Abstract
We have developed and validated a methodology for determining the antibody composition of the polyclonal serum response after immunization. Pepsin-digested serum IgGs were subjected to standard antigen-affinity chromatography, and resulting elution, wash, and flow-through fractions were analyzed by bottom-up, liquid chromatography–high-resolution tandem mass spectrometry. Identification of individual monoclonal antibodies required the generation of a database of IgG variable gene (V-gene) sequences constructed by NextGen sequencing of mature B cells. Antibody V-gene sequences are characterized by short complementarity determining regions (CDRs) of high diversity adjacent to framework regions shared across thousands of IgGs, greatly complicating the identification of antigen-specific IgGs from proteomically observed peptides. By mapping peptides marking unique VH CDRH3 sequences, we identified a set of V-genes heavily enriched in the affinity chromatography elution, constituting the serum polyclonal response. After booster immunization in a rabbit, we find that the antigen-specific serum immune response is oligoclonal, comprising antibodies encoding 34 different CDRH3s that group into 30 distinct antibody VH clonotypes. Of these 34 CDRH3s, 12 account for ∼60% of the antigen-specific CDRH3 peptide mass spectral counts. For comparison, antibodies with 18 different CDRH3s (12 clonotypes) were represented in the antigen-specific IgG fraction from an unimmunized rabbit that fortuitously displayed a moderate titer for BSA. Proteomically identified antibodies were synthesized and shown to display subnanomolar affinities. The ability to deconvolute the polyclonal serum response is likely to be of key importance for analyzing antibody responses after vaccination and for more completely understanding adaptive immune responses in health and disease.
Keywords: antibody proteomics, antibody repertoire, serum immunoprofiling, B-cell response, humoral response
The first Nobel Prize in Medicine was awarded to Emil von Behring, who in collaboration with Kitasato Shibasaburo and Paul Ehrlich discovered serum antitoxins (1, 2). Remarkably, after more than 100 y of intense research in immunology, little is known about the clonality, relative concentrations, and binding properties of the monoclonal antibodies that constitute the antigen-specific Ig pool in serum.
At steady state, circulating antibodies are produced by terminally differentiated B lymphocytes (plasma cells) within the bone marrow, and thus cannot be accessed in living individuals (3). Although recent single B-cell cloning methods (4, 5) have led to the identification of peripheral antigen-specific B memory and/or antibody-secreting cells (plasmablasts), it is generally unknown whether the Igs encoded by peripheral blood B cells correspond to the antibodies present in circulation and especially whether they are present at physiologically relevant levels (i.e., at serum concentrations above KD corresponding to >1 μg/mL for an average affinity of individual antibodies of 5 nM).
The proteomic deconvolution of serum Igs presents two major technical challenges: first, antibody genes in antigen stimulated B-lymphocytes are not simply encoded in the germline but are extensively diversified by somatic recombination, revision, and/or mutation. Therefore, the sequence database required for the interpretation of mass spectra is not available a priori (6, 7) and is completely different for each individual. Second, the antigen-specific antibody pool comprises a wide variety of Igs that display very high levels of amino acid identity within the framework regions. As a result, standard approaches for proteomic analysis by MS are confounded by this exceptionally high rate of identical sequence shared among Ig-derived peptides, which greatly complicates the task of confidently identifying individual variable (V) genes through peptide mapping. Advancements in sequencing and MS technologies have shown some success against these challenges. MS-based de novo sequencing approaches have been used for the identification of purified monoclonal antibodies (8). More recently the identification of a limited subset of antigen-specific antibodies in serum after very stringent enrichment to reduce the complexity of the antigen-specific polyclonal antibody pool to a limited set of Igs from humans and animals was reported (9–12). However, because of the inherent difficulties associated with the proteomic analysis of complex mixtures of antibodies, these studies had focused on the identification of only a small subset of the antigen-specific serum IgGs present in a fraction isolated after stringent affinity chromatography.
In contrast, complete understanding of how B-cell differentiation ultimately shapes humoral immunity requires addressing the more difficult problem of how to deconvolute the entire repertoire of antigen-specific antibodies in serum or in other secretions.
Here we describe the proteomic deconvolution of the serum-derived antigen-specific polyclonal antibody pool by combining NextGen sequencing of the immunoglobulin heavy chain variable region (VH gene) repertoire with liquid chromatography–high-resolution tandem mass spectrometry (LC-MS/MS) (Fig. 1). Proteomic identifications of unique VH-derived peptides (overwhelmingly from the CDR3 region of the VH sequences) were used to determine the VH repertoire of circulating antigen-specific antibodies, and identified VH genes were shown to encode antibodies with subnanomolar antigen affinity.
Fig. 1.
Schematic of the workflow for serum Ig deconvolution. (Left) V gene repertoire sequencing pipeline: total RNA from desired B-cell subpopulations is reverse transcribed and amplified by 5′ RACE with IgG-specific (VH) or Igκ/Igλ-specific (VL) primers and sequenced by Roche 454 sequencing. Reads are processed bioinformatically to obtain a database of unique V genes and their relative transcript abundances. The V gene database is used to interpret the MS spectra. (Right) F(ab)2 purification and proteomic pipeline: F(ab)2 fragments from IgG are prepared and subjected to antigen-affinity chromatography. Proteins in the eluent, flow-through, and wash buffer are denatured, alkylated, proteolyzed, and resolved by high-resolution LC-MS/MS. Full-length V genes containing the identified iCDR3 peptides are then determined from the repertoire database. (Lower) Antibody production and validation: synthetic VH genes are assembled into an scFv library using the VL cDNA, then antigen-specific antibodies are isolated by two to three rounds of phage panning and characterized for antigen affinity.
Results
Overview of Experimental Approach.
The first step for serum antibody deconvolution is the high-throughput sequencing of B lymphocyte cDNAs to generate a database of class-switched antibody VH sequences in a particular individual (Fig. 1). In parallel, full-length IgGs are purified by protein A affinity chromatography and treated with pepsin to prepare F(ab)2 fragments. Antigen-specific F(ab)2 fragments are then isolated by affinity chromatography on immobilized antigen. Bound antibodies are eluted using standard immunoaffinity chromatography conditions (elution with pH 2.7 buffer), and the flow-through and the wash buffer from the column are also collected to identify weakly antigen-binding antibodies. The F(ab)2 fragments are proteolytically digested, and the resulting peptides are resolved by high-resolution LC-MS/MS analysis on an Orbitrap Velos (Thermo Scientific). To ensure comprehensive coverage of the antibody repertoire, we performed three replicate injections per sample, using LC elution with a long, shallow gradient of 5–40% acetonitrile over 245 min, yielding an average of >100,000 MS2 fragmentation spectra per sample. Peptides found to be enriched >10-fold in the elution fraction compared with wash and flow-through were considered as corresponding to IgGs exhibiting a high degree of antigen specificity.
By far the highest diversity in antibodies occurs within the CDR3 region of the VH (CDRH3), which is overwhelmingly responsible for antigen recognition (13) (Fig. 2A). Composed of the V(D)J join with its inherent junctional diversity, the CDRH3 specifies the VH clonotype. The VH clonotype is defined as the group of VH sequences that share germ-line V and J segments, have identical CDRH3 lengths, and exhibit greater than 80% amino acid identity in the CDRH3 sequences (14, 15). The VH clonotype is an important immunological concept because it accounts for antibodies that likely originate from a single B-cell lineