Abstract
The importance of gene gain through duplication has long been appreciated. In contrast, the importance of gene loss has only recently attracted attention. Indeed, studies in organisms ranging from plants to worms and humans suggest that duplication of some genes might be better tolerated than that of others. Here we have undertaken a large-scale study to investigate the existence of duplication-resistant genes in the sequenced genomes of 20 flowering plants. We demonstrate that there is a large set of genes that is convergently restored to single-copy status following multiple genome-wide and smaller scale duplication events. We rule out the possibility that such a pattern could be explained by random gene loss only and therefore propose that there is selection pressure to preserve such genes as singletons. This is further substantiated by the observation that angiosperm single-copy genes do not comprise a random fraction of the genome, but instead are often involved in essential housekeeping functions that are highly conserved across all eukaryotes. Furthermore, single-copy genes are generally expressed more highly and in more tissues than non–single-copy genes, and they exhibit higher sequence conservation. Finally, we propose different hypotheses to explain their resistance against duplication.
Keywords: evolution, gene duplication, polyploidy
Following Ohno (1), gene duplication has been repeatedly reported to play an important role in evolution. For instance, mechanisms such as sub- or neofunctionalization underlie the evolution of many novel gene functions. Conversely, gene duplication can also be strongly deleterious (2, 3) and has been associated with diseases such as Parkinson (4) and cancer (5). The full complement of genes for which duplication is not tolerated, to the extreme that some genes occur as one single copy in any genome, is currently unknown. Such a set of genes might, however, reveal whether general evolutionary and functional characteristics could explain the deleterious effects of their duplication. Obtaining such genes from individual genomes is problematic due to the difficulty of discerning neutral from selective gene loss. Therefore, pioneering studies in identifying single-copy genes have used comparative genomics approaches across large evolutionary time scales. For instance, Paterson et al. (6) identified a set of genes present in Arabidopsis thaliana, Oryza sativa, Saccharomyces cerevisiae, and Tetraodon that were convergently restored to single-copy status following independent whole-genome duplication (WGD) events in each of these four organisms. Based on this observation, these authors argued that there was strong selection pressure to preserve these genes as singletons and they coined them as “duplication-resistant” genes. Single-copy genes were also observed in a comparative study of four angiosperm species (7), in a comparative analysis spanning seven eukaryotic genomes (8), and in a study covering 40 vertebrates, 23 arthropods, and 32 fungal genomes (9).
With the exception of refs. 9 and 8, most studies cited above were based on only a few genomes, thus restricting the statistical power of the observations made. Therefore, it is not clear whether the presence of single-copy genes is the outcome of selection or whether stochastic processes of gene loss dynamics can explain the presence of shared sets of single-copy genes. To assess selection against retention of certain gene duplicates and to study the relationship between gene duplicability and gene function, it is necessary to study a large number of genomes. The angiosperm lineage is especially well-suited to study duplication resistance because all angiosperm genes have repeatedly undergone duplication through multiple shared and independent WGD events (10, 11). Moreover, these WGDs have occurred over different time scales, with ancient events dating back to the origin of the seed and flowering plants (12), the monocots (13), and the (core) eudicots (14), whereas other WGDs have occurred more recently in many independent plant lineages (10, 15). In addition to WGDs, single gene duplications in angiosperms are also prevalent and have played a considerable role in shaping plant genomes (16, 17). For instance, the estimated rate of single gene duplications lies between 300 and 500 genes per 10 million years (18, 19). As the angiosperm lineage spans ∼150–200 million years of evolution, we expect there to be a high recurrence of gene duplication by both large- and small-scale duplication (SSD) events, which enables investigating the existence of selection for gene loss (in case of WGD) or selection against fixation of duplicates (in case of SSD).
Here, we take advantage of the increasing number of available sequenced angiosperm genomes to identify single-copy genes at high resolution. In particular, we identified single-copy genes in 20 sequenced angiosperm genomes, comprising six monocots and 14 eudicots and spanning multiple shared and independent WGD events (Fig. 1). We describe the functional and evolutionary characteristics of these genes and provide hypotheses to explain their single-copy status.
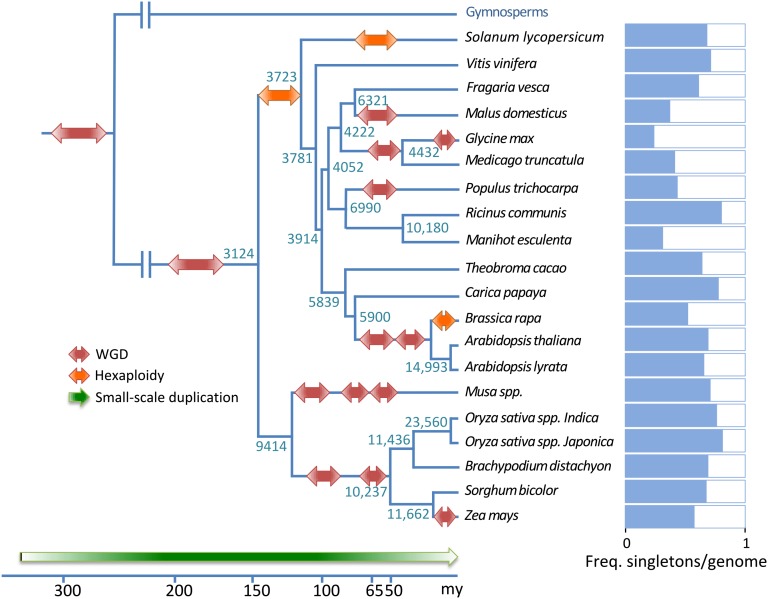
Fig. 1.
