Abstract
Dynamic epigenetic modifications play a key role in mediating the expression of genes required for neuronal development. We previously identified nitric oxide (NO) as a signaling molecule that mediates S-nitrosylation of histone deacetylase 2 (HDAC2) and epigenetic changes in neurons. Here, we show that HDAC2 nitrosylation regulates neuronal radial migration during cortical development. Bead-array analysis performed in the developing cortex revealed that brahma (Brm), a subunit of the ATP-dependent chromatin-remodeling complex BRG/brahma-associated factor, is one of the genes regulated by S-nitrosylation of HDAC2. In the cortex, expression of a mutant form of HDAC2 that cannot be nitrosylated dramatically inhibits Brm expression. Our study identifies NO and HDAC2 nitrosylation as part of a signaling pathway that regulates cortical development and the expression of Brm in neurons.
Keywords: neural development, transcription, nitric oxide synthase, polar morphology, schizophrenia
Neuronal development relies on a number of sequential events whereby a pool of undifferentiated embryonic progenitor cells gives rise to a variety of highly specialized postmitotic neurons and glia. In the cortex, neuronal and glial cells derive from multipotent neural progenitor cells (NPCs) generated in the ventricular zone (VZ) of the developing brain. NPCs either undergo self-renewal or exit the cell cycle, differentiate into postmitotic neurons, and migrate to the external layers of the cortex (1). Cortical layers are formed between embryonic day 11 (E11) and E18 in an inside-out manner with deep layers formed first, followed by the more superficial layers, and the formation depends on radial neuron migration. Extracellular cues that regulate this process include brain-derived neurotrophic factor (BDNF) (2). Although BDNF and TrkB signaling are not required for neuronal survival in the embryonic cortex, they have been implicated in the regulation of NPC proliferation (3) and radial and tangential neuronal migration (2).
In addition to the coordinated expression of nuclear factors that regulate, first, neurogenesis and neuronal migration and, later, promote gliogenesis, epigenetic modifications of both DNA and histones represent fundamental mechanisms by which neurons adapt their transcriptional response to corticogenic cues (4). Histone acetylases and deacetylases (HDACs) are nuclear enzymes that maintain chromatin acetylation in balance, thereby contributing to both transcriptional activation and repression. All HDACs identified so far are present in the nervous system and are often developmentally regulated. For example, the highly homologous HDAC1 and HDAC2 are detected at different stages of neuronal development, with HDAC1 confined to neural stem cells and glia and HDAC2 predominantly expressed in postmitotic neuroblasts and differentiated neurons (5).
Although it is now established that many posttranslational modifications of HDACs impact on their activity, there is little evidence directly linking extrinsic stimuli to changes of nuclear HDAC functions and histone acetylation in neurons. One exception is represented by NO, a signaling molecule that plays a pivotal role in regulating NPC proliferation in vitro and neurogenesis in the adult hippocampus (6) and granule cell migration in the cerebellum (7). NO is a gaseous molecule capable of modifying thiol groups of cysteines by means of cysteine nitrosylation (S-nitrosylation), which often affects protein function, including protein–protein interaction, catalytic activity, or subcellular localization. A vast number of proteins have been shown to undergo stimulus-dependent S-nitrosylation, both in the nucleus (8–10) and in the cytoplasm (11). We have recently demonstrated in cortical neurons that the nuclear factor HDAC2 undergoes S-nitrosylation at two cysteine residues, Cys262 and Cys274 (9). This results in HDAC2 dissociation from gene promoters, histone acetylation, and transcriptional activation of specific genes. Importantly, HDAC2 bearing a mutation of Cys262 and Cys274 to alanine (HDAC2C262/274A) cannot be nitrosylated and acts as a powerful transcriptional repressor. When expressed in cortical neurons, HDAC2C262/274A induced a remarkable decrease of both dendritic growth and branching (9). However, whether nitrosylation of HDAC2 regulates the expression of genes necessary for neuronal development remains unknown.
Results
Defects of Neuronal Migration in E14.5 Cortex of nNOS−/− Mice.
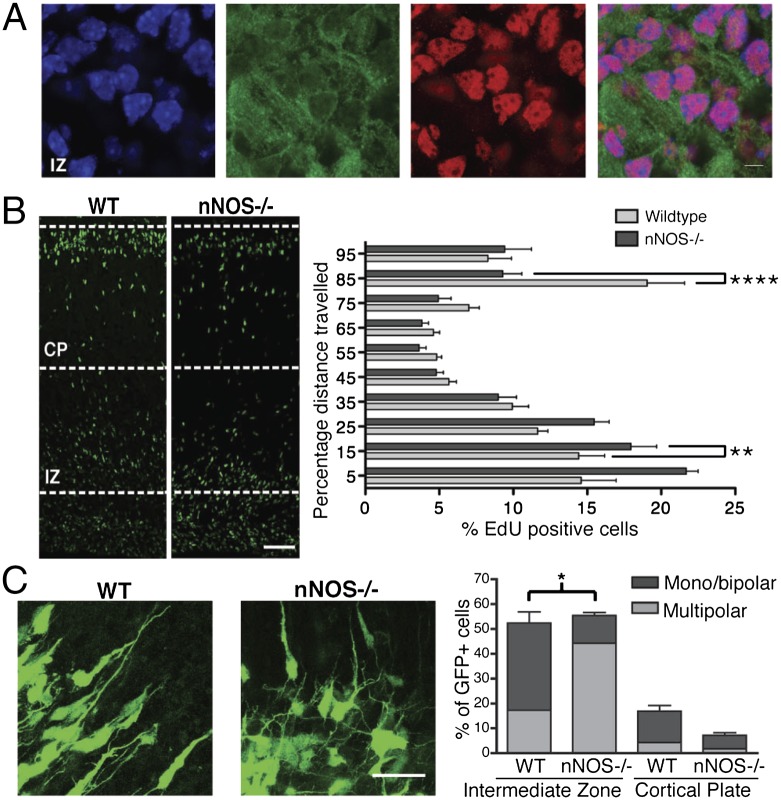
To examine whether neuronal nitric oxide synthase (nNOS) may be involved in regulating neuronal migration in the mammalian cortex, we first performed nNOS immunostaining at different embryonic stages (Fig. S1A). The nNOS antibody specificity was confirmed by performing Western blotting and immunostaining of brains obtained from either wild type (WT) or mice lacking neuronal nitric oxide synthase (nNOS−/−) (12) (Fig. S1 B and C). Although at E12.5, little or no nNOS staining was detected, nNOS expression steadily increased, and at E15.5, nNOS was clearly detectable in both the intermediate zone (IZ) and the cortical plate (CP) (Fig. 1A and Fig. S1A). In agreement with previous studies (13, 14), NPCs within the VZ did not show any significant expression of nNOS at any embryonic stage. Similarly, immunostaining of HDAC2 in mouse cortex at different developmental stages showed that little HDAC2 was detected at E12.5 and that its levels progressively increased at E15.5 and E18.5 (Fig. S1A). A similar developmental regulation of HDAC2 and nNOS was observed by quantitative RT-PCR (qRT-PCR) and RT-PCR and Western blotting of E12.5, E15.5, and E18.5 cortex (Figs. S1C and S2). In E15.5 cortex, nNOS and HDAC2 colocalized in neurons within the IZ and the CP albeit in distinct cellular compartments; nNOS protein was localized mainly in the cytoplasm whereas HDAC2 was confined to the nucleus (Fig. 1A).
