Abstract
Recent reports have demonstrated that somatic cells can be directly converted to other differentiated cell types through ectopic expression of sets of transcription factors, directly avoiding the transition through a pluripotent state. Our previous experiments generated induced neural progenitor-like cells (iNPCs) by a novel combination of five transcription factors (Sox2, Brn2, TLX, Bmi1 and c-Myc). Here we demonstrated that the iNPCs not only possess NPC-specific marker genes, but also have qualities of primary brain-derived NPCs (WT-NPCs), including tripotent differentiation potential, mature neuron differentiation capability and synapse formation. Importantly, the mature neurons derived from iNPCs exhibit significant physiological properties, such as potassium channel activity and generation of action potential-like spikes. These results suggest that directly reprogrammed iNPCs closely resemble WT-NPCs, which may suggest an alternative strategy to overcome the restricted proliferative and lineage potential of induced neurons (iNCs) and broaden applications of cell therapy in the treatment of neurodegenerative disorders.
Selective degeneration of functional neurons is associated with the pathogenesis of neurodegenerative disorders, such as degeneration of midbrain dopaminergic neurons in Parkinson's disease1, forebrain cholinergic neurons in Alzheimer's disease2 and cortical GABAergic neurons in schizophrenia3. One promising strategy to treat these neurodegenerative diseases is cell transplantation therapy, which uses cells derived from embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs). Recent studies have demonstrated that derivations of these neuronal subtypes from ESCs and iPSCs can be used to treat neurodegenerative disorders in rodent models4,5,6. Although various strategies have been developed to derive neuron subtypes, including direct differentiation from ESCs and iPSCs4,5,6 and direct reprogramming from somatic cells by forced expression of developmental genes7,8,9,10, the major obstacles that limit the broad application in cell transplantation therapy are the heterogeneous populations generated from neuronal differentiation protocols and low yield of neurons derived from direct reprogramming for conducting clinical trials.
Derivation of somatic stem cells, such as neural progenitor cells (NPCs), is highly desirable because NPCs are self-renewing, tripotent cells which can give rise to neurons, astrocytes and oligodendrocytes11. Although NPCs can be derived from either embryonic or adult tissue, the use and the accessibility of tissues source will limit the application of NPCs in research and clinics. Recent studies have shown that a combination of NPC fate determinants and selected transcription factors responsible for cell self-renewal and maintenance can directly convert both mouse and human fibroblasts into NSCs/NPCs with proliferation, self-renewal and tripotent differentiation capabilities12,13,14,15,16. These advances suggest that direct reprogramming of NPCs from differentiated somatic cells, instead of neurons, will not only provide an alternative to derivation from fetal tissue or pluripotent cells, but also provide a potentially unlimited source of neurons.
We previously showed that adult mouse skin fibroblasts could be successfully reprogrammed into iNPCs using our novel combination of transcription factors12. In this study, we have further identified the characteristics and potential functionalities of iNPCs, and our results demonstrate that directly reprogrammed iNPCs closely resemble WT-NPCs in several characteristics, including specific marker gene expression, tripotent differentiation potential, and neuroelectrophysiological properties. These findings further validate that iNPCs derived using our novel cocktail of transcription factors will be a promising candidate for cell transplantation therapy of neurodegenerative diseases.
Results
Marker gene expression and differentiation potential of iNPCs
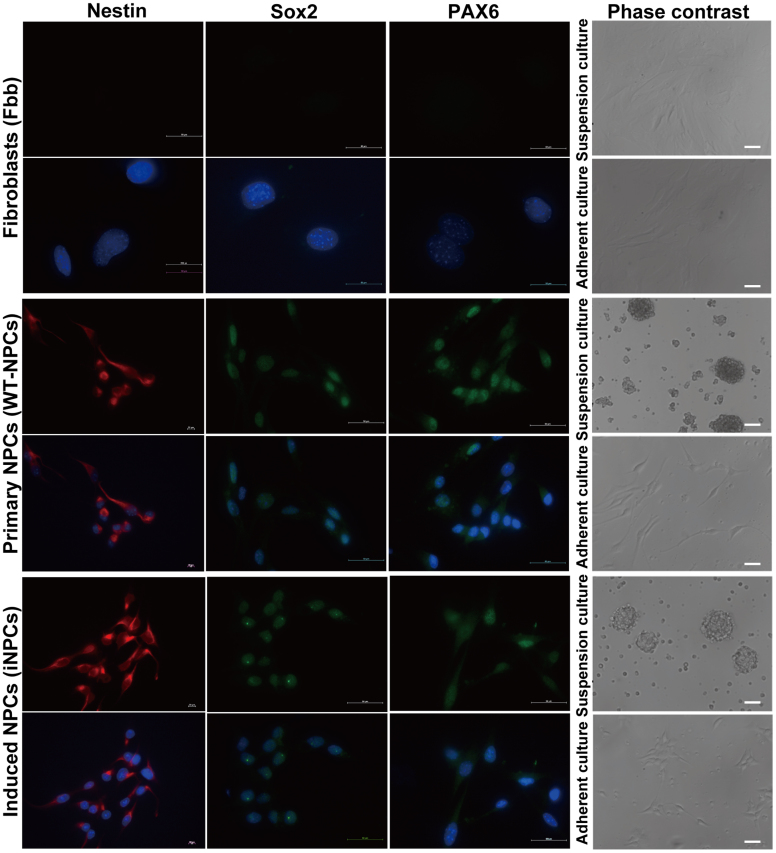
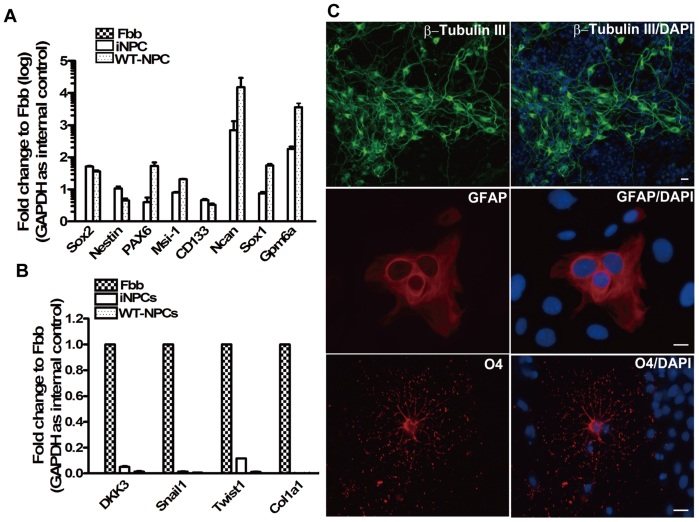