Phylogenetic tree showing the relationships between the 20 angiosperm species used in this study. Genome duplications are denoted in red (ancient tetraploidy) and orange (ancient hexaploidy). Numbers at nodes refer to (strictly and mostly) single-copy genes. Bars at the right represent the fraction of singletons in each genome. Placing of duplication events was based on refs. 12, 13, 22, 24, and 59.
Results
Identification of Single-Copy Genes.
To identify genes that are single copy in a large number of angiosperm genomes, we used the orthologous groups (OGs), predicted by the OrthoMCL method (20), from the PLAZA 2.5 database (21). These OGs span 17 angiosperm genomes and were augmented with the recently published genomes of Solanum lycopersicum (tomato), Brassica rapa (Chinese cabbage), and Musa acuminata (banana), to independently verify single-copy status (SI Appendix, section SI1 and Fig. S1). Of these, the former two both underwent a hexapolyploidization event (22, 23), whereas banana probably underwent three rounds of WGD (24). None of these events have been shared with any of the species in the PLAZA database. In total, for all further analyses, we considered 9,513 OGs that represent gene evolution dating back to the common ancestor of the 17 angiosperm genomes in PLAZA. Single-copy genes sensu stricto are defined as genes that are conserved in all angiosperm genomes and with a one-to-one orthology relationship in these genomes; that is, they have remained single-copy since the angiosperm common ancestor or have consistently been restored to single-copy status following duplication. To accommodate potential problems with genome annotation and the presence of recent duplicates, alleles, or pseudogenes, we slightly relaxed these criteria and defined single-copy OGs as the OGs that are present in all angiosperm genomes, yet tolerating missing copies in up to two species and allowing for duplicates in up to three species. OGs that are truly single copy for all species are further referred to as “strictly single-copy” OGs, whereas OGs with duplicates for one up to three species are referred to as “mostly single-copy” OGs. In total, 392 OGs are strictly single copy and 3,488 OGs are mostly single copy.
Although OrthoMCL performs reasonably well to predict OGs in different benchmark studies (25, 26), it also has some drawbacks that can lead to misinterpretation of the data (27). In addition, the outcome might be susceptible to the choice of the inflation parameter, which controls the size of the OGs. Therefore, we developed a phylogenetic approach (SI Appendix, section SI2 and Fig. S3) to validate the single-copy status of the obtained OGs. In particular, OGs were expanded with mutual best BLAST hits followed by a construction of the corresponding gene tree to assess whether the additional genes might represent missed inparalogs. Using this approach, the single-copy status of 2,840 (2,663 mostly single-copy + 177 strictly single-copy) of the 3,880 initially observed (strictly and mostly) single-copy OGs could be confirmed.
The majority of the “mostly single-copy” OGs shows a species bias for the presence of extra copies: they have extra (inparalog) duplicates for soybean (Glycine max), poplar (Populus trichocarpa), and apple (Malus domestica) (SI Appendix, Fig. S6). Indeed, of the 2,663 mostly single-copy OGs, there are only 621 OGs that have duplicates for species other than soybean, poplar, and apple. The presence of extra copies in soybean is not surprising given that this species underwent a relatively recent polyploidization event ∼13 million years ago (mya), and the genome is still highly duplicated (Fig. 1) (28). The high incidence of duplicates in apple and poplar might be explained by slower rates of evolution in these species, with many duplicates still on the route to pseudogenization following independent genome duplications that occurred ∼65 mya (Fig. 1) (29–31). Alternatively, the high frequency of duplicates for the apple genome could be partially explained by misclassification of alleles as duplicates (30). These observations further motivate the inclusion of OGs with duplicates for a limited number of species (mostly single-copy OGs) into our set of “single-copy groups.” It should be noted that from now on, when using the term “single-copy genes,” we actually refer to the joint set of “strictly” and “mostly” single-copy OGs.
Number of Identified Single-Copy OGs Exceeds Random Expectation.
Considering the large number of duplication events that have occurred within the angiosperm lineage, it seems surprising that there exists a large set of genes that have consistently been restored to single-copy status in all species. A potential explanation for this observation is that duplication is deleterious for this particular set of genes and hence is selected against. Alternatively, single-copy OGs might arise through the effects of random duplicate loss in different lineages. To distinguish between these possibilities, we performed simulations of gene evolution to estimate the probability that the observed number of single-copy OGs might have arisen by chance (SI Appendix, section SI3). In particular, we used parsimony-based gene-tree species-tree reconciliation (32) to obtain estimates of net duplications, losses, and unchanged copy numbers (including cases in which genes are first duplicated and then lost on the same branch) that occur along each branch of the angiosperm species tree. Next, copy-number changes of a single ancestral angiosperm gene along this tree were simulated (i.e., genes were duplicated/lost/kept unchanged according to the numbers predicted for each branch) under the assumption that the duplication/loss dynamics of the gene family along the separate branches of the tree are independent, reflecting a scenario in which OG sizes are randomly assorted. The outcome of this simulation strongly supports a pattern of convergent gene loss, with the number of observed single-copy OGs significantly exceeding the expectation under a scenario of independent gene duplication/loss across lineages (P < 0.00001, based on 100,000 simulations) (SI Appendix, section SI3 and Fig. S4).
