | Groove width (nm) | Depth (nm) |
|---|---|---|---|
| 400 nm | 200 | 200 | 300 |
| 800 nm | 400 | 400 | 300 |
| 1200 nm | 600 | 600 | 300 |
| 1600 nm | 800 | 800 | 300 |
| 2000 nm | 1000 | 1000 | 300 |
| 4000 nm | 2000 | 2000 | 300 |
Preparation of substrates for cell culture
In preparation for cell seeding, the topographically patterned substrates were soaked in 70% ethanol for 10 minutes, rinsed 3 times with sterile 1x phosphate buffered saline (PBS, pH 7.2, Invitrogen, CA), air dried and exposed to 280 nm UV light for 30 minutes in a laminar flow hood.
Primary human corneal epithelial cell harvest and culture
Human corneoscleral rims with no history of ocular diseases that were unsuitable for transplant were used to establish epithelial cultures. All experiments were conducted in accord with the Declaration of Helsinki. The primary HCECs were harvested as described previously30. HCECs from 2–4 corneas were plated in either epithelial medium (EP) or in EpiLife® medium with 60 μM calcium (Invitrogen, CA). EP contained a 3:2 ratio of Ham’s F12 and Dulbelco’s Modified Eagles medium (DMEM) (Invitrogen, CA), supplemented with 2.5 % (v/v) fetal bovine serum (FBS), 0.4 μg/ml hydrocortisone, 8.4 ng/ml cholera toxin, 5 μg/ml insulin, 24 μg/ml adenine, 10 ng/ml epidermal growth factor, 100 units penicillin, and 100 μg/ml streptomycin43,44. HCECs in EP were plated onto 100 mm tissue culture plates containing a mitomycin-c treated Swiss 3T3 mouse fibroblast layer. EpiLife® medium was supplemented with a proprietary combination of bovine serum albumin, bovine transferrin, hydrocortisone, recombinant human insulin-like growth factor type-1, prostaglandin, and recombinant human epidermal growth factor (EpiLife® Defined Growth Supplement, Invitrogen, CA). Cells in EpiLife® medium were plated into 100 mm tissue culture plates coated with fibronectin and collagen (FNC coating mix, AthenaES, MD). All HCECs were incubated at 37°C and 5% CO2 until they reached approximately 70% confluence. Cells were used before passage 3. For all experiments, 3–5 replicates of each of six different topographic surfaces (six packs) were used and the cells were plated at a density of 10,000 cells per cm2 in the absence of any FNC coating. All cells were incubated for 12 hours after plating to allow for attachment and cell spreading.
Immunocytochemistry
To determine HCEC nucleus and cytoskeleton alignment and elongation response to topographically patterned substrates, cytoskeletal actin-filaments and the cell nucleus were stained. Following the 12 hours of incubation, the surfaces were rinsed with 1x PBS. The surfaces were then fixed with 4% paraformaldehyde-PBS (Electron Microscopy Sciences, PA) at room temperature for 20 minutes. Following a 1x PBS wash, the cells were permeabilized with 0.1% (v/v) Triton X –100 (Sigma-Aldrich, MO) in 1x PBS for 7 minutes, washed in 1x PBS for 10 minutes, and then immersed in 1% (w/v) bovine serum albumin (Sigma-Aldrich, MO) in 1x PBS for 20 minutes to block non-specific binding. Cells were rinsed with 1x PBS for 10 minutes, then incubated with 5 μg/ml of TRITC-phalloidin (Sigma-Aldrich, MO) and 0.1 μg/ml 4′,6-Diamidino-2-phenylindole (DAPI) (Invitrogen, CA) in 1x PBS for 40 minutes, to label both filamentous actin (red), and the nucleus (blue). Following a final rinse with 1x PBS, each substrate was mounted between a glass slide and a glass coverslip using DABCO® (Fluka, Switzerland), an anti-fading mounting medium.
Quantification of cell and nuclear shape and orientation
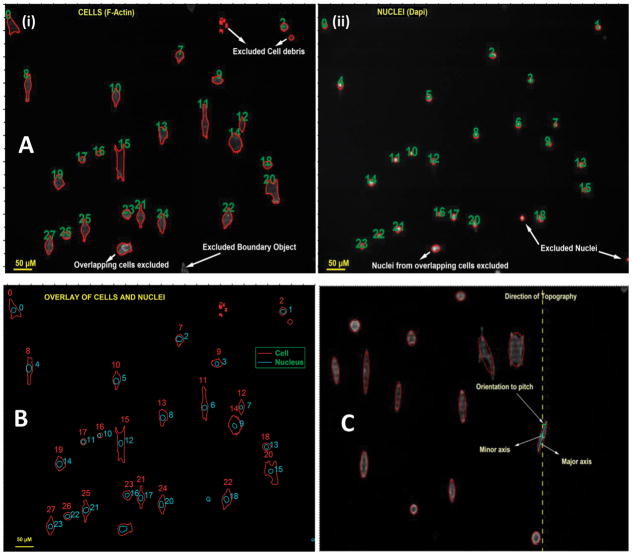
Samples were imaged using a Zeiss Axiovert 200M fluorescent microscope (Zeiss, Germany). Images of fluorescent cytoskeleton and nulei on each of the six packs were obtained using a 10x objective lens. At least four separate images were taken from each patterned pitch and flat areas separating the topographies. A total of eight individual experiments were performed for each pitch resulting in analysis of approximately 300–400 cells and their respective nuclei for a given topography. Image analysis was performed using built-in functions in Igor Pro 6.1 (Wavemetrics Inc., Portland, OR) along with user-defined functions to ensure that orientation and elongation data from an individual cell body (as reflected by imaging of the cytoskeleton) could be compared directly to its nucleus42,45. Not all cells were included in our analysis. For inclusion in this study, the cell body had to be fully attached and contained within the borders of the image, not in physical contact with another cell and not undergoing mitosis (e.g. cell bodies with more than one nucleus). The nuclei inclusion criteria specified that the numbers of nuclei and cells were identical. Examples of images based on these criteria are shown in figures 1A (i), (ii) and B. The cell and nucleus object numbers in Figure 1B correspond to row locations in a generated data table that necessitated a user-defined function to overlay cells and nuclei to eliminate cell debris or nuclei not apparently within the cellular boundaries.