Fig. 1.
Lack of nitric oxide signaling impairs radial neuron migration. (A) Images of the IZ of E15.5 sagittal sections stained with DAPI (blue), anti-nNOS (green), and anti-HDAC2 (red). (Scale bar, 30 µm.) n = 3. (B) Images of WT and nNOS−/− cortices from embryos of E15.5 mice injected with EdU, killed 72 h after injection. Quantitative analysis of EdU-positive neurons as percentage distance from VZ. Shown are averages and SEM; **P < 0.01, ****P < 0.0001, two-way ANOVA, Bonferroni post hoc test. (Scale bar, 50 µm.) WT embyos, n = 10; nNOS−/− embryos, n = 9. (C) (Left) Embryos were ex vivo electroporated at E14.5, and organotypic slices were cultured for 3 d in vitro. The images show eGFP-expressing neurons within the IZ. (Right) Quantitative analysis was performed by calculating the percentage of GFP cells within either the IZ or the CP with monopolar/bipolar or multipolar morphology. (Scale bar, 50 µm.) Averages and SEM, n = 3; *P < 0.001, two-way ANOVA, Bonferroni post hoc test.
The influence of nNOS and NO signaling on radial migration of cortical neurons was analyzed in vivo by performing (5-ethynyl-2′-deoxyuridine) (EdU)-based birthdating of E15.5 pregnant mice. EdU-positive cell positioning was analyzed at 48 and 72 h after EdU injection. In the nNOS−/− mice, cortical migration was disrupted, as indicated by the higher percentage of EdU-positive cells observed within the subventricular zone (SVZ) and IZ at 48 and 72 h after EdU administration (Fig. 1B and Fig. S3A). After 72 h, a remarkably larger number of EdU-positive cells reached the CP in WT mice compared with nNOS−/− embryos. Neuronal distribution in nNOS−/− mice was analyzed by immunostaining for upper-layer and deep-layer cortical neurons using the layer-specific markers Cux1 and Ctip2, respectively (15). At E15.5, nNOS−/− embryos showed a disorganization of cortical layer markers, with expansion of deep-layer Ctip2-positive neurons at the expense of upper-layer Cux1-positive neurons (Fig. S3 B and C). Although NO affects proliferation of NPCs in vitro and adult neurogenesis in vivo (6, 13), the high number of cells observed within the deep layers of the nNOS−/− cortex was not due to increased proliferation of NPCs. E13.5 nNOS−/− and WT pregnant female mice were injected with EdU and, after 24 h, 10-μm cryostat sections of embryos were immunostained for Ki-67, a nuclear marker of cell proliferation. Quantitative analysis of EdU and Ki-67–positive cells indicated that, in the embryonic cortex in vivo, the absence of nNOS did not affect the exit of NPCs from the cell cycle (Fig. S4A).
In the cortex, postmitotic neurons initially grow a number of immature neurites; this multipolar morphology is transient and precedes the establishment of neuronal polarity that is essential for radial migration (16). To test whether abnormal neuronal polarization was responsible for the defects of radial migration, we analyzed the morphology of postmitotic neurons within the IZ and the CP of nNOS−/− and WT embryos. Brains were ex vivo electroporated at E14.5, and organotypic slices were analyzed after 3 d in culture. Strikingly, in the IZ, absence of NO signaling increased the number of multipolar neurons compared with WT brains (Fig. 1C). As expected, most neurons that reached the CP were bipolar in both WT and nNOS−/− mice, although in the absence of NO signaling the total number of neurons in the CP was significantly lower. Nestin immunostaining showed no radial glia defects in brains of nNOS−/− mice (Fig. S4B). Taken together, these findings indicate that NO regulates radial migration of postmitotic cortical neurons, possibly by contributing to the establishment of neuronal polarity.
S-Nitrosylation of HDAC2 Is Required for Radial Migration of Cortical Neurons.