Characterization of Single-Copy OGs.
Because the number of single-copy OGs greatly exceeds random expectation (SI Appendix, Fig. S4), we hypothesize that there is a selective force that works to restore duplicated genes to single-copy status in these OGs. To investigate the factors that may underlie the duplication resilience of single-copy OGs, we investigated their functional and evolutionary characteristics.
Functional bias.
First, we assessed enrichment of single-copy OGs for certain Gene Ontology (GO) categories (33). Because A. thaliana is the best annotated of all angiosperm genomes, we restricted this and further analyses to the A. thaliana genes within the single-copy OGs (see Dataset S1). Of the 2,986 A. thaliana genes in the single-copy OGs, 2,313 had a GO annotation.
Functional enrichment analysis reveals that single-copy genes are biased toward conserved processes such as DNA repair, recombination, and DNA damage response and also to organelle-related functions such as photosynthesis, whereas genes involved in regulation of transcription and phosphorylation are underrepresented (Table 1; see SI Appendix for a full list of significant GO terms). Because single-copy genes are by definition conserved across the 20 profiled species (see above), we assessed whether functional overrepresentation is specifically linked to single-copy status rather than phylogenetic conservation. Therefore, we also calculated the functional enrichment of single-copy genes relative to the set of A. thaliana genes that are conserved across angiosperms (i.e., all genes in the 9,513 OGs considered, both single copy as non–single copy), rather than all A. thaliana genes. This analysis yielded similar results, suggesting that overrepresentation is not only due to conservation but is specifically related to single-copy status (SI Appendix, Table S1).
Table 1.
Significantly over- and underrepresented GO categories among A. thaliana single-copy OG member genes
| GO term | Ontology | Adjusted P value (FDR < 0.05) | No. of A. thaliana single-copy genes | Total no. of A. thaliana genes | |
| Overrepresented | DNA repair | BP | 1.8137E-21 | 64 | 154 |
| Response to DNA damage stimulus | BP | 1.8137E-21 | 66 | 163 | |
| DNA recombination | BP | 1.3949E-12 | 28 | 52 | |
| DNA metabolic process | BP | 6.3775E-33 | 114 | 311 | |
| DNA replication | BP | 4.9269E-8 | 32 | 94 | |
| Photosynthesis | BP | 1.2060E-10 | 40 | 113 | |
| Plastid organization | BP | 1.5284E-11 | 39 | 102 | |
| Cofactor metabolic process | BP | 4.0235E-10 | 59 | 219 | |
| Embryonic development | BP | 1.0020E-6 | 83 | 429 | |
| Meiosis I | BP | 3.0321E-6 | 15 | 30 | |
| Chloroplast | CC | <1.0000E-99 | 538 | 2,070 | |
| Underrepresented | Regulation of transcription | BP | 2.2454E-15 | 63 | 1,468 |
| Regulation of gene expression | BP | 1.8458E-13 | 82 | 1,604 | |
| Phosphorylation | BP | 1.9760E-5 | 51 | 910 |
P values shown were obtained by hypergeometric test and FDR adjusted. FDR, false discovery rate; BP, biological process; CC, cellular component.
We also assessed whether genes that are strictly single copy show a different functional enrichment pattern than the mostly single-copy genes. Strictly single-copy genes are also enriched for functions related to DNA damage and repair, but not photosynthesis (SI Appendix, Table S2). This might indicate that duplicates of genes involved in the former processes are more resistant against SSD and/or removed faster after large-scale duplication events. However, in each of the significant categories, the absolute numbers increase when taking into account the mostly single-copy families, suggesting that even for these categories duplication resistance is not absolute and/or duplicate loss is not instantaneous.
We observed that single-copy genes, except for the ones involved in chloroplast functions, are in general well-conserved in metazoans (i.e., they are not specific to the plant lineage) (SI Appendix, section SI4 and Table S3). This observation, combined with the GO enrichment results, suggests that many single-copy genes are involved in core cellular processes. To further test this hypothesis, we assessed the dispensability of single-copy genes in a dataset of 5,360 A. thaliana mutants (34), which contained 771 single-copy genes. A significant portion of our single-copy genes (316 genes) overlapped with the 1,742 mutants showing a visible phenotype (P value = 5.35e-08, hypergeometric test).
Expression bias in single-copy genes.
