Abstract
Many studies have utilized the Inbred Long Sleep and Inbred Short Sleep mouse strains to model the genetic influence on initial sensitivity to ethanol. The mechanisms underlying this divergent phenotype are still not completely understood. In this study, we attempt to identify genes that are differentially expressed between these two strains and to identify baseline networks of co-expressed genes, which may provide insight regarding their phenotypic differences. We examined the whole brain and striatal transcriptomes of both strains, using next generation RNA sequencing techniques. Many genes were differentially expressed between strains, including several in chromosomal regions previously shown to influence initial sensitivity to ethanol. These results are in concordance with a similar sample of striatal transcriptomes measured using microarrays. In addition to the higher dynamic range, RNA-Seq is not hindered by high background noise or polymorphisms in probesets as with microarray technology, and we are able to analyze exome sequence of abundant genes. Furthermore, utilizing Weighted Gene Co-expression Network Analysis (WGCNA) we identified several modules of co-expressed genes corresponding to strain differences. Several candidate genes were identified, including protein phosphatase 1 regulatory unit 1b (Ppp1r1b), prodynorphin (Pdyn), proenkephalin (Penk), ras association (RalGDS/AF-6) domain family member 2 (Rassf2), myosin 1d (Myo1d), and transthyretin (Ttr). In addition, we propose a role for potassium channel activity as well as map kinase signaling in the observed phenotypic differences between the two strains.
Keywords: RNA-Sequencing, Tophat, Cufflinks, WGCNA, Loss-of-righting-reflex, Inbred Short Sleep, Inbred Long Sleep, eQTL, Lore QTL, alcohol
Introduction
The heritability of alcohol use disorders, estimated to be approximately 0.5, suggests that genetics plays an important role in determining an individual’s risk (Dick et al. 2009). One possibility for how this risk manifests itself is in first response to alcohol (Schuckit 1980), where it was demonstrated that a low level of response to alcohol is a strong predictor of future alcohol use disorders (Schuckit 1994; Schuckit 2000). In animals, measures of acute ethanol response from a single intra-peritoneal injection include: ethanol-stimulated activity, metabolism, hypothermia, ataxia, and loss of righting reflex (LORR). The Inbred Long Sleep (ILS) and Inbred Short Sleep (ISS) mouse strains were selected for differences in LORR and show a large phenotypic divergence (McClearn & Kakihana 1981). Since this phenotype is present in ethanol-naïve animals, it is likely that genetically mediated differences in baseline gene expression could account for much of this phenotypic difference.
The ILS and ISS mice have been extensively studied, and are phenotypically different beyond ethanol-induced LORR (DeFries et al. 1989; Erwin et al. 1990; Gehle & Erwin 2000), for example, the strains differ in ethanol preference with the ISS mice consuming more ethanol than the ILS mice (Saba et al. 2011). The underlying genetics of these quantitative traits have been explored successfully using recombinant panels of mice to identify regions of interest likely involved in LORR (Lore QTLs) on chromosomes 1, 2, 3, 8, 11, and 15 (Bennett et al. 2002; Bennett et al. 2008; Bennett et al. 2006; Christensen et al. 1996; Markel et al. 1997; Markel et al. 1996). Genes in these regions were sequenced to find polymorphisms that may contribute to the observed phenotypes, and fifteen genes with coding sequence differences were identified (Ehringer et al. 2001). Further, gene expression studies, in both whole brain (Xu et al. 2001), and cerebellum (Maclaren & Sikela 2005) identified many differentially expressed genes (DEGs) between the strains. Maclaren identified several DEGs within Lore QTL regions with promoter region sequence differences (Maclaren et al. 2006).
The current study utilized Next Generation RNA Sequencing (RNA-Seq) technology to investigate baseline gene expression differences between these two strains. RNA-Seq produces millions of short reads which, when mapped back to the genome, provide a measure of gene expression as well as strain-specific sequence, at least for abundantly expressed genes. It provides a higher level of resolution of gene expression than is possible with hybridization microarrays. A high level of background noise, typical with microarrays, does not limit RNA-Seq (Marguerat & Bähler 2010; Wang et al. 2009). RNA-Seq has also been shown to improve network characteristics compared to microarrays (Iancu et al. 2012). The purpose of this study is to identify both DEGs and networks of co-expressed genes for future study of initial response to alcohol and risk of alcohol use disorders. While priority will be given to genes previously identified in alcohol or drug studies, we will use multiple bioinformatics resources to filter candidate genes depending on differential expression, sequence differences, genome locations, and co-expression with other candidate genes.
Materials and Methods
Statement on animal care
This study was conducted with approval from the Institutional Animal Care and Use Committee at the University of Colorado Health Sciences Center (Denver, Colorado) following guidelines established by the Office of Laboratory Animal Welfare. Measures were taken to minimize discomfort.
RNA extraction
Mice were bred and housed at the specific pathogen free facility at the Institute for Behavioral Genetics (University of Colorado, Boulder) under a 12-hour light/dark cycle with ad libitum access to food and water. On post-natal day 60, twelve ethanol-naïve adult male mice (n=6/strain) were sacrificed by cervical dislocation and whole brains were removed. Six brains (n=3/strain) were further dissected to isolate the striatum. Total RNA, from whole brains (WB, n=6, 3/strain) and striatum (ST, n=6, 3/strain) was extracted using RNeasy midi kits (Qiagen, Valencia, CA), and quantity and quality were determined using a NanoDrop™ spectrophotometer (Thermo Fisher Scientific, Wilmington, DE) and Agilent 2100 BioAnalyzer™ (Agilent Technologies, Santa Clara, CA). Ratios of absorbance at 260nm and 280nm were shown to be excellent (>1.8). RNA Integrity scores were also shown to be excellent (>8.0).
Library preparation
The preparation of the cDNA library for RNA-Sequencing was conducted according to Illumina (San Diego, California) protocol for quantitative RNA Sequencing on the Genome Analyzer II (GAII) platform. Starting with 10 μg total RNA for each RNA sample, the samples were enriched for poly-A RNA using Sera Mag Magnetic Oligo(dT) Beads™. The poly-A enriched RNA samples were then fragmented with a 3M NaOAc solution at 94°C for 5 minutes. The samples were reverse transcribed with random primers, and end repair was performed with T4 and Klenow DNA polymerase. Double stranded Illumina adaptors, with a single thymine overhang, were ligated to the ends of the cDNA fragments by first adding a single adenine to each 3′ end of the cDNA. Next, 200bp fragments were selected by agarose gel electrophoresis and subsequent gel extraction with Qiagen Gel Purification kits. Libraries were enriched with 15 cycles of PCR, and purified using QIAquick PCR Purification kits (Qiagen). Each cDNA library was run on one GAII lane sequencing to 36bp.
Alignment
Raw 36 nucleotide reads were trimmed to 28nt due to inherent decrease in quality score toward the 3′ end (Shendure & Ji 2008). Reads were mapped to the mouse reference genome (mm9, Ensembl) using TopHat (v1.2.0, http://tophat.cbcb.umd.edu) (Trapnell et al. 2009). TopHat first maps reads using Bowtie (v0.12.7, http://bowtie-bio.sourceforge.net/) (Trapnell & Salzberg 2009) alignment software, which utilizes a Burrows-Wheeler index of the mouse genome (obtained from Bowtie source webpage, http://bowtie-bio.sourceforge.net/) to rapidly align short reads. TopHat then uses the resulting read pileup to deduce likely exon/intron boundaries, and identifies reads aligning across boundaries. Reads with up to 2 mismatches were allowed, and reads were removed if they aligned to more than 10 places in the genome. Visualization of read pileups was done using the Integrated Genomics Viewer (IGV v2.1, www.broadinstitute.org/igv) (Robinson et al. 2011).