Figure 1.
(A) Figure illustrates the representative raw data from individual channels images of cells (actin) and nuclei (dapi), grown in EpiLife medium on 400 nm topography, with their outline identified. (B) Representative image overlay of the outline of cells and their corresponding nucleus. Figure demonstrates accurate matching of a cell to its nucleus. (C) Defining the axes of an object. Figure illustrates a representative of an ellipse defined around a cell to determine its major and minor axes, and hence its orientation to the underlying topography.
For each cell body and nucleus, an ellipse was created based on the defined boundary of the object that conserved the total area of the defined object (Figure 1C). From this ellipse, major and minor axes were defined. The cytoskeleton and nucleus alignment angle was defined as the angle between the major axis of the defined object and the long axis of the underlying topographic pattern. The cell body or nucleus was considered aligned parallel if the angle between the object and the topography was less than 10° and perpendicular if the angle was between 80° and 90°. From this information we then described the “net orientation’ as the difference between the number of objects aligned parallel to those aligned perpendicular. For a more detailed analysis, objects were then sorted into 10 degree increments (0–10, 10–20 etc.). The elongation of both the cell body and the nucleus were quantified by the ratio of the major axis to the minor axis (axis ratio).
Data Analysis Criteria
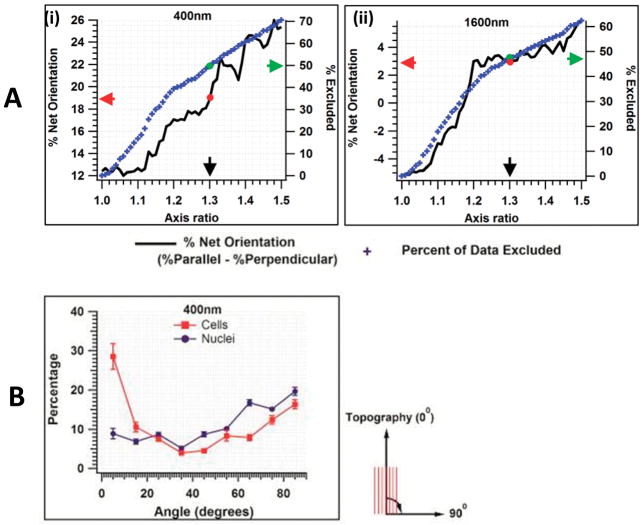
The alignment of a cytoskeleton or nucleus is defined by measuring the angle between the major axis of the cytoskeleton or nucleus (Figure 1C) and the underlying topographic ridge and groove. As the axis ratio increases, there is increased certainty that the orientation angle has been correctly determined. However as the axis ratio of the cytoskeleton or nucleus approaches a value of one (a round object), the likelihood of incorrectly assigning the orientation angle increases; for example, assigning an orientation of parallel when the correct orientation is perpendicular. This has the potential to influence the interpretation of the net orientation of the cytoskeleton or nucleus to a topographic feature as well as errors in the relationship between the orientation of the cytoskeleton and nucleus within the same cell. To try and overcome this potential error, our lab has previously excluded all objects that were less than 30% elongated, i.e. an axis ratio of 1.3. We have not employed this criterion to these data for the following reason. Figure 2A demonstrates the effect of applying an axis ratio cutoff on the net orientation (number of parallel cytoskeleton minus number of perpendicular cytoskeleton) of HCECs on our 400nm pitch substrate. Below an axis ratio cutoff of 1.12 there was no effect on net orientation i.e. there are a similar number of perpendicular objects improperly labeled parallel as for parallel objects improperly labeled as perpendicular. However, above 1.12, the effect of an arbitrary cutoff alters the interpretation of the cell body’s net orientation towards increasing net parallel orientation. The right hand axis of the figure demonstrates the percentage of data that was removed as a consequence of any cutoff value chosen. In figure 2B the same analysis was applied to the cytoskeleton of HCECs on a 1600nm pitch patterned substrates. In this case, a clear boundary did not exist. More importantly, applying the cutoff in this case alters the interpretation of the net cell orientation from net perpendicular to net parallel demonstrating that data eliminated arbitrarily on different pitches did not result in the identical effect on net orientation.
Figure 2.
A) Representative figure illustrating the effect of a ‘cut-off’ applied for data exclusion, on % net orientation, based on aspect ratio of cells. The black curve depicts the % net orientation, blue symbols the percent of data excluded. On the 400 nm pitch, if a cut-off for 1.3 axis ratio is applied; we observe the net orientation (red arrow) to be 19% parallel with 50% of the data excluded (green arrow) from analysis. B) Representative histogram of cell and nuclear orientation to underlying topography (400 nm) for the complete data set. Nuclei are predominantly oriented perpendicular to the underlying ridges and grooves while the cytoskeleton is aligned parallel. Results are mean ± SEM, n=8 individual experiments. A zero angle indicates orientation is parallel to the topography.
The above analysis demonstrated that it was not necessary to exclude data based on an arbitrary ratio to determine the number of cells aligned to a topographic feature. Also, removing data within the lower limits, which might be considered as noise, had no effect on interpretation of net orientation. However, applying a constant cutoff outside the noise limit to all substrates might inadvertently result in exclusion of data that is essential to understanding the orientation response of the cytoskeleton to an underlying topographic cue. A representative histogram of nuclear and cytoskeleton orientation to underlying topography (400 nm) is illustrated in figure 2B.
In addition to orientation response, we were also interested in the effect of orientation on elongation. The minimum axis ratio for these HCECs that could reliably be used was 12% elongation. If a cell could not reliably be assigned an orientation, it was removed from analysis of elongation. All subsequent results presented in the paper were based on these parameters.
Results
Alignment of the cell body and nucleus depends on pitch size
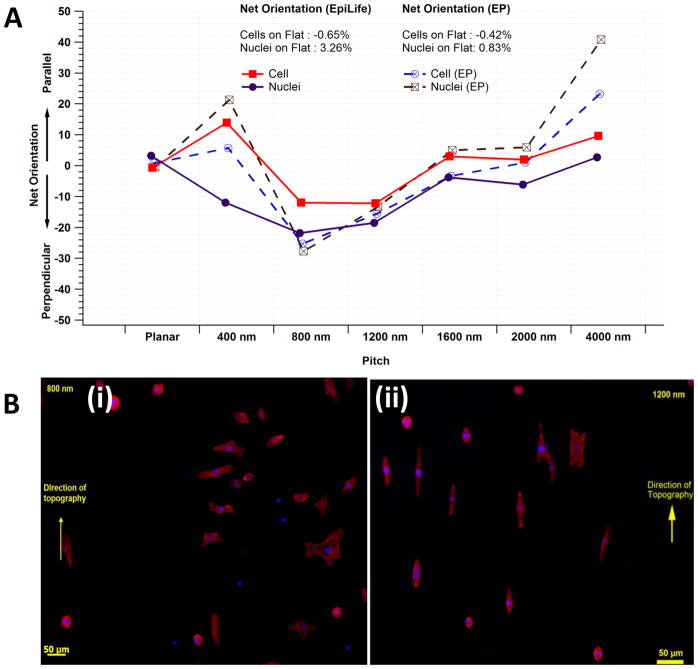
The alignment of the cell body and nucleus were dependent on pitch size. The trends in orientation were independent of the type of growth medium; however, the net orientation of both the nuclei and cytoskeleton were more pronounced in EP medium compared with EpiLife medium. Figure 3A illustrates the net alignment of cell bodies and nuclei in response to topographic cues in both EP and EpiLife medium. Approximately 10% of all cell bodies or nuclei, per ten degree increment, were randomly oriented on flat surfaces and there was no apparent preferential net alignment. On the 400 nm pitch, approximately 30–35% of all cell bodies were aligned parallel in both EpiLife and EP media. As the pitch increased from 400 nm to 800 nm, the percentage of perpendicular nuclei and cytoskeleton increased, with a maximum perpendicular percentage of 30.17 ± 2.05 % in EpiLife medium and 37.68 ± 16.72 % in EP medium for alignment of cell bodies. As the pitch increased from 800 to 4000nm the number of parallel cell bodies and nuclei continued to increase, until nuclei and cytoskeletal elements were preferentially aligned parallel at the 4000 nm pitch substrate. The resulting net orientation of cells are listed in table 2. For cell bodies at 4000 nm pitch, parallel alignment was 23.88 ± 2.08 % of the population in EpiLife medium and 41.70 ± 2.96 % of the population in EP medium. The incomplete response of a cellular population of HCECs is a well-reported phenomenon28–31,38,40,42. A representative image illustrating the differential alignment of cells is depicted in figure 3B. A relatively equal distribution of perpendicular and parallel nuclei/cells was observed on the intermediate pitch sizes of 1600 nm and 2000 nm. The absolute orientation of nuclei and cells in both media is illustrated in supplementary figures 1A and B. The absolute orientation of cell bodies and nuclei with an arbitrary cell aspect ratio cut off of 1.3 is illustrated in supplementary figure 2. The relative trends for alignment of cells and nuclei are similar to those when no cut-off is applied; however the interpretation of net orientation (perpendicular or parallel) for 1600nm was modified, as discussed previously.
Figure 3.
A) Net orientation of cells and nuclei when cultured in EpiLife and EP media. Net orientation was determined as the difference between total number of objects aligned parallel to those aligned perpendicular to the substratum. Nuclei of cells cultured on the 400 nm pitch in EpiLife medium were predominantly net-perpendicular (~10%) while their cell bodies were aligned net-parallel (~15%). However, in EP medium both cell and nuclei had a net parallel orientation to the topography. Results are Mean of n=8 individual experiments. B) Representative images of primary corneal epitheial cells aligned (i) perpendicular to the underlying surface (800 nm) and (ii) parallel to the topography (1200 nm).
Table 2.
Net orientation of cells on 800 nm and 4000 nm pitch in EP and EpiLife media
| Net Orientation (% of all cells) | EP medium | EpiLife medium |
|---|---|---|
| 800 nm | 21.30 % net perpendicular | 13.89% net perpendicular |
| 4000 nm | 40.81 % net parallel | 9.65 % net parallel |
Nuclei and cells cultured on topographically patterned substrates are more elongated than cells on planar substrates
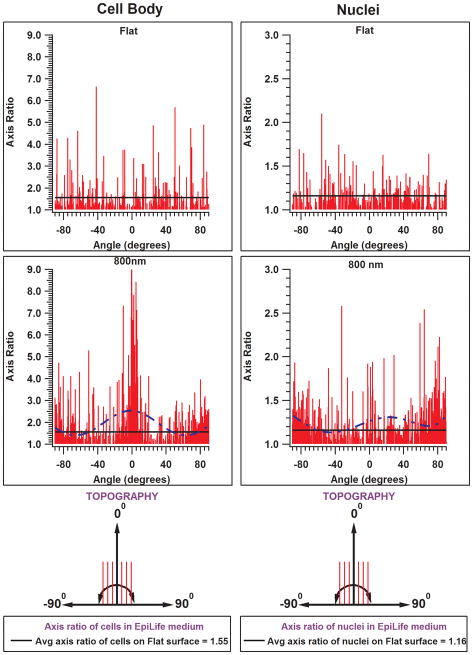
The influence of nucleus and cytoskeleton orientation on elongation is demonstrated as a histogram in Fig 4. Both the nuclei and cell bodies were significantly more elongated when cultured on substrates patterned with ridges and grooves as compared to those on planar substrates. Cells that aligned parallel to the long axis of the underlying features were also significantly longer than those aligned perpendicular. Curiously, the nuclei of cells cultured on 800 nm and 1200 nm pitches were longer when aligned perpendicular to the topography than those aligned parallel. This trend is evident with a polynomial curve fitted to the data. At other pitches (400, 1600, 2000, and 4000 nm; data not shown) greater numbers of nuclei were elongated when aligned parallel to the topography. The trends for elongation of cell body were more evident for cells cultured in EpiLife than in EP medium.