We previously used a retroviral delivery system to ectopically express two neural progenitor fate determinants (Brn2 and TLX) with three neural progenitor-related transcription factors (Sox2, Bmi1 and c-Myc) in mouse skin fibroblasts and successfully converted skin fibroblasts derived from E/Nestin: EGFP transgenic mice into neural progenitor cell line12. During the culture process, we found that iNPCs showed higher proliferation rates than WT-NPCs, and can be maintained with similar proliferation rates, self-renewal and differentiation potentials for over fifty cell passages (data not shown). iNPCs between passages 5 to 10 were used for all experiments. To identify similarities between iNPCs and WT-NPCs, we cultured and expanded iNPCs under standard NPC growth conditions, and performed the immunostaining analysis with antibodies specific for NPC marker genes. Our results demonstrated that iNPCs and WT-NPCs share similar cell morphology, whether cultured in adherent (biopolar) or suspension (neurosphere) culture condition (Fig. 1, right panel). Additionally, immunochemistry data also revealed that the expression level and distribution of NPC marker genes including Sox2, Nestin and Pax6 in iNPCs were similar to those in WT-NPCs (Fig. 1, left three panels). To further confirm the similarities, we used SYBR-Green-based quantitative Real-Time PCR with specific primer pairs (Table 1) to detect Nestin and other NPC specific marker gene expression levels, as well as the fibroblast marker gene expression in fibroblasts, WT-NPCs and iNPCs. Our results confirmed the transcription of Nestin, Sox2, Musashi-1 (Msi-1), CD133, Ncan, Sox1 and Gpm6a, all of which are characteristic markers of NPCs16 (Fig. 2A). Importantly, iNPCs from our cocktail of transcription factors showed a reduction in the fibroblast-specific genes including DKK3, Snail1, Twist1 and Col1α1 to levels similar to WT-NPCs when compared to skin fibroblasts (Fbb) (Fig. 2B), suggesting that iNPCs share similar marker gene expression profile with WT-NPCs, and that the fibroblast-specific transcriptional program is down-regulated in iNPCs. In addition, we observed that the transcription levels of Sox1, Msi-1 and Pax6 in iNPCs were lower than that in WT-NPCs, and that the transcription level of Pax6 was consistent with its protein level in iNPCs (Fig. 1 lower panel), indicating the possibility that exogenous genes may compensate for the functions of endogenous genes with lower expression levels in iNPCs. Our real-time analyses for transcription levels of transgenic and endogenous genes including Brn2, Sox2, Bmi1, Nr2e1 and c-Myc in iNPCs supported this possibility (Fig. S1A and S1B). Furthermore, we compared the differentiation potential and transgenic and endogenous gene expression profiles in iNPCs between early and late passages. Our results showed that late passaged iNPCs still possessed tripotent differentiation potentials similar to that of early passaged iNPCs (Fig. S5A), and most transgenes used for direct reprogramming were down-regulated except for c-Myc, whereas endogenous Brn2, Sox2, Bmi1, and Nr2e1 were significantly up-regulated in late passages compared to those in early passages (Fig. S5C and Fig. S5D). In addition, some NPC specific marker gene expression levels were significantly up-regulated in late passage, such as Sox1 and Gpm6a, some of them had no significant changes, such as Nestin and CD133, and other genes including Msi-1, Pax6 and Ncan were down-regulated (Fig. S5B). These results suggest that the endogenous gene networks have been activated in the late passage, and iNPCs will be gradually independent on the expression of transgenes.
Figure 1. Light image and Immunostaining analyses of iNPCs, WT-NPCs and Fbb.
iNPCs, WT-NPCs and Fbb were subjected to suspension culture or adherent culture (coated with Poly-D-Lysine/Fibronectin) and morphology of cells were assessed by bright-field microscopy (right panel). Cells (5 × 104) cultured in 24-well plate containing coverslips coated with Poly-D-Lysine/Fibronectin, were fixed with 4% PFA and permeabilized with 0.2% Triton X-100 in PBS and then subjected to immunostaining with Nestin (red), Sox2 and Pax6 (green) antibodies, and nuclear staining with DAPI (blue) (left panel). (Scale bars: 50 μm).
Table 1. Primer pairs used for SYBR-Green-based quantitative Real-Time RT-PCR.
| Target (mouse) | Forward Primer | Reverse Primer | Annealing Temp |
|---|---|---|---|
| Nestin | 5′GTCTCAGGACAGTGCTGAGCCTTC3′ | 5′TCCCCTGAGGACCAGGAGTCTC3′ | 60°C |
| Pax6 | 5′GCGGAGTTATGATACCTACACC3′ | 5′GAAATGAGTCCTGTTGAAGTGG3′ | 60°C |
| Musashi1 | 5′GATGGCTCCCCCTCCAGGTT3′ | 5′CATTGGTGAAGGCTGTGGCA3′ | 60°C |
| CD133 | 5′TCGTACTGGTGGCTGGGTGGC3′ | 5′ACCACAAGGATCATCAATATCCAG3′ | 60°C |
| Sox2 | 5′CCTCCGGGACATGATCAGCATGTA3′ | 5′GCAGTGTGCCGTTAATGGCCGTG3′ | 60°C |
| Ncan | 5′GCACCGTGTATGGCTGTAGT3′ | 5′ATTCTCGCAAGGGCTGCATA3′ | 60°C |
| Sox1 | 5′GAGATGATCAGCATGTACCTGCC3′ | 5′GTAGTGCTGTGGCAGCGAGT3′ | 60°C |
| Gpm6a | 5′GCTTGGACAAGTGGACTGC3′ | 5′TAGGGAATACCTCCCAGGCA3′ | 60°C |
| DKK3 | 5′ACAGTGAGTGCTGTGGAGACCAGC3′ | 5′TCCCTCTGGTTGTCACAGATGGTC3′ | 60°C |
| Snail1 | 5′GTCTGCACGACCTGTGGAAA3′ | 5′AGCCAGACTCTTGGTGCTTG3′ | 60°C |
| Twist1 | 5′GAGGTCTTGCCAATCAGCCA3′ | 5′CCAGTTTGATCCCAGCGTTT3′ | 60°C |
| Col1a1 | 5′CTGACGCATGGCCAAGAAGA3′ | 5′ATACCTCGGGTTTCCACGTC3′ | 60°C |
| GAPDH | 5′AAGGGCTCATGACCACAGTC3′ | 5′GGATGACCTTGCCCACAG3′ | 60°C |
Figure 2. Marker gene expression of neural progenitor cells in iNPCs and their differentiation potential.
(A) Transcriptional profiling of NPC-specific genes in iNPCs, Fbb as well as WT-NPCs was analyzed by SYBR-Green based quantitative RT-PCR with specific primer pairs (see Table 1). Error bars correspond to SEM. (B) The expression levels of fibroblast-specific genes in Fbb, iNPCs as well as WT-NPCs was analyzed by SYBR-Green based quantitative RT-PCR with specific primer pairs (see Table 1). GAPDH was used as an internal control. (C) iNPCs cultured on collagen IV-coated coverslips were incubated under neuronal, astrocyte and oligodendrocyte differentiation conditions, respectively, and then subjected to immunostaining with polyclonal β-tubulin III (upper panel), monoclonal GFAP (middle panel) and polyclonal O4 (lower panel) antibodies, and nuclear staining with DAPI (blue). (Scale bars: 50 μm).
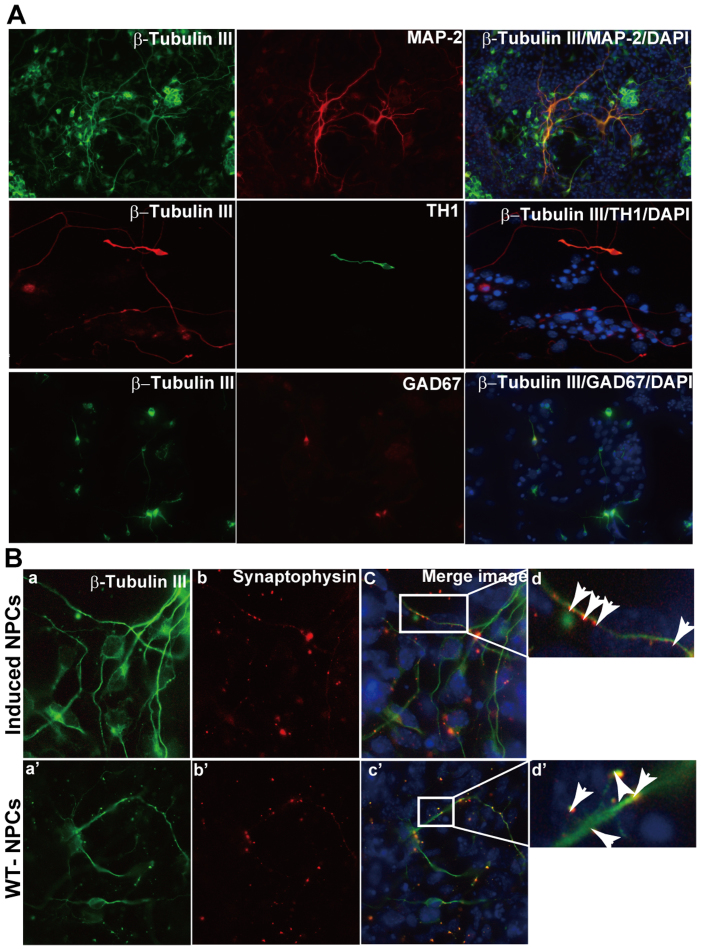
To investigate the tripotent potential of iNPCs, we examined the capability of iNPCs to differentiate into the three main neural lineages. Astrocyte differentiation was induced by culturing cells with 10% FBS. After 7 days differentiation, approximately 30–40% of cells in the culture system showed typical astrocyte morphology that uniformly stained for GFAP (Fig. 2C middle panel). When iNPCs were cultivated in Neurobasal medium supplemented with 2% B27 for 7 days, approximately 80% β-Tubulin III positive cells were observed in culture (Fig. 2C, upper panel). Although iNPCs have been shown to readily generate neurons and astrocytes in our previous study12, their oligodendroglial differentiation potential has not yet been explored, which raises concerns as to their tripotency. Here, our data further validate that the iNPCs derived from mouse skin fibroblasts are indeed tripotent. Upon proliferation with FGF2, platelet-derived growth factor (PDGF) and forskolin, followed by differentiation in the presence of triiodothyronine (T3) and ascorbic acid, iNPCs can generate oligodendrocytes (Fig. 2C, lower panel). Moreover, for mature neuronal differentiation, iNPCs were cultivated without EGF but in the presence of BDNF with increasing concentrations (20 ng/ml to 30 ng/ml) and FGF-2 with gradually decreasing concentrations (10 ng/ml to 6.7 ng/ml and 5 ng/ml). Most differentiated cells showed neuronal morphology and expression of the neuronal marker β-Tubulin III, with MAP-2+ (microtubule-associated protein) cell population accounting for 50% (Fig. 3A, upper panel). To further identify the subtypes of neurons, we detected the expression of the subtype neuron markers choline acetyltransferase (ChAt) (cholinergic neurons), serotonin (5-HT) (serotonergic neurons), GAD67 (GABAergic neurons) and tyrosine hydroxylase (TH, dopaminergic neurons). The results demonstrated that a relatively low percentage of iNPCs differentiated into TH+ and GAD67+ cells in the presence of BDNF (Fig. 3A, middle and lower panel), but no ChAt and 5-HT positive neurons were observed in culture conditions with BDNF (data not shown). These results suggest that artificial iNPCs are indeed tripotent cells and can be differentiated into selected subtypes of neurons depending on the differentiation procedure.
Figure 3. Mature neuronal differentiation and synapse formation between neurons derived from iNPCs.
iNPCs were cultured on collagen IV-coated glass coverslips and subjected to subtype neuronal differentiation. After three weeks differentiation, cells were fixed with 4% PFA and (A) then subjected to double immunostaining with β-tubulin III and MAP-2 (upper panel), β-tubulin III and TH1 (middle panel) and β-tubulin III and GAD67 (low panel). (B) Double immunostaining with β-tubulin III and Synaptophysin antibodies. (a, a′) β-tubulin III (green); (b, b′) Synaptophysin (red); (d, d′) shows a magnification of (c, c′), and arrows indicate the formation of synapses between neurons. Nuclear staining with DAPI (blue). (Scale bars: 50 μm).
Synapse formation and electrophysiological properties of functional neurons derived from iNPCs
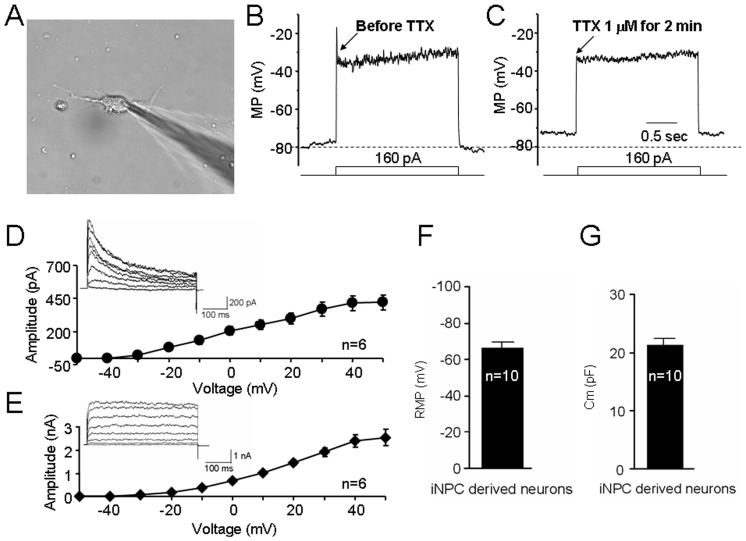
Synapses are connections between neurons that transmit electrical or chemical signals within the nervous system. To evaluate the ability of iNPC-derived neurons to form connections with each other, we differentiated neurons from iNPCs under a condition conducive to primary neuron culture, and observed that neurons derived from iNPCs expressed Synaptophysin with punctate distribution (Fig. 3B, upper panel: b and d) similar to the expression pattern and distribution of Synaptophysin observed in WT-NPCs (Fig. 3B, lower panel: b′ and d′), suggesting synapse formation of neurons from iNPCs in vitro. To analyze whether the resulting neurons from iNPCs exhibit functional membrane properties of neurons from NPCs, we performed electrophysiological recording. In current-clamp mode, injection of depolarizing current (160 pA) induced an action potential-like spike (Fig. 4A and 4B) in 20% (12/60) recorded neuron-like cells. Additionally, this spike could be blocked by 1 μM TTX, a Na+ channel blocker (Fig. 4C). In voltage-clamp mode, voltage-dependent outward K+ currents generated by voltage steps from −60 mV to +30 mV were recorded in iNPC-derived neurons after 7–10 days in culture. 10 mM TEA and 1 mM 4-AP were added to the bath solutions to record A-type (Fig. 4D) and D-type (Fig. 4E) K+ currents, respectively. Moreover, whole-cell patch-clamp recording revealed neuron-like resting membrane potentials (−66.0 ± 1.4 mV, n = 10; Fig. 4F) and membrane capacitances (21.3 ± 1.1 pF, n = 10; Fig. 4G) in iNPC-derived neurons, further suggesting similar functional membrane properties and activities compared to primary brain NPCs.
Figure 4. Electrophysiological properties of iNPC-derived neurons assessed by whole-cell patch-clamp recordings.
A phase contrast image of a patch pipette attached to the membrane of a cultured iNPC-differentiated neuron (A). Typical traces of action potential-like spike induced by current injection steps before (B) and after (C) 1 μM TTX treatment. (D) A-type K+ currents were induced by an initial hyperpolarization (voltage steps from the holding potential of −60 mV to −90 mV and return to −60 mV in the first step), and then stepped to +30 mV in increments of 10 mV (500 ms duration) in the presence of 10 mM TEA. (E) D-type K+ currents were induced by an initial depolarization (voltage steps from the holding potential of −60 mV to −30 mV and return to −60 mV in the first step), and then stepped to +30 mV in increments of 10 mV (500 ms duration) in the presence of 1 mM 4-aminopyridine. Bar graphs show the resting membrane potentials (F) and membrane capacitances (G) of iNPC-derived neurons. (Scale bars: 50 μm).
Discussion
Recent studies have shown that a combination of defined transcription factors or microRNA can directly convert somatic cells into glutamatergic, dopaminergic, GABAergic, and motor neurons7,8,10,17,18,19. The direct conversion of these somatic cells into subtypes of induced neuronal cells (iNCs) or self-renewable neural progenitor cells (iNPCs) has major applications in the treatment of neurodegenerative disorders. However, iNCs are not self-renewing, which limits the amount of cells available for research and clinical studies. In this regard, iNPCs are more attractive than that of iNCs.
Although various strategies have been developed to derive NPCs or regionally specified neural progenitors from embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs)20,21,22,23, the ethical issues of ESCs and the risk for teratoma formation from iPSCs limit their clinical and research applications. It has recently been reported that fibroblasts can be directly reprogrammed into NPCs through either Yamanaka factors in combination with NPC culture conditions24 or with a pool of several factors13,15,16. The critical pluripotency factor Oct4 is lacking in the latter method, which may decrease the risk of teratoma formation. Additionally, Sox2, a critical regulator gene for neural progenitor cell identity and maintenance, has also been successfully used for the direct conversion of fibroblasts into neural progenitor cells14. These studies mentioned above are consistent with our findings that mouse fibroblasts can be directly converted into iNPCs that exhibit typical primary NPC properties and differentiation abilities. However, our earlier study employed a novel pool of five factors (Brn2, TLX, Sox2, Bmi1 and c-Myc) and successfully converted adult skin fibroblast into iNPCs12. Additionally, compared to other studies, our system and Dr. Wernig's system16 provide a valuable tool to monitor the kinetics of the direct conversion process toward iNPC through observing the formation of GFP+ colonies (Fig. S2). Importantly, our iNPC reprogramming procedure shortened the process of iNPC generation, by which GFP+ colonies show up at day 9, rather than day 2516 or longer15. In addition, our procedure is also independent of culture conditions compared to that with a single factor system14. In this study, we further demonstrate that our iNPC line not only shows tripotent differentiation potentials and abilities to differentiate into mature neurons with synapse formation, but also exhibits characteristic electrophysiologic features (Fig. 2, Fig. 3 and Fig. 4).
Nuclear orphan receptor TLX determines the neural progenitor cell fate through a negative regulatory loop with miR-9, but also contributes to the proliferation and self-renewal of NPCs25,26,27. Brn2 also plays an important role in determining nestin gene expression by collaborating with Sox2 or Sox1128,29,30. Therefore, in our study, the application of Brn2 and TLX as NPC fate determinants and other factors as regulators of cell proliferation and self-renewal may offer better iNPC reprogramming. However, our previous study did not demonstrate detailed properties of these iNPCs. Here we show that the iNPCs from the combination of five factors are tripotent, self-renewing iNPCs, which can give rise to neurons, astrocytes and oligodendrocytes (Fig. 2C). In addition, iNPCs here come from skin fibroblasts (E/Nestin:EGFP transgenic mouse), which initially express EGFP upon transformation and keep green fluorescence until passage 5. After that, we observed that EGFP disappeared consistent with that in WT-NPCs (Fig. S3A), however, the disappearance of EGFP in iNPCs does not affect the expression of specific NPC marker genes, such as Sox2, Nestin and Pax6 (Fig. 1, Fig. 2 and Fig. S4), and differentiation potentials of iNPCs (Fig. 2 and Fig. S5A). Transgene silencing may contribute to the EGFP disappearance. To validate this possibility, we analyzed the exogenous EGFP expression by RT-PCR with EGFP specific primer in early and late passages of iNPCs and also detected the endogenous Nestin expression by real-time RT-PCR analysis. The results showed that EGFP expression disappears in late passage. However, endogenous Nestin expression still maintains high level (Fig. S3 B and C), indirectly supporting the possibility of transgene silencing. It has been proposed that DNA methylation can cause transcriptional repression by directly interfering with the binding of transcription factors to DNA, which has been confirmed by the identification of a number of transcriptional regulators that cannot