Transcript assembly, quantification, and differential expression testing
To assemble transcripts and estimate abundance, output from TopHat and the annotated reference genome (mm9, Ensembl) was analyzed using Cufflinks (v2.0.2, http://cufflinks.cbcb.umd.edu/) (Trapnell et al. 2010) to construct the minimum number of transcripts that explain the maximum number of reads. Since the sequenced sample had been enriched for poly-A mRNA transcripts, a mask file was used to discriminate against alignments in rRNA, tRNA, and small RNA genes. Once transcripts were assembled, their abundances were estimated by counting the number of aligned reads contained in the transcript, and normalizing both to the size of transcript and to the total number of aligned reads in the sample (fragments per kilobase exon per million mapped fragments, FPKM). Cuffcompare was then used to compile the set of transcripts from each group, and each transcript was tested for differential expression using Cuffdiff. Data for the four groups of three samples (ILS/WB, ILS/ST, ISS/WB, and ISS/ST) were input into Cuffdiff to calculate each pairwise comparison of gene expression. Cuffdiff outputs estimates of the Jensen-Shannon divergence of each pair to determine statistical significance. Due to the exploratory nature of this study, we applied a less stringent correction for multiple testing, using a False Discovery Rate (FDR=0.1). Since the Cuffdiff minimum threshold of 1000 reads allows inclusion of intronic reads, reads aligning to close neighbors, and/or genes contained within an intron, we wanted to ensure that we only included reads which aligned within the exon structure, therefore we set a minimum expression level FPKM of at least 1 for genes to be included in subsequent analyses. Minimum thresholds have been employed in previous studies, and a minimum FPKM of 1 is consistent (Brooks et al. 2011; Graveley et al. 2010). In addition, using the Ensembl annotation information, we identified expressed genes (FPKM>1) with overlapping features, i.e. un-translated regions on opposite strands. Visual examination of each of these cases resulted in removal of 139 genes from further analysis.
Weighted Gene Co-expression Network Analysis (WGCNA)
Weighted gene co-expression networks were generated using the statistical program R (v2.11.1, www.r-project.org) and the WGCNA package (http://www.genetics.ucla.edu/labs/horvath/CoexpressionNetwork/). Cufflinks output from all twelve samples were used for a single WGCNA. Data were merged based on unique Ensembl Gene Id, and genes were excluded if no group reached an average FPKM≥1. Briefly, WGCNA first attempted to impute missing data using a k-nearest neighbors algorithm, then removed genes where imputation was impossible, and removed genes with no variance in expression values. Next, a signed similarity matrix was constructed by taking each pairwise gene-gene Pearson correlation, adding 1 then dividing by 2. This was converted to a weighted adjacency matrix by a power function, determined by a scale-free topology model (β=4). Therefore, the adjacency matrix contained values from 0 to 1 for each gene, with 0, 0.5, and 1; signifying negative correlation (0–0.5), no correlation (0.5), and positive correlation (0.5–1). Genes were clustered based on hierarchical clustering of topological overlap matrix-based dissimilarity, with the dynamic tree cutting algorithm cutreeDynamic, and the deepSplit option set to 4. Gene clusters with a minimum of 20 genes were identified using a dynamic tree-cutting algorithm, which identified 21 gene clusters (modules). Similar gene modules were merged using the mergeCloseModules command, with a dissimilarity threshold of 0.1 (Pearson correlation greater than 0.9). Merging similar modules resulted in 16 remaining modules used in downstream analysis. Hub genes in each module were determined by ranking each gene by its module membership, calculated by WGCNA. Module robustness was tested in two ways. First, average module adjacencies were calculated and compared to the average adjacencies of randomly sampled “modules” of the same size. One thousand permutations of randomly sampled modules were generated. Modules were considered robust if average module adjacencies were significantly higher than the randomly generated modules. Second, the intramodular and extramodular connectivity of each module was calculated and scaled according to module size. Modules with higher scaled intramodular connectivity were considered robust.
Identification of relevant co-expression modules
To identify biologically relevant co-expression modules, we took the first principle component of each module, or module eigengene, using the moduleEigengenes command from the WGCNA R-package. Each module eigengene is representative of the gene expression levels for each module, if the module were reduced to a single gene. An analysis of variance of the resulting module eigengene values was used to identify module eigengenes different due to strain, region, or an interaction. Significant p-values were less than 0.05/16=0.003125. Each module was tested for enrichment of differentially expressed genes using a hypergeometric distribution function in R, and p-values were corrected using the p.adjust function in R, utilizing the Benjamini-Hochberg method (Benjamini & Hochberg 1995). The set of differentially expressed genes had been determined using the Cufflinks package as described above, and genes were included if significant at FDR=0.1.
Bioinformatics analyses
The set of differentially expressed genes were tested for functional group over-representation with the Web-based gene set analysis toolkit (WebGestalt, http://bioinfo.vanderbilt.edu/webgestalt) (Duncan et al. 2010; Zhang et al. 2005). Functional groups based on Gene Ontology (GO) (Ashburner et al. 2000), Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al. 2011; Kanehisa & Goto 2000), and WikiPathways (Kelder et al. 2012; Pico et al. 2008). Over-represented Lore QTL regions were identified using a hypergeometric distribution function in R. Cis-regulation of differentially expressed and WGCNA module Lore QTL hub genes was determined using publicly available datasets at www.genenetwork.org. Expression QTLs were identified using two LxS datasets, hippocampus (Aug07) and prefrontal cortex (Aug06), as well as two BxD datasets, striatum (Dec10v2) and whole brain (Nov06). Peak LOD score for expression must occur within 10Mb of gene locus to have been considered cis-regulated. Furthermore, since multiple datasets were used to interrogate regulation of expression, and most datasets contained multiple probes for each gene, cis- peaks had to occur in the majority of all the probes and at least once in each dataset to be considered having evidence of cis-regulation. MicroRNA binding sites were identified from www.microrna.org, visualizing all miRNAs with good mirSVR scores. In addition, sets of differentially expressed genes and co-expression modules were tested for over-representation of genes previously identified as being significantly differentially expressed (at least 3-fold higher) by cell type—neuron, astrocyte, or oligodendrocyte (Cahoy et al. 2008).
Identifying gene sequence differences
Cis-regulated differentially expressed genes in Lore QTL regions, as well as Lore QTL hub genes from WGCNA modules were visualized in IGV to identify sequence differences between strains. IGV incorporates annotated SNP information from dbSNP (build 128), which we used to classify SNPs as known or novel. In addition, genes sequenced previously (Ehringer et al. 2001) were visualized for confirmation of previous results.
Affymetrix microarray analysis
A reanalysis of previously published ILS/ISS striatal Affymetrix microarray results (Radcliffe et al. 2006) was conducted as a validation study of the current RNA-seq DEG results. Briefly, striatal tissue was dissected and total RNA was isolated from 15 naïve mice from each strain. RNA was quantitatively pooled from 3 mice for a total of 5 microarray samples for each strain. RNA preparation, array hybridization (Affymetrix 430 v2.0), and array scanning was performed using standard procedures; details can be found in Radcliffe et al. (2006).
Two probe masks were created and implemented to eliminate erroneous probes from calculations of transcript expression, thereby, increasing accuracy of expression estimates. Probe sequences were obtained directly from Affymetrix and aligned to the mouse genome (mm9) using BLAT (Kent 2002). First, individual probes that aligned to more than one location or did not perfectly align were removed. Second, probes that targeted regions of the genome harboring SNPs were eliminated because an “expression” difference detected from these probes was more likely to represent differences in hybridization efficiency rather than true differences in RNA expression levels (Walter et al. 2007). SNPs were identified from the current RNA-seq data using Partek Genomics Suite (v6.6; St. Louis, MO). We were less concerned about keeping probesets as ensuring that the retained probesets were of the highest quality possible. A liberal statistical criterion was thus used to test for significance of the SNPs (LOD>5.0) at the risk of increased type I errors for SNP identification, but at the same time, increased type II errors for probe removal, which we felt was acceptable in this case. Finally, probesets were required to consist of at least five probes. Following a global scaling procedure (average signal intensity of each array was set to a default target signal of 500), probe level normalization was performed using the Robust Multi-array Average method (RMA). Any RMA value that was less than 0.01 was converted to 0.01.