Figure 4.
Representative histograms of axis ratio of cells, and nuclei on flat surfaces and 800 nm pitches in EpiLife medium. A zero angle indicates cells/nuclei are aligned parallel to the topography. Positive angles (0° to 90°) indicate cells/nuclei oriented at an angle to the right of the ridges and grooves and negative angles (−90° to 0°) indicate orientation to the left. On 800 nm pitch, it can be observed that the extend of cell elongation is greatest when aligned parallel to the topography. In contrast, a greater number of nuclei oriented perpendicular were significantly elongated than those aligned parallel to the topography when cultured in EpiLife medium. Dashed lines are a result of curve fitting highlighting the trend in axis ratio over the entire range of angle measurements.
Correlating cell body and nuclear alignment
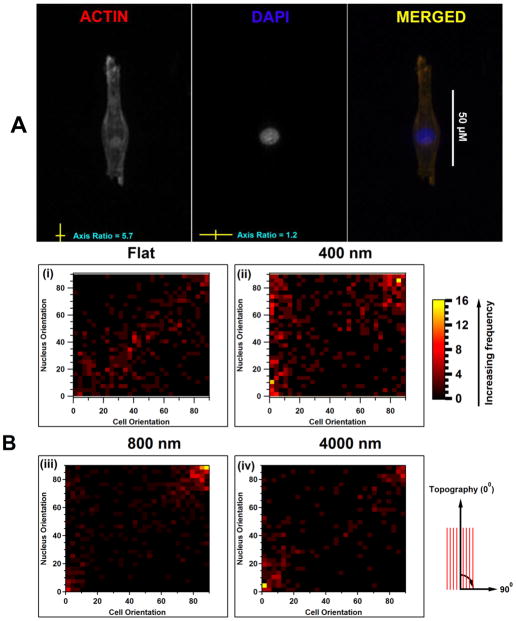
The relationship between nuclear orientation with respect to orientation of the cell body was determined to ascertain whether the nuclear orientation in response to topographic cues was independent of the cytoskeletal alignment (figure 5). The density maps in figure 5B represent the frequency of nuclear orientation to the orientation of its cytoskeleton. The orientation of nuclei was predominantly dependent on the orientation of the cytoskeleton; however this was not always the case e.g., on 400 nm. Interestingly, subsets of cell nuclei on the 400 nm pitch in EpiLife medium did not follow their cytoskeletal orientation (Figure 5B). A number of cells aligned parallel to the underlying topography had nuclei that were rotated 90° to the cytoskeleton (Figure 5B; approximately 10% of all nuclei had a net perpendicular orientation while the cell body was aligned net-parallel as demonstrated with an increased density on top left of the heat map). This striking feature was found to be dependent on soluble factors; nuclei followed the orientation of their cell body on the 400 nm pitches when cultured in EP medium. In both growth media, on flat surfaces, nuclei predominantly correlated with cytoskeletal orientation (linear distribution of frequency from bottom left to top right). On 800nm pitch, the density maps demonstrate that orientation of cells and nuclei were highly correlated and that they reversed their preferred orientation from net parallel to net perpendicular (increased frequency in top right corner). In addition, the shift from perpendicular nuclei/cells from 800 nm to both parallel on 4000 nm surfaces was significantly more evident when the HCECs were cultured in EP medium than in EpiLife medium. Approximately 21% of the cells cultured in EP medium had a net orientation perpendicular to the 800 nm anisotropic topography compared with 14% of cells cultured in EpiLife medium (Table 2) In contrast, on 4000 nm ridges and grooves 40% of the cells cultured in EP medium had a net parallel orientation in comparison with 9.65% in EpiLife medium.
Figure 5.
A) Figure illustrates the orientation of nucleus (DAPI) and its cell body (F-actin) on 400 nm topography. The cell body is observed to align parallel to the topography while its nucleus is perpendicular. The orientation was determined by monitoring the axis ratio; the ratio of the long axis of the cell body to its short axis. B) Heat maps are frequency of nuclear orientation expressed as a function of cell orientation. (i) On flat surfaces the cell body and nucleus are oriented randomly and follow each other (a linear relation). (ii) On 400 nm surfaces, there is a subset of cells aligned parallel to the substratum with their nuclei oriented perpendicular (Top left of the heat map). (iii) On 800 nm pitch, both cellbody and nuclei are perpendicularly aligned and follow each other (high frequency on top right). (iv) On the 4000 nm pitch, both cells and nuclei follow each other in orientation (top right and bottom left.)
Discussion
Topographic cues in the form of parallel ridges and grooves influence the morphology of primary HCECs. Conventionally, alignment studies have used either the cytoskeleton or the nucleus to determine the orientation. Integration of the alignment and morphology of both the cell body and nucleus documents that for the majority of pitches studied, cell body and nuclear alignment is coincident. But that is not always the case. There are topographical cues such as the 400nm pitch substrates which can influence the nucleus and cytoskeleton differentially and this may also be facilitated by soluble factors in the growth medium. These results are particularly important to consider when designing biomaterials to elicit a desired cellular response. The mechanisms by which these materials influence nuclear shape, geometry, alignment, positioning and mechanics could have a dominant role on fundamental behaviors, including: RNA diffusion, signaling, chromosomal localization, gene expression, cell migration and division46–49. We have previously demonstrated in HUVECs that submicron topographic features alter the expression of a large number of genes that regulate these basic cellular processes32.