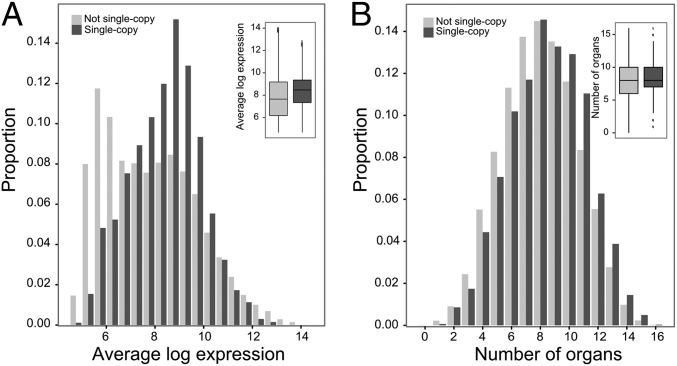
Previous research has established a clear relationship between gene duplicability and gene expression levels (35, 36), but was mainly focused on the relationship between duplicate retention and gene expression, and it remains unknown whether duplication-resistant genes show similar biases. Therefore, we analyzed expression levels of single-copy genes in an A. thaliana expression compendium, which contains expression measurements for 20,777 genes in 425 different conditions, representing different plant organs and developmental time points (37). Expression measurements are available for 2,654 of the 2,986 A. thaliana single-copy genes. Single-copy genes show a clear bias toward higher absolute expression levels, calculated as the gene expression level averaged across all samples, compared with A. thaliana genes that are not single-copy (P < 2.2e-16, one-sided Mann–Whitney U test) (Fig. 2). Since photosynthesis genes are generally highly expressed (38), we removed genes belonging to this functional category and reassessed absolute gene expression levels for single-copy and non–single-copy genes to make sure that the observations are not due to the overrepresentation of photosynthesis genes in our single-copy OGs (SI Appendix, Fig. S7) (P < 2.2e-16, one-sided Mann–Whitney U test). A possible explanation for the on average higher expression levels of single-copy genes could be that potential high needs for certain proteins cannot be provided through duplication because duplication would be disadvantageous (Discussion).
Fig. 2.
Expression analysis of single-copy genes. (A) Absolute expression levels of A. thaliana genes in single-copy and non–single-copy OGs. The figure shows the proportion of genes (y axis) that has a certain absolute expression level (x axis), calculated as the geometric mean of all expression measurements of a certain gene. (B) Expression breadth of A. thaliana genes in single-copy and non–single-copy OGs calculated as the number of organs in which a gene is expressed.
We also assessed the expression breadth of the genes over 16 organs (SI Appendix, section SI4). We observed a higher expression breadth for single-copy genes compared with non–single-copy genes (P = 3.681e-15, one-sided Mann–Whitney U test), which is consistent with the observation that many single-copy genes seem to be involved in housekeeping functions. A separate analysis for strictly single-copy genes gave borderline significant results for gene expression breadth and no significant distinction for the gene expression level comparison (SI Appendix, Table S4). The limited sample size of the set of strictly single-copy genes (177 genes), however, complicates the identification of significant patterns.
Sequence conservation.
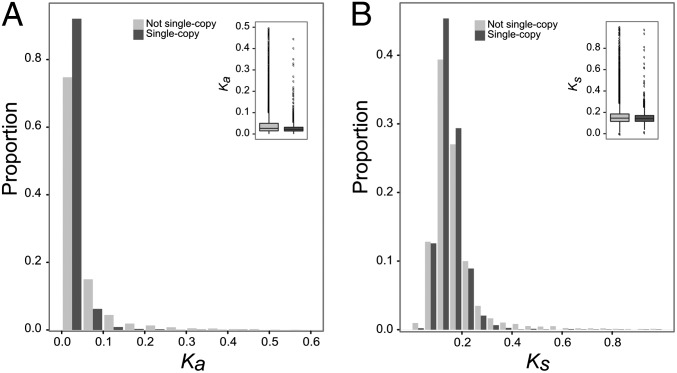
Similar to gene expression level, sequence conservation has previously been associated with gene duplicability (9, 39). We estimated sequence conservation of the single-copy genes by calculating the number of synonymous substitutions per synonymous site (Ks) and the number of nonsynonymous substitutions per nonsynonymous site (Ka) between strictly one-to-one orthologs in A. thaliana and Arabidopsis lyrata. Upon comparison of the Ka and Ks distributions of single-copy genes versus non–single-copy genes (also here strictly one-to-one orthologs were used), we observed that both Ks (P < 2.2e-16, one-sided Mann–Whitney U test) and Ka (P < 2.2e-16, one-sided Mann–Whitney U test) values were significantly lower for the single-copy genes (Fig. 3).
Fig. 3.
Sequence conservation of the single-copy genes. (A) The number of nonsynonymous substitutions per nonsynonymous sites (Ka) plotted against the proportion of orthologous pairs with a certain Ka value. (B) The number of synonymous substitutions per synonymous sites (Ks) plotted against the proportion of orthologous pairs with a certain Ks value.
Hence, while synonymous substitutions seem to be mainly neutral in non–single-copy genes, they are, to some extent, selected against in single-copy genes. In accordance with this, we find that codon use, as assessed by the Codon Adaptation Index (40), is more biased in single-copy genes than in non–single-copy genes (P = 1.33e-14, one-sided Mann–Whitney U test) (SI Appendix, Fig. S8). Because single-copy genes are also often highly expressed, the skewed codon bias observed for these genes could be explained by the “translational accuracy” hypothesis (41, 42), which states that codons are adapted to reduce mistranslation costs that are potentially higher for highly expressed genes. We also observe increased purifying selection for the single-copy genes versus non–single-copy genes at the level of nonsynonymous substitutions; that is, there seems to be a higher selection pressure for single-copy genes to both maintain their function (lower Ka) and high expression levels (lower Ks). When analyzed separately, strictly single-copy genes also have significantly lower Ka- and Ks- values than non–single-copy genes (SI Appendix, Table S4).