Results
Illumina GAII sequencing
Quantitative RNA Sequencing was completed on an Illumina GAII platform. Twelve samples total were sequenced, 6 each of whole brain (WB) and striatum (ST). Three samples from each region were from ILS mice, three from ISS mice. Whole brain data yielded short-read libraries of 12.7 and 13.1 million reads on average in ILS and ISS strains respectively. Striatum sequencing produced libraries of 26.9 and 26.5 million reads on average in ILS and ISS strains (Table 1). Differences in library size are due to updates in Illumina software occurring between sequencing dates.
Table 1.
Alignment statistics for each strain and region.
| Region | Strain | # mice | Total readsa | Reads removedb | Unique hitsc | Parameter hitsd | # Exon junctionse |
|---|---|---|---|---|---|---|---|
| Striatum | ILS | 3 | 26927097 ± 882830 | 4923 (0.0184%) | 72.45% | 88.89% | 80214 |
| ISS | 3 | 26466323 ± 1020682 | 5122 (0.0195%) | 73.97% | 89.63% | 83274 | |
| Whole Brain | ILS | 3 | 12786365 ± 1355373 | 3301 (0.0273%) | 74.78% | 89.17% | 72643 |
| ISS | 3 | 13130036 ± 481554 | 1853 (0.0141%) | 74.37% | 88.31% | 71220 |
Total number of short reads generated per group with standard deviation.
Low complexity reads are filtered prior to any attempt to align.
Percent of reads aligned to exactly one region of the genome.
Percent of reads aligned when allowing for up to 10 alignments.
Number of unique exon boundaries identified.
Alignment
Approximately 0.02% of low-complexity reads were discarded prior to alignment. Of the remaining reads, when alignment was constrained to 2 mismatches and 1 alignment, between 72 and 75% of reads aligned to the mouse genome. When constraints were relaxed to allow for up to 10 alignments, ~89% of reads were aligned. Over 70,000 (WB) and 80,000 (ST) unique exon-exon boundaries were identified (Table 1).
Differential expression
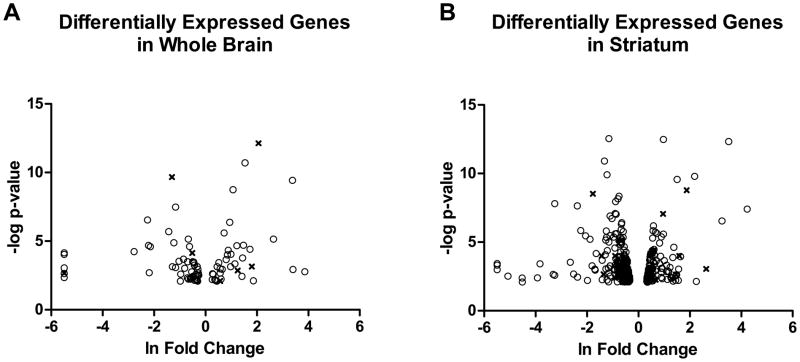
Using a minimum expression threshold of FPKM≥1 (in at least one sample) and a false discovery rate (FDR=0.1), 90 genes were differentially expressed between strains in the whole brain. In striatum, 336 genes were differentially expressed (Figure 1). Fifty-three genes were identified as differentially expressed in both data sets. Of those, 52 were differentially expressed in the same direction, while only one was higher in one strain compared to the other depending on region. Eight WB DEGs and 31 ST DEGs reside in previously identified Lore QTL regions. A complete list of all analyzed genes is given in supplemental files (Supplemental Table S1). Noteworthy differences include 14 potassium channel subunit ST DEGs, previously identified candidate genes—ras association (RalGDS/AF-6) domain family member 2 (Rassf2) and myosin 1d (Myo1d), and genes previously implicated in alcohol/drug response phenotypes—protein phosphatase 1 regulatory unit 1b (Ppp1r1b), opioid peptide precursor genes prodynorphin (Pdyn) and proenkephalin (Penk), and transthyretin (Ttr).
Figure 1.
Differentially expressed genes in whole brain and striatum.
Figure 1 displays the distribution of differentially expressed genes between strain in Whole Brain (A) and Striatum (B) samples. The x-axis represents the natural log of the fold change, with positive values corresponding to higher expression in ILS mice, and negative values corresponding to higher expression in ISS mice. The y-axis represents the negative log of the p-value of the difference in expression, with more significant differences corresponding to higher numbers. Open circles (82 WB, 305 ST) represent genes significant at a False Discovery Rate (FDR) of 0.1. X’s (8 WB, 31 ST) represent genes lying in Lore QTL regions.
Of the 336 DEG from the striatum, 297 had one or more valid probesets represented on the Affymetrix array. These Affymetrix probesets were tested for DE using one-way ANOVA (uncorrected; one-tail test). Over 90% of the Affymetrix probesets were expressed in the same direction as the RNA-seq DEG (Supplemental Figure S1). Of these, 65.7% were DE at p<0.05, 10.8% were DE at a p value between 0.05 and 0.1, and the remainder were DE at p>0.1 (Supplemental Figure S1). A complete list of the Affymetrix probesets and their expression levels is shown in supplemental table S2.
Over-representation analysis of differentially expressed genes
Utilizing the online resource WebGestalt, GO and KEGG functional group, and chromosomal region over-representation was determined on the set of 90 differentially expressed genes in WB, and the 336 differentially expressed genes in ST, with the reference set of genes based on the total number of genes detected at FPKM≥1 and tested for differential expression (12,678 genes in WB, 12,395 in ST). The results are shown in Table 2. Briefly, the most significant functional groups represented in whole brain include groups related to ribosomes, extracellular regions, and the major histocompatibility protein complex (corrected group p-values range from 9.19×10−6 − 0.0285). In striatum, the most significant functional groups include those related to ribosomes, potassium channel activity, and signal transduction (corrected group p-values range from 3.45×10−6 − 0.0482).
Table 2.
Over-representation analysis for DEGs in whole brain and striatum.
| Brain Region | General category | Classification terma | Resourceb | # genesc | p-valued | Corrected p-valuee |
|---|---|---|---|---|---|---|
| Striatum | Synapse/Signaling | Potassium channel activity | Gene Ontology | 14 | 9.92E-08 | 1.01E-05 |
| G-protein coupled receptor signaling pathway | Gene Ontology | 23 | 3.01E-06 | 5.00E-04 | ||
| Signal transduction | Gene Ontology | 68 | 1.00E-03 | 2.00E-03 | ||
| Neuron development | Gene Ontology | 17 | 7.15E-05 | 2.00E-03 | ||
| Non-odorant G-protein coupled receptors | Wikipathways | 13 | 2.00E-04 | 3.40E-03 | ||
| Calcium signaling pathway | KEGG | 12 | 2.00E-04 | 5.60E-03 | ||
| Dopamine receptor activity | Gene Ontology | 2 | 9.00E-04 | 6.10E-03 | ||
| Beta-adrenergic receptor kinase activity | Gene Ontology | 2 | 9.00E-04 | 6.10E-03 | ||
| Neuroactive ligand-receptor interation | KEGG | 11 | 4.00E-04 | 7.50E-03 | ||
| Negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | Gene Ontology | 4 | 5.00E-04 | 8.80E-03 | ||
| Synapse | Gene Ontology | 15 | 4.20E-03 | 3.65E-02 | ||
| Opioid peptide activity | Gene Ontology | 2 | 8.40E-03 | 4.18E-02 | ||
| Gap junction | KEGG | 7 | 4.30E-03 | 4.82E-02 | ||
| Behavior | Response to amphetamine | Gene Ontology | 4 | 1.00E-04 | 2.00E-03 | |
| Ribosome | Cytoplasmic ribosomal proteins | Wikipathways | 13 | 1.22E-07 | 4.15E-06 | |
| Ribosome | KEGG | 13 | 2.61E-07 | 1.46E-05 | ||
| Ribosome | Gene Ontology | 13 | 1.10E-03 | 1.41E-02 | ||
| Cell types | Neuron | Cahoy et al | 96 | 1.00E-16 | ||
| Oligodendrocyte | Cahoy et al | 26 | 2.50E-03 | |||
| Lore QTL | Lore4 Chr11:79000000-108000000 | Bennett et al | 13 | 4.10E-02 | ||
|
| ||||||
| Whole Brain | Ribosome | Cytoplasmic ribosomal proteins | Wikipathways | 6 | 1.04E-05 | 7.28E-05 |
| Ribonucleoprotein complex | Gene Ontology | 8 | 4.20E-03 | 2.85E-02 | ||
| Ribosome | KEGG | 7 | 9.19E-07 | 9.19E-06 | ||
| Cell membrane | Extracellular region | Gene Ontology | 13 | 7.00E-04 | 1.86E-02 | |
| Metabolic pathway | Retinol metabolism | Wikipathways | 2 | 1.17E-02 | 4.10E-02 | |
| Immune | MHC protein complex | Gene Ontology | 2 | 3.80E-03 | 2.85E-02 | |
| Cell types | Astrocyte | Cahoy et al | 10 | 2.60E-02 | ||
| Lore QTL | LoreChr3 Chr3:130000000-155000000 | Bennett et al | 2 | 2.70E-02 | ||
Term used to classify related genes.
Resource used for classification, Gene Ontology, KEGG, NCBI Entrez Gene, Wikipathways, Cahoy et al (2008), or Bennett et al (2006)/personal communication with Dr. Bennett.
Number of differentially expressed genes in each category.
Uncorrected hypergeometric p-value testing whether number of DEGs in each term more than expected.
Benjamini-Hochberg corrected p-values.
Additionally, LoreChr3 on chromosome 3 was enriched with WB DEGs (2 genes, hypergeometric p=0.027). In striatum, Lore4 on chromosome 11 (13 genes, hypergeometric p=0.041) was enriched (Table 2). The set of ST DEGs was also enriched for genes previously shown to be at least 3-fold over-expressed in oligodendrocytes (26 genes, hypergeometric p=0.0025) and neurons (96 genes, hypergeometric p<1×10−16). The set of WB DEGs was enriched for astrocyte-related genes (10 genes, hypergeometric p=0.026).
Weighted gene co-expression network analysis (WGCNA)
A single WGCNA of all 12 samples produced 16 distinct clusters (modules) of similarly expressed genes. The number of genes in each module ranged from 24 to 8,288. Each gene was assigned to a colored module, and no grey module (representing non co-expressed genes) was created (Supplementary Figure S2). Module robustness was tested using two methods. First, in each module, permutation testing confirmed that average module adjacency was always greater than the mean of 1000 randomly sampled “modules” of equal size (all modules p<0.001). Second, all modules were shown to display higher scaled intramodular connectivity compared to scaled extramodular connectivity (Supplemental Figure S3).
WGCNA gene modules enriched with differentially expressed genes
To determine whether each module contained more differentially expressed genes than expected, the number of observed differentially expressed genes in each module was compared to the hypergeometric distribution of the expected number of differentially expressed genes. Six modules were enriched with striatum DEGs (blue, cyan, green, greenyellow, magenta, and yellow) (Table 3). Of the 336 striatal DEGs, 96 out of 3211 in the blue module were differentially expressed (hypergeometric p=0.025), 8 of 76 in the cyan module (hypergeometric p=3.67×10−4), 12 of 123 in the green module (hypergeometric p=1.48×10−4), 12 of 171 in the greenyellow module (hypergeometric p=9.1×10−4), 9 of 87 in the magenta module (hypergeometric p=2.59×10−4), and 19 of 299 in the yellow module (hypergeometric p=2.59×10−4). Four modules were enriched with whole brain DEGs (darkred, green, magenta, and yellow, Table 3). Of the 90 whole brain DEGs, 1 of 24 in darkred were differentially expressed (hypergeometric p=0.042), 7 of 123 in green (hypergeometric p=7.1×10−6), 10 of 87 in magenta (hypergeometric p=1.45×10−10), and 12 of 299 in yellow (hypergeometric p=5.27×10−7). All p-values have been adjusted for multiple corrections according to the Benjamini-Hochberg method, using the p. adjust function in R. A complete table of the WGCNA modules can be found in the Supplemental Table S3).
Table 3.
WGCNA Co-expression module characteristics.
| Modulea | #genes | Module eigengene significantb | DEG enrichmentc | Cell type enrichmentd | Top genese | Lore QTLf | DEGg |
|---|---|---|---|---|---|---|---|
| black | 165 | Region (2.13E-05) | Matk | ||||
| Kcnh3 | |||||||
| Psd | |||||||
| Tmem191c | |||||||
| Ppp2r2c | |||||||
|
| |||||||
| yellow | 299 | Strain (4.12E-04) | ST (2.60E-04) WB (5.26E-07) |
Ptprn | Lore1 | ||
| Eif3k | |||||||
| Glt25d1 | |||||||
| Tnip1 | |||||||
| Tpd52 | |||||||
|
| |||||||
| brown | 1082 | Region (1.11E-03) | Oligodendrocyte (2.31E-07) | Rps6ka4 | |||
| Gsn | |||||||
| Rbx1 | |||||||
| Ephb1 | |||||||
| Icam5 | |||||||
|
| |||||||
| cyan | 76 | ST (3.66E-04) | Robo3 | ST | |||
| Kalrn | |||||||
| Kcns1 | Lore2b | ST | |||||
| Cacnb3 | |||||||
| Sytl2 | |||||||
|
| |||||||
| greenyellow | 171 | Strain (6.63E-06) Region (2.37E-04) |
ST (9.10E-04) | 6030458C11Rik | ST | ||
| Selplg | ST, WB | ||||||
| 4933439F18Rik | ST | ||||||
| 4632428N05Rik | |||||||
| Gm10116 | |||||||
|
| |||||||
| magenta | 87 | Strain (2.26E-07) | ST (2.60E-04) WB (1.45E-10) |
Gm10516 | ST, WB | ||
| Folh1 | WB | ||||||
| Prss50 | WB | ||||||
| Rnasel | ST, WB | ||||||
| 4930452B06Rik | |||||||
|
| |||||||
| grey60 | 36 | Strain (1.39E-04) | 2610002J02Rik | ||||
| Polr1b | Lore2a | ||||||
| Lama2 | |||||||
| Chi3l1 | |||||||
| Adi1 | |||||||
|
| |||||||
| darkred | 24 | WB (4.23E-02) | Rnd2 | Lore4 | |||
| Uchl1 | |||||||
| Add1 | |||||||
| Ncan | |||||||
| Tstd2 | |||||||
|
| |||||||
| green | 123 | Strain (1.78E-07) | ST (1.48E-04) WB (7.10E-06) |
Tmem181a | |||
| A530054K11Rik | ST, WB | ||||||
| Copb1 | ST | ||||||
| Tmem181b-ps | ST | ||||||
| Trmt6 | Lore2a | ||||||
|
| |||||||
| blue | 3211 | Strain (2.20E-03) Region (2.01E-06) |
ST (2.51E-02) | Rasgrp1 | |||
| Ppp1r9a | |||||||
| Pde7b | ST | ||||||
| Nexn | LoreChr3 | ST | |||||
| Rgs4 | ST | ||||||
|
| |||||||
| turquoise | 8288 | Region (1.63E-06) | Neuron (5.45E-04) Astrocyte (2.07E-02) |
Gm672 | |||
| Kndc1 | ST | ||||||
| Pcdh1 | |||||||
| Plxna1 | ST | ||||||
| Slc20a2 | |||||||
Gene co-expression module produced by WGCNA. Modules not significant for eigengene difference, DEG or cell-type enrichment are not shown.
Results of Analysis of Variance of the first principle component, or module eigengene, of expression values for each module. Each module eigengene was tested across strain and region.
Module over-representation of DEGs from either ST or WB.
Module over-representation of genes expressed at least 3-fold higher in specific cell types, neurons, astrocytes, or oligodendrocytes (Cahoy et al, 2008).
The top five most interconnected genes in each module.
Lore QTL region containing corresponding top gene.
Region of differential expression of corresponding top gene.
Module eigengenes associated with strain/region differences
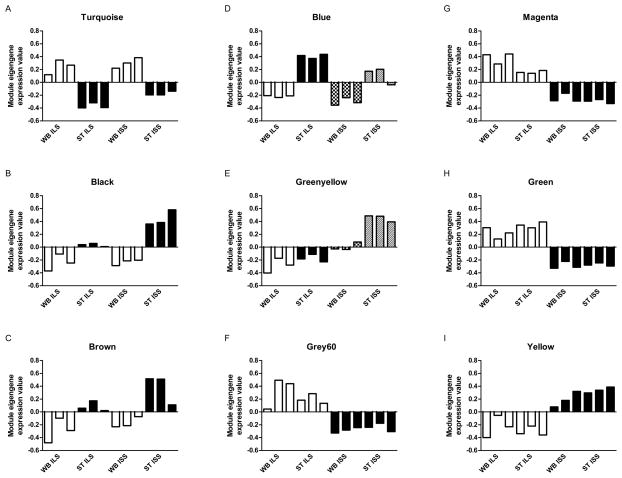
We calculated the 1st principle component (PC) of each module using the moduleEigengenes command from the WGCNA R-package. The 1st PC, or module eigengene, represents the sample-specific expression levels if each module were reduced to a single gene (Hierarchical clustering of module eigengenes is shown in Supplemental Figure S4). An analysis of variance (ANOVA) of the module eigengenes (Figure 2) resulted in strain differences in four modules: green (F1,8=274.6, p=1.78×10−7), grey60 (F1,8=46.11, p=1.39×10−4), magenta (F1,8=258.3, p=2.26×10−7) and yellow (F1,8=65.06, p=4.12×10−5). Three modules were different by region—black (F1,8=78.03, p=2.13×10−5),, brown (F1,8=24.62, p=1.11×10−3), and turquoise (F1,8=154.6, p=1.63×10−6). Two modules were different for both strain and region: blue (F1,8=19.61, p=0.0022, strain; F1,8=146.43, p=2.01×10−6, region) and greenyellow (F1,8=106.8, p=6.63×10−6, strain; F1,8=39.49, p=2.37×10−3, region), No module eigengenes had significant strain × region interaction effects. P-values were considered significant when less than 0.05/16=0.003125.
Figure 2.
WGCNA module eigengene expression levels.
Figure 2 displays the calculated expression level of module eigengenes, the first principle component of each module expression pattern. Individual mouse samples (bars) are in groups of 3 for each set of whole brain ILS, striatum ILS, whole brain ISS and striatum ISS. Only module eigengenes significant for strain or region differences are shown. Module eigengenes reduce the expression value of all genes in the module to one value per sample. An ANOVA of each module eigengene reveals modules different by region (A–C), both region and strain (D–E), and strain (F–I). No module eigengenes had significant strain × region interactions.
Cell type over-representation in WGCNA modules
Using genes identified as being significantly over-expressed, by at least 3-fold, in neurons, astrocytes, or oligodendrocytes, we tested whether modules were enriched for these sets of genes (Table 3 and Figure 2) (Cahoy et al. 2008). Of the 13,802 genes used in the WGCNA, 1,099 (neuron), 803 (astrocyte), and 556 (oligodendrocyte) had been identified as being over-expressed by at least 3-fold in each cell type. The turquoise module was enriched with 721 neuron genes (hypergeometric p=5.45×10−4) and 522 astrocyte genes (hypergeometric p=0.021). The brown module was enriched with 81 oligodendrocyte genes (hypergeometric p=2.31×10−7). All p-values were adjusted for the Benjamini-Hochberg false discovery rate.
Gene module hub gene identification
WGCNA identifies networks of interconnected genes, and it is possible to further identify the most interconnected genes in each module. The top five most interconnected genes (hub genes) in the eleven modules either enriched for DEGs or different across strain or region are listed in Table 3. Seventeen DEGs were identified as hub genes, 11 ST DEGs, 2 WB DEGs, and 4 DEGs from both ST and WB. Nine genes located within Lore QTLs were also hub genes. Of the six modules identified as different across strain, four had DEGs as hub genes. In the blue module, phosphodiesterase 7b (Pde7b), nexilin, F-actin binding protein (Nexn), and regulator of G-protein signaling 4 (Rgs4), all ST DEGs, are hub genes. Three ST DEGs in the greenyellow module were hub genes, 6030458C11Rik, 4933439F18Rik, and selectin P ligand (Selplg). Additionally, three genes in the green module, A530054K11Rik, coatamer protein complex subunit beta 1 (Copb1), and transmembrane protein 181b pseudogene (Tmem181b-ps), along with four genes in the magenta module, Gm10516, folate hydrolase (Folh1), protease, serine 50 (Prss50), and ribonuclease A, family 1 (Rnasel), are differentially expressed in either ST, WB, or both.
Functional group over-representation in WGCNA modules
Co-expression modules were analyzed using WebGestalt to test for functional group over-representation (Supplemental Table S4). In the six modules modules differing by strain, several signaling pathways were over-represented, including mitogen-activated protein kinase (MAPK) signaling (blue, yellow), peroxisome proliferator activated protein (PPAR) signaling (blue, greenyellow), transforming growth factor (TGF) beta signaling (blue, greenyellow), nuclear factor κB (NF-κB) signaling (blue, magenta, yellow), and toll-like receptor (TLR) signaling (blue, yellow). Genes involved in regulating the actin cytoskeleton were enriched in blue, green, and yellow. Complement and coagulation cascades were enriched in the magenta module. All group p-values range from 1.1 × 10−38 − 0.048 and have been corrected for multiple testing and were significant at <5% false discovery rate.
Identification of cis-regulated Lore QTL genes
Utilizing publicly accessible databases of recombinant inbred gene expression data from the online WebQTL tool (www.genenetwork.org), we identified differentially expressed genes from both striatum and whole brain, as well as hub genes, in Lore QTL regions that have evidence of cis-regulation. Each hub gene and DEG lying in Lore QTL regions was interrogated. A total of 11 genes showed evidence of cis-regulation. Three DEGs, alanine-glyoxylate aminotransferase 2-like 1 (Agxt2l1) located in LoreChr3, ras association (RalGDS/AF-6) domain family member 2 (Rassf2) located in Lore2a and keratin 12 (Krt12) located in Lore4 were differentially expressed in both WB and ST, and show strong evidence of cis-regulation. Six genes differentially expressed in the ST, Lore1 genes regulated endocrine-specific protein 18 (Resp18) and serine peptidase inhibitor, clade E, member 2 (Serpine2), Lore3 gene centromere protein t (Cenpt), Lore4 genes Rap guanine nucleotide exchange factor GEF-like 1 (Rapgefl1), myosin light chain 4 (Myl4), and keratin 9 (Krt9), Lore5 all show evidence of cis-regulation. The WB DEG and LoreChr3 gene DNA-damage-inducible transcript 4-like (Ddit4l), as well as the grey60 module hub gene, polymerase (RNA) I polypeptide B (Polr1b) also could be cis-regulated.
Sequence differences
Of the Lore QTL genes with evidence of cis-regulation, only Resp18 and Agxt2l1 did not have any detectable sequence differences (Supplemental Table S5). Of note, an unnamed missense single nucleotide polymorphism (SNP) in Serpine2, resulting in an isoleucine to valine substitution (I313V) in both ILS and ISS mice was observed. Four missense SNPs in Cenpt, three of which were unnamed were only observed in ISS. More unnamed SNPs were observed in Myl4, Polr1b, and Ddit4l. Also notable are the multitude of polymorphisms in 3′ UTR of Rassf2. According to www.microrna.org, these polymorphisms could potentially disrupt the binding sites of multiple miRNAs.
Fifteen genes previously reported to contain coding sequence differences were examined, and each polymorphism was confirmed in twelve of the genes (Ehringer et al. 2001). Low expression levels in Tgfb1 and Pth2r (named Pthr in original paper) made it impossible to identify polymorphisms. Znf133 has since been classified as a pseudogene, although it is expressed in our sample, and several single nucleotide polymorphisms are confirmed; however, frame shift mutations could not be confirmed. Although there are numerous sequence differences between the two strains, complete identification and classification of polymorphisms was beyond the scope of the study.
Discussion
Loss of righting reflex in response to acute ethanol has been well studied in the ILS and ISS strains, and respective QTLs have been identified and replicated using recombinant panels, both LSxSS and LxS (Bennett et al. 2002; Bennett et al. 2008; Bennett et al. 2006; Christensen et al. 1996; Maclaren & Sikela 2005; Markel et al. 1997; Markel et al. 1996). The goal of this study was to identify baseline differences in gene expression and co-expression between these two selected inbred strains, which will provide insight into the underlying biology that contributes to their differential sensitivity to alcohol. While previous studies have identified candidate genes based on expression differences, this study uses multiple methods, differential expression, Weighted Gene Co-expression Network Analysis, identification of cis-regulated Lore QTL genes, and identification of sequence differences in coding and un-translated regions. The use of RNA-Seq technology, as opposed to previous use of microarray, provides higher dynamic range, lower background noise, improved network characteristics, and the elimination of hybridization issues due to polymorphisms and annotation (Iancu et al. 2012; Marguerat & Bähler 2010; Wang et al. 2009). In this study, 90 genes in WB and 336 in ST samples were differentially expressed. We prioritize genes that are located in previously identified Lore QTL regions for future study. Eight WB and 31 ST DEGs are located in Lore QTL regions. While the total number of QTL genes is no different than chance, two Lore QTL regions were enriched for DEGs, LoreChr3 on chromosome 3 was enriched with WB DEGs and Lore4 on chromosome 11 was enriched for ST DEGs. This could potentially signify regional differences in gene expression, and future transcriptome examinations may identify regions enriched with other Lore QTL genes.
Two previously identified candidate genes (Maclaren & Sikela 2005), Rassf2 (ras association (RalGDS/AF-6) domain family member 2), located in Lore2a, and Myo1d (Lore4 gene myosin 1d) were identified by our analysis as differentially expressed in both ST and WB. MacLaren sequenced the promoter region of Rassf2, finding several polymorphisms (Maclaren et al. 2006). One advantage of RNA-Seq is the acquisition of the genetic sequence of exons and untranslated regions (UTRs). Examination of the 3′ UTR of Rassf2 shows distinct genotypes. ISS mice have the C57Bl/6J haplotype, while the ILS 3′ UTR shows many SNPs, several unnamed in dbSNP. Since the 3′ UTR is implicated in post-transcriptional regulation, including microRNA binding sites, the polymorphisms could account for some of the previously observed differences in expression. The observed ILS polymorphisms disrupt the consensus sequences for binding sites of 9 miRNAs (www.microrna.org). We were unable to detect expression levels for these miRNAs, so whether they affect expression levels of Rassf2 remains to be seen. We present evidence that several genes, including Rassf2, are cis-regulated, meaning that polymorphisms in gene regions between the two strains could contribute to differences in gene expression. If these are cis-regulated, it is likely that differences in gene expression could be explained by genetic polymorphisms in either coding regions or UTRs. Furthermore, while synonymous polymorphisms in exons may not affect protein function, they are indicative of distinct haplotypes between strains and of possible polymorphisms in intergenic or intronic regions that could affect expression. It is not clear how Rassf2 and Myo1d could influence ethanol-related behavior. Rassf2 has been characterized as a pro-apoptotic gene, residing in the nucleus and binding K-Ras, inducing apoptosis (Donninger et al. 2010). Differences could also arise from the role of Myo1d in the development of the nervous system (Benesh et al. 2012). Taken together, it is possible that strain specific neural development could lead to phenotypic differences.
Located in Lore2a is the DEG prodynorphin, Pdyn. More highly expressed in ST of ILS mice, Pdyn is differentially expressed in other animal models of ethanol behaviors. Consistent with our findings, low drinking ANA rats have increased levels of striatal Pdyn compared to higher drinking AA rats (Nylander et al. 1994). Another opioid precursor gene, proenkephalin, Penk, is also more highly expressed in the ST of ILS mice. While the difference between strains in opioid signaling has not been explored in depth, it has been shown that SS and LS mice differ in response to morphine injection and withdrawal (Brick & Horowitz 1983). Another QTL gene, in Lore4, Ppp1r1b, which codes for protein phosphatase 1 regulatory unit 1b, also known as DARPP-32, has been implicated in the neurobiological response to many drugs of abuse (Yger & Girault 2011). Ppp1r1b is expressed in striatal medium spiny neurons (MSNs), and plays a large role in the cellular response to dopaminergic signaling.
In addition to genes from Lore QTL regions, transthyretin (Ttr) on chromosome 19 was also identified in both samples as being differentially expressed. Gamma-protein kinase C (PKC-γ) null mutant mice and their wild-types have similar ethanol-related behaviors as the ISS and ILS mice, and these differences were correlated with baseline Ttr expression, which is higher in mutant mice (Smith et al. 2006). Similar to the ISS mice, PKC-γ null mutants are less sensitive to acute ethanol than their wild-type littermates (Harris et al. 1995), and voluntarily consume more ethanol (Bowers & Wehner 2001). Likewise, baseline expression of Ttr in ISS mice is higher relative to ILS mice. While it is unknown whether a chronic ethanol diet would increase expression of Ttr in the ISS mice, as in the PKC-γ null mutants, future confirmation would further implicate Ttr in ethanol behavior. Also of interest are the 14 potassium channel subunits differentially expressed in the striatum; as potassium channels have been implicated in responses to ethanol (Brodie et al. 1999; Hopf et al. 2011; Hopf et al. 2010; Mulholland 2012) and the cumulative effect of differential expression of all of these channels could contribute to the difference in ethanol sensitivity between the strains.
While RNA-Seq is thought to offer several advantages over microarrays, it still suffers a problem inherent to any massively parallel method: finding the appropriate statistical balance between type I and type II errors. Validation by an independent method is one approach and here we have used microarray data to validate the RNA-Seq DEGs. The results are similar, perhaps slightly better, to a comprehensive comparison of RNA-Seq to hybridization microarrays conducted by Bottomly et al. (2011); i.e., they found that 48.4% of genotype-dependent RNA-Seq DEG were also DE on the Affymetrix platform and we found that this was true for 65.7% of our RNA-Seq DEG, although our statistical criteria was somewhat less stringent. In addition to the possibility of statistical errors, reasons for less than perfect consistency between RNA-Seq and microarrays probably include the broader dynamic range of RNA-Seq and, more importantly, the likelihood of genotype effects on transcript isoform abundance meaning for microarrays, quantification of a given transcript is dependent on probeset location (Bottomly et al. 2011). Indeed, we have seen hints of evidence for strain-by-isoform interactions for some of the microarray probesets that were not significant, although this particular RNA-Seq dataset is not ideal for a comprehensive splice variant analysis.
Using WebGestalt to identify over-represented groups in our sets of DEGs, we identified several distinct groups of differentially regulated gene systems. In ST, there were many DEGs involved in signal transduction and synaptic signaling. In addition to functional groups, we identified cell type specific (neurons, astrocytes, and oligodendrocytes) genes over-represented in each set of DEGs. The set of ST DEGs was enriched for neuron and oligodendrocyte genes. Specifically, the set of 127 DEGs up-regulated in ILS mice was only enriched for neuronal genes, while the set of 209 DEGs up-regulated in ISS mice was enriched for all three types of cells. This suggests that while there are differences in neuronal processes between the two strains, there may be more important differences in glial related processes. This holds up when looking at WB DEGs, as the set of WB DEGs is enriched only for astrocyte related genes.