Relative dimensions of topographic features modulate cell and nuclear orientation
The predominant substratum biophysical cues affecting epithelial cells are topography and stiffness of the native basement membrane. Our results confirm that the nucleus and cytoskeleton of primary HCECs respond to anisotropic topographies (with features in the biological range) with ridges and grooves of equal width and a fixed depth of 300 nm. Fluorescence microscopy observations have revealed that these topographies altered the shape and alignment of both the cell body and nucleus in a pitch dependent manner such that, both cells and nuclei on topographies were more aligned and longer than those on planar substrates.
We previously reported that HCECs aligned perpendicular to topography on a 400 nm pitch patterned substrate (silicon as substratum) with unequal ridge-groove width (70 nm ridge width/330 nm groove width and a depth of 600 nm) and became increasingly aligned parallel to the topography as pitch size increased to 4000 nm (1900 nm ridge/2100 groove) in EpiLife medium29. In contrast, we demonstrated here that there were a significantly larger number of cells aligned parallel to topography on 400 nm pitch (NOA81 as substratum) of equal ridge and groove width and 300 nm depth, and they maximally oriented perpendicular to the topography on 800 nm pitch in both EP and EpiLife media. Interestingly, we observed that HCECs switch their preferential alignment from parallel to perpendicular, between 400 nm and 800 nm pitches; and from perpendicular to parallel between 800 nm and 4000 nm pitches. This suggests that the response of HCEC is impacted by the dimensional and spatial attributes of the underlying topographic as well as potentially by the substrate chemistry. It is known that HCECs are responsive to anisotropic topographies at groove depths greater than 150 nm30. However, there is little evidence in the literature that suggests topography alone dictates differential orientation of cells between the 400 nm – 4000 nm range. More recently, we demonstrated an increase in nuclear volume, area and elastic modulus of immortalized human corneal epithelial cells, cultured in EpiLife medium, on larger pitches while the nuclear area, volume and elastic modulus was lowered on the smallest pitch (400 nm) compared with planar substrates42. It is now known that changes in nuclear lamina50,51 can alter nuclear shape by association of the cytoskeleton to peri-nuclear proteins. Other factors such as surface chemistry and/or soluble factors may indeed play an additional role. The trends in the switch from perpendicular (on 800 nm) to parallel (on 4000 nm) alignment remained the same in both media. Although relatively greater number of cells oriented to/against topography when cultured in the presence of serum (EP medium) in comparison to the serum free EpiLife medium. Also, the elongation of cells was more pronounced in EpiLife medium than in EP medium. Results from our study further highlight that biophysical cues in the form of topography are capable of altering nuclear morphology. These results are particularly important to consider when designing biomaterials to elicit a desired cellular response.
Alignment response of the cell body and nucleus
Translation of mechanical cues from the substratum to the nucleus is mediated by the cytoskeletal network52,53. Various studies have quantified the orientation of cells based on either actin, or DAPI or phase contrast images. While Manwaring et al27, and Biela et al54 quantified cell alignment based on the outline of cells (phase contrast), Heydarkhan-Hagvall et al55 qualitatively observed orientation of fibroblasts on nanotopography following actin staining. In another study, a particular cell’s alignment was determined by its nuclear alignment26. Nathan et al56 recently demonstrated that the actin network predominantly mediates nuclear deformation in mesenchymal cells on nanofibrous scaffolds. This is balanced somewhat by non-compressive forces of microtubules and intermediate filaments. Their results suggest the cytoskeleton controls the alignment of nuclei. However, our results suggest that this is not always the case. Correlating nucleus and cytoskeletal alignment demonstrated that the nuclei of HCECs, on 400 nm surfaces in EpiLife medium, predominantly aligned perpendicular to the underlying topography, while the cell body aligned parallel. This observation would have been missed had we chosen to determine orientation based on either actin or DAPI staining by itself. The observed differential alignment response is novel and is dependent on the soluble microenvironment of the cell as well as the size scale of the cues presented. This investigation demonstrates that methodologically it is important to ascertain both nuclear and cellular orientation in studies of topographic cueing. Further study on the interplay between the nucleus and cytoskeleton in response to substrate topography based on size range may yield other cases of misaligned nuclei with cells. The biological consequence of such a differential alignment of the cytoplasm and its nucleus is currently not known. Clearly, further work needs to be done to determine if the observed differences have significant consequences on gene and protein expression. Our results suggest that a more thorough understanding of the processes that enable a differential response could disclose novel pathways and/or cytoskeletal processes that could inform evolving strategies cell, stem cell and tissue engineering and contribute to the design and fabrication of improved prosthetic devices.
Supplementary Material
Supplementary fig 1. The plots show the percentage of cells and nuclei within 10-degree angle intervals indicated on the x-axis. A): Alignment of cells and nuclei in EP medium. Cells and nuclei predominantly oriented parallel to topography on 400 nm and 4000 nm pitches. Significant number of cells and nuclei oriented perpendicular on 800 nm pitch. Cells aligned both parallel and perpendicular to the ridges and grooves on patterns with 1200 nm, 1600 nm and 2000 nm pitches. B) Alignment of cells and nuclei in EpiLife medium. Cells predominantly oriented parallel while nuclei oriented perpendicular to topography on 400 nm pitch. Significant number of cells and nuclei oriented perpendicular on 800 nm pitch. Cells aligned both parallel and perpendicular to the ridges and grooves on patterns with 1200 nm, 1600 nm and 2000 nm pitches.
Supplementary fig 2. Alignment of cells and nuclei with an arbitrary cut off of 1.3 applied for cell elongation in A) in EP medium. The trends for cell and nuclear orientation are comparable to those observed without any cut off applied (supplementary figure 1A). B) in EpiLife medium. Note that the orientation of cells is relatively pronounced on topography when compared to those observed without application of a cut-off (supplementary figure 1B).