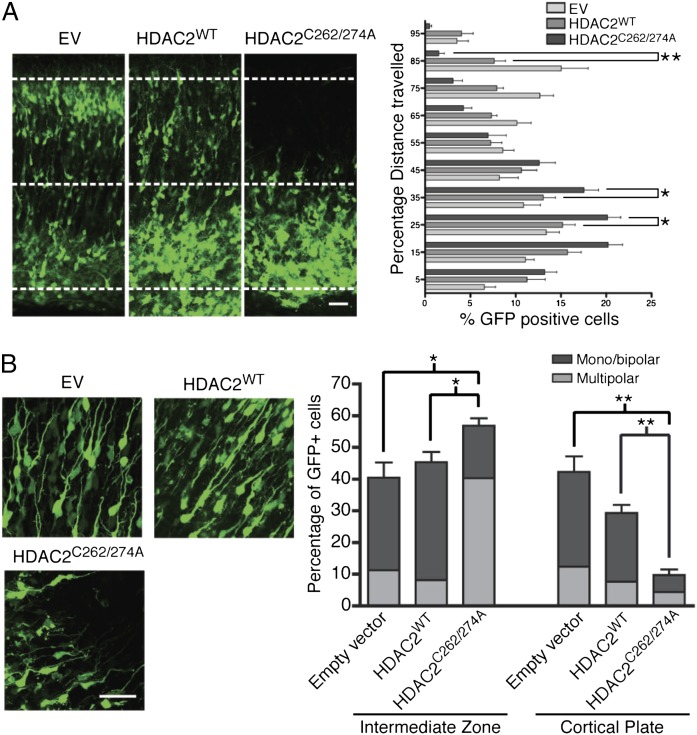
In the brain, HDAC2 levels are developmentally regulated (5, 17). Although in NPCs both HDAC2 and the highly homologous HDAC1 are present, in postmitotic neurons, HDAC2 levels are much higher than HDAC1 (5). To ask whether NO signaling and S-nitrosylation of HDAC2 influenced radial migration of cortical neurons, E14.5 brains were ex vivo electroporated with vectors expressing either empty vector (EV), HDAC2WT, or HDAC2C262/274A. The mutant HDAC2C262/274A cannot undergo S-nitrosylation and acts as a powerful transcriptional repressor. In all experiments, endogenous HDAC2 levels were greatly reduced by cotransfection of siRNA that specifically targeted mouse HDAC2 (Fig. S5 A and B). In slices of electroporated brains, expression levels of both HDAC2WT and HDAC2C262/274A were comparable (Fig. S5C). Cells electroporated with HDAC2C262/274A strikingly accumulated within the IZ and failed to reach the CP (Fig. 2A). In contrast, expression of HDAC2WT had little effect on radial migration. In accordance with our findings observed in nNOS−/− cortex, quantitative analysis of cells labeled with EdU indicated that expression of HDAC2C262/274A did not influence proliferation of NPCs (Fig. S6A). Morphological analysis of neurons electroporated with EV, HDAC2WT, or HDAC2C262/274A showed alterations that closely resembled the defects observed in the brains of nNOS−/− mice (Fig. 2B). Similarly, electroporation of either HDAC2WT or HDAC2C262/274A did not disrupt the radial glial scaffold of the cortex (Fig. S6B).
Fig. 2.
S-nitrosylation of HDAC2 regulates radial neuron migration. (A) Images of cortical slices obtained from E14.5 embryos ex vivo electroporated with HDAC2 siRNA and pCIG-IRES-eGFP, pCIG-HDAC2WT, or pCIG-HDAC2C262/274A and cultured for 5 d. Quantitative analysis of eGFP-positive cells as percentage distance from VZ. (Scale bar, 50 µm.) Shown are averages and SEM. *P < 0.05, two-way ANOVA, Bonferroni post hoc test; EV, n = 7; HDAC2WT and HDAC2C262/274A, n = 8. (B) (Left) E14.5 embryos were ex vivo electroporated, and organotypic slices were cultured for 5 d in vitro. The images show eGFP-expressing neurons within the IZ. (Right) Quantitative analysis was performed by calculating the percentage of GFP cells within either the IZ or the CP with monopolar/bipolar or multipolar morphology. (Scale bar, 50 µm.) Averages and SEM, n = 3. *P < 0.001 for empty vector and HDAC2WT versus HDAC2C262A/C274A multipolar cells and HDAC2WT versus HDAC2C262A/C274A monopolar/bipolar cells; *P < 0.005 for empty vector versus HDAC2C262A/C274A monopolar/bipolar cells in IZ; **P < 0.001 for empty vector and HDAC2WT versus HDAC2C262A/C274A monopolar/bipolar cells in CP; two-way ANOVA, Bonferroni post hoc test.
Characterization of the Transcriptional Program Regulated by HDAC2 S-Nitrosylation.
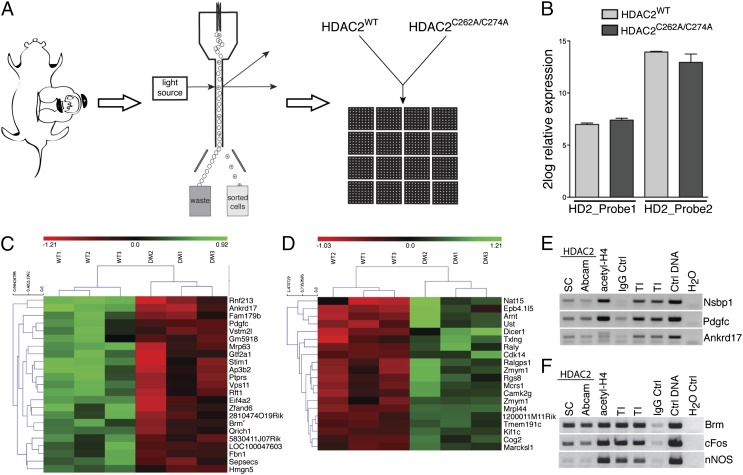
To identify genes that were regulated by HDAC2 nitrosylation in the cortex and that may regulate embryonic cortical development, we performed a genome-wide bead-array screen of NPCs electroporated with either pCIG-HDAC2WT-IRES-eGFP (HDAC2WT) or pCIG-HDAC2C262/274A-IRES-eGFP (HDAC2C262/274A). E14.5 mouse embryos were subjected to in utero electroporation, and, after 18 h, cortices were acutely dissociated, GFP-positive cells were isolated by FACS, and mRNA was subjected to genome-wide bead-array screens (Fig. 3A). Two probes for HDAC2 were included in the microarray chip; one probe bound the coding region of HDAC2, recognizing both endogenous HDAC2 and overexpressed HDAC2 (HDAC2probe_2); the second probe spanned the 3′ UTR of HDAC2, thereby recognizing only endogenous HDAC2 transcripts (HDAC2probe_1). The transcript levels of probe_2 were significantly higher than probe_1 in both the HDACWT and HDAC2C262A/C274A samples, and, importantly, the transcript levels of probe_2 were similar for the samples electroporated with either HDAC2WT or HDAC2C262A/C274A (Fig. 3B). A multicenter large-scale quality control analysis recently showed that fold cutoff is a more reliable statistical tool than false discovery rate (FDR) cutoffs (18), and a 1.3-fold cutoff has been previously used