To further characterize strain specific differences in gene expression, we employed the agnostic network analysis tool WGCNA, which clustered genes based on topological overlap dissimilarity. The results of the WGCNA display its usefulness at analyzing large expression datasets. Gene modules were enriched for cell specific genes, and module eigengenes highlight strain- and region-specific differences. However, there is a limitation on the interpretations due to the small sample size in our study, even though each module passed strict robustness testing. No hub genes were immediately identifiable as strong candidate genes, however it is important to acknowledge that the WGCNA identifies networks of related genes, and the effect of any single gene could be minimal. It differs in this way from the differential expression analysis, where the genes with the largest differences in expression, and possibly having larger effects, are identified. In this analysis, we were less confident in some of the smaller modules where some samples appeared to be outliers, but more confident of modules showing consistent expression levels within groups (either regional or strain). These patterns of expression are striking, and show that genes can be consistently co-expressed at different levels depending on region or strain.
Of the 16 gene co-expression networks (modules), three were enriched for ST DEGs, one for WB DEGs, and three were enriched for both ST and WB DEGs. This made it possible to identify not only DEGs, but also gene networks in which those DEGs reside. Functional group over-representation of DEG-enriched modules revealed many genes related to neuronal structure and function, as well as transcriptional regulation. Interestingly, these modules were enriched for several signaling pathways, including MAP Kinase signaling pathways, previously shown to regulate ethanol behaviors (Carnicella et al. 2008).
One module, turquoise, was enriched with neuron genes. Since this module eigengene differed across region, and not strain, this module is most likely composed of neuronal genes differentially expressed due to regional differences, and given that this is the largest module, most of the co-expression differences can likely be due to brain regional differences. Of the six modules different across strain, five were enriched for ST DEGs, while three of those were also enriched for WB DEGs.
Utilizing RNA-Seq technology to identify gene expression differences and gene co-expression networks has provided insight into the differences between ILS and ISS mice. Genes previously identified as candidates from expression/QTL studies, Rassf2, Myo1d, and drug response studies, Pdyn, Penk, Ppp1r1b, and Ttr are again implicated. While these differences exist, this study is not designed to specify causal differences. Therefore, it is important for future research to focus on manipulation, genetic or pharmacological, of genes and gene networks to further elucidate the differences between these strains, in order to understand the cause of ethanol-related behaviors.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Dr. Katerina Kechris and Dr. Laura Saba for the development of the Affymetrix probe mask. This research was supported by National Institutes of Health grants T32 DA017637 (TMD), R01 AA017889 (MAE, salary support), and R01 AA016957 (RAR).
Footnotes
The authors have no conflicts of interest to declare.
Contributor Information
Todd Darlington, Institute for Behavioral Genetics, University of Colorado, Boulder and Department of Integrative Physiology, University of Colorado, Boulder.
Marissa Ehringer, Institute for Behavioral Genetics, University of Colorado, Boulder and Department of Integrative Physiology, University of Colorado, Boulder.
Colin Larson, Department of Pharmaceutical Sciences, University of Colorado Health Sciences Center, Denver.
Tzu Phang, Division of Pulmonary Sciences and Critical Care Medicine, University of Colorado Health Sciences Center, Denver.
Richard Radcliffe, Institute for Behavioral Genetics, University of Colorado, Boulder and Department of Pharmaceutical Sciences, University of Colorado Health Sciences Center, Denver.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, et al. Gene ontology: tool for the unification of biology. The GeneOntology Consortium. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benesh AE, Fleming JT, Chiang C, Carter BD, Tyska MJ. Expression and localization of myosin-1d in the developing nervous system. Brain research. 2012;1440:9–22. doi: 10.1016/j.brainres.2011.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- Bennett B, Beeson M, Gordon LG, Carosone-Link P, Johnson TE. Genetic dissection of quantitative trait loci specifying sedative/hypnotic sensitivity to ethanol: mapping with interval-specific congenic recombinant lines. Alcoholism: Clinical and Experimental Research. 2002;26:1615–1624. doi: 10.1097/01.ALC.0000037136.49550.B3. [DOI] [PubMed] [Google Scholar]
- Bennett B, Carosone-Link P, Beeson M, Gordon LG, Phares-Zook N, Johnson TE. Genetic dissection of quantitative trait locus for ethanol sensitivity in long-and short-sleep mice. Genes, Brain and Behavior. 2008;7:659–668. doi: 10.1111/j.1601-183X.2008.00403.x. [DOI] [PubMed] [Google Scholar]
- Bennett B, Carosone-Link P, Zahniser NR, Johnson TE. Confirmation and fine mapping of ethanol sensitivity quantitative trait loci, and candidate gene testing in the LXS recombinant inbred mice. Journal of Pharmacology and Experimental Therapeutics. 2006;319:299–307. doi: 10.1124/jpet.106.103572. [DOI] [PubMed] [Google Scholar]
- Bottomly D, Walter NAR, Hunter JE, Darakjian P, Kawane S, Buck KJ, Searles RP, Mooney M, McWeeney SK, Hitzemann RJ. Evaluating Gene Expression in C57BL/6J and DBA/2J Mouse Striatum Using RNA-Seq and Microarrays. PLoS ONE. 2011;6:e17820. doi: 10.1371/journal.pone.0017820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers BJ, Wehner JM. Ethanol consumption and behavioral impulsivity are increased in protein kinase Cgamma null mutant mice. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2001;21:RC180–RC180. doi: 10.1523/JNEUROSCI.21-21-j0004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick J, Horowitz GP. Tolerance and cross-tolerance to morphine and ethanol in mice selectively bred for differential sensitivity to ethanol. Journal of studies on alcohol. 1983;44:770–779. doi: 10.15288/jsa.1983.44.770. [DOI] [PubMed] [Google Scholar]
- Brodie MSM, McElvain MAM, Bunney EBE, Appel SBS. Pharmacological reduction of small conductance calcium-activated potassium current (SK) potentiates the excitatory effect of ethanol on ventral tegmental area dopamine neurons. The Journal of pharmacology and experimental therapeutics. 1999;290:325–333. [PubMed] [Google Scholar]
- Brooks MJ, Rajasimha HK, Roger JE, Swaroop A. Next-generation sequencing facilitates quantitative analysis of wild-type and Nrl−/− retinal transcriptomes. Molecular Vision. 2011;17:3034. [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. Journal of Neuroscience. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnicella S, Kharazia V, Jeanblanc J, Janak PH, Ron D. GDNF is a fast-acting potent inhibitor of alcohol consumption and relapse. Proceedings of the National Academy of Sciences. 2008;105:8114–8119. doi: 10.1073/pnas.0711755105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SC, Johnson TE, Markel PD, Clark VJ, Fulker DW, Corley RP, Collins AC, Wehner JM. Quantitative Trait Locus Analyses of Sleep-Times Induced by Sedative-Hypnotics in LSXSS Recombinant Inbred Strains of Mice. Alcoholism: Clinical and Experimental Research. 1996;20:543–550. doi: 10.1111/j.1530-0277.1996.tb01090.x. [DOI] [PubMed] [Google Scholar]
- DeFries JC, Wilson JR, Erwin VG, Petersen DR. LS × SS recombinant inbred strains of mice: initial characterization. Alcoholism: Clinical and Experimental Research. 1989;13:196–200. doi: 10.1111/j.1530-0277.1989.tb00310.x. [DOI] [PubMed] [Google Scholar]
- Dick DM, Prescott C, McGue M. The Genetics of Substance Use and Substance Use Disorders. Handbook of Behavior Genetics. 2009:433–453. [Google Scholar]