Primary corneal epithelial cells do not migrate in the absence of FNC coating. Primary HCECs were seeded onto topographically patterned surfaces and imaged by time lapse microscopy over 24 h using a Zeiss Axiomat 200M inverted microscope, a 10X objective. Minimal or no migration by HCECs on a 2000 nm surface was observed over a 24 h period.
Acknowledgments
This project was supported by the National Institute of Health through grants from the National Eye Institute R01EY01613404, and P30EY12576 and unrestricted funds from Research to Prevent Blindness.
References
- 1.Resnikoff SPD, Etya’ale D, Kocur I, Pararajasegaram R, Pokharel GP, Mariotti SP. Global data on visual impairment in the year 2002. Bulletin of the World Health Organization. 2004;82:844–851. [PMC free article] [PubMed] [Google Scholar]
- 2.Nakamura K, Kurosaka D, Bissen-Miyajima H, Tsubota K. Intact corneal epithelium is essential for the prevention of stromal haze after laser assisted in situ keratomileusis. British Journal of Ophthalmology. 2001;85(2):209–213. doi: 10.1136/bjo.85.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Discher DE, Janmey P, Wang Y-l. Tissue Cells Feel and Respond to the Stiffness of Their Substrate. Science. 2005;310(5751):1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 4.Engler AJ, Richert L, Wong JY, Picart C, Discher DE. Surface probe measurements of the elasticity of sectioned tissue, thin gels and polyelectrolyte multilayer films: Correlations between substrate stiffness and cell adhesion. Surface Science. 2004;570(1–2):142–154. [Google Scholar]
- 5.Saha K, Keung AJ, Irwin EF, Li Y, Little L, Schaffer DV, Healy KE. Substrate Modulus Directs Neural Stem Cell Behavior. Biophysical Journal. 2008;95(9):4426–4438. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nemir S, West JL. Synthetic materials in the study of cell response to substrate rigidity. Annals of Biomedical Engineering. 2010;38(1):2–20. doi: 10.1007/s10439-009-9811-1. [DOI] [PubMed] [Google Scholar]
- 7.Healy KE, Thomas CH, Rezania A, Kim JE, McKeown PJ, Lom B, Hockberger PE. Kinetics of bone cell organization and mineralization on materials with patterned surface chemistry. Biomaterials. 1996;17(2):195–208. doi: 10.1016/0142-9612(96)85764-4. [DOI] [PubMed] [Google Scholar]
- 8.Anselme K, Linez P, Bigerelle M, Le Maguer D, Le Maguer A, Hardouin P, Hildebrand HF, Iost A, Leroy JM. The relative influence of the topography and chemistry of TiAl6V4 surfaces on osteoblastic cell behaviour. Biomaterials. 2000;21(15):1567–1577. doi: 10.1016/s0142-9612(00)00042-9. [DOI] [PubMed] [Google Scholar]
- 9.Mrksich M. A surface chemistry approach to studying cell adhesion. Chemical Society Reviews. 2000;29(4):267–273. [Google Scholar]
- 10.Stevens MM, George JH. Exploring and Engineering the Cell Surface Interface. Science. 2005;310(5751):1135–1138. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- 11.MAROIS Y, MARIE-FRANCOISE, SUGOT-LUIZARD, GUIDOIN R. Endothelial Cell Behavior on Vascular Prosthetic Grafts: Effect of Polymer Chemistry, Surface Structure, and Surface Treatment. ASAIO Journal. 1999;45(4):272–280. [PubMed] [Google Scholar]
- 12.Ponsonnet L, Reybier K, Jaffrezic N, Comte V, Lagneau C, Lissac M, Martelet C. Relationship between surface properties (roughness, wettability) of titanium and titanium alloys and cell behaviour. Materials Science and Engineering: C. 2003;23(4):551–560. [Google Scholar]
- 13.Deligianni DD, Katsala ND, Koutsoukos PG, Missirlis YF. Effect of surface roughness of hydroxyapatite on human bone marrow cell adhesion, proliferation, differentiation and detachment strength. Biomaterials. 2000;22(1):87–96. doi: 10.1016/s0142-9612(00)00174-5. [DOI] [PubMed] [Google Scholar]
- 14.Hallab NJ, Bundy KJ, O’Connor K, Moses RL, Jacobs JJ. Evaluation of Metallic and Polymeric Biomaterial Surface Energy and Surface Roughness Characteristics for Directed Cell Adhesion. Tissue Engineering. 2001;7(1):55–71. doi: 10.1089/107632700300003297. [DOI] [PubMed] [Google Scholar]
- 15.Xu C, Yang F, Wang S, Ramakrishna S. In vitro study of human vascular endothelial cell function on materials with various surface roughness. Journal of Biomedical Materials Research Part A. 2004;71A(1):154–161. doi: 10.1002/jbm.a.30143. [DOI] [PubMed] [Google Scholar]
- 16.Dupps WJ, Jr, Wilson SE. Biomechanics and wound healing in the cornea. Experimental Eye Research. 2006;83(4):709–720. doi: 10.1016/j.exer.2006.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zieske JD. Extracellular matrix and wound healing. Current Opinion in Ophthalmology. 2001;12(4):237–41. doi: 10.1097/00055735-200108000-00001. [DOI] [PubMed] [Google Scholar]
- 18.Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Biol. 2002;3(5):349–363. doi: 10.1038/nrm809. [DOI] [PubMed] [Google Scholar]
- 19.Abrams GA, Goodman SL, Nealey PF, Franco M, Murphy CJ. Nanoscale topography of the basement membrane underlying the corneal epithelium of the rhesus macaque. Cell and Tissue Research. 2000;299(1):39–46. doi: 10.1007/s004419900074. [DOI] [PubMed] [Google Scholar]
- 20.Abrams GA, Schaus SS, Goodman SL, Nealey PF, Murphy CJ. Nanoscale Topography of the Corneal Epithelial Basement Membrane and Descemet’s Membrane of the Human. Cornea. 2000;19(1):57–64. doi: 10.1097/00003226-200001000-00012. [DOI] [PubMed] [Google Scholar]
- 21.Bruder JM, Lee AP, Hoffman-Kim D. Biomimetic materials replicating Schwann cell topography enhance neuronal adhesion and neurite alignment in vitro. Journal of Biomaterials Science --Polymer Edition. 2007;18(8):967–982. doi: 10.1163/156856207781494412. [DOI] [PubMed] [Google Scholar]
- 22.Charest JL, García AJ, King WP. Myoblast alignment and differentiation on cell culture substrates with microscale topography and model chemistries. Biomaterials. 2007;28(13):2202–2210. doi: 10.1016/j.biomaterials.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 23.Curtis A, Wilkinson C. Topographical control of cells. Biomaterials. 1997;18(24):1573–1583. doi: 10.1016/s0142-9612(97)00144-0. [DOI] [PubMed] [Google Scholar]
- 24.Fujita S, Ohshima M, Iwata H. Time-lapse observation of cell alignment on nanogrooved patterns. J R Soc Interface. 2009;6:S269–S277. doi: 10.1098/rsif.2008.0428.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersson A-S, Olsson P, Lidberg U, Sutherland D. The effects of continuous and discontinuous groove edges on cell shape and alignment. Experimental Cell Research. 2003;288(1):177–188. doi: 10.1016/s0014-4827(03)00159-9. [DOI] [PubMed] [Google Scholar]
- 26.Dalby MJ, Riehle MO, Yarwood SJ, Wilkinson CDW, Curtis ASG. Nucleusalignment and cell signaling in fibroblasts: response to a micro-grooved topography. Experimental Cell Research. 2003;284(2):272–280. doi: 10.1016/s0014-4827(02)00053-8. [DOI] [PubMed] [Google Scholar]
- 27.Manwaring ME, Walsh JF, Tresco PA. Contact guidance induced organization of extracellular matrix. Biomaterials. 2004;25(17):3631–3638. doi: 10.1016/j.biomaterials.2003.10.043. [DOI] [PubMed] [Google Scholar]
- 28.Tocce EJ, Smirnov VK, Kibalov DS, Liliensiek SJ, Murphy CJ, Nealey PF. The ability of corneal epithelial cells to recognize high aspect ratio nanostructures. Biomaterials. 2010;31(14):4064–4072. doi: 10.1016/j.biomaterials.2010.01.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teixeira AI, McKie GA, Foley JD, Bertics PJ, Nealey PF, Murphy CJ. The effect of environmental factors on the response of human corneal epithelial cells to nanoscale substrate topography. Biomaterials. 2006;27(21):3945–3954. doi: 10.1016/j.biomaterials.2006.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraser SA, Ting Y-H, Mallon KS, Wendt AE, Murphy CJ, Nealey PF. Sub-micron and nanoscale feature depth modulates alignment of stromal fibroblasts and corneal epithelial cells in serum-rich and serum-free media. Journal of Biomedical Materials Research Part A. 2008;86A(3):725–735. doi: 10.1002/jbm.a.31519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teixeira AI, Abrams GA, Bertics PJ, Murphy CJ, Nealey PF. Epithelial contact guidance on well-defined micro-and nanostructured substrates. J Cell Sci. 2003;116(10):1881–1892. doi: 10.1242/jcs.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gasiorowski JZ, Liliensiek SJ, Russell P, Stephan DA, Nealey PF, Murphy CJ. Alterations in gene expression of humanvascular endothelial cells associated with nanotopographic cues. Biomaterials. 2010;31(34):8882–8888. doi: 10.1016/j.biomaterials.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang S, Chen CS, Ingber DE. Control of Cyclin D1, p27Kip1, and Cell Cycle Progression in Human Capillary Endothelial Cells by Cell Shape and Cytoskeletal Tension. Molecular Biology of the Cell. 1998;9(11):3179–3193. doi: 10.1091/mbc.9.11.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Houtchens GR, Foster MD, Desai TA, Morgan EF, Wong JY. Combined effects of microtopography and cyclic strain on vascular smooth muscle cell orientation. J Biomech. 2008;41(4):762–9. doi: 10.1016/j.jbiomech.2007.11.027. Epub 2008 Jan 28. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biggs MJP, Richards RG, Gadegaard N, Wilkinson CDW, Oreffo ROC, Dalby MJ. The use of nanoscale topography to modulate the dynamics of adhesion formation in primary osteoblasts and ERK/MAPK signalling in STRO-1+ enriched skeletal stem cells. Biomaterials. 2009;30(28):5094–5103. doi: 10.1016/j.biomaterials.2009.05.049. [DOI] [PubMed] [Google Scholar]
- 36.Dahl KN, Kahn SM, Wilson KL, Discher DE. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J Cell Sci. 2004;117(20):4779–4786. doi: 10.1242/jcs.01357. [DOI] [PubMed] [Google Scholar]
- 37.Thomas CH, Collier JH, Sfeir CS, Healy KE. Engineering gene expression and protein synthesis by modulation of nuclear shape. Proceedings of the National Academy of Sciences. 2002;99(4):1972–1977. doi: 10.1073/pnas.032668799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karuri NW, Liliensiek S, Teixeira AI, Abrams G, Campbell S, Nealey PF, Murphy CJ. Biological length scale topography enhances cell-substratum adhesion of human corneal epithelial cells. J Cell Sci. 2004;117(15):3153–3164. doi: 10.1242/jcs.01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Odom TW, Love JC, Wolfe DB, Paul KE, Whitesides GM. Improved Pattern Transfer in Soft Lithography Using Composite Stamps. Langmuir. 2002;18(13):5314–5320. [Google Scholar]
