Abstract
Multidrug-resistant Staphylococcus aureus infections pose a significant threat to human health. Antibiotic resistance is most commonly propagated by conjugative plasmids like pLW1043, the first vancomycin-resistant S. aureus vector identified in humans. We present the molecular basis for resistance transmission by the nicking enzyme in S. aureus (NES), which is essential for conjugative transfer. NES initiates and terminates the transfer of plasmids that variously confer resistance to a range of drugs, including vancomycin, gentamicin, and mupirocin. The NES N-terminal relaxase–DNA complex crystal structure reveals unique protein–DNA contacts essential in vitro and for conjugation in S. aureus. Using this structural information, we designed a DNA minor groove-targeted polyamide that inhibits NES with low micromolar efficacy. The crystal structure of the 341-residue C-terminal region outlines a unique architecture; in vitro and cell-based studies further establish that it is essential for conjugation and regulates the activity of the N-terminal relaxase. This conclusion is supported by a small-angle X-ray scattering structure of a full-length, 665-residue NES–DNA complex. Together, these data reveal the structural basis for antibiotic multiresistance acquisition by S. aureus and suggest novel strategies for therapeutic intervention.
Keywords: MRSA, USA300, F-plasmid, pSK41, hairpin
Antibiotic resistance, which arises in bacterial pathogens through conjugative plasmid DNA transfer, is a well-established threat to global health. For example, whereas vancomycin has been essential in treating recalcitrant Staphylococcus aureus infections for decades, vancomycin-resistant S. aureus (VRSA) strains have now appeared in clinical settings worldwide (1–3). VRSA first arose in the United States through the interplay of conjugative DNA transfer and resistance-determinant transposition. The resulting plasmid, pLW1043, has been sequenced and contains not only a vanHAX vancomycin-resistance transposon, but also a cadre of putative DNA transfer genes (4). It was recently shown that the S. aureus plasmid pSK41, which is closely related to pLW1043, mediates the transfer of vancomycin resistance from Enterococcus faecalis into strains of methicillin resistant S. aureus (MRSA) (5). Conjugative bacterial plasmids use almost exclusively plasmid-encoded factors that work in concert to coordinate the cell-to-cell transfer of one strand of the duplex plasmid (6, 7). An element common to all conjugative processes is the plasmid-encoded relaxase enzyme that initiates and terminates transfer by creating a transient single-strand DNA break and covalent protein–DNA intermediate (8, 9).
The vancomycin-resistance plasmid pLW1043 (4) and related plasmids from S. aureus (10, 11), including pSK41 and pGO1 (12–14) as well as plasmids from streptococcal, lactococcal, and clostridial strains, encode a relaxase enzyme termed nicking enzyme in Staphylococcus (NES) that exhibits a unique full-length sequence (Fig. S1). It is 665 residues in length, confines its relaxase motifs to its N-terminal ∼220 aa, and encodes a “single tyrosine”-type relaxase like MobA from plasmid R1162 (15), although the NES and MobA relaxases share only 28% sequence identity. NES is distinct from the second major class of conjugative relaxases, like those from the F and R388 plasmids, which contain more than one catalytic tyrosine (7). The C-terminal region (residues 221–665) of NES is unique to pLW1043 and related S. aureus plasmids, and its function has remained unclear. Most plasmid relaxases are fused to catalytic C-terminal domains like a primase (e.g., R1162) or helicase (e.g., F, R388) that are essential to DNA transfer. In contrast, pLW1043's NES C-terminal region shares no sequence similarity with other bacterial proteins (13).
Here, we present crystal structures of the N- and C-terminal domains of NES, as well as a small-angle X-ray scattering (SAXS) structure of the full-length NES–DNA complex. We show the two domains of NES work in concert to process DNA, design a minor-groove targeted polyamide that inhibits NES at low micromolar concentrations, and establish specific regions of NES essential for plasmid transfer in S. aureus. Together, these data unravel the molecular basis for antibiotic resistance acquisition by S. aureus and suggest potential new avenues for disrupting the spread of multidrug resistance.
Results
NES Crystal Structures.
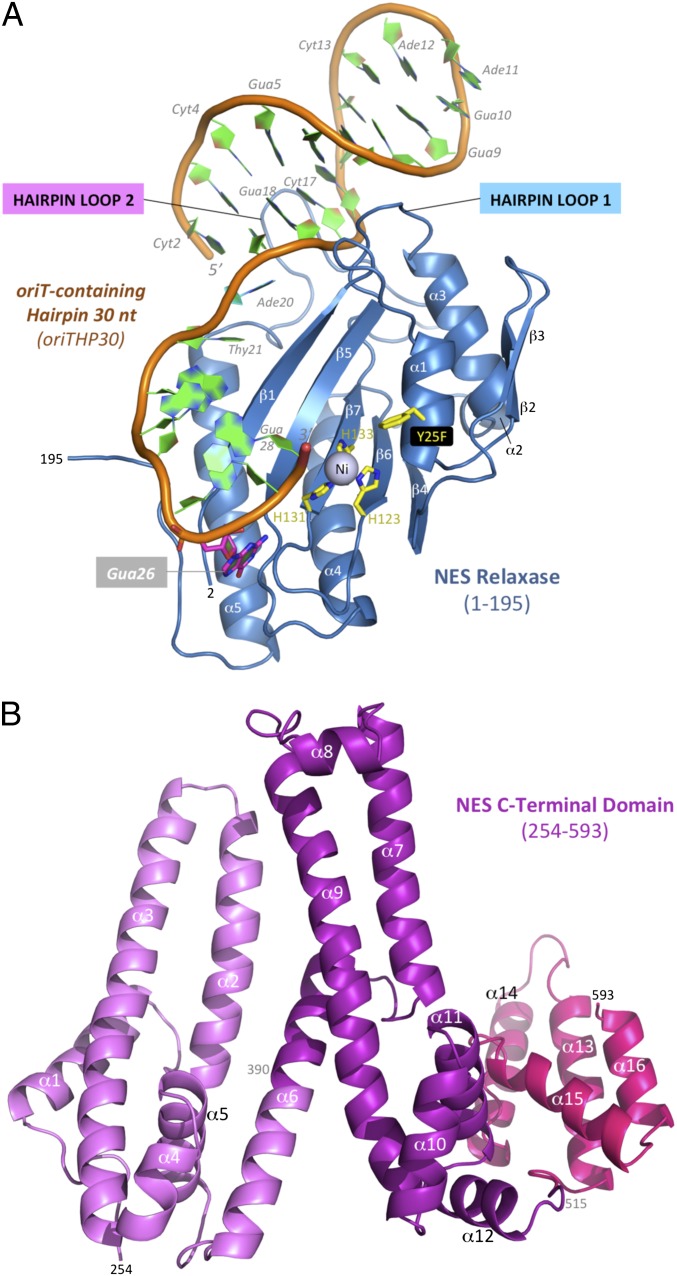
The pLW1043 nes gene was subcloned into Escherichia coli expression vectors used to produce full-length, as well as N-terminal and C-terminal forms of NES. The crystal structure of the NES relaxase (residues 2–195) was determined in complex with a 30-nt DNA sequence to 2.9 Å resolution (Fig. 1A and Table S1). The NES relaxase exhibits a structure similar overall to that observed for the other conjugative relaxases resolved to date (sharing 2.2, 2.9, 3.1, and 3.1 Å Cα rmsd with the MobA, F, R388, and pCU1 relaxases, respectively) (15–18) and contains a metal ion coordinated by three histidines and an adjacent catalytic tyrosine 25 (mutated to phenylalanine in this structure) (Fig. 1A). Mass spectrometry data indicate that the bound metal ion is nickel (Table S2), which is one of the ions that we show supports NES relaxase activity in vitro (Fig. S2). The bound DNA oligonucleotide contains a 7-bp duplex hairpin followed by a 9-nt ordered single-stranded region that extends into the relaxase’s active site.
Fig. 1.
NES structure from S. aureus pLW1043. (A) Crystal structure of the NES N-terminal relaxase (1–195) bound to a 30-nt oriT-containing hairpin (OriTHP30). (B) Crystal structure of the C-terminal domain (254–593) with domains highlighted in different shades of magenta.
The crystal structure of the pLW1043 NES C-terminal domain (residues 254–593) was determined to 3.0 Å resolution and exhibits an all α-helical fold (α1–α16) with no structural homologs identified using the Dali matrix-alignment method (Fig. 1B and Table S1). Furthermore, it bears no structural similarity to a helicase or a primase, like other relaxase-fused C-terminal domains examined previously. Together these structures provide unique atomic-level information on NES, an enzyme that drives drug resistance propagation in S. aureus.
DNA Binding.