to perform network analyses in a similar experimental setup (19). We have combined a more stringent fold change of >1.6 with an uncorrected P value ≤ 0.01 to identify possible target genes of HDAC2 S-nitrosylation. This analysis generated 23 transcripts that were decreased and 20 transcripts that were increased in neurons expressing HDAC2C262/274A compared with neurons expressing HDAC2WT (Fig. 3 C and D and Fig. S7 A and B). A second analysis excluding sample DM2, which displayed a lower degree of HDAC2 overexpression, revealed an FDR ≤ 0.2 for most of the identified targets (Table S1). To obtain a broader view of the biological processes possibly affected by HDAC2 S-nitrosylation and NO signaling, a GO analysis using all transcripts that displayed P ≤ 0.05 and a fold-change of >1.3 in HDAC2C262/274A vs. HDAC2WT samples identified biological processes possibly affected by S-nitrosylation of HDAC2 such as neural development, differentiation, and migration (Stim1, Ptprs, Dlg1, Marcksl1), posttranslation protein modification, chromatin modification and histone methylation (Nsbp1, Arnt1), metabolism, and cell cycle (Fig. S7C). Brm, Nspb1, Pdgfc, and Ankrd17, which were identified as targets of S-nitrosylated HDAC2, were validated by chromatin immunoprecipitation (ChIP) assay using chromatin from the E14.5 cortices (Fig. 3 E and F). Brm is part of the mammalian Brm/Brg1 (BAF) complex that belongs to the evolutionarily conserved SWI/SNF family of ATPase-dependent chromatin-remodeling factors. There are at least 30 genes encoding BAF proteins; during neuronal development, BAF subunits that are encoded by homologous gene families and have similar functions undergo a switch in subunit composition, resulting in remodeling complexes with high affinity for specific DNA motifs within gene promoters (20, 21). Importantly, Brm levels are tightly developmentally regulated in the brain (22).
Fig. 3.
Characterization of the transcriptional program regulated by HDAC2 S-nitrosylation. (A) Embryos were in utero electroporated at E14.5, and after 18 h cortices were microdissected and dissociated. GFP-expressing cells were FAC-sorted, and RNA was extracted and hybridized to genome-wide bead arrays. (B) Relative expression of endogenous HDAC2 (HDAC2_Probe1) and overexpressed HDAC2WT or HDAC2C262A/C274A (HDAC2_Probe2). HDAC2 transcript levels were comparable between HDAC2C262A/C274A and HDAC2WT samples. The y axis represents 2log relative expression; n = 3. (C and D) Heat maps generated using the 43 transcripts potentially regulated by HDAC2 S-nitrosylation. Shown are 2log relative transcript levels in HDAC2WT and HDAC2C262/274A electroporation conditions. Samples were clustered based on euclidean distance (clustering of samples is indicated at the top; clustering of target genes is indicated at the left). Color scale bar represents 2log relative expression [2log(expression sample) – 2log(average expression all samples)]; green, up-regulation; red, down-regulation; black, no change; n = 3. (E and F) ChIP validation of target transcripts in E14.5 cortical tissue. Immunoprecipitation against HDAC2 (Santa Cruz and Abcam), acetylated histone H4, and rabbit IgG was followed by PCR for Nsbp1, Pdgfc, Ankrd17, Brm, nNOS, and cFos promoters. n = 3.
NO Signaling and HDAC2 Nitrosylation Regulate Brm Levels in the Cortex.
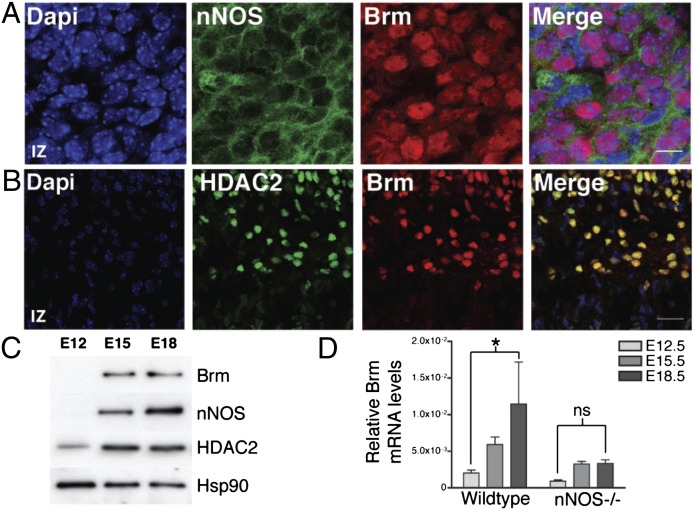
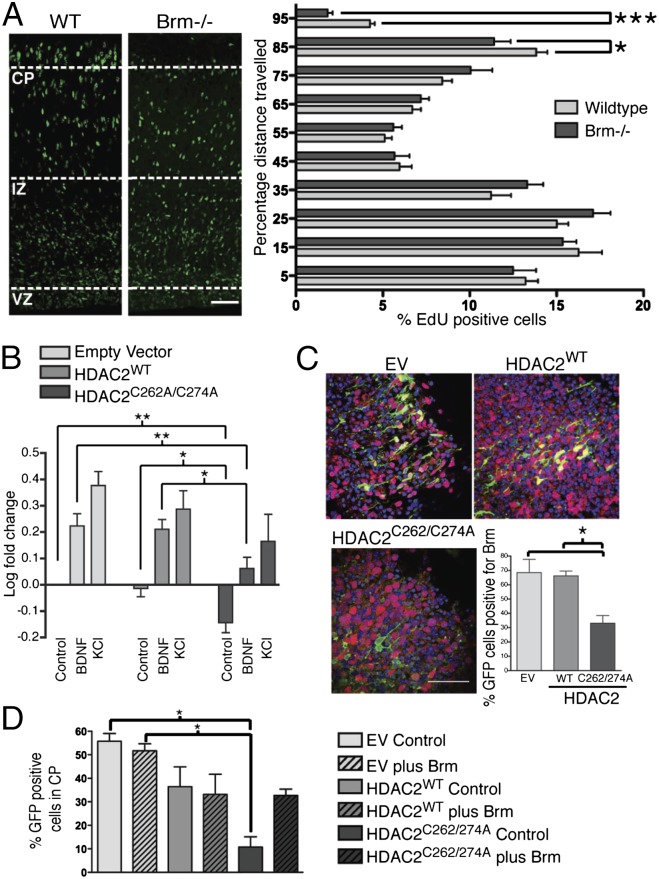
Microarray analysis indicated that Brm expression was strongly decreased in NPCs electroporated with only HDAC2C262/274A [1.8-fold down-regulation in NPCs electroporated with HDAC2C262/274A compared with HDAC2WT (P = 0.01)]. Importantly, HDAC2 and other class I HDACs are recruited to the Brm promoter GATA-binding sites in undifferentiated cells, and their binding progressively decreases during cell differentiation (23). At E12.5, few cells showed detectable Brm expression; however, at E15.5 and E18.5, most neurons within the CP coexpressed Brm with both nNOS and HDAC2 (Fig. 4 A–D; Figs. S8 and S9A). This striking colocalization pattern, together with the evidence that HDAC2 directly binds the Brm promoter, prompted us to study whether absence