- 40.Liliensiek SJ, Campbell S, Nealey PF, Murphy CJ. The scale of substratum topographic features modulates proliferation of corneal epithelial cells and corneal fibroblasts. J Biomed Mater Res A. 2006;79(1):185–92. doi: 10.1002/jbm.a.30744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liliensiek SJ, Wood JA, Yong J, Auerbach R, Nealey PF, Murphy CJ. Modulation of human vascular endothelial cell behaviors by nanotopographic cues. Biomaterials. 2010;31(20):5418–26. doi: 10.1016/j.biomaterials.2010.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKee Clayton T, Raghunathan Vijay K, Nealey Paul F, Russell P, Murphy Christopher J. Topographic Modulation of the Orientation and Shape of Cell Nuclei and Their Influence on the Measured Elastic Modulus of Epithelial Cells. Biophysical Journal. 2011;101(9):2139–2146. doi: 10.1016/j.bpj.2011.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen-Hoffmann BL, Rheinwald JG. Polycyclic aromatic hydrocarbon mutagenesis of human epidermal keratinocytes in culture. Proceedings of the National Academy of Sciences of the United States of America. 1984;81(24):7802–7806. doi: 10.1073/pnas.81.24.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sabatini L, Allen-Hoffmann B, Warner T, Azen E. Serial cultivation of epithelial cells from human and macaque salivary glands. In Vitro Cellular & Developmental Biology -Animal. 1991;27(12):939–948. doi: 10.1007/BF02631121. [DOI] [PubMed] [Google Scholar]
- 45.McKee CT, Raghunathan VK, Russel P, Murphy CJ. Probing Effect of Biophysical Cues on Cell Behavior with Atomic Force Microscopy. Microscopy and Analysis-UK. 2011;(147) [Google Scholar]
- 46.Friedl P, Wolf K, Lammerding J. Nuclear mechanics during cell migration. Current Opinion in Cell Biology. 2010 doi: 10.1016/j.ceb.2010.10.015. In Press, Corrected Proof. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cremer T, Cremer C. Chromosome territories, nucleararchitecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2(4):292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 48.Jaalouk DE, Lammerding J. Mechanotransduction gone awry. Nat Rev Mol Cell Biol. 2009;10(1):63–73. doi: 10.1038/nrm2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taddei A, Hediger F, Neumann FR, Gasser SM. THE FUNCTION OF NUCLEAR ARCHITECTURE: A Genetic Approach. Annual Review of Genetics. 2004;38(1):305–345. doi: 10.1146/annurev.genet.37.110801.142705. [DOI] [PubMed] [Google Scholar]
- 50.Webster M, Witkin KL, Cohen-Fix O. Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. Journal of Cell Science. 2009;122(10):1477–1486. doi: 10.1242/jcs.037333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dahl KN, Ribeiro AJS, Lammerding J. Nuclear Shape, Mechanics, and Mechanotransduction. Circ Res. 2008;102(11):1307–1318. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol. 2009;10(1):75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 53.Roca-Cusachs P, Alcaraz J, Sunyer R, Samitier J, Farré R, Navajas D. Micropatterning of Single Endothelial Cell Shape Reveals a Tight Coupling between Nuclear Volume in G1 and Proliferation. Biophysical Journal. 2008;94(12):4984–4995. doi: 10.1529/biophysj.107.116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Biela SA, Su Y, Spatz JP, Kemkemer R. Different sensitivity of human endothelial cells, smooth muscle cells and fibroblasts to topography in the nano-micro range. Acta Biomaterialia. 2009;5(7):2460–2466. doi: 10.1016/j.actbio.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 55.Heydarkhan-Hagvall S, Choi C-H, Dunn J, Heydarkhan S, Schenke-Layland K, MacLellan WR, Beygui RE. Influence of Systematically Varied Nano-Scale Topography on Cell Morphology and Adhesion. Cell Communication and Adhesion. 2007;14(5):181–194. doi: 10.1080/15419060701755594. [DOI] [PubMed] [Google Scholar]
- 56.Nathan AS, Baker BM, Nerurkar NL, Mauck RL. Mechano-topographic modulation of stem cell nuclear shape on nanofibrous scaffolds. Acta Biomaterialia. 2011;7(1):57–66. doi: 10.1016/j.actbio.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary fig 1. The plots show the percentage of cells and nuclei within 10-degree angle intervals indicated on the x-axis. A): Alignment of cells and nuclei in EP medium. Cells and nuclei predominantly oriented parallel to topography on 400 nm and 4000 nm pitches. Significant number of cells and nuclei oriented perpendicular on 800 nm pitch. Cells aligned both parallel and perpendicular to the ridges and grooves on patterns with 1200 nm, 1600 nm and 2000 nm pitches. B) Alignment of cells and nuclei in EpiLife medium. Cells predominantly oriented parallel while nuclei oriented perpendicular to topography on 400 nm pitch. Significant number of cells and nuclei oriented perpendicular on 800 nm pitch. Cells aligned both parallel and perpendicular to the ridges and grooves on patterns with 1200 nm, 1600 nm and 2000 nm pitches.
Supplementary fig 2. Alignment of cells and nuclei with an arbitrary cut off of 1.3 applied for cell elongation in A) in EP medium. The trends for cell and nuclear orientation are comparable to those observed without any cut off applied (supplementary figure 1A). B) in EpiLife medium. Note that the orientation of cells is relatively pronounced on topography when compared to those observed without application of a cut-off (supplementary figure 1B).
Primary corneal epithelial cells do not migrate in the absence of FNC coating. Primary HCECs were seeded onto topographically patterned surfaces and imaged by time lapse microscopy over 24 h using a Zeiss Axiomat 200M inverted microscope, a 10X objective. Minimal or no migration by HCECs on a 2000 nm surface was observed over a 24 h period.