Fluorescence anisotropy studies established that DNA oligonucleotides ∼30 nt in length containing duplex hairpin bound the NES relaxase domain with affinities as low as 4 nM (Tables S3 and S4). From this set, oligonucleotide oriTHP30 (30 nt, with the origin of transfer, “oriT”, and a hairpin, “HP”; 8 nM affinity, Table S3) produced the cocrystal structure with the NES relaxase outlined above (Fig. 1A). In the structure, 12 nt contact 21 residues of the NES relaxase, forming 17 base-specific interactions and 9 nonspecific phosphate interactions (Fig. 2A). Of particular note, DNA hairpin-binding loop 1 (residues 78–82) interacts with the DNA hairpin duplex minor groove and forms two base-specific interactions and one phosphate contact (Fig. 2B). DNA hairpin-binding loop 2 works in concert with loop 1 to close around the DNA duplex (Figs. 1A and 2A). Loop 2 (residues 150–156) binds to the major groove of the hairpin, forming 6 base-specific interactions and 4 phosphate contacts (Fig. 2C).
Fig. 2.
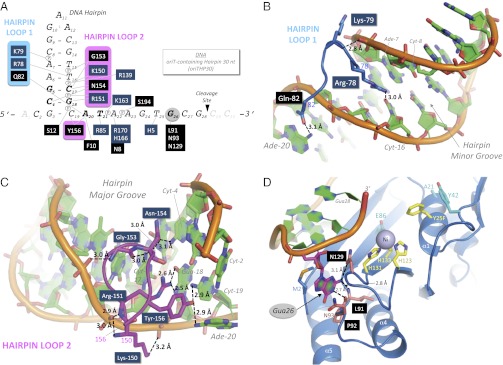
DNA binding by S. aureus NES. (A) Schematic of the NES relaxase–DNA contacts with bases examined using variant oligos in boldface type. (B) Minor groove-binding hairpin loop 1 of NES. (C) Major groove-binding hairpin loop 2 of NES. (D) Active site region of NES, with the buried Gua26 base highlighted.
The single-stranded nine ordered nucleotides of the oriTHP30 oligonucleotide form 12 base-specific and 2 nonspecific phosphate interactions with the relaxase (Fig. 2D). The nucleotide base of guanine 26 is buried in a cavity created by several residues, including two making hydrogen-bonding contacts. Fluorescence anisotropy polarization studies show that eliminating this region of the oligonucleotide reduces relaxase binding by 20-fold (oriT27 vs. oriT24(-3); Table S3). Thus, NES employs specific major- and minor-groove contacts around the DNA hairpin duplex, as well as backbone and unique base-specific interactions, to align the DNA substrate for catalysis.
NES Catalysis.
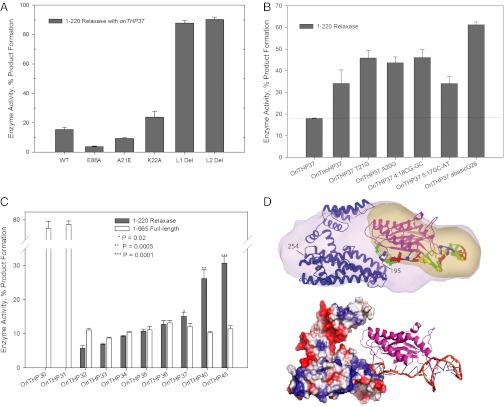
DNA cleavage assays using a 5′-6-FAM–labeled substrate (oriTHP37; Table S4) revealed the NES relaxase domain (residues 1–220) produces an equilibrium level of 15% (±1%) cleaved DNA product (Fig. 3A), a value similar to that of relaxases examined previously (18, 19). In contrast to the related S. aureus pGO1 plasmid, which was reported to have a cleavage site at GCC∧C (where ∧ indicates the putative nic site) (14), we found that pLW1043 NES cleaves 2 nt upstream, at G∧CCC (where residues in boldface type indicate the validated nic site; Fig. S3 and Tables S3 and S4). NES relaxase mutants were then designed to test the role that specific regions of the enzyme play in catalytic function (Fig. 3A). An E86A substitution was found to reduce the cleavage-religation equilibrium significantly; in contrast, a K22A mutation increases cleavage-religation equilibrium, whereas the addition of an adjacent charge (Ala-21-Glu) has no impact on the enzyme (Fig. 3A). The K22A and A21E mutant proteins show wild-type DNA binding; the E86A variant, however, exhibits only nonspecific binding, indicating the important role coordinated metal binding plays in substrate association (Table S5 and Fig. 2). K22 may interact with the DNA region downstream of the cleavage site, as its mutation to an alanine produces an equilibrium shift toward cleaved product (Fig. 3A; see also Fig. 2D).
Fig. 3.
Functional analysis of NES. (A) NES wild-type (WT) and variant relaxase activities, including that of loop 1 (L1) and loop 2 (L2) deletions. (B) Activity of NES relaxase with variant oligos, including an abasic G26 site. (C) Effect of DNA oligo length on NES relaxase and full-length activity. (D) Upper, SAXS envelope shape (pink) representing averaged and filtered dummy atom model of the NES–DNA complex, with the relatively electron-rich region indicated (yellow) and NES crystal structures docked by SUPCOMB. Lower, a positively charged region of the NES C-terminal domain is in proximity to the DNA-bound active site of the NES relaxase in the docked model.
The elimination of DNA-binding loop 1 or 2 exerted dramatic impacts on NES activity. Removal of either of these regions shifted the cleavage-religation equilibrium of the enzyme to ∼90% cleaved DNA, close to trapping the enzyme in its covalent protein–DNA complex state (Fig. 3A). This occurs despite the fact that the loop 1 or loop 2 deletion mutants exhibit either nonspecific or poor DNA binding relative to that in wild-type NES (Table S5). Similarly, changes in specific DNA nucleotides also shift the NES cleavage-religation equilibrium toward cleavage, particularly the elimination of Gua-26 (Fig. 3B). Again, this occurs despite the impact these changes have on DNA binding. The shifts in cleavage-religation equilibrium are reminiscent of specific topoisomerase mutants as well as topoisomerase-targeted drugs that stabilize the covalent enzyme–DNA complex (20–22), and they establish that loop 1, loop 2, and the Gua-26 binding pocket play important roles in NES relaxase catalysis.
The isolated NES relaxase (residues 1–220) exhibited no DNA cleavage until 4 nt were present past the nic site (oriTHP32; Fig. 3C). After that, the stepwise addition of nucleotides produced a stepwise increase in 1–220 relaxase activity up to 35% product formation (oriTHP33-45; Fig. 3C). Intriguingly, when the full-length NES enzyme (1–665) was used, product equilibrium does not exceed ∼10% regardless of the length of oligonucleotide assayed (Fig. 3C). Thus, the presence of the C-terminal domain significantly impacts the catalytic activity of the N-terminal relaxase. Furthermore, full-length NES is able to cleave DNA substrates with only 2- to 3-nt 3′ extensions, in contrast to 1–220 NES (Fig. 3C). Taken together, these data indicate that the unique C-terminal domain of NES regulates the relaxase cleavage-religation equilibrium of the intact enzyme.
SAXS Structure of Full-Length NES with DNA.
Full-length NES alone and in complex with oriTHP30 DNA were examined by SAXS for low-resolution structure determination. NES alone produced SAXS data with clear concentration-dependent radius of gyration (Rg) values, indicating the formation of dynamic complexes not amenable to interpretation. The complex of full-length NES with DNA was well behaved, however, and gave a concentration-independent 38-Å Rg in solution (Fig. S4 A and B). A dummy atom bead model and associated molecular envelope of the NES–DNA complex were calculated using MONSA (23) and Chimera (24), respectively. These revealed the overall shape of the complex (Fig. 3D, top, pink) and the location of the relatively electron-rich DNA (Fig. 3D, top, yellow). SASREF (25) was used for rigid-body fitting of the structures of the N-terminal relaxase–DNA complex and C-terminal domain crystal structures into this SAXS envelope. A solution was chosen that both accommodated the SAXS envelope and placed the DNA of the relaxase complex within the electron-rich region (Fig. 3D, top). In addition, it positioned a positively charged region of the C-terminal domain in close proximity to the DNA-bound NES relaxase active site (Fig. 3D, bottom). Thus, this SAXS-based structural model appears to corroborate the functional observation regarding the impact of the C-terminal domain on NES relaxase activity (Fig. 3C). Together, they suggest a putative model by which the C-terminal and N-terminal regions work in concert to regulate the DNA cleavage-religation equilibrium of NES.
Regions Essential for DNA Transfer in S. aureus.
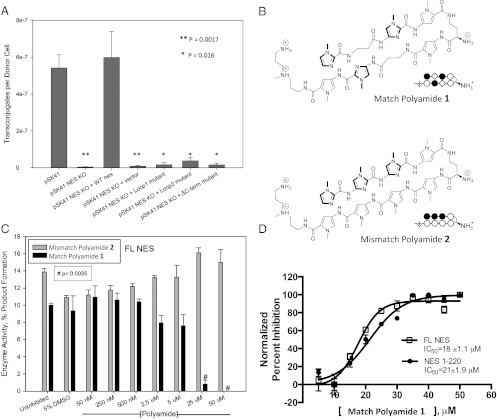
Conjugative transfer was examined in S. aureus, using the staphylococcal conjugative plasmid family prototype pSK41 because its conjugative transfer genes are nearly identical to pLW1043 but it lacks the dangerous vancomycin-resistance transposon. Disruption of the nes gene in pSK41 led to the elimination of plasmid transfer, as measured by the number of DNA transfer events (the formation of “transconjugants”) per donor S. aureus cell (Fig. 4A). This result confirms the essential role of NES in the transmission of pSK41 and related staphylococcal plasmids, including pLW1043 (14). Importantly, whereas complementation with wild-type nes returns conjugation to wild-type levels, complementation with specific mutant nes genes designed on the basis of the NES crystal structures dramatically impacted plasmid transfer (Fig. 4A). Variant nes genes containing the deletion of either loop1 or loop2, both of which significantly disrupted the DNA cleavage-religation equilibrium of NES in vitro (Figs. 2 and 3A), significantly reduced conjugative DNA transfer (Fig. 4A). Thus, the specific major- and minor-groove DNA contacts formed by NES loops 1 and 2 are essential for conjugative DNA transfer between S. aureus cells. Furthermore, the C-terminal domain of NES is also required for conjugation. Introduction of a stop codon after the nucleotides encoding residue 303 in nes eliminated plasmid transfer in S. aureus (Fig. 4A). This residue was chosen because the nes gene in the MRSA plasmid pUSA03 is truncated at this position (26), as discussed below. Taken together, these data establish that the loop 1 and 2 regions of the NES relaxase domain, and the C-terminal domain of NES, are required for the transfer of plasmids in S. aureus cells.
Fig. 4.
NES in S. aureus and polyamide inhibition. (A) Effect of nes deletion (KO) and complementation with WT and designed NES loop 1 (L1), loop 2 (L2), and C-terminal deletion (ΔC-term) mutants on conjugation in S. aureus. (B) Structures of the Match Polyamide 1 and Mismatch Polyamide 2. (C) Inhibition of full-length NES with Match 1 (solid bars) and Mismatch 2 (shaded bars) polyamides. (D) Eighteen- and 21-μM IC50 values of the Match Polyamide 1 for the full-length and relaxase forms of NES, respectively.
Polyamide Inhibition.
The dependence of conjugative plasmid transfer on an intact DNA-binding domain suggests the potential for inhibiting relaxase function and limiting the spread of antibiotic resistance genes through disruption of NES–DNA interactions. As a proof-of-concept, we examined the ability of synthetic minor-groove binding ligands to inhibit NES activity. The GC-rich hairpin sequence recognized by NES represents a low-affinity target for traditional minor-groove binding of natural products such as distamycin and netropsin, which preferentially bind AT tracts (27, 28). Indeed, in our hands we observed no significant change in NES relaxase activity when netropsin was added up to concentrations of 1 mM. This led us to investigate inhibition of NES by Py-Im polyamides, a class of sequence-specific minor-groove binding compounds that can be programmed to bind GC-rich sequences with high affinity (Fig. 4B) (29). Match polyamide 1 was designed to bind selectively to the 5′-GCGAA-3′ sequence contacted by the essential loop 1 of NES (Figs. 1A and 2B). As a control the activity of this compound was compared with that of a mismatch polyamide 2, which shows lower affinity for the 5′-GCGAA-3′ sequence (Fig. S5). The DNA cleavage activity of the relaxase 1–220 region of NES was eliminated by match polyamide 1 at 50 μM (Fig. S6). Interestingly, 2.5- to 50-μM concentrations of the mismatch control polyamide 2 caused a significant increase in product formation by the relaxase (Fig. S6). This result is similar to those obtained when the loop 1 and 2 regions of the relaxase were deleted (Fig. 3A). Taken together, these data indicate that, for the isolated relaxase domain, disrupting the protein–DNA interactions around the DNA hairpin can significantly shift the enzyme’s cleavage-religation equilibrium. This effect, however, is not seen when intact, 1–665 NES is used, as outlined below.