of Brm affected cortical development in vivo. E15.5 Brm−/− and WT mice were subjected to EdU-based birthdating, and cell positioning of EdU-positive neurons was performed 72 h after injection. A lower number of EdU-positive cells reached the CP of Brm−/− mice compared with WT (Fig. 5A). Analysis of the lower CP marker showed that the number of Cux1-positive neurons was reduced in Brm−/− mice compared with WT control (Fig. S9B). Importantly, both abnormal cortical development and reduced number of Cux1-expressing neurons observed in Brm−/− embryos closely mirrored that observed in nNOS−/− mice. It should be noted that for both nNOS−/− and Brm−/− mice compensatory mechanisms occurring in vivo may have blunted the phenotype observed in vitro. A residual nNOS activity (about 5%) is present in the brain of nNOS−/− mice due to the expression of a truncated form of nNOS that has residual enzymatic activity (25). Moreover, deletion of Brm results in overexpression of Brg1, the functional homolog of Brm present in the BAF complex that is capable of compensating, at least in part, Brm functions (26). To determine whether NO signaling and HDAC2 nitrosylation regulated transcriptional activation of Brm in cortical neurons, analysis of the Brm promoter was performed using plasmids that encoded EV, HDAC2WT, or HDAC2C262/274A together with a vector that contained the Brm promoter (−1 to –1872) fused to a luciferase reporter gene (23) (Fig. 5B). Cortical neurons transfected with HDAC2 siRNA and either EV or HDAC2WT and stimulated with BDNF or depolarized with KCl showed increased activity of the Brm promoter. In contrast, in cortical neurons expressing HDAC2C262/274A, stimulation with BDNF and KCl did not induce activation of the Brm promoter to the same extent, suggesting that NO signaling and HDAC2 nitrosylation regulate Brm expression. Importantly, Brm mRNA was strikingly reduced in brains of nNOS−/− mice (Fig. 4D).
Fig. 4.
Expression of Brm is regulated during cortical development. (A and B) Images of sagittal sections of E18.5 cortex stained with DAPI (blue), anti-Brm (red), anti-nNOS (green; A), or anti-HDAC2 (green; B). (Scale bars, 10 µm in A and 30 µm in B.) n = 3. (C) Western blot analysis of Brm, nNOS, HDAC2, and Hsp90 in cortical lysates from E12.5, E15.5, and E18.5 embryos. n = 3. (D) qRT-PCR analysis of Brm mRNA levels normalized to Rpl11. mRNA was isolated from cortices of either wild-type or nNOS−/− embryos at the indicated stages. Averages are SEM; *P < 0.05; ns, nonsignificant; two-way ANOVA, Bonferroni post hoc test; n = 4.
Fig. 5.
NO and S-nitrosylation of HDAC2 regulates Brm cortical expression. (A) Confocal images of WT and Brm−/− cortices from embryos of E15.5 mice injected with EdU, killed 72 h after injection. Quantitative analysis of distance traveled of EdU-positive neurons. Values are expressed as the percentage distance from VZ. Shown are averages and SEM; *P < 0.05, ***P < 0.0002, unpaired t test. (Scale bar, 50 µm.) WT embyos, n = 9; Brm−/− embryos, n = 9. (B) Luciferase assay of Brm promoter using cortical neurons transfected with HDAC2 siRNA and EV, pCIG-HDAC2WT, or pCIG-HDAC2C262/274A. Neurons were stimulated with either vehicle, BDNF (75 ng⋅ml−1) or KCl (50 mM), for 24 h. Averages and SEM; *P < 0.05, **P < 0.01; one-way ANOVA, Bonferroni post hoc test; n = 5. (C) Expression of Brm in cortices ex vivo electroporated with HDAC2 siRNA and EV, pCIG-HDAC2WT, or pCIG-HDAC2C262/274A. Images of the CP from slices cultured for 5 d and stained for DAPI (blue), GFP (green), and Brm (red). (Scale bar, 50 µm.) n = 5. Quantitative analysis of GFP-positive cells expressing Brm in the CP. Shown are averages and SEM; *P < 0.05; one-way ANOVA, Bonferroni post hoc test; n = 3. (D) Images of cortex from slices of E14.5 embryos ex vivo electroporated with siHDAC2 and pCIG-IRES-eGFP, pCIG-HDAC2WT, or pCIG-HDAC2C262/274A and cultured for 5 d. Brains were coelectroporated with either pDEST26-hBrm (24) (Brm) or EV. Quantitative analysis of eGFP-expressing neurons expressed as percentage distance from VZ. (Scale bar, 50 µm.) Shown are averages and SEM; *P < 0.05; one-way ANOVA, Bonferroni post hoc test; n = 3.
Finally, the effect of HDAC2 nitrosylation on Brm expression was studied during radial neuronal migration in the cortex. E14.5 brains were electroporated with EV, HDAC2WT, or HDAC2C262/274A together with siRNA that targeted endogenous HDAC2. Cortical slices were maintained in culture for 5 d and immunostained for GFP and Brm. As expected, in brains expressing EV or HDAC2WT, Brm was clearly detectable in neurons that had migrated to the CP (Fig. 5C). However, when brains were electroporated with HDAC2C262A/C274A, little or no Brm was detected in neurons at the CP. Importantly, coelectroporation of a Brm expression vector completely rescued the migration defects observed in neurons expressing HDAC2C262/274A (Fig. 5D and Fig. S9C). These findings demonstrate that HDAC2 nitrosylation regulates Brm expression in the cortex, possibly promoting a subunit switch of BAF complexes that is necessary for neuronal radial migration.
Discussion
Epigenetic modifications are emerging as essential mechanisms regulating a host of developmental processes, including neurogenesis, differentiation of NPCs, and neuronal migration (20, 27). HDAC1 and HDAC2 control differentiation of NPCs into mature neurons (17). Mice lacking both HDAC1 and HDAC2 are characterized by severe developmental defects that include abnormal cardiac morphogenesis, hippocampal abnormalities, and aberrant organization of cortical layers (17). Although these findings suggest that HDAC1 and HDAC2 may be functionally redundant during development, their expression in the brain is tightly regulated and compartmentalized (5). Indeed, lack of either HDAC1 or HDAC2 is often associated with defects that may not be functionally compensated by the other, indicating that they may have nonredundant functions.
We previously demonstrated that NO induces S-nitrosylation of HDAC2 and that this posttranslational modification mediates BDNF- and synaptic activity-dependent epigenetic changes in developing neurons (9). NO-dependent inhibition of HDAC2 activity and histone acetylation is not confined to neurons and has also been demonstrated in muscle cells (28, 29). Thus, S-nitrosylation of HDAC2 represents a fundamental and conserved regulatory mechanism that controls gene transcription in mammalian cells. Here, we show that HDAC2 nitrosylation is necessary for neuronal morphology and radial neuron migration in the cortex. Importantly, the abnormal radial migration and impaired neuronal morphology observed in nNOS−/− cortex are strikingly similar to the defects observed in brain slices expressing the NO-insensitive HDAC2C262A/C274A. Expression of both nNOS and HDAC2 is developmentally regulated (5, 14). At E12.5, nNOS is virtually absent in the cortex, whereas at this stage HDAC2 can be detected in glial cells (5) and at much lower levels in NPCs (5). As NPCs differentiate into neurons and migrate toward the external layers of the cortex, both nNOS and HDAC2 expression were dramatically increased and colocalized in most neurons within the CP. The distribution of nNOS in the developing cortex is in agreement with the antiproliferative and prodifferentiation role of NO previously demonstrated in vitro (7) and in vivo (6).
How does NO and HDAC2 regulate radial neuron migration? Our genome-wide screen of embryonic cortices identified a number of genes involved in chromatin modification, neural migration, and axonogenesis that were specifically affected in NPCs expressing HDAC2C262A/C274A compared with HDAC2WT, indicating that S-nitrosylation of HDAC2 regulates these developmental processes. Changes in expression of some of these genes may account for the morphological defects and abnormal polarity observed in neurons lacking nNOS or expressing HDAC2C262A/C274A. For example, Ptprs (2.2-fold down-regulated) is involved in neurite growth, axonal growth and pathfinding, and neurogenesis. Ptprs−/− mice display developmental delays and neurological and neuroendocrine defects (30, 31). Many genes that were changed in NPCs expressing HDAC2C262A/C274A were also regulated by NO signaling, including Pdgfc (1.9-fold down-regulated) (32). Interestingly, Stim1 (2.4-fold down-regulated), an endoplasmic reticulum Ca2+ sensor that influences neuronal development and growth cone turning, also regulates NO synthesis (33, 34). Genes involved in transcriptional regulation and chromatin modification, such as Brm, the ATPase subunit of the ATP-dependent chromatin-remodeling complex BAF, were also significantly inhibited (1.8-fold) in NPCs expressing HDAC2C262A/C274A compared with HDAC2WT.
In mammals, a switch of BAF subunit composition takes place during the differentiation of stem cells into NPCs and eventually into postmitotic neurons. BAF45a and BAF53a subunits, for example, are highly expressed in neuronal progenitors; however, in differentiated neurons, the levels sharply decrease, and they are replaced by BAF45b/c and BAF53b (21). Similarly, Brm levels progressively increase during neuronal differentiation, whereas the homologous ATPase subunit Brg1 is already present in ES cells, and its levels remain unchanged during neuronal development (22). Importantly, the subunit composition of BAF complexes may influence their ability to interact with specific nuclear factors. Brg1, for example, binds to the corepressor neural restrictive silencing complex known as Neuron-Restrictive Silencer Factor/RE1-silencing transcription factor (NRSF/REST), and its depletion in nonneuronal cells results in increased expression of NRSF/REST-dependent, neuron-specific genes (35). It is conceivable that BAF complexes containing subunits specific to either NPCs or neurons will bind and regulate distinct sets of genes. In neurons (or in any other cell type), the genomic occupancy of both Brm and Brm-dependent gene transcription are unknown; thus it is difficult to assess which proportion of the genes whose expression is affected by S-nitrosylation of HDAC2 is directly regulated by Brm. Given that our bead-array analysis was performed 18 h after brain electroporation, it is possible that genes that are directly regulated by Brm or other chromatin-remodeling factors identified with the screen may represent secondary targets of HDAC2 nitrosylation.
One fundamental question is to understand how the switch of BAF complex subunits is regulated during neuronal development. Because the expression of functionally homologous subunits of BAF is temporally and spatially regulated, it is likely that relative nuclear levels of homologous subunits play an essential role in determining BAF complex composition. Indeed, the transition of BAF53a to BAF53b in neuronal progenitors is mediated by miR-9 and miR-124, two microRNAs that target the 3′ UTR of BAF53a, thereby inhibiting its translation (20). Our study identifies NO-dependent HDAC2 S-nitrosylation as the first signaling pathway by which the BAF subunit Brm is regulated in the developing cortex. HDAC2 binds the Brm promoter and inhibits its transcription. Both in the absence of NO signaling and in cortices expressing the NO-insensitive HDAC2C262A/C274A, Brm transcript and protein levels are dramatically decreased. Importantly, we found that inhibition of Brm expression results in defective radial migration. Brm−/− mice show an increased body weight and defects of cell proliferation (26). The lack of severe developmental abnormalities may be due to the compensatory up-regulation of Brg1 that can functionally replace, at least in part, Brm within the BAF complexes (26).
Our findings that NO-dependent expression of Brm influences radial neuron migration may have implications that extends beyond the mechanisms of cortical development. A genome-wide screen of single nucleotide polymorphisms recently identified Brm as a gene associated with schizophrenia (24). Although behavioral analyses of Brm−/− mice have not been performed, nNOS−/− mice display a host of abnormalities that span from aggressive behavior to impaired cognitive performance (36). Given that NO signaling has been implicated in many developmental events that are altered in schizophrenia, including cell migration and synaptogenesis (37), it is conceivable that disregulation of NO and HDAC2 signaling may play an essential role in the pathogenesis of this and other neurodegenerative diseases.
Materials and Methods
EdU Migration Assay.
All animal studies were approved by the Institutional Animal Care and Use Committees at University College London. E15.5 mice were injected with EdU (5 µg/g of mouse weight) and culled at either E17.5 or E18.5. Tissues were flash-frozen in optimal cutting temperature (OCT) medium with isopentane on dry ice. Ten-micrometer sections were collected using a cryostat (Leica) and stored at −20 °C. Sections were stained for EdU incorporation using the Click-iT EdU mixture with AlexaFlour 594 (Invitrogen).
Ex Vivo Electroporation.
The ex vivo protocol was developed by Franck Polleux (38). DNA (1 µg⋅µL−1) and siRNA (10 µM) diluted in 0.05% Fast Green Dye (Sigma) were microinjected into the lateral ventricles, and brains were subjected to electroporation. Brains were embedded in 3% (wt/vol) low-melting-point agarose dissolved in HBSS medium and sliced. Slices were cultured on semipermeable organotypic membranes (VWR) in slice culture medium and cultured for 5 d in vitro.
In Utero Electroporation and FACS Sorting.
DNA (2 μg⋅μL−1) in 10 mM Tris (pH 8.0) with 0.01% Fast Green Dye was microinjected into the lateral ventricles, and brains were electroporated using an ECM 830 Electro-square-porator (Harvard Apparatus). Embryos were electroporated in utero at E14.5, and brains were removed after 18 h. Live cells were sorted using forward scatter versus side scatter, and clumps were discarded by combining the forward scatter versus pulse width. GFP fluorescent cells were sorted on forward scatter versus fluorescence channel 1 (528/38 filter) using a 100-µm nozzle at a pressure of 15 psi with an average speed of 7,000 cells/sec−1. mRNA was isolated and subjected to bead-array analysis.
A full description of the methods used in this study can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Catia Andreassi, Rejji Kuruvilla, and Alison Lloyd for insightful comments on the manuscript and Franck Polleux for helping with the ex vivo electroporation technique and for providing pCIG vector. The Brm-luciferase expression vector was kindly provided by K. Miyake and the pDEST26-hBrm vector by Tadao Arinami. This work was supported by Medical Research Council Research Grant G0500/792 (to A.R.), Netherlands Organization for Health Research and Development Grants ZonMW-VIDI and ZonMW-TOP (to R.J.P.), and by Netherlands Organization for Scientific Research VICI Grant 865.09.002 (to M.P.S.). A.R. is the recipient of Medical Research Council Senior Non Clinical Fellowship G0802010.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218126110/-/DCSupplemental.
References
- 1.Gupta A, Tsai LH, Wynshaw-Boris A. Life is a journey: A genetic look at neocortical development. Nat Rev Genet. 2002;3(5):342–355. doi: 10.1038/nrg799. [DOI] [PubMed] [Google Scholar]
- 2.Medina DL, et al. TrkB regulates neocortex formation through the Shc/PLCgamma-mediated control of neuronal migration. EMBO J. 2004;23(19):3803–3814. doi: 10.1038/sj.emboj.7600399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartkowska K, Paquin A, Gauthier AS, Kaplan DR, Miller FD. Trk signaling regulates neural precursor cell proliferation and differentiation during cortical development. Development. 2007;134(24):4369–4380. doi: 10.1242/dev.008227. [DOI] [PubMed] [Google Scholar]
- 4.Riccio A. Dynamic epigenetic regulation in neurons: Enzymes, stimuli and signaling pathways. Nat Neurosci. 2010;13(11):1330–1337. doi: 10.1038/nn.2671. [DOI] [PubMed] [Google Scholar]
- 5.MacDonald JL, Roskams AJ. Histone deacetylases 1 and 2 are expressed at distinct stages of neuro-glial development. Dev Dyn. 2008;237(8):2256–2267. doi: 10.1002/dvdy.21626. [DOI] [PubMed] [Google Scholar]
- 6.Packer MA, et al. Nitric oxide negatively regulates mammalian adult neurogenesis. Proc Natl Acad Sci USA. 2003;100(16):9566–9571. doi: 10.1073/pnas.1633579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka M, Yoshida S, Yano M, Hanaoka F. Roles of endogenous nitric oxide in cerebellar cortical development in slice cultures. Neuroreport. 1994;5(16):2049–2052. doi: 10.1097/00001756-199410270-00015. [DOI] [PubMed] [Google Scholar]
- 8.Hara MR, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7(7):665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 9.Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S-nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455(7211):411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- 10.Kornberg MD, et al. GAPDH mediates nitrosylation of nuclear proteins. Nat Cell Biol. 2010;12(11):1094–1100. doi: 10.1038/ncb2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sha Y, Marshall HE. S-nitrosylation in the regulation of gene transcription. Biochim Biophys Acta. 2012;1820(6):701–711. doi: 10.1016/j.bbagen.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang PL, Dawson TM, Bredt DS, Snyder SH, Fishman MC. Targeted disruption of the neuronal nitric oxide synthase gene. Cell. 1993;75(7):1273–1286. doi: 10.1016/0092-8674(93)90615-w. [DOI] [PubMed] [Google Scholar]
