Abstract
Much of the knowledge about cell differentiation and function in the immune system has come from studies in mice, but the relevance to human immunology, diseases, and therapy has been challenged, perhaps more from anecdotal than comprehensive evidence. To this end, we compare two large compendia of transcriptional profiles of human and mouse immune cell types. Global transcription profiles are conserved between corresponding cell lineages. The expression patterns of most orthologous genes are conserved, particularly for lineage-specific genes. However, several hundred genes show clearly divergent expression across the examined cell lineages, and among them, 169 genes did so even with highly stringent criteria. Finally, regulatory mechanisms—reflected by regulators’ differential expression or enriched cis-elements—are conserved between the species but to a lower degree, suggesting that distinct regulation may underlie some of the conserved transcriptional responses.
Keywords: comparative transcriptomics, evolution, gene expression, microarray
The immune system is extensively studied in both human and mouse. Studies of human immune cells are restricted largely to in vitro or ex vivo assays, whereas studies in mice allow manipulation and monitoring of the immune system in an organismal setting. Although the mouse is an invaluable model for studies of immune function, there are substantial differences between the two species because of both biological and experimental factors, and studies have repeatedly suggested caution when translating findings from mouse to human (1, 2).
Charting the similarities and differences between immune cell lineages of human and mouse can provide a reference map that will help translate mouse findings to human and determine when (and why) the mouse immune response is likely to diverge from the human immune response. On the one hand, many orthologous transcription factors play conserved roles in both human and mouse immune system differentiation. On the other hand, studies reported prevalent variation between human and mouse (3–5), but it is hard to distinguish real differences from differences caused by the conditions compared, representation of cell populations per se, and their ability to interact and communicate with each other. Genome-wide profiling of mRNA levels in each lineage opens the way for such a comprehensive comparison.
Here, we compare two compendia of transcriptional profiles collected from human and mouse cell lineages during immune system differentiation. As expected, we show that the transcriptional program is extensively conserved from global transcriptional profiles to lineage-specific gene expression to the underlying regulatory mechanisms. Extensive conservation is also observed even when comparing activated immune T cells. We highlight genes with different expression patterns in human and mouse that were not previously reported and validate a few of them experimentally. We provide our data and analysis in a web portal that will serve as a reference map for future immunology studies.
Results
Transcriptional Maps of the Human and Mouse Immune Systems.
We compared two compendia of the human and mouse mRNA expression profiles measured using microarrays from immune cells at different differentiation states. The human D-MAP compendium (6) consists of 38 cell types (Fig. S1A). The mouse ImmGen compendium (7) consists of 244 cell types (Fig. S1B). Both span a wide range of myeloid and lymphoid cells of the innate and adaptive immune systems. Using the Ensembl COMPARA database, we mapped 10,248 one-to-one orthologs between the two species that were measured in both studies. Although many of the major cell lineages are common to both compendia (Fig. 1A), there are several important distinctions in the design of the two studies in the measured cell types, which were more finely parsed in the mouse data, and the tissue source (only blood for human; all lymphoid organs for mouse) (Dataset S1, Note 1). In addition, different samples of the same cell type in human were typically from unrelated healthy donors, whereas mouse data were almost exclusively derived from inbred C57BL/6 (B6) mice raised in a barrier facility.
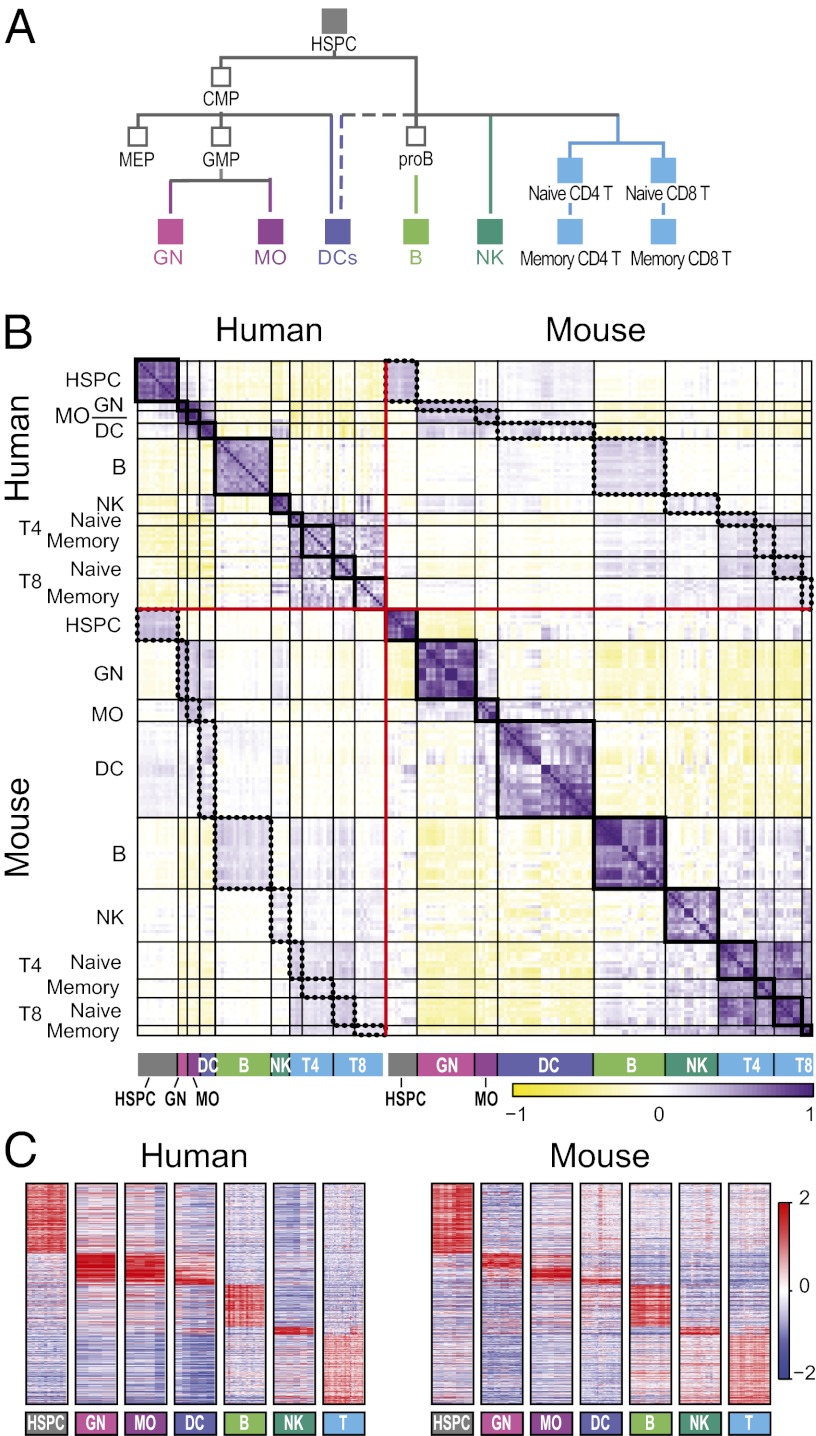
Fig. 1.