With full-length NES, mismatch polyamide 2 showed no effect, whereas the match polyamide 1 significantly reduced DNA cleavage activity at 25 μM (Fig. 4C). A closer examination of NES inhibition by concentrations of polyamide 1 between 5 and 50 μM produced inhibition curves and IC50 values of 18 and 21 μM for the full-length and relaxase regions of NES, respectively (Fig. 4D). Thus, full-length NES is inhibited by a polyamide targeted to the hairpin minor-groove DNA sequence and is resistant to the effects of the nonspecific polyamide. Taken together, these results confirm that full-length NES acts in a manner distinct from that of the isolated relaxase with respect to catalytic activity and suggest that with further development small molecules capable of inhibiting NES–DNA interactions could disrupt NES function to prevent antibiotic resistance transfer.
Discussion
Antibiotic resistance propagation is largely mediated by the conjugative DNA transfer of plasmids encoding resistance genes, as well as nearly all of the factors required for DNA transmission from donor to recipient cells (7, 9, 30). We focused on the NES relaxase enzyme from plasmid pLW1043, the first vancomycin-resistance vector identified in S. aureus (4). pLW1043 and related plasmids like pSK41 variously harbor resistance to key antibiotics like vancomycin, aminoglycosides, β-lactams, and mupirocin (6). The NES relaxase was shown to contain two essential loops (1, 2) that close around the bound DNA hairpin (Figs. 1A and 2). We establish that the disruption of these loops, using either mutagenesis or a polyamide, inhibits NES activity in vitro (Figs. 3A and 4 B–D). Furthermore, the deletion of either of these loops eliminates plasmid DNA transfer in S. aureus (Fig. 4A).
Minor-groove–targeted Py-Im Polyamide 1, designed to bind to the GC-rich DNA minor groove of the NES substrate hairpin, demonstrated IC50 values of 20 μM with respect to DNA cleavage. Although optimization of polyamide architecture and functionalization will be necessary to promote organism-specific uptake in S. aureus, these results suggest a future approach for targeted disruption of antibiotic resistance transfer. Minor-groove binders similar to Py-Im polyamides have shown activity against S. aureus in vitro and in animal models of infection (31, 32). Interestingly, there is a natural precedent for targeting NES activity as a means to limit DNA transfer, in the form of a functional CRISPR containing a nes-specific spacer that is present in the genome of Staphylococcus epidermidis strain RP62a (33).
This is a unique single-tyrosine relaxase structure resolved in complex with DNA, and DNA binding by NES is clearly distinct relative to the “multityrosine” relaxase–DNA complex structures elucidated previously. The cavity that holds the buried Gua-26 nucleotide base in NES is conserved in the single-tyrosine relaxase structure of plasmid R1162 MobA determined without DNA (Fig. S7A and Fig. 2D) (15). In contrast, the multityrosine relaxases from plasmids pCU1, R388, and F all position a helix in place of the short loop in NES (Fig. S7 B–D) (16–18), and these helices occlude the Gua-26 binding pocket. Thus, whereas the multityrosine relaxases use a “thumb” region not present in NES or MobA (Fig. S7 C and D), the shorter single-tyrosine relaxases likely rely on burying a nucleotide adjacent to its active site to align the substrate DNA. Recall that deletion of Gua-26 significantly impacted the equilibrium cleavage activity of NES (Fig. 3B). These data suggest the Gua-26 binding pocket in NES-like relaxases may be targeted in future studies to disrupt enzyme activity.
The structural and biochemical data presented here, together with in vivo observation that the deletion of the NES C-terminal domain significantly reduces conjugation, suggest that the C-terminal domain regulates the NES processing of the DNA substrate during conjugative transfer. The 11 deposited S. aureus nes genes are identical in amino acid sequence within their conserved N-terminal relaxase domains (residues 1–220). In addition, 7 encode 665-residue proteins with complete C-terminal domains. In contrast, 4 exhibit significantly shorter C-terminal regions, including one truncated at residue 555 (accession no. EIK19451) and another that ends at position 482 (ADM29113) (34) (Fig. S8). The 2 remaining sequenced S. aureus nes genes have severely limited C-terminal domains, terminating their coding regions at residues 300 or 303 (CCJ09841, YP_492718) (Fig. S8). The 303-residue NES (YP_492718) is produced by a frameshift in the nes gene on plasmid pUSA03 in some USA300 strains of MRSA (26). The truncated NES we used was created to mimic this 303-residue protein. Our finding that the C-terminal 362 residues of NES were required for efficient conjugation strongly suggests that pUSA03 plasmids with truncated nes genes would be unable to conjugate on their own and may require factors present on coresident plasmids.
Methods
Crystal Structure Determination.