- Donninger H, Hesson L, Vos M, Beebe K, Gordon L, Sidransky D, Liu JW, Schlegel T, Payne S, Hartmann A, Latif F, Clark GJ. The Ras effector RASSF2 controls the PAR-4 tumor suppressor. Molecular and cellular biology. 2010;30:2608–2620. doi: 10.1128/MCB.00208-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan D, Prodduturi N, Zhang B. WebGestalt2: an updated and expanded version of the Web-based Gene Set Analysis Toolkit. BMC bioinformatics. 2010;11:10. [Google Scholar]
- Ehringer MA, Thompson J, Conroy O, Xu Y, Yang F, Canniff J, Beeson M, Gordon LG, Bennett B, Johnson TE, Sikela JM. High-throughput sequence identification of gene coding variants within alcohol-related QTLs. Mammalian genome. 2001;12:657–663. doi: 10.1007/s00335-001-1001-x. [DOI] [PubMed] [Google Scholar]
- Erwin VG, Jones BC, Radcliffe RA. Further characterization of LSxSS recombinant inbred strains of mice: activating and hypothermic effects of ethanol. Alcoholism: Clinical and Experimental Research. 1990;14:200–204. doi: 10.1111/j.1530-0277.1990.tb00472.x. [DOI] [PubMed] [Google Scholar]
- Gehle V, Erwin V. The genetics of acute functional tolerance and initial sensitivity to ethanol for an ataxia test in the LSxSS RI strains. Alcoholism: Clinical and Experimental Research. 2000;24:579–587. [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, Brown JB, Cherbas L, Davis CA, Dobin A, Li R, Lin W, Malone JH, Mattiuzzo NR, Miller D, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2010;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RA, McQuilkin SJ, Paylor R, Abeliovich A, Tonegawa S, Wehner JM. Mutant mice lacking the gamma isoform of protein kinase C show decreased behavioral actions of ethanol and altered function of gamma-aminobutyrate type A receptors. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:3658–3662. doi: 10.1073/pnas.92.9.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf FW, Simms JA, Chang S-J, Seif T, Bartlett SE, Bonci A. Chlorzoxazone, an SK-Type Potassium Channel Activator Used in Humans, Reduces Excessive Alcohol Intake in Rats. Biological Psychiatry. 2011;69:618–624. doi: 10.1016/j.biopsych.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf FWF, Bowers MSM, Chang S-JS, Chen BTB, Martin MM, Seif TT, Cho SLS, Tye KK, Bonci AA. Reduced Nucleus Accumbens SK Channel Activity Enhances Alcohol Seeking during Abstinence. Neuron. 2010;65:13–13. doi: 10.1016/j.neuron.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu OD, Kawane S, Bottomly D, Searles R, Hitzemann RJ, McWeeney S. Utilizing RNA-Seq data for de novo coexpression network inference. Bioinformatics (Oxford, England) 2012;28:1592–1597. doi: 10.1093/bioinformatics/bts245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Research. 2011;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa MM, Goto SS. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelder T, van Iersel MP, Hanspers K, Kutmon M, Conklin BR, Evelo CT, Pico AR. WikiPathways: building research communities on biological pathways. Nucleic Acids Research. 2012;40:D1301–1307. doi: 10.1093/nar/gkr1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT---The BLAST-Like Alignment Tool. Genome research. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclaren EJ, Bennett B, Johnson TE, Sikela JM. Expression profiling identifies novel candidate genes for ethanol sensitivity QTLs. Mammalian genome : official journal of the International Mammalian Genome Society. 2006;17:147–156. doi: 10.1007/s00335-005-0065-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclaren EJ, Sikela JM. Cerebellar gene expression profiling and eQTL analysis in inbred mouse strains selected for ethanol sensitivity. Alcoholism: Clinical and Experimental Research. 2005;29:1568–1579. doi: 10.1097/01.alc.0000179376.27331.ac. [DOI] [PubMed] [Google Scholar]
- Marguerat S, Bähler J. RNA-seq: from technology to biology. Cellular and Molecular Life Sciences. 2010;67:569–579. doi: 10.1007/s00018-009-0180-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markel PD, Bennett B, Beeson M, Gordon LG, Johnson TE. Confirmation of quantitative trait loci for ethanol sensitivity in long-sleep and short-sleep mice. Genome research. 1997;7:92–99. doi: 10.1101/gr.7.2.92. [DOI] [PubMed] [Google Scholar]
- Markel PD, Fulker DW, Bennett B, Corley RP, DeFries JC, Erwin VG, Johnson TE. Quantitative trait loci for ethanol sensitivity in the LS × SS recombinant inbred strains: interval mapping. Behavior Genetics. 1996;26:447–458. doi: 10.1007/BF02359489. [DOI] [PubMed] [Google Scholar]
- McClearn G, Kakihana R. Selective Breeding for Ethanol Sensitivity: Short-Sleep and Long-Sleep Mice. In: McClearn G, Deitrich RA, Erwin V, editors. Development of Animal Models as Pharmacogenetic Tools. Washington, DC: US Government Printing Office; 1981. pp. 147–159. DHHS Publication No [ADM] 81–113. [Google Scholar]
- Mulholland PJP. K(Ca)2 channels: Novel therapeutic targets for treating alcohol withdrawal and escalation of alcohol consumption. Alcohol. 2012;46:309–315. doi: 10.1016/j.alcohol.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander I, Hyytia P, Forsander O, Terenius L. Differences between Alcohol-Preferring (AA) and Alcohol-Avoiding (ANA) Rats in the Prodynorphin and Proenkephalin Systems. Alcoholism: Clinical and Experimental Research. 1994;18:1272–1279. doi: 10.1111/j.1530-0277.1994.tb00118.x. [DOI] [PubMed] [Google Scholar]
- Pico AR, Kelder T, van Iersel MP, Hanspers K, Conklin BR, Evelo C. WikiPathways: pathway editing for the people. PLoS Biology. 2008;6:e184–1407. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe RA, Lee MJ, Williams RW. Prediction of cis-QTLs in a pair of inbred mouse strains with the use of expression and haplotype data from public databases. Mammalian genome : official journal of the International Mammalian Genome Society. 2006;17:629–642. doi: 10.1007/s00335-005-0178-9. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nature biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba LM, Bennett B, Hoffman PL, Barcomb K, Ishii T, Kechris K, Tabakoff B. A systems genetic analysis of alcohol drinking by mice, rats and men: influence of brain GABAergic transmission. Neuropharmacology. 2011;60:1269–1280. doi: 10.1016/j.neuropharm.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA. Self-rating of alcohol intoxication by young men with and without family histories of alcoholism. Journal of studies on alcohol. 1980;41:242–249. doi: 10.15288/jsa.1980.41.242. [DOI] [PubMed] [Google Scholar]
- Schuckit MA. Low level of response to alcohol as a predictor of future alcoholism. American Journal of Psychiatry. 1994;151:184–189. doi: 10.1176/ajp.151.2.184. [DOI] [PubMed] [Google Scholar]
- Schuckit MA. Genetics of the risk for alcoholism. The American journal on addictions / American Academy of Psychiatrists in Alcoholism andAddictions. 2000;9:103–112. doi: 10.1080/10550490050173172. [DOI] [PubMed] [Google Scholar]
- Shendure J, Ji H. Next-generation DNA sequencing. Nature biotechnology. 2008;26:1–11. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Smith AM, Bowers BJ, Radcliffe RA, Wehner JM. Microarray analysis of the effects of a gamma-protein kinase C null mutation on gene expression in striatum: a role for transthyretin in mutant phenotypes. Behavior Genetics. 2006;36:869–881. doi: 10.1007/s10519-006-9083-6. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Salzberg SL. How to map billions of short reads onto genomes. Computation Biology. 2009;27:1–3. doi: 10.1038/nbt0509-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter NA, McWeeney SK, Peters ST, Belknap JK, Hitzemann RR, Buck KJ. SNPs matter: impact on detection of differential expression. Nature Methods. 2007;4:679–680. doi: 10.1038/nmeth0907-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ehringer MA, Yang F, Sikela JM. Comparison of global brain gene expression profiles between inbred long-sleep and inbred short-sleep mice by high-density gene array hybridization. Alcoholism: Clinical and Experimental Research. 2001;25:810–818. [PubMed] [Google Scholar]
- Yger M, Girault J-A. DARPP-32, jack of all trades... master of which? Frontiers in Behavioral Neuroscience. 2011;5:1–14. doi: 10.3389/fnbeh.2011.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Kirov S, Snoddy JR. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic AcidsResearch. 2005;33:W741–748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.