Conserved transcriptional profiles in matched immune lineages between human and mouse. (A) Simplified lineage tree for the immune system cell populations measured in both human and mouse compendia. CMP, common myeloid progenitor; GMP, granulocyte–monocyte progenitor; MEP, megakaryocyte–erythroid progenitor. Filled rectangles, cell types included in the common lineages. Colors mark those lineages in all figures. (B) Global correlation matrix of the Pearson coefficients (yellow/purple color scale on the bottom) between every pair of human and mouse samples in the common lineages. Samples are in rows and columns sorted by the lineage tree. Red lines separate species, black lines separate lineages, thick black rectangles mark correlations within each lineage within a species, and dotted rectangles mark correlations between samples of the same lineage between species. (C) Mean-centered expression values (red/blue color scale on the right) of the genes shared between matching lineage signatures in human (Left) and mouse (Right). Signatures were defined in each species separately as composed of genes that are expressed in a lineage-specific pattern in that species. Every pair of matching signatures was compared between the species to identify all those pairs of orthologs that were members of both signatures. Genes are sorted by human lineage with maximal expression level.
Because the differences between the compendia may confound subsequent comparisons, we first identified matching cell types in the two studies. We reasoned that broad comparisons are likely to be more robust, because individual cell types are harder to map between the species, and there are many more cell types in the mouse compendium. We, therefore, focused on comparing at the level of seven broad cell groups: hematopoietic stem and progenitor cells (HSPCs), granulocytes (GNs), monocytes (MOs), dendritic cells (DCs), B cells, natural killer cells (NKs), and T cells (Fig. 1A, filled rectangles). Using other groupings, such as separating CD8 and CD4 naïve and memory T cells, adding progenitors, or combining all myeloid cells into one group, did not qualitatively change the results. Within each group, we selected the specific cell types to include based on known biology (Dataset S2). For example, we excluded one human mature NK cell population (CD56− CD16+ CD3− cells), which is more similar to human monocytes than to NK cells, and mouse plasma cells, whose equivalent is missing from the human compendium. Many of the relevant cell surface markers are similarly expressed in the corresponding groups in both species, confirming their comparability (Fig. S2A). Overall, we compared seven groups of cell types, consisting of 80 human and 137 mouse samples.
Global Expression Profiles and Lineage-Specific Signatures Are Conserved.
The global expression profiles of matching lineages are similar between the two species for both individual cell types and average group profiles (Fig. 1B, dotted rectangles; and Fig. S2B, thick black rectangles). The correlation between a pair of mouse/human profiles from the same lineage group (mean correlation = 0.2) is lower than the correlation between two cell types of the same lineage within a species (mean correlation in human = 0.51, mouse = 0.58) or between profiles of the exact same cell type in different human donors (mean correlation = 0.58) or different inbred mouse strains (mean correlation = 0.79). Finally, the correlation between matching immune cell groups is comparable with the correlation between two matching solid tissues (8) (Fig. S2C).
Similarly, signatures of lineage-specific gene expression are conserved between the species. For each lineage in each species, we defined two signatures (induced and repressed) as the set of genes with significantly high or low expression, respectively (Bonferroni-corrected t test < 0.05), in this lineage compared with the other lineages. For all lineages, there was a significant overlap in corresponding gene signatures between species (Fig. 1C and Dataset S2). Overall, of 5,588 orthologous gene pairs that belong to a signature in at least one species, 22% (1,227 orthologs) also belong to the corresponding signature in the other species (i.e., induced or repressed in the same lineage). Because the signatures were defined strictly, the 22% estimation is most likely an underestimate of the level of conservation.
Majority of Genes Have Conserved Expression.
More broadly, there is a significant similarity in the expression of individual pairs of orthologous genes between the species. To test this similarity, we first defined, for each gene in each species, an immune expression profile based on its median expression in each of the seven groups. We then calculated a conservation of expression (COE) measure as the correlation between the immune expression profiles of each pair of one-to-one human and mouse orthologs in our set (Fig. 2A). COE values are significantly higher compared with a null distribution [P < 10−10, Kolmogorov–Smirnov (KS) test against a permutation of ortholog assignments] (Fig. 2B). A similar COE distribution is obtained when using a tissue atlas of human and mouse gene expression (8) (Fig. 2B).
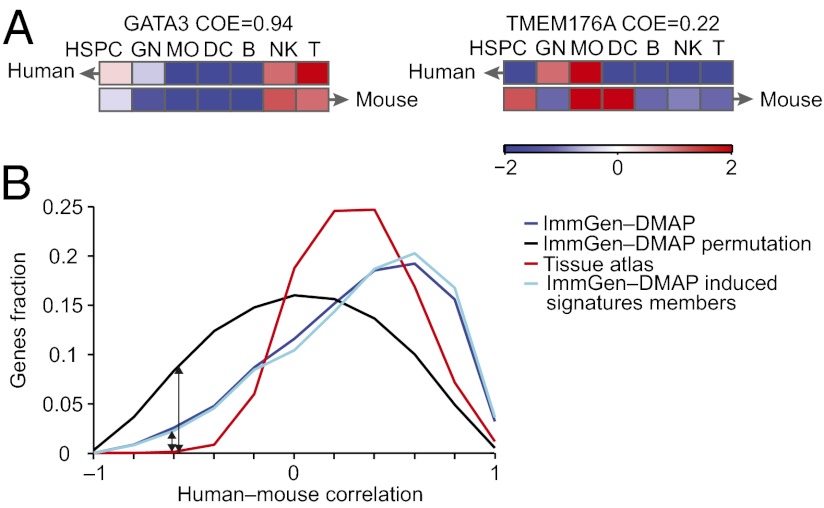
Fig. 2.
COE between human–mouse orthologs. (A) COE is calculated for a pair of human–mouse orthologs as the Pearson correlation coefficient between their median expression profiles (color scale on the bottom) in each lineage. Shown are examples for (Left) highly conserved orthologs and (Right) lowly conserved orthologs. (B) Significantly high COE values in human and mouse. Shown is a distribution of human and mouse COE values (blue), only lineage signature genes (light blue), a tissue atlas (8) (red), and a null distribution created by random pairings of one-to-one orthologs (black).
We next used the COE distributions to estimate the proportion of conserved genes. We assume that the observed COE distribution is a mixture of two distributions: one reflecting the conserved genes and the other reflecting the nonconserved genes. The COE distribution of nonconserved genes should be similar to the ImmGen-DMAP permutation distribution (Fig. 2B, black), whereas the COE distribution of conserved genes should be concentrated in the positive correlation region of the overall observed COE distribution. Thus, the observed total distribution of COE values for all genes, denoted Fall-genes, is obtained as a mixture of the COE distribution of nonconserved genes F0 (estimated by the permutation-based distribution) and the (unknown) COE distribution of conserved genes Fconserved: Fall-genes = α Fconserved + (1 − α) F0, where the mixture parameter α is the fraction of genes with conserved COE.
We next sought to estimate the mixture parameter α representing the fraction of genes with conserved COE. We first used a nonparametric method applicable for any distribution. Because Fall-genes(x) ≥ (1 − α) F0(x) for any x, we can estimate the maximal possible fraction of the nonconserved genes (1 − α) by calculating the ratio between the ImmGen-DMAP and ImmGen-DMAP permutation distributions: Fall-genes(x)/F0(x) for any value of x (Fig. 2B, black and dark blue curves, respectively). To avoid errors resulting from inaccurate estimates of Fall-genes(x) and F0(x) from data that may skew our estimator for 1 − α, we took the minimal ratio Fall-genes(x)/F0(x) for values of x on the negative region (x < 0) and excluded the left tail (x > −0.8), resulting in a minimal ratio of 1 − α = 0.3 (Fig. 2B, black arrows) and indicating that at least α = 1 − 0.3 = 70% of the genes have conserved COE scores. As an alternative strategy, we fit the COE data with a mixture of Gaussians model (Fig. S2D), where the nonconserved genes Gaussian mean and SD are taken from the permuted data. In the resulting model, the conserved genes Gaussian (black) contained α = 51% of the genes (Fig. S2D). Thus, both the parametric and nonparametric estimators for the fraction of conserved genes indicated that the majority of genes (51–70%) had conserved COE score.
Genes with high COE (Fig. S2E) share several transcriptional characteristics, including higher maximal expression (P = 2.7 × 10−9 and 2.9 × 10−24, human and mouse, respectively) (Dataset S1, Note 2 discusses alternative explanations of this phenomenon and an analysis of their likelihood), membership in lineage-specific induced signatures (KS test, P = 5.6 × 10−20) (Fig. 2B), and a presence of TATA box in their promoter (t test P = 0.04). The latter finding is in contrast to previous reports in yeast and other species of enhanced expression divergence of TATA-containing genes (9).
Known and Previously Undescribed Differences in Individual Genes.