Purified NES constructs were crystallized using hanging drop vapor diffusion, and data for the relaxase–DNA complex and the C-terminal domain, respectively (Table S1), were collected at the National Institute of General Medical Sciences and National Cancer Institute Collaborative Access Team (GM/CA-CAT). Structures were determined and refined using Phenix and manually adjusted with Coot (SI Methods).
DNA-Binding Studies.
Direct binding to relaxase and full-length constructs were measured using fluorescence polarization anisotropy and a 6FAM-oriT31 (Table S3). Competition binding studies used fluorescence polarization anisotropy and titrations of unlabeled DNA (SI Methods).
DNA Cleavage Assays.
Equilibrium DNA cleavage was examined using 6FAM-labeled oligos with product formation monitored on PAGE gels. Percentage of product formation was quantified using samples in triplicate (SI Methods).
Small-Angle X-ray Scattering.
Full-length NES was examined in complex with oriTHP30 DNA, using SAXS on a Rigaku Bio-SAXS 1000 instrument with 60-min collection scans at three concentrations. Concentration-independent Rg values were obtained, and averaged and filtered dummy atom models were created and used for docking (25) (SI Methods).
Conjugation Assays in S. Aureus.
An nes gene knockout S. aureus strain was generated using the Lactococcus lactis Ll.LtrB group II intron (35). WT and mutant nes complementation plasmids were generated in pSK5632 in which nes transcription was driven by a lac promoter (36). Matings were performed on solid surfaces for 20 h, followed by cell recovery and plating with antibiotic selection to determine conjugation frequency (SI Methods).
Polyamide Inhibition Studies.
Polyamides were synthesized and characterized, and their effects on full-length and relaxase domain NES constructs were determined as described in SI Methods.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants AI078924 (to M.R.R.) and GM51747 (to P.B.D.), by National Medical Research Council of Australia Grant 457454 (to S.M.K. and N.F.), by American Cancer Society Fellowship PF-10-015-01-CDD (to J.L.M.), and by the APS and GM/CA CAT.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Database deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4HT4 and 4HTE).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219701110/-/DCSupplemental.
References
- 1.Chang S, et al. Vancomycin-Resistant Staphylococcus aureus Investigative Team Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N Engl J Med. 2003;348(14):1342–1347. doi: 10.1056/NEJMoa025025. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention (CDC) Vancomycin-resistant Staphylococcus aureus—Pennsylvania, 2002. MMWR Morb Mortal Wkly Rep. 2002;51(40):902. [PubMed] [Google Scholar]
- 3.Werner G, Strommenger B, Witte W. Acquired vancomycin resistance in clinically relevant pathogens. Future Microbiol. 2008;3(5):547–562. doi: 10.2217/17460913.3.5.547. [DOI] [PubMed] [Google Scholar]
- 4.Weigel LM, et al. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science. 2003;302(5650):1569–1571. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 5.Zhu W, Clark N, Patel JB. pSK41-like plasmid is necessary for Inc18-like vanA plasmid transfer from Enterococcus faecalis to Staphylococcus aureus in vitro. Antimicrob Agents Chemother. 2013;57(1):212–219. doi: 10.1128/AAC.01587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grohmann E, Muth G, Espinosa M. Conjugative plasmid transfer in gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67(2):277–301. doi: 10.1128/MMBR.67.2.277-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EP, de la Cruz F. Mobility of plasmids. Microbiol Mol Biol Rev. 2010;74(3):434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la Cruz F, Frost LS, Meyer RJ, Zechner EL. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev. 2010;34(1):18–40. doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- 9.Wong JJ, Lu J, Glover JN. Relaxosome function and conjugation regulation in F-like plasmids - A structural biology perspective. Mol Microbiol. 2012;85(4):602–617. doi: 10.1111/j.1365-2958.2012.08131.x. [DOI] [PubMed] [Google Scholar]