bind methylated recognition elements31. In addition, transgene silencing was found to be associated with the onset of DNA methylation in the CpG island of gene promoter32. Comparing to transgene promoter, in normal cells, methylation mainly involves CpG-poor regions, while CpG islands seem to be protected from this inhibitory modification33. Our DNA methylation analysis clearly revealed that the higher levels of DNA methylation in transgenic promoter of late passaged iNPCs than that in early passaged iNPCs may contribute to EGFP disappearance (Fig. S6). Consistent with the DNA methylation analysis, we found that DNA methylation inhibitor (5-AZa), but not histone deacetylase inhibitor (TSA) reversed the EGFP expression in both early and late passaged iNPC cultures (data not shown). In this study, we employed two NPC fate determinants Brn2 (Pou3f2) and Sox2, which have been reported to determine Nestin expression by synergic interactions between Sox2 and Brn2 within nestin enhancer29. The high levels of those genes by transgene expression in early passage and by endogenous gene expression in late passage may contribute to the Nestin expression. Moreover, we speculated that exogenous genes may be silenced and lose their binding activity to the second-intronic enhancer of nestin after direct reprogramming, and that endogenous expression of NPC-related genes including Pax6, Msi-1 Ncan, Sox1, and Gpm6a responsible for maintaining a stable cell fate were turned on. However, the transcription analyses of transgenic and endogenous genes in iNPCs revealed that transgenes used for the direct conversion of fibroblasts were not silenced after reprogramming (Fig. S1A), but some transgenes will be down-regulated in iNPCs after long-term passages (Fig. S5C and S5D) and selected endogenous genes responsible for maintaining NPC properties were up-regulated such as Sox1 and Gpm6a (Fig. S5B). In addition, the fibroblast-specific genes Dkk3, Snail1, Twist1, and Col1α1 were significantly down-regulated in both iNPCs and WT-NPCs compared to fibroblasts (Fig. 2A and B), suggesting that the reprogrammed iNPCs suppress the fibroblast-specific transcription network and establish a new transcriptional network. Additionally, we observed that endogenous expression levels of NPC-related genes Pax6, Msi-1 and Sox1 were lower than that in WT-NPCs. It was reported that Pax6 is a key regulator in the neuronal fate determination and proliferation of NPCs through direct modulation of Sox2 expression34, and Msi-1 and Sox1 also contribute to the maintenance of the immature state and self-renewal activity of neural progenitor cells35,36. Therefore, it is possible that more exogenous expression of Sox2 and other genes (Fig. S1) compensates for the lower expression of Pax6, Msi-1, and Sox1 in iNPCs. Until now, it remains unknown whether the epigenetic memory of the starting cells determines the efficiency of iNPC generation and their differentiation potential in the future. To address these questions, we have developed iNPCs from other starting cells, such as astrocytes (unpublished data) and will do the comparison between iNPCs whose starting cells are from different germ layers.
Besides our report12, four research teams using a combination of different factors reported inducible production of neural progenitor cells13,14,15,16, which may suggest that the transcription factors are not the only elements that determine induction of NPCs from fibroblasts. Rather, these studies suggest that if cell fate determinants work coordinately with related regulators, the direct conversion of NPCs will be achieved. Future studies will need to determine whether the use of different reprogramming cocktails will alter features in iNPC lines. Long-term goals should remain focused on developing the procedures for clinical applications of human neural progenitor cells in the treatment of neurodegenerative disorders.
Methods
Cell culture
iNPCs have been successfully converted from mouse skin fibroblasts by retroviral transduction with Brn2, Sox2, Bmi1, Nr2e1, and c-Myc12, and WT-NPCs were isolated from embryonic day 14.5 (E14.5) C57BL/6J mouse embryos. iNPCs and WT-NPCs were maintained in NPC medium (NeuroCult® NSC Basal Medium (Mouse) supplemented with NeuroCult® NSC Proliferation Supplement (STEMCELL Technologies Inc.), 10 ng/ml of bFGF and EGF (R&D Systems, Minneapolis, MN)). iNPC line was used for all experiments at passage 5 to 10, and all experiments involving animals were done following protocols approved previously by the University of Nebraska Medical Center Institutional Animal Care and Use Committee and utilizing National Institutes of Health (NIH) ethical guidelines.
Differentiation of iNPCs
For general neural differentiation, WT-NPCs and iNPCs were cultured on collagen type IV-coated coverslips in NPC medium. The next day, the medium was replaced by neural differentiation medium (Neurobasal medium with 2% B27, 2 mM glutamine). For generation of glial fibrillary acidic protein (GFAP)-expressing astrocytes, the NPC medium was replaced by DMEM/F-12 [1:1] with 10% FBS, and cultured for 7 days, with a medium change every other day. On day 7 of differentiation, cells were immunostained. For oligodendrocyte differentiation, iNPCs were cultivated in DMEM/F12 with 1× N2, 10 ng/ml PDGF (R&D Systems, Minneapolis, MN), 10 ng/ml FGF-2 and 10 μM forskolin (Sigma-Aldrich, Saint Louis, MO) for 4 days. Afterwards, PDGF and forskolin were replaced by 30 ng/ml 3, 3, 5-triiodothyronine (T3) hormone and 200 mM ascorbic acid (all from Sigma-Aldrich, Saint Louis, MO) for another 7 days.