- 13.Cheng A, Wang S, Cai J, Rao MS, Mattson MP. Nitric oxide acts in a positive feedback loop with BDNF to regulate neural progenitor cell proliferation and differentiation in the mammalian brain. Dev Biol. 2003;258(2):319–333. doi: 10.1016/s0012-1606(03)00120-9. [DOI] [PubMed] [Google Scholar]
- 14.Bredt DS, Snyder SH. Transient nitric oxide synthase neurons in embryonic cerebral cortical plate, sensory ganglia, and olfactory epithelium. Neuron. 1994;13(2):301–313. doi: 10.1016/0896-6273(94)90348-4. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Ge G, Uchida Y, Luu B, Ahn S. 2011. Gli3 is required for maintenance and fate specification of cortical progenitors. J Neurosci 31(17):6440–6448. [DOI] [PMC free article] [PubMed]
- 16.Barnes AP, Polleux F. Establishment of axon-dendrite polarity in developing neurons. Annu Rev Neurosci. 2009;32:347–381. doi: 10.1146/annurev.neuro.31.060407.125536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montgomery RL, Hsieh J, Barbosa AC, Richardson JA, Olson EN. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc Natl Acad Sci USA. 2009;106(19):7876–7881. doi: 10.1073/pnas.0902750106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo L, et al. Rat toxicogenomic study reveals analytical consistency across microarray platforms. Nat Biotechnol. 2006;24(9):1162–1169. doi: 10.1038/nbt1238. [DOI] [PubMed] [Google Scholar]
- 19.Gohlke JM, et al. Characterization of the proneural gene regulatory network during mouse telencephalon development. BMC Biol. 2008;6:15. doi: 10.1186/1741-7007-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460(7255):642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lessard J, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55(2):201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machida Y, Murai K, Miyake K, Iijima S. Expression of chromatin remodeling factors during neural differentiation. J Biochem. 2001;129(1):43–49. doi: 10.1093/oxfordjournals.jbchem.a002834. [DOI] [PubMed] [Google Scholar]
- 23.Itoh T, Miyake K, Iijima S. Differentiation-specific expression of chromatin remodeling factor BRM. Biochem Biophys Res Commun. 2008;366(3):827–833. doi: 10.1016/j.bbrc.2007.12.026. [DOI] [PubMed] [Google Scholar]
- 24.Koga M, et al. Involvement of SMARCA2/BRM in the SWI/SNF chromatin-remodeling complex in schizophrenia. Hum Mol Genet. 2009;18(13):2483–2494. doi: 10.1093/hmg/ddp166. [DOI] [PubMed] [Google Scholar]
- 25.Eliasson MJ, Blackshaw S, Schell MJ, Snyder SH. Neuronal nitric oxide synthase alternatively spliced forms: Prominent functional localizations in the brain. Proc Natl Acad Sci USA. 1997;94(7):3396–3401. doi: 10.1073/pnas.94.7.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reyes JC, et al. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) EMBO J. 1998;17(23):6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirabayashi Y, Gotoh Y. Epigenetic control of neural precursor cell fate during development. Nat Rev Neurosci. 2010;11(6):377–388. doi: 10.1038/nrn2810. [DOI] [PubMed] [Google Scholar]
- 28.Cacchiarelli D, et al. MicroRNAs involved in molecular circuitries relevant for the Duchenne muscular dystrophy pathogenesis are controlled by the dystrophin/nNOS pathway. Cell Metab. 2010;12(4):341–351. doi: 10.1016/j.cmet.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 29.Colussi C, et al. HDAC2 blockade by nitric oxide and histone deacetylase inhibitors reveals a common target in Duchenne muscular dystrophy treatment. Proc Natl Acad Sci USA. 2008;105(49):19183–19187. doi: 10.1073/pnas.0805514105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirkham DL, et al. Neural stem cells from protein tyrosine phosphatase sigma knockout mice generate an altered neuronal phenotype in culture. BMC Neurosci. 2006;7:50. doi: 10.1186/1471-2202-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson KM, et al. Receptor protein tyrosine phosphatase sigma inhibits axonal regeneration and the rate of axon extension. Mol Cell Neurosci. 2003;23(4):681–692. doi: 10.1016/s1044-7431(03)00120-9. [DOI] [PubMed] [Google Scholar]
- 32.Kourembanas S, McQuillan LP, Leung GK, Faller DV. Nitric oxide regulates the expression of vasoconstrictors and growth factors by vascular endothelium under both normoxia and hypoxia. J Clin Invest. 1993;92(1):99–104. doi: 10.1172/JCI116604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell CB, Gasperini RJ, Small DH, Foa L. STIM1 is necessary for store-operated calcium entry in turning growth cones. J Neurochem. 2012;122(6):1155–1166. doi: 10.1111/j.1471-4159.2012.07840.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhang W, et al. Regulation of STIM1, store-operated Ca2+ influx, and nitric oxide generation by retinoic acid in rat mesangial cells. Am J Physiol Renal Physiol. 2007;292(3):F1054–F1064. doi: 10.1152/ajprenal.00286.2006. [DOI] [PubMed] [Google Scholar]
- 35.Ooi L, Belyaev ND, Miyake K, Wood IC, Buckley NJ. BRG1 chromatin remodeling activity is required for efficient chromatin binding by repressor element 1-silencing transcription factor (REST) and facilitates REST-mediated repression. J Biol Chem. 2006;281(51):38974–38980. doi: 10.1074/jbc.M605370200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nelson RJ, et al. Behavioural abnormalities in male mice lacking neuronal nitric oxide synthase. Nature. 1995;378(6555):383–386. doi: 10.1038/378383a0. [DOI] [PubMed] [Google Scholar]
- 37.Bernstein HG, Bogerts B, Keilhoff G. The many faces of nitric oxide in schizophrenia. A review. Schizophr Res. 2005;78(1):69–86. doi: 10.1016/j.schres.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 38.Polleux F, Ghosh A. The slice overlay assay: A versatile tool to study the influence of extracellular signals on neuronal development. Sci STKE. 2002;2002(136):pl9. doi: 10.1126/stke.2002.136.pl9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.