- 10.McDougal LK, et al. Emergence of resistance among USA300 methicillin-resistant Staphylococcus aureus isolates causing invasive disease in the United States. Antimicrob Agents Chemother. 2010;54(9):3804–3811. doi: 10.1128/AAC.00351-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shearer JE, et al. Major families of multiresistant plasmids from geographically and epidemiologically diverse staphylococci. Genes Genomes Genet. 2011;1:581–591. doi: 10.1534/g3.111.000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berg T, et al. Complete nucleotide sequence of pSK41: Evolution of staphylococcal conjugative multiresistance plasmids. J Bacteriol. 1998;180(17):4350–4359. doi: 10.1128/jb.180.17.4350-4359.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caryl JA, O’Neill AJ. Complete nucleotide sequence of pGO1, the prototype conjugative plasmid from the Staphylococci. Plasmid. 2009;62(1):35–38. doi: 10.1016/j.plasmid.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Climo MW, Sharma VK, Archer GL. Identification and characterization of the origin of conjugative transfer (oriT) and a gene (nes) encoding a single-stranded endonuclease on the staphylococcal plasmid pGO1. J Bacteriol. 1996;178(16):4975–4983. doi: 10.1128/jb.178.16.4975-4983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monzingo AF, Ozburn A, Xia S, Meyer RJ, Robertus JD. The structure of the minimal relaxase domain of MobA at 2.1 A resolution. J Mol Biol. 2007;366(1):165–178. doi: 10.1016/j.jmb.2006.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guasch A, et al. Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC. Nat Struct Biol. 2003;10(12):1002–1010. doi: 10.1038/nsb1017. [DOI] [PubMed] [Google Scholar]
- 17.Larkin C, et al. Inter- and intramolecular determinants of the specificity of single-stranded DNA binding and cleavage by the F factor relaxase. Structure. 2005;13(10):1533–1544. doi: 10.1016/j.str.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 18.Nash RP, Habibi S, Cheng Y, Lujan SA, Redinbo MR. The mechanism and control of DNA transfer by the conjugative relaxase of resistance plasmid pCU1. Nucleic Acids Res. 2010;38(17):5929–5943. doi: 10.1093/nar/gkq303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nash RP, Niblock FC, Redinbo MR. Tyrosine partners coordinate DNA nicking by the Salmonella typhimurium plasmid pCU1 relaxase enzyme. FEBS Lett. 2011;585(8):1216–1222. doi: 10.1016/j.febslet.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bax BD, et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature. 2010;466(7309):935–940. doi: 10.1038/nature09197. [DOI] [PubMed] [Google Scholar]
- 21.Champoux JJ. DNA topoisomerases: Structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 22.Chrencik JE, et al. Mechanisms of camptothecin resistance by human topoisomerase I mutations. J Mol Biol. 2004;339(4):773–784. doi: 10.1016/j.jmb.2004.03.077. [DOI] [PubMed] [Google Scholar]
- 23.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76(6):2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 25.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89(2):1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diep BA, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367(9512):731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 27.Luck G, Triebel H, Waring M, Zimmer C. Conformation dependent binding of netropsin and distamycin to DNA and DNA model polymers. Nucleic Acids Res. 1974;1(3):503–530. doi: 10.1093/nar/1.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wartell RM, Larson JE, Wells RD. Netropsin. A specific probe for A-T regions of duplex deoxyribonucleic acid. J Biol Chem. 1974;249(21):6719–6731. [PubMed] [Google Scholar]
- 29.Dervan PB, Edelson BS. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr Opin Struct Biol. 2003;13(3):284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 30.Malachowa N, DeLeo FR. Mobile genetic elements of Staphylococcus aureus. Cell Mol Life Sci. 2010;67(18):3057–3071. doi: 10.1007/s00018-010-0389-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anthony NG, et al. Antimicrobial lexitropsins containing amide, amidine, and alkene linking groups. J Med Chem. 2007;50(24):6116–6125. doi: 10.1021/jm070831g. [DOI] [PubMed] [Google Scholar]
- 32.Kaizerman JA, et al. DNA binding ligands targeting drug-resistant bacteria: Structure, activity, and pharmacology. J Med Chem. 2003;46(18):3914–3929. doi: 10.1021/jm030097a. [DOI] [PubMed] [Google Scholar]
- 33.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322(5909):1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kennedy AD, et al. Complete nucleotide sequence analysis of plasmids in strains of Staphylococcus aureus clone USA300 reveals a high level of identity among isolates with closely related core genome sequences. J Clin Microbiol. 2010;48(12):4504–4511. doi: 10.1128/JCM.01050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao J, et al. Use of targetrons to disrupt essential and nonessential genes in Staphylococcus aureus reveals temperature sensitivity of Ll.LtrB group II intron splicing. RNA. 2006;12(7):1271–1281. doi: 10.1261/rna.68706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grkovic S, Brown MH, Hardie KM, Firth N, Skurray RA. Stable low-copy-number Staphylococcus aureus shuttle vectors. Microbiology. 2003;149(Pt 3):785–794. doi: 10.1099/mic.0.25951-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.