For subtype neuronal differentiation, iNPCs were plated at 5 × 104 cells/cm2 on collagen IV-coated glass coverslips in fresh pre-warmed medium A, composed of a 1:3 mix of DMEM/F12 and Neurobasal media (Invitrogen) containing 0.5% N2 and 1% B27 supplements, bFGF-2 10 ng/ml and BDNF 20 ng/mL for 3 days. Cells were then shifted to medium B composed of a 1:3 mix of DMEM/F12 and Neurobasal media containing 0.5% N2 and 1% B27 supplements, FGF-2 6.7 ng/mL, and BDNF 30 ng/mL. After 3 days, medium B was replaced with fresh medium B with a reduced amount of FGF-2 5 ng/ml. The cells were maintained under these conditions for an additional 10-15 days; the medium was partially changed every 3 days.
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde, washed 3 times with PBS, and then incubated in PBS containing 0.1% Triton X-100, 1% BSA (both Sigma-Aldrich), and 10% FBS (Invitrogen) for 45 min at room temperature. The cells were then incubated with the primary antibodies anti-Nestin (Millipore, 1:200), mouse anti-Tuj1 (Sigma-Aldrich, 1:1000), rabbit anti-Tuj1 (Sigma-Aldrich, 1:1000), rabbit anti-GFAP IgG (Sigma-Aldrich, 1:1000), mouse anti-O4 (R&D Systems, 1:100), mouse anti-Synaptophysin (Abcam, ab32127, 1:250), rabbit anti-CHAT (Millipore, AB144P, 1:100), mouse anti-Sox2 (Millipore, MAB4343, 1:1000), mouse anti-Pax6 (Developmental Studies Hybridoma Bank (DSHB), Iowa City, IA, 1:100) and mouse anti-Tyrosine Hydroxylase (TH) (Covance, MMS-503P, 1:1000) overnight at 4°C. One day after incubation with primary antibodies, the cells were washed with PBS containing 0.1% BSA and further incubated with secondary antibodies for 60 min at room temperature. Nuclei were detected by DAPI (1:1000) staining.
SYBR-Green-based quantitative Real-Time RT-PCR
Total mRNA was isolated with TRIzol Reagent (Invitrogen) and RNeasy Mini Kit (QIAGEN Inc., Valencia, CA) using the manufacturer's recommendations. The reverse transcription was performed using Transcription 1st Strand cDNA Synthesis Kit (Roche, USA). The RT-PCR analyses for the detection of neural progenitor cell specific mRNAs was performed using SYBR® Select Master Mix (Life Technologies, Los Angeles, CA) with 0.5 μl of cDNA, corresponding to 1 μg of total RNA in a 15 μl final volume, 1.5 μl H2O, 7.5 μl SYBR Green, 5.5 μl oligonucleotide primer pairs (synthesized at Fisher) at 1 mM (Table 1 and Table S1). PCR-program: 1. 50°C for 2 min, 2. 95°C for 2 min; 3. 95°C for 15 sec, 4. specific annealing temperature for 15 sec, 5. 72°C for 1 min. Steps 2-4 were repeated 40 times. All samples were amplified in duplicates and the mean was used for further analysis.
Whole cell Patch-Clamp analysis
Electrophysiological properties of neurons derived from iNPCs were analyzed 15 days after neuronal differentiation using standard whole-cell patch-clamp technique at room temperature. Cells with mature neuronal morphology were selected for whole-cell-patch-clamp recordings at the indicated time points. Cells were cultured on coverslips and placed in a submerged recording chamber and constantly perfused with oxygenated bath solution containing (in mmol l-1): 142 NaCl, 8.1 KCl, 1 CaCl2, 6 MgCl2, 10 HEPES, 10 D-Glucose (pH 7.4). Visualization of cells was done using an upright microscope equipped with near-infrared differential interference contrast and a 60 ×water immersion objective (Nikon). Whole-cell current-clamp and voltage-clamp recordings were carried out with an Axopatch-200B amplifier (Axon Instruments) that was interfaced by an A/D-converter (Digidata 1320; Axon Instruments,) to a PC running PClamp software (version 9; Axon Instruments). Recording pipette was filled with internal solution containing (in mmol l-1): 153 KCl, 1 MgCl2, 5 EGTA, 2.5 MgATP and 0.3 NaGTP, 10 HEPES. For measurements of action potentials and voltage responses, cells were current-clamped between −20 mV and −80 mV (depending on resting membrane potential), and 500-ms hyperpolarizing and depolarizing current steps were delivered in 160-pA increments through the whole-cell pipette. Voltage-gated sodium channels were blocked in the presence of 1 μM TTX (TOCRIS bioscience).
Author Contributions
C.T. designed the experiments, performed all the experiments and wrote the manuscript; Q.L. and J.W. performed the electrophysiological recording and data analysis; K.M, R.A and Y.W. helped with constructs, real-time analysis and immunochemistry assay; K.M.,Y.L., Y.H. and Q.C. helped with supplemental data; D.K. helped with DNA methylation analysis; J.D. helped in data discussion; J.Z. oversaw the project and wrote the manuscript.
Supplementary Material
Supplemental information
Acknowledgments
We kindly thank Drs. Yulong Li, Jinxu Liu and Lie Gao, and Mrs. Kristin M. Leland Wavrin, who provided technical support for this work. Julie Ditter, Lenal Bottoms, Johna Belling, and Robin Taylor provided outstanding administrative and secretarial support. Monoclonal antibody against PAX6 was obtained from the Developmental Studies Hybridoma Bank maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. This work was supported in part by research grants by the National Institutes of Health: R01 NS 41858-01, R01 NS 061642-01, 3R01NS61642-2S1, P01 NS043985, and P20 RR15635-01 (JZ), the State of Nebraska, DHHS-LB606 Stem Cell 2009-10 (JZ), LB606 Stem Cell-2010-10 (SD and CT), National Natural Science Foundation of China (NSFC) # 81028007 (JZ) and National Natural Science Foundation of China (NSFC) #81271419 (CT).