Discussion
By comparing 20 sequenced angiosperm genomes, we show that, despite the large number of SSD and large-scale duplication events that have taken place, there exists a set of genes that has been repeatedly restored to single-copy status. Because the observed number of single-copy families greatly exceeds what can be expected from random gene loss effects, selection likely promotes convergent evolution of these genes to single-copy status across angiosperms.
In agreement with previous work on single-copy genes (8, 9, 43–45), we found this set of genes not to be a random fraction of the genome but to encode housekeeping and other essential functions, as suggested by their functional enrichment, conservation throughout the eukaryotic tree, and expression breadth. Striking is the overrepresentation of genes targeted to the chloroplast and genes functioning in DNA repair and replication. As for the former, Duarte et al. (7) observed a similar overrepresentation for a set of single-copy genes identified in four angiosperm genomes. The overrepresentation of DNA repair and replication genes is in line with previous findings that cancer “caretakers”—that is, genes that are involved in maintaining genome stability—are often ancient singletons in the human genome (44). This remarkable congruence of different studies (7–9, 43, 44), based on different species and different methods, on the involvement of single-copy genes in ancient, well-conserved processes suggests that this is a robust pattern. Hence, it can be argued that single-copy genes form a well-conserved core that is sensitive to either mutation or duplication. In contrast, we observed that other highly conserved proteins, such as ribosomal proteins, are not biased toward singleton status. In addition, high duplicability of conserved genes was observed in vertebrates (9), yeast (39), and Paramecium (35). Hence, conservation alone seems not to be a sufficient explanation for single-copy status. Below we present hypotheses that might explain why genes would be preferentially retained as singletons.
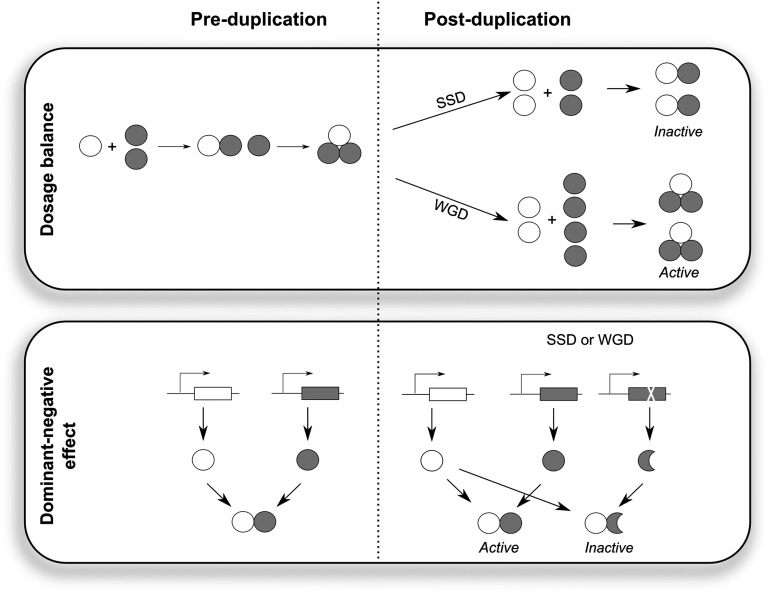
First, single-copy genes might be dosage balance sensitive; that is, it is important that they are maintained in the correct relative dose because an increase in copy number might unbalance their interactions with other proteins within the cell (Fig. 4, Upper). We indeed observe that many of the single-copy genes encode subunits of protein complexes, such as the photosynthesis light harvesting complexes, the chloroplast NADH dehydrogenase complex, the Origin Recognition Complex, and the switch/sucrose nonfermentable (SWI/SNF) chromatin remodeling complex. However, previously it was suggested that duplication of dosage-sensitive genes is tolerated in case of WGDs, as such events duplicate entire complexes and pathways and hence conserve the stoichiometric relations among the individual gene components (3, 16, 46–52). The genes identified in our analysis are, in contrast, duplication-resistant under both SSDs and WGD scenarios. A variant of the dosage balance hypothesis might, however, still apply to single-copy genes that encode proteins involved in organellar processes such as photosynthesis, one of the clearly overrepresented functional classes in our analysis, as was also suggested by ref. 47. Many of the photosynthesis-related processes and protein complexes are mosaics of nuclear- and chloroplast-encoded proteins, and communication between the nuclear and chloroplast genome is tightly regulated to maintain balance between proteins encoded by the separate genomes (53, 54). WGDs only duplicate the nuclear genome and not the organellar genomes, which might upset the stoichiometric balance between the nuclear- and organelle-encoded subunits or might disrupt the intricate signaling that exists between nucleus and chloroplast. On the other hand, it has been observed that the number of chloroplasts increases with ploidy level (55, 56). Hence, it is possible that the ratio of nuclear-encoded genes versus chloroplast-encoded genes remains relatively constant following WGD, in which case WGD would not result in an imbalance. However, the exact effects of WGD on the nuclear/chloroplast gene dosage ratio remain to be ascertained.
Fig. 4.
Two hypotheses to explain single-copy status. (Upper) The dosage balance hypothesis, which predicts that stoichiometric imbalance among protein complex subunits is harmful. WGD ensures that relative ratios among subunits are maintained, whereas this is not necessarily the case for SSD (e.g., when the white subunit is duplicated). (Lower) The dominant-negative mutation hypothesis, which can explain single-copy status under both scenarios of SSD and WGD. In this hypothesis, gene duplication can result in an extra mutational target, in which mutations can occur that interfere with wild-type function. This is, for instance, possible if a mutation occurs in a protein complex subunit, in such a sense that protein interaction capabilities are maintained and hence the mutant protein competes with the wild-type protein for forming complexes.
Alternatively, duplication of some genes might not be tolerated because mutations in one of the copies produce dominant-negative phenotypes (55–57); that is, the expression of the mutant variant disrupts wild-type function when occurring simultaneously in one cell (Fig. 4, Lower). In this case, because duplications increase the mutational target, there might be selection for removal of the extra gene copy such that it cannot acquire dominant-negative mutations. Several of our observations are indeed in support of this hypothesis. First, we observe a high sequence conservation for single-copy genes, which suggests that most mutations in these proteins are detrimental. This is further substantiated by the observation that single-copy genes are involved in processes essential for plant viability, such as photosynthesis and DNA damage response, and that mutants of single-copy genes produce strong phenotypic effects. Secondly, different single-copy genes encode subunits of protein complexes (see above), which are especially prone to dominant-negative mutations because it might result in wild-type proteins being trapped in inactive protein complexes due to binding with mutant subunits or the production of toxic aggregates (41, 55, 56). Hence, although the gene balance and the dominant-negative effects hypotheses might produce similar effects in certain instances (e.g., a reduction in active protein complex levels), the underlying mechanisms and the predictions they make are different. Noteworthy, according to Herskowitz (56), large protein complexes, such as the DNA replication machinery, are especially sensitive to dominant-negative effects. In contrast to the mainstream version of the aforementioned dosage balance hypothesis, the dominant-negative hypothesis may help explain why dosage-sensitive single-copy genes are also preferentially lost after WGD. In contrast, others suggested that genes sensitive to dominant-negative effects are generally retained in duplicate after WGD (35, 57), based on the fact that pseudogenization would involve the accumulation of mutations that could lead to such effects. We propose that the expression of one of the duplicates may also rapidly be silenced epigenetically or through permissible mutations in the promoter region, so that it can pseudogenize without deleterious effects.
In the absence of large-scale experimental studies that, for instance, investigate the effect of gene overexpression (dosage balance hypothesis) or heterozygosity (dominant-negative effect) of single-copy genes on organism phenotype, it is hard to establish with certainty the underlying mechanisms that might explain the deleterious effects of duplication in a defined set of genes. Nevertheless, with this study we hope to have shown that there exists a substantial number of genes for which duplication seems to be deleterious. Both the specific characteristics of these single-copy genes and the hypotheses put forward to explain their single-copy status will hopefully encourage further research to elucidate the evolutionary forces acting on these genes.
Materials and Methods
Full Methods.
Details regarding the simulation study and functional and evolutionary characterization of single-copy genes can be found in SI Appendix.
Obtaining Single-Copy Genes.
We extracted all OGs constructed for 17 angiosperm genomes and seven outgroup species from the PLAZA 2.5 database (21). These OGs were constructed using OrthoMCL and augmented with all TribeMCL families in the PLAZA database that did not show overlap with the OrthoMCL families. Single-copy OGs were defined as those with copies present in at least 15 out of the 17 genomes and with duplicates in maximum three species. Afterward, as an independent verification of single-copy status, the OGs were expanded with genes from three additional angiosperm genomes (SI Appendix, section SI1).
To validate the single-copy status of each of the obtained single-copy OGs, we expanded each of these OGs with the best BLAST hits that were not included in the OG. To this end, for each species in the OG, the two top-scoring (E-value) reciprocal best BLAST hits not in the OG, obtained from an all-against-all BLASTP analysis, were selected. For each OG we analyzed the topology of the corresponding rooted gene tree (created as described in SI Appendix, section SI2). For this we only considered trees with a multiple sequence alignment (MSA) length exceeding 100 amino acids, as for other trees the bootstrap support of the branches was often too low. Ideally, in these trees the genes in the original PLAZA OG all cluster together, with the expanded BLAST hits being outparalogous to the original OG genes (SI Appendix, section SI2 and Fig. S3). However, this is often not the case, and hence tree topologies were analyzed in more detail. For each gene tree we identified the subtree that contained a high coverage for OG genes and a low coverage for BLAST hit genes. To identify this subtree we introduced the “purity measure,” which at each node compares the number of OG genes to the total number of genes (including both OG genes and BLAST hit genes) in the subtree:
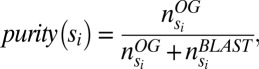 |
with 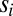 being the subtree at node
being the subtree at node 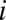 and
and 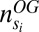 the number of OG genes in the subtree and
the number of OG genes in the subtree and 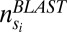 the number of BLAST hits in the subtree.
the number of BLAST hits in the subtree.
In a breadth-first traversal of the tree topology, the subtree with the maximal purity-score, further referred to as the “core set,” was retrieved. Only subtrees that contained at least 85% of the OG genes were considered. By setting this threshold lower than 100%, potential mispredictions of orthology relationships by OrthoMCL could be eliminated. Next, phylogenetic profiles, detailing the number of gene copies per species, were calculated for each core set to assess whether the core set conformed to the initial criteria for (mostly) single-copy families: duplicates for maximum three species and missing copies for maximum two species. For further downstream analysis, the genes within each OG were replaced by the genes in the core set obtained from the associated phylogenetic tree. The pipeline for this analysis was programmed in Perl, using the Bio::Phylo package (58), and is available upon request.
Supplementary Material
Acknowledgments
The authors thank Sebastian Proost and Michiel Van Bel for their assistance with the PLAZA data and Stefanie De Bodt for providing the gene expression data. The authors also thank the reviewers for their useful comments. This work was supported by Research Foundation–Flanders (FWO) Project G.0059.08. K.L.A. was supported in part by a Killam Research Fellowship from the University of British Columbia. K.V. acknowledges the Multidisciplinary Research Partnership “Bioinformatics: From Nucleotides to Networks” Project (01MR0310W) of Ghent University. S.M. is a fellow of the FWO.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300127110/-/DCSupplemental.
References
- 1.Ohno S. Evolution by Gene Duplication. New York: Springer; 1970. p. 160. [Google Scholar]
- 2.Conrad B, Antonarakis SE. Gene duplication: A drive for phenotypic diversity and cause of human disease. Annu Rev Genomics Hum Genet. 2007;8:17–35. doi: 10.1146/annurev.genom.8.021307.110233. [DOI] [PubMed] [Google Scholar]
- 3.Makino T, McLysaght A. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Natl Acad Sci USA. 2010;107(20):9270–9274. doi: 10.1073/pnas.0914697107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singleton AB, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302(5646):841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 5.Seeger RC, et al. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med. 1985;313(18):1111–1116. doi: 10.1056/NEJM198510313131802. [DOI] [PubMed] [Google Scholar]
- 6.Paterson AH, et al. Many gene and domain families have convergent fates following independent whole-genome duplication events in Arabidopsis, Oryza, Saccharomyces and Tetraodon. Trends Genet. 2006;22(11):597–602. doi: 10.1016/j.tig.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Duarte JM, et al. Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol Biol. 2010;10:61. doi: 10.1186/1471-2148-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koonin EV, et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5(2):R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waterhouse RM, Zdobnov EM, Kriventseva EV. Correlating traits of gene retention, sequence divergence, duplicability and essentiality in vertebrates, arthropods, and fungi. Genome Biol Evol. 2011;3:75–86. doi: 10.1093/gbe/evq083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van de Peer Y, Fawcett JA, Proost S, Sterck L, Vandepoele K. The flowering world: A tale of duplications. Trends Plant Sci. 2009;14(12):680–688. doi: 10.1016/j.tplants.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16(7):1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiao Y, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473(7345):97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 13.Tang H, Bowers JE, Wang X, Paterson AH. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc Natl Acad Sci USA. 2010;107(1):472–477. doi: 10.1073/pnas.0908007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vekemans D, et al. Gamma paleohexaploidy in the stem lineage of core eudicots: Significance for MADS-box gene and species diversification. Mol Biol Evol. 2012;29(12):3793–3806. doi: 10.1093/molbev/mss183. [DOI] [PubMed] [Google Scholar]
- 15.Proost S, Pattyn P, Gerats T, Van de Peer Y. Journey through the past: 150 million years of plant genome evolution. Plant J. 2011;66(1):58–65. doi: 10.1111/j.1365-313X.2011.04521.x. [DOI] [PubMed] [Google Scholar]
- 16.Freeling M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol. 2009;60:433–453. doi: 10.1146/annurev.arplant.043008.092122. [DOI] [PubMed] [Google Scholar]
- 17.Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytol. 2009;183(3):557–564. doi: 10.1111/j.1469-8137.2009.02923.x. [DOI] [PubMed] [Google Scholar]
- 18.Moore RC, Purugganan MD. The early stages of duplicate gene evolution. Proc Natl Acad Sci USA. 2003;100(26):15682–15687. doi: 10.1073/pnas.2535513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanneste K, Van de Peer Y, Maere S. Inference of genome duplications from age distributions revisited. Mol Biol Evol. 2013;30(1):177–190. doi: 10.1093/molbev/mss214. [DOI] [PubMed] [Google Scholar]
- 20.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Bel M, et al. Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. 2012;158(2):590–600. doi: 10.1104/pp.111.189514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomato Genome Consortium The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485(7400):635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, et al. Brassica rapa Genome Sequencing Project Consortium The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2011;43(10):1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 24.D’Hont A, et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature. 2012;488(7410):213–217. doi: 10.1038/nature11241. [DOI] [PubMed] [Google Scholar]