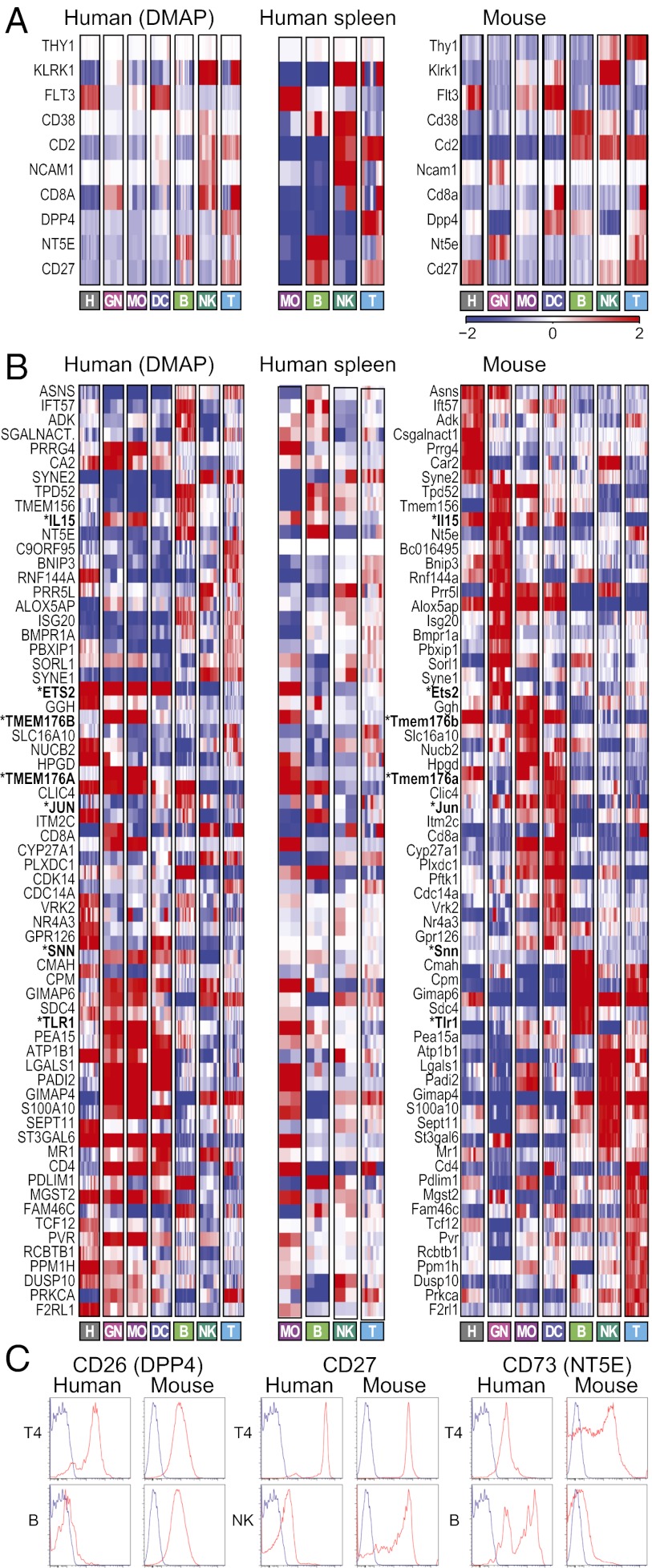
On the backdrop of the global conservation pattern, there are differences in the expression of certain genes, some consistent with previous reports and others identified in this study. First, we found several differences already noted previously (Fig. 3A, Fig. S3A, and Dataset S3), including in THY1, KLRK1, FLT3, CD38, CD2 (reviewed in ref. 10), NCAM1 (CD56) (11), and CD8A (12). Importantly, most of the human–mouse differences are also observed when using data from human monocytes and B, NK, and T cells isolated from spleen (Fig. 3A, Center and Fig. S3A), ruling out the possibility that the differences are merely caused by different tissue sources in the human and mouse compendia. The few previously reported differences that we did not observe in our analysis are typically in cells or expression levels that we do not expect to detect in our data (Dataset S3).
Fig. 3.
Differentially expressed genes between human and mouse. (A) Expression profiles (color scale on the bottom) in (Left) the human blood cell compendium, (Center) human splenocytes, and (Right) mouse of genes that were previously reported to be differentially expressed between human and mouse and are consistently different in our datasets or different in our datasets and validated below. (B) Selected genes with different expression patterns between human (Left) blood, (Center) spleen, and (Right) mouse immune cell types that were not previously reported. Shown are mean-centered expression values of genes sorted by mouse lineage with maximal expression level. Genes discussed in the text are marked with bold and asterisks. (C) Flow cytometry analysis of predicted human/mouse differences. Several cell surface markers were chosen (based on reagent availability) among differentially expressed genes and analyzed by staining of human peripheral blood mononuclear cells (PBMCs) or B6 splenocytes. Profiles in Upper and Lower depict cell populations for which expression was predicted to be shared or divergent in both species, respectively.
Next, we identified cell surface markers that display different expression patterns and systematically identified another 169 orthologous genes with expression that differs between human and mouse and were not previously reported (using stringent criteria of COE < 0.25 and at least a fourfold change between the lineage means in each species). These genes include IL15, ETS2, TMEM176A, TMEM176B, SNN, DPP4, NT5E, and CD27 (Fig. 3B, Fig. S3B, and Dataset S3). For example, IL15, a trophic cytokine for NK cells and memory T cells (13), is expressed in human in granulocytes, monocytes, and B cells, but in mouse, it is expressed in HSPC and some myeloid cells only. Some differences do not agree with previous findings (e.g., TLR1) (14) or may result from distinct immune activation states (e.g., JUN). Importantly, the differences are also detected comparing with cells isolated from human spleen (Fig. 3B, Center and Fig. S3B). We confirmed by flow cytometry a few of the differences in cell surface markers (Fig. 3C), staining human blood cells and mouse splenocytes in parallel. CD26 (DPP4) was, indeed, expressed in mouse but not human B cells; the opposite held for CD73 (NT5E), although human B cells include a CD27lo subset that may match with the mouse; CD27 was high in mouse but not human NK cells.
Contribution of Gene Duplication to Transcriptional Divergence.
The comparison above focused on one-to-one orthologs (a major portion of the genes in the human and mouse genomes), but gene duplication, loss, and appearance events were previously shown to play a major role in divergence between species in other systems (15). To explore the contribution of gene duplication to divergence here, we considered 66 cases where one ortholog in one species has many paralogs in the other. There are 16 conserved cases, in which expression of all paralogs is conserved with the expression of their ortholog (COE > 0.5) (Dataset S4). For example, the human gene FTL and its two mouse orthologs Ftl1 and Ftl2 are all expressed in GNs, MOs, and DCs (Fig. S4). Another 17 cases show potential evidence for neofunctionalization, with at least one of the paralogs having COE > 0.5 and another paralog having COE < 0.5 (Dataset S4). For example, human OAS1 and its mouse orthologs Oas1a and Oas1g are expressed in myeloid cells (GNs, MOs, and DCs), whereas two other mouse orthologs, Oas1b and Oas1c, are also expressed in T cells. In another example, human SCD is induced in HSPC along with one of its mouse orthologs, Scd2. The other mouse ortholog, Scd1, is induced in B cells, suggesting neofunctionalization (Fig. S4). Finally, in 33 groups, all paralogs have diverged in expression compared with their one ortholog (COE ≤ 0.5) (Dataset S4): in some cases, because of subfunctionalization and in others cases, because of neofunctionalization of all paralogs. One example of subfunctionalization is human myeloid cell nuclear differentiation antigen (MNDA; expressed in all myeloid cells) and its mouse orthologs, Ifi204 and Ifi205 (each induced in different myeloid cell subtypes) (Fig. S4).
Conservation and Divergence in T-Cell Activation.
The two compendia analyzed above primarily profiled resting cell states, and it was of interest to ask whether the balance of substantial conservation and limited divergence would shift during cell activation, especially because immune responses are under strong evolutionary selection. To test this hypothesis, we compared expression profiles generated under parallel conditions at early (1–4 h) and late (20–48 h) times of activation of purified CD4+ T cells by anti-CD3/CD28. We found that the human and mouse responses are substantially conserved by several measures to an extent comparable with or higher than the conservation observed for the resting cell populations in the main compendia. COE measures calculated based on the three time points are significantly higher than observed for random permutations (KS test, P < 10−300) (Fig. S5A) and the COE levels calculated based on any three lineages chosen from the seven lineages analyzed above (KS test, P < 2.7 × 10−71) (Fig. S5A). Using our nonparameteric approach, we estimate that at least 64% of genes have conserved COE in this response. Finally, there is a substantial overlap between the response patterns in the two species when partitioning the differentially expressed genes into distinct patterns in one species and examining their expression in the other species (Fig. 4A and Fig. S5B).
Fig. 4.
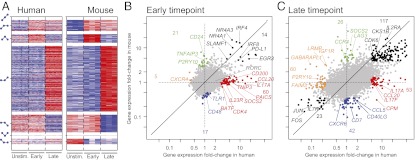
Transcriptional conservation in human- and mouse-activated CD4+ T cells. (A) Conservation of activation patterns. (Right) The significantly differentially expressed mouse genes were sorted by their expression patterns (schematically showing change over time on the left of the heat maps) and (Left) the expression of their orthologous genes in human-activated T cells. (B and C) Comparison of differential expression. Shown are scatter plots comparing the change in expression in (B) early and (C) later time points relative to prestimulation in human (x axis) and mouse (y axis) CD4+ T cells. The global off-diagonal skew reflects the slightly weaker activation of mouse CD4+ T cells under these conditions. The black points denote transcripts that are induced in both species or repressed in both species. The colored points denote transcripts that are affected in only one species (red, unchanged in mouse and induced in human; orange, unchanged in mouse and repressed in human; green, induced in mouse and unchanged in human; blue, repressed in mouse and unchanged in human) (Dataset S3).
A close examination of the response patterns showed that some segments of the responses were quantitatively or qualitatively different between the two species (Fig. 4 B and C and Dataset S3). Strikingly, these segments include much of the canonical Th17 differentiation signature (IL17A,F, IL23R, RORC, BATF, and CCL20) because of either an inherently higher responsiveness of the Th17 module in human or perhaps, a higher proportion of quickly reactive memory T cells in the human cells tested. Conversely, activation of CD24 or Lag3 seemed unique to mouse cells under these conditions. Some of the expression changes likely reflect functional differences: the NF-κB inhibitor TNIP3 is induced in human but not mouse, consistent with the fact that the mouse ortholog is nonfunctional because of lack of an essential homology domain (16). Most differences reflect lack of activation in one species, whereas in a few others, genes are either already fully expressed in the prestimulated state in mouse (e.g., CDK4 and PAICS) (Fig. 4B, red) or induced earlier in human than in mouse (e.g., SOCS2) (Fig. 4 B, red and C, green). Some respond in opposite ways in each species (e.g., LRMP, IGF1R, and GABARAPL1) (Fig. 4C, orange).
Comparison of Regulatory Mechanisms in Immune Cells.
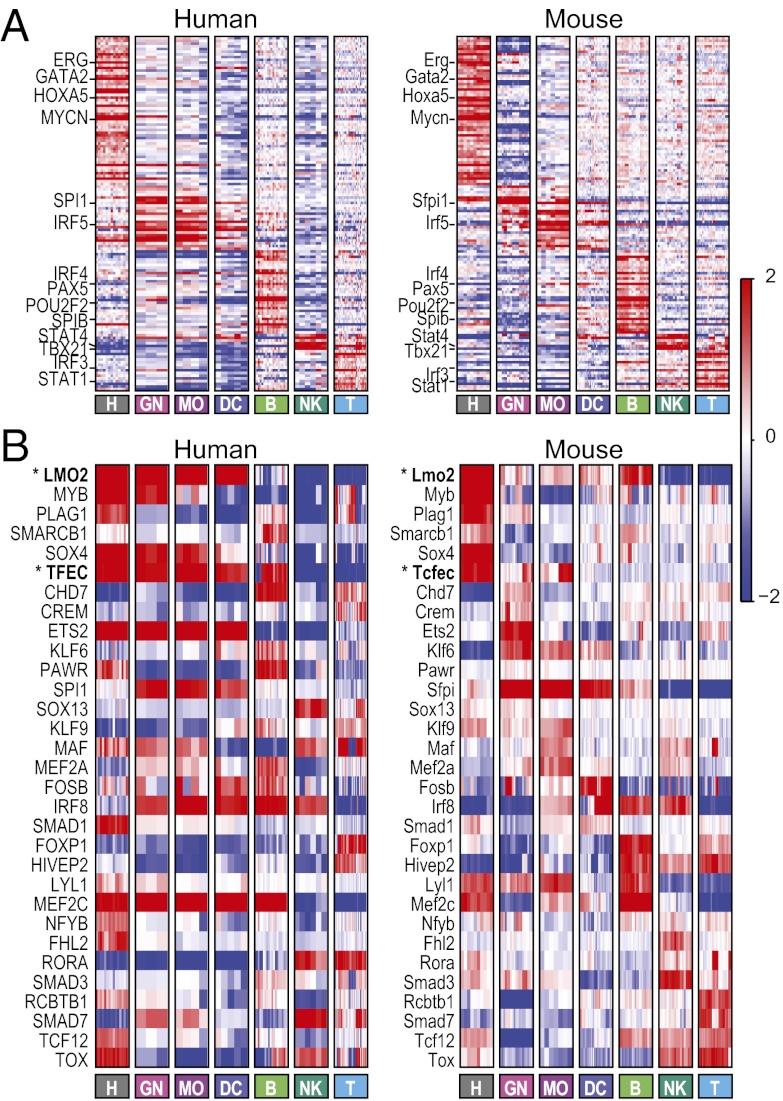
We next asked how conservation and/or changes in regulatory mechanisms relate to expression patterns between human and mouse. As expected, there is a significant overlap in the cis-regulatory elements enriched in the set of all lineage-specific genes in the two species (187 motifs, hypergeometric P < 10−10) (Dataset S2) and the motifs enriched in a pair of matching human and mouse signatures (Dataset S2). This overlap suggests that similar regulatory elements (and cognate transcription factors) are often used to control immune system differentiation and immune cells in both species. Furthermore, the expression patterns of genes encoding known transcriptional regulators, including known master regulators, were partially conserved between mouse and human, parallel to the genome-wide conservation of expression. The 353 regulators that are expressed in each species in at least one lineage have significantly higher COE values than the COE values of the rest of the one-to-one filtered orthologs set (KS test, P = 5.27 × 10−12); 131 of them (37%) are maximally expressed in the same lineage in both species (Fig. 5A) (e.g., GATA2 and HOXA5 in HSPCs and PAX5 and SPIB in B cells).
Fig. 5.
Regulation of human and mouse immune system differentiation is largely conserved. (A) Conserved lineage-specific expression of regulators. Shown are mean-centered expression values (red/blue color bar on the right) of regulators with maximal expression in the same lineage in (Left) human and (Right) mouse. Genes are sorted by mouse lineage with maximal expression. Selected gene names are highlighted on the left. (B) Divergent expression of regulators. Shown are mean-centered expression values of selected regulators with coherent expression patterns within each species that differ between the two species (Left, human; Right, mouse). Genes are sorted by mouse lineage with maximal expression. Genes mentioned in the text are marked with bold and asterisks.
However, some of the annotated regulators expressed in both species show divergent expression patterns (Fig. 5B), and other regulators are expressed only in human lineages and not others. Intriguing examples of divergent expression include transcription factor EC (TFEC) [human: HSPCs, myeloid cells, and B cells; mouse (Tcfec): HSPCs and some myeloid cells but not B cells] and LIM domain only 2 (LMO2) (human: HSPCs and myeloid cells; mouse: HSPCs and B cells). Furthermore, 16 regulators are expressed in human but not mouse common samples (3 regulators not expressed in any mouse sample) (Dataset S3). For example, MYBL1, not mentioned previously in immune context, is expressed in human NK and T cells but not expressed in mouse common lineages. LDB2, not mentioned previously in immune context, is expressed in human HSPCs and NK cells but not expressed in mouse common lineage. Similarly, there are 76 regulators expressed in mouse but not human common lineages (37 regulators not expressed in any human sample) (Dataset S3). For example, Nfix is expressed in mouse HSPCs and monocytes but not the human common lineages, and Prdm1 is expressed in mouse monocytes and NK cells but not the common human lineages. These differences must be interpreted with caution, because some may reflect technical probeset issues and not real biological differences.
Finally, we identified nine instances of putative divergent regulation based on cis-regulatory elements enriched in one species’ signature but not its counterpart signature in the other species (Dataset S3), and no corresponding change in expression pattern was found. Among those instances are ZFP161 (only enriched in the mouse NK signature), PAX3 (only enriched in human T-cell signature), and HIF1 (only enriched in mouse repressed HSPC signature).
Discussion
The laboratory mouse has been an important model from which many current paradigms of immunology are derived. Despite the many features conserved between the human and mouse systems, there are important known differences between them (10). Systematically determining these differences is critical for translating findings from mouse immunology to the human setting. Here, we have begun the development of such a reference map by comparing two extensive transcriptional compendia (6, 7).
We find an extensive conservation of the transcriptional program at several levels—global profiles, individual genes, and lineage-specific gene signatures. Gene and signature conservation is often reflected by concomitant conservation of the associated regulatory mechanisms with conservation of most regulators and cis-regulatory elements. Nevertheless, in many cases, lineage-specific genes are associated with different cis-regulatory elements. Although some of these discrepancies are likely related to the lower reliability of cis-regulatory predictions, other cases may reflect real divergence, as observed elsewhere (17–19).
Recent findings that the rate of gene expression evolution varies between mammalian organs, lineages, and chromosomes (20) may create the expectation that such rates will be particularly high in the immune system, because it is demonstrably under strong evolutionary pressure (as observed at the sequence level) (21–25). Our analysis suggests, however, that, for both steady expression states and during CD4 T-cell activation, overall gene expression conservation level in our compendia is comparable with the conservation in a solid tissue atlas (8), although different genes have highly conserved expression in each context. One intriguing possibility is that some of the transcriptional innovation in the immune system is mediated through divergence after gene duplication and not the change in regulation of one-to-one orthologs.
On the overall background of conservation, we identified cell surface markers, regulators, and 169 one-to-one orthologs with highly distinct expression patterns between human and mouse, more than tripling the number of previously reported differences (10). We tested and validated a few of these differences at the protein level. However, these results must be interpreted with some caution, because some distinctions may stem from probeset design, differences in the sampled cell types, or population and environmental variation in the human samples, but they should serve as a flag for future analyses.
Our analysis provides a valuable resource for future comparison of the human and mouse immune systems and studies of evolution of gene regulation. To facilitate this comparison and other uses, we provide all of our data, analyses, and results on a web portal (http://rstats.immgen.org/comparative/comparative_search.php). These results should help to identify when and where the mouse can, in fact, be considered predictive of human immunology.
Materials and Methods
Datasets.
Gene expression in mouse cell types from the ImmGen March 2011 release robust multichip average (RMA)-normalized data was used, including 802 arrays (Affymetrix MoGeneST1.0) of 244 cell types, essentially all in triplicate (7). Gene expression in human cell types was measured on 211 Affymetrix array U133A from 38 cell types, with four to seven replicates from different donors. Data were normalized and batch-corrected as described (6). Profiles from human-activated CD4+ T cells and splenocytes on the HuGene 1.0ST platform were obtained from the ImmVar project: 14 replicates from different human subjects per time points. Profiles from mouse-activated T cells from Wakamatsu et al. (26) were three replicates per time point from different pools of mice. Human and mouse normal tissue datasets (32 comparable tissues) were downloaded from http://biogps.org/downloads/, files U133AGNF1B.gcrma.avg_ann and GNF1M_plus_macrophage_small.bioGPS_ann.
We used Ensembl COMPARA rel 63 to map orthologs between mouse and human ENSEMBL gene IDs, yielding 10,248 one-to-one ortholog pairs measured in both compendia. Only 5,841 genes with expression levels above 120 (recommended ImmGen threshold) in more than three cell types of lineages common to both species were included in the filtered one-to-one orthologs set. When a given gene was represented by more than one probeset, the probeset with highest mean expression was used.
Mapping Human–Mouse Samples.
For each of the seven common lineages, comparable cell types were selected based on markers used for sorting and expert knowledge in the ImmGen Consortium. For 18 human selected cell types, we removed 23 outlier human samples (of 103 samples of those cell types) compared with all the samples from the same cell type. This removal resulted in 80 human samples of 18 cell types and 137 mouse samples of 44 cell types (Dataset S2).
Global Samples Correlation.
Expression values of all the genes in the filtered set of one-to-one orthologs were standardized per species, and Pearson correlation coefficient was calculated between the samples.
Differentially Expressed Gene Signatures.
Using the same procedure described in ref. 6, we defined for each of the seven lineages (HSPC, GN, MO, DC, NK, T cell, and B cell) in each species signatures of significantly induced and repressed genes in that lineage (14 signatures per species) (Dataset S2) based on a t test between a gene's expression level in a particular lineage’s samples and the samples from all other lineages [Bonferroni corrected P < 0.05 and the mean of the lineage-specific expression is higher (lower) than the global mean]. We test the significance of the overlap of two signatures using a hypergeometric test (cutoff false discovery rate < 10%).
COE.
The COE of a gene is a measure of agreement of its expression in comparable lineages between two species. For each gene in each species, we first computed its median expression in a lineage. We then calculated the COE of the gene as the Pearson correlation coefficient between these group-level summaries. The significance of the COE was estimated by a KS test compared with a computed null distribution of COE (computing COE after permuting ortholog assignments, 1,000 random permutations). Matlab function MixtureOfGaussiansGivenInit (O.Z.; http://www.broadinstitute.org/~orzuk/matlab/libs/stats/mog/MixtureOfGaussiansGivenInit.m) was used to estimate the mixture of Gaussians.
Activation of T cells.
Naive CD4+ T cells from 14 human donor blood or pooled spleen and lymph nodes of male C57BL/6foxp3-gfp mice were enriched by negative selection and stimulated in vitro with anti-CD3 and -CD28–conjugated beads; human total CD4+ T cells were then purified using positive magnetic selection, and mouse CD4+ GFP− T cells were flow sorted. Microarray profiling was performed on unstimulated cells and at early (4 h for human cells and pooled RNA at 1 and 4 h for mouse cells) and late (48 h for human cells and pooled 24 and 48 h for mouse cells) time points.