References
- Dauer W. & Przedborski S. Parkinson's disease: mechanisms and models. Neuron 39, 889–909 (2003). [DOI] [PubMed] [Google Scholar]
- Auld D. S., Kornecook T. J., Bastianetto S. & Quirion R. Alzheimer's disease and the basal forebrain cholinergic system: relations to beta-amyloid peptides, cognition, and treatment strategies. Prog Neurobiol 68, 209–245 (2002). [DOI] [PubMed] [Google Scholar]
- Lewis D. A., Hashimoto T. & Volk D. W. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci 6, 312–324 (2005). [DOI] [PubMed] [Google Scholar]
- Daadi M. M., Grueter B. A., Malenka R. C., Redmond D. E., Jr. & Steinberg G. K. Dopaminergic neurons from midbrain-specified human embryonic stem cell-derived neural stem cells engrafted in a monkey model of Parkinson's disease. PLoS One 7, e41120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gonzalez R. & Velasco I. Therapeutic potential of motor neurons differentiated from embryonic stem cells and induced pluripotent stem cells. Arch Med Res 43, 1–10 (2012). [DOI] [PubMed] [Google Scholar]
- Karumbayaram S. et al. Directed differentiation of human-induced pluripotent stem cells generates active motor neurons. Stem Cells 27, 806–811 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazzo M. et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 476, 224–227 (2011). [DOI] [PubMed] [Google Scholar]
- Pang Z. P. et al. Induction of human neuronal cells by defined transcription factors. Nature 476, 220–223 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T. et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfisterer U. et al. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A 108, 10343–10348 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage F. H. Mammalian neural stem cells. Science 287, 1433–1438 (2000). [DOI] [PubMed] [Google Scholar]
- Tian C. et al. Direct conversion of dermal fibroblasts into neural progenitor cells by a novel cocktail of defined factors. Curr Mol Med 12, 126–137 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thier M. et al. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell 10, 473–479 (2012). [DOI] [PubMed] [Google Scholar]
- Ring K. L. et al. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell 11, 100–109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D. W. et al. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell 10, 465–472 (2012). [DOI] [PubMed] [Google Scholar]
- Lujan E., Chanda S., Ahlenius H., Sudhof T. C. & Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc Natl Acad Sci U S A 109, 2527–2532 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo A. S. et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228-231 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son E. Y. et al. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell 9, 205–218 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marro S. et al. Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell 9, 374–382 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkeby A. et al. Generation of Regionally Specified Neural Progenitors and Functional Neurons from Human Embryonic Stem Cells under Defined Conditions. Cell Rep 1, 703–714 (2012). [DOI] [PubMed] [Google Scholar]
- Fujimoto Y. et al. Treatment of a mouse model of spinal cord injury by transplantation of human induced pluripotent stem cell-derived long-term self-renewing neuroepithelial-like stem cells. Stem Cells 30, 1163–1173 (2012). [DOI] [PubMed] [Google Scholar]
- Nori S. et al. Grafted human-induced pluripotent stem-cell-derived neurospheres promote motor functional recovery after spinal cord injury in mice. Proc Natl Acad Sci U S A 108, 16825–16830 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S. et al. ES cell-derived renewable and functional midbrain dopaminergic progenitors. Proc Natl Acad Sci U S A 108, 9703–9708 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. et al. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci U S A 108, 7838–7843 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G., Yu R. T., Evans R. M. & Shi Y. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc Natl Acad Sci U S A 104, 15282–15287 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Sun G., Li S. & Shi Y. A feedback regulatory loop involving micro RNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol 16, 365–371 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Q. et al. Orphan nuclear receptor TLX activates Wnt/beta-catenin signalling to stimulate neural stem cell proliferation and self-renewal. Nat Cell Biol 12, 31–40; sup pp 31–39 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi S. et al. The Sox-2 regulatory regions display their activities in two distinct types of multipotent stem cells. Mol Cell Biol 24, 4207–4220 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S. et al. Interplay of SOX and POU factors in regulation of the Nestin gene in neural primordial cells. Mol Cell Biol 24, 8834–8846 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi S. et al. The Sox2 regulatory region 2 functions as a neural stem cell-specific enhancer in the telencephalon. J Biol Chem 281, 13374–13381 (2006). [DOI] [PubMed] [Google Scholar]
- Eden S. & Cedar H. Role of DNA methylation in the regulation of transcription. Curr Opin Genet Dev 4, 255–259 (1994). [DOI] [PubMed] [Google Scholar]
- O'Hagan H. M., Mohammad H. P. & Baylin S. B. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet 4, e1000155 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross S. H. & Bird A. P. CpG islands and genes. Curr Opin Genet Dev 5, 309–314 (1995). [DOI] [PubMed] [Google Scholar]
- Wen J. et al. Pax6 directly modulate Sox2 expression in the neural progenitor cells. Neuroreport 19, 413–417 (2008). [DOI] [PubMed] [Google Scholar]
- Elkouris M. et al. Sox1 maintains the undifferentiated state of cortical neural progenitor cells via the suppression of Prox1-mediated cell cycle exit and neurogenesis. Stem Cells 29, 89–98 (2011). [DOI] [PubMed] [Google Scholar]
- Kawahara H. et al. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J Cell Biol 181, 639–653 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental information