Purpose and Appropriate Sample Types
This panel was developed, optimized and validated for assessment of CD4+ and CD8+ T-cell responses to various peptide pools for antigens of interest in cryopreserved peripheral blood mononuclear cells (PBMC) from adult humans. The panel has been used to evaluate HIV- and TB-specific responses to candidate vaccines for these pathogens, although the panel can be used with peptide pools for any proteins. The panel has not been tested with freshly-isolated PBMC or with whole blood.
Background
The focus of this panel is functional T-cell characterization and therefore only a minimum number of phenotyping markers are included in order to identify the CD4+ and CD8+ T cell lineages (CD3, CD4, CD8). This allows for a large number of functional markers, which include functions common to both CD4+ and CD8+ T cells and also functions more likely to be associated with a single T-cell lineage. IFN-γ, IL-2, and TNF-α are considered key cytokines for both lineages and are commonly examined in intracellular cytokine staining (ICS) assays (1); thus, these were given high priority. MIP-1β is a chemokine that can be produced by both CD4+ and CD8+ T cells and hasbeen shown to be useful in identifying polyfunctional T cells (2). It has relatively higher background when examined individually in terms of cells staining in the unstimulated control condition, and thus we mainly examine it in the context of co-expression with other functional markers. A fifth function commonly included when examining polyfunctionality is CD107a (3), a marker of degranulation, which may be a surrogate for cytotoxic potential. Although traditionally considered a function of cytolytic CD8+ T cells, some CD4+ T cells can degranulate.
We included two additional markers of interest for CD4+ T cells: IL-4 (a representative Th2 cytokine) and CD40 ligand (CD40L, also known as CD154). For IL-4, a bright fluorochrome was chosen since IL-4-producing cells are difficult to detect due to the low intensity of staining. Cells expressing CD40L interact with B cells expressing CD40; CD40L thus likely identifies T cells (mainly CD4+ T cells) that can provide help to B cells (4). CD40L is expressed on the surface of cells and can be detected either by surface staining in a co-culture assay where the fluorochrome-conjugated antibody is included during the ex vivo antigen stimulation (4), or through intracellular staining. We used the latter method since the CD40L co-culture assay is incompatible with use of Brefeldin A, which is essential for the most sensitive detection of some cytokines, in particular TNF-α. In situations where sensitivity is not as critical, monensin alone can be used to allow for co-culture surface staining of CD40L. Note that both Brefeldin A and monensin were used in our assay, since monensin was required for the CD107a co-culture assay.
Finally, a viability marker is considered essential for any assay identifying cells present at low frequency, and CD14 was included to exclude monocytes in order to improve the specificity of the assay. This could have been included in the same channel as the viability marker, but we chose to use another unused channel.
Similarity to Published OMIPs
OMIP-001 (5), OMIP-008 (6) and OMIP-009 (7), since these OMIPs examine antigen-specific human T cells by ICS. Our OMIP is focused on multiple functions rather than a combination of functions and memory markers (OMIP-001, OMIP-009) and includes a larger variety of functions compared with all these OMIPs. Additionally, our panel was developed for use in a good clinical laboratory practices (GCLP) setting and has been validated.
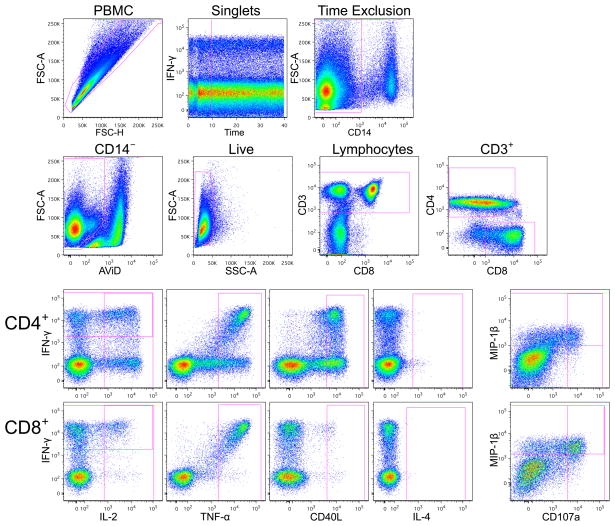
Figure 1.