Multiple Comparison Control.
The Benjamini Hochberg False Discovery Rate (27) procedure was used to control the false discovery rate at 5% or 10% whenever stated.
Supplementary Material
Acknowledgments
We thank the members of the ImmGen Consortium for discussions and eBioscience, Affymetrix, and Expression Analysis for support of the ImmGen Project. We thank L. Lanier, R. Gazit, P. A. Monach, N. Novershtern, B. L. Ebert, and M. Kowalczyk for helpful discussions. We also thank L. Gaffney and S. Hart for help with graphics. Work was supported by National Institutes of Health Grants R24 AI072073, RC2-GM093080, U54-CA149145, and 149644.0103 (to V.J. and D.K.), National Science Foundation Grant DBI-0345474, the Howard Hughes Medical Institute, and the Merkin Foundation for Stem Cell Research at the Broad Institute (A.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222738110/-/DCSupplemental.
References
- 1.Payne KJ, Crooks GM. Immune-cell lineage commitment: Translation from mice to humans. Immunity. 2007;26(6):674–677. doi: 10.1016/j.immuni.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 2.Davis MM. A prescription for human immunology. Immunity. 2008;29(6):835–838. doi: 10.1016/j.immuni.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dermitzakis ET, Clark AG. Evolution of transcription factor binding sites in Mammalian gene regulatory regions: Conservation and turnover. Mol Biol Evol. 2002;19(7):1114–1121. doi: 10.1093/oxfordjournals.molbev.a004169. [DOI] [PubMed] [Google Scholar]
- 4.Odom DT, et al. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet. 2007;39(6):730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ravasi T, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140(5):744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Novershtern N, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144(2):296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heng TSP, Painter MW. Immunological Genome Project Consortium The Immunological Genome Project: Networks of gene expression in immune cells. Nat Immunol. 2008;9(10):1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 8.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101(16):6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tirosh I, Weinberger A, Carmi M, Barkai N. A genetic signature of interspecies variations in gene expression. Nat Genet. 2006;38(7):830–834. doi: 10.1038/ng1819. [DOI] [PubMed] [Google Scholar]
- 10.Mestas J, Hughes CCW. Of mice and not men: Differences between mouse and human immunology. J Immunol. 2004;172(5):2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 11.Lanier LL, Hemperly JJ. NK3 CD56 and CD57 cluster workshop report. In: Schlossman SF, et al., editors. Leucocyte Typing V: White Cell Differentiation Antigens. Vol 2. Oxford: Oxford Univ Press; 1995. pp. 1398–1400. [Google Scholar]
- 12.Kieffer LJ, et al. Human CD8 α expression in NK cells but not cytotoxic T cells of transgenic mice. Int Immunol. 1996;8(10):1617–1626. doi: 10.1093/intimm/8.10.1617. [DOI] [PubMed] [Google Scholar]
- 13.Ma A, Koka R, Burkett P. Diverse functions of IL-2, IL-15, and IL-7 in lymphoid homeostasis. Annu Rev Immunol. 2006;24(1):657–679. doi: 10.1146/annurev.immunol.24.021605.090727. [DOI] [PubMed] [Google Scholar]
- 14.Dasari P, Nicholson IC, Hodge G, Dandie GW, Zola H. Expression of toll-like receptors on B lymphocytes. Cell Immunol. 2005;236(1–2):140–145. doi: 10.1016/j.cellimm.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449(7158):54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- 16.Weaver BK, Bohn E, Judd BA, Gil MP, Schreiber RD. ABIN-3: A molecular basis for species divergence in interleukin-10-induced anti-inflammatory actions. Mol Cell Biol. 2007;27(13):4603–4616. doi: 10.1128/MCB.00223-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanay A, Regev A, Shamir R. Conservation and evolvability in regulatory networks: The evolution of ribosomal regulation in yeast. Proc Natl Acad Sci USA. 2005;102(20):7203–7208. doi: 10.1073/pnas.0502521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuch BB, Galgoczy DJ, Hernday AD, Li H, Johnson AD. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 2008;6(2):e38. doi: 10.1371/journal.pbio.0060038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hogues H, et al. Transcription factor substitution during the evolution of fungal ribosome regulation. Mol Cell. 2008;29(5):552–562. doi: 10.1016/j.molcel.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478(7369):343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 21.Schlenke TA, Begun DJ. Natural selection drives Drosophila immune system evolution. Genetics. 2003;164(4):1471–1480. doi: 10.1093/genetics/164.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McTaggart SJ, Obbard DJ, Conlon C, Little TJ. Immune genes undergo more adaptive evolution than non-immune system genes in Daphnia pulex. BMC Evol Biol. 2012;12(1):63. doi: 10.1186/1471-2148-12-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fumagalli M, et al. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet. 2011;7(11):e1002355. doi: 10.1371/journal.pgen.1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen R, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3(6):e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Downing T, Cormican P, O’Farrelly C, Bradley DG, Lloyd AT. Evidence of the adaptive evolution of immune genes in chicken. BMC Res Notes. 2009;2(1):254. doi: 10.1186/1756-0500-2-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wakamatsu E, Mathis D, Benoist C. Convergent and divergent effects of costimulatory molecules in conventional and regulatory CD4+ T cells. Proc Natl Acad Sci USA. 2012;110(3):1023–1028. doi: 10.1073/pnas.1220688110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B. 1995;57(1):289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.