- 25.Chen F, Mackey AJ, Vermunt JK, Roos DS. Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS ONE. 2007;2(4):e383. doi: 10.1371/journal.pone.0000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altenhoff AM, Dessimoz C. Phylogenetic and functional assessment of orthologs inference projects and methods. PLOS Comput Biol. 2009;5(1):e1000262. doi: 10.1371/journal.pcbi.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trachana K, et al. Orthology prediction methods: A quality assessment using curated protein families. Bioessays. 2011;33(10):769–780. doi: 10.1002/bies.201100062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmutz J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463(7278):178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 29.Tuskan GA, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313(5793):1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 30.Velasco R, et al. The genome of the domesticated apple (Malus × domestica Borkh.) Nat Genet. 2010;42(10):833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- 31.Smith SA, Donoghue MJ. Rates of molecular evolution are linked to life history in flowering plants. Science. 2008;322(5898):86–89. doi: 10.1126/science.1163197. [DOI] [PubMed] [Google Scholar]
- 32.Durand D, Halldórsson BV, Vernot B. A hybrid micro-macroevolutionary approach to gene tree reconstruction. J Comput Biol. 2006;13(2):320–335. doi: 10.1089/cmb.2006.13.320. [DOI] [PubMed] [Google Scholar]
- 33.Ashburner M, et al. The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanada K, et al. Evolutionary persistence of functional compensation by duplicate genes in Arabidopsis. Genome Biol Evol. 2009;1:409–414. doi: 10.1093/gbe/evp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gout JF, Kahn D, Duret L. Paramecium Post-Genomics Consortium The relationship among gene expression, the evolution of gene dosage, and the rate of protein evolution. PLoS Genet. 2010;6(5):e1000944. doi: 10.1371/journal.pgen.1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang L, Gaut BS. Factors that contribute to variation in evolutionary rate among Arabidopsis genes. Mol Biol Evol. 2011;28(8):2359–2369. doi: 10.1093/molbev/msr058. [DOI] [PubMed] [Google Scholar]
- 37.De Bodt S, Hollunder J, Nelissen H, Meulemeester N, Inzé D. CORNET 2.0: integrating plant coexpression, protein-protein interactions, regulatory interactions, gene associations and functional annotations. New Phytol. 2012;195(3):707–720. doi: 10.1111/j.1469-8137.2012.04184.x. [DOI] [PubMed] [Google Scholar]
- 38.Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37(5):501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 39.Davis JC, Petrov DA. Preferential duplication of conserved proteins in eukaryotic genomes. PLoS Biol. 2004;2(3):E55. doi: 10.1371/journal.pbio.0020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharp PM, Li WH. The codon Adaptation Index—A measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134(2):341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akashi H. Synonymous codon usage in Drosophila melanogaster: Natural selection and translational accuracy. Genetics. 1994;136(3):927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Armisén D, Lecharny A, Aubourg S. Unique genes in plants: Specificities and conserved features throughout evolution. BMC Evol Biol. 2008;8:280. doi: 10.1186/1471-2148-8-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D’Antonio M, Ciccarelli FD. Modification of gene duplicability during the evolution of protein interaction network. PLOS Comput Biol. 2011;7(4):e1002029. doi: 10.1371/journal.pcbi.1002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449(7158):54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- 46.Maere S, et al. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA. 2005;102(15):5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edger PP, Pires JC. Gene and genome duplications: The impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 2009;17(5):699–717. doi: 10.1007/s10577-009-9055-9. [DOI] [PubMed] [Google Scholar]
- 48.Birchler JA, Veitia RA. Gene balance hypothesis: Connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci USA. 2012;109(37):14746–14753. doi: 10.1073/pnas.1207726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Birchler JA, Veitia RA. The gene balance hypothesis: From classical genetics to modern genomics. Plant Cell. 2007;19(2):395–402. doi: 10.1105/tpc.106.049338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Papp B, Pál C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424(6945):194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- 51.Aury JM, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444(7116):171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 52.Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16(7):1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pogson BJ, Woo NS, Förster B, Small ID. Plastid signalling to the nucleus and beyond. Trends Plant Sci. 2008;13(11):602–609. doi: 10.1016/j.tplants.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 54.Kleine T, Voigt C, Leister D. Plastid signalling to the nucleus: Messengers still lost in the mists? Trends Genet. 2009;25(4):185–192. doi: 10.1016/j.tig.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 55.Veitia RA. Exploring the molecular etiology of dominant-negative mutations. Plant Cell. 2007;19(12):3843–3851. doi: 10.1105/tpc.107.055053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987;329(6136):219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- 57.Gibson TJ, Spring J. Genetic redundancy in vertebrates: polyploidy and persistence of genes encoding multidomain proteins. Trends Genet. 1998;14(2):46–49. doi: 10.1016/s0168-9525(97)01367-x. discussion 49–50. [DOI] [PubMed] [Google Scholar]
- 58.Vos RA, Caravas J, Hartmann K, Jensen MA, Miller C. BIO:Phylo-phyloinformatic analysis using perl. BMC Bioinformatics. 2011;12:63. doi: 10.1186/1471-2105-12-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fawcett JA, Maere S, Van de Peer Y. Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc Natl Acad Sci USA. 2009;106(14):5737–5742. doi: 10.1073/pnas.0900906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.