Example staining profile for PBMC stimulated with Staphylococcal enterotoxin B (SEB). The upper two rows show the gating hierarchy to identify CD4+ and CD8+ T cells. Due to pressure fluctuations during the first 10 seconds of collection of the sample using the high-throughput sampler (HTS), events collected during this time are excluded. The lower two rows show expression of the functional markers for CD4+ and CD8+ T cells. A gate is applied for each functional marker, not accounting for co-expression of other markers. Boolean gates are later created based on the gates shown to identify cells expressing various combinations of markers. Note that the functional gates for many markers are placed relatively high in reference to the negative cells to lower the number of cells falling into these gates for the unstimulated samples (not shown).
Table 1.
Summary table for application of OMIP-014.
| Purpose | T-cell cytokine production and function after in vitro stimulation |
| Species | Human |
| Cell types | Cryopreserved PBMC |
| Cross references | OMIP-001, OMIP-008, OMIP-009 |
Table 2.
Reagents used for OMIP-014.
| Specificity | Clone | Fluorochrome | Purpose |
|---|---|---|---|
| IFN-γ | B27 | V450 | Function |
| IL-2 | MQ1-17H12 | PE | |
| TNF-α | MAb11 | FITC | |
| IL-4 | MP4-25D2 | APC | |
| MIP-1β | D21-1351 | Ax700 | |
| CD40L (CD154) | TRAP1 | PE-Cy5 | |
| CD107a | H4A3 | PE-Cy7 | |
| CD3 | UCHT1 | PE-TR | Lineage |
| CD4 | 13B8.2 | APC-Ax750 | |
| CD8 | SK1 | PerCP-Cy5.5 | |
| CD14 | TüK4 | QD655 | Dump |
| Dead cells | - | AViD |
APC, allophycocyanin; Ax, Alexa; AViD, LIVE/DEAD fixable aqua dead cell stain; Cy, cyanine; FITC, fluorescein isothiocyanate; PE, R-phycoerythrin; PerCP, peridinin chlorophyll protein; QD, quantum dot; TR, Texas Red.
Acknowledgments
Funding support:
This work was supported by the HIV Vaccine Trials Network Laboratory Program, a cooperative agreement with the National Institutes of Health Division of AIDS (National Institute of Allergy and Infectious Diseases) (UM1 AI068618). This work was also supported through the University of Washington Center for AIDS Research, a National Institutes of Health-funded program (P30 AI027757).
We thank the James B. Pendleton Charitable Trust for their generous equipment donation. The authors thank the laboratory technicians who performed the assays and Stephen Voght for help with editing.
Literature Cited
- 1.Horton H, Thomas EP, Stucky JA, Frank I, Moodie Z, Huang Y, Chiu YL, McElrath MJ, De Rosa SC. Optimization and validation of an 8-color intracellular cytokine staining (ICS) assay to quantify antigen-specific T cells induced by vaccination. J Immunol Methods. 2007;323:39–54. doi: 10.1016/j.jim.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–9. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betts MR, Brenchley JM, Price DA, De Rosa SC, Douek DC, Roederer M, Koup RA. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J Immunol Methods. 2003;281:65–78. doi: 10.1016/s0022-1759(03)00265-5. [DOI] [PubMed] [Google Scholar]
- 4.Chattopadhyay PK, Yu J, Roederer M. A live-cell assay to detect antigen-specific CD4+ T cells with diverse cytokine profiles. Nat Med. 2005;11:1113–7. doi: 10.1038/nm1293. [DOI] [PubMed] [Google Scholar]
- 5.Mahnke YD, Roederer M. OMIP-001: Quality and phenotype of Ag-responsive human T-cells. Cytometry A. 2010;77A:819–20. doi: 10.1002/cyto.a.20944. [DOI] [PubMed] [Google Scholar]
- 6.Zuleger CL, Albertini MR. OMIP-008: measurement of Th1 and Th2 cytokine polyfunctionality of human T cells. Cytometry A. 2012;81A:450–2. doi: 10.1002/cyto.a.22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamoreaux L, Koup RA, Roederer M. OMIP-009: Characterization of antigen-specific human T-cells. Cytometry A. 2012;81A:362–3. doi: 10.1002/cyto.a.22042. [DOI] [PubMed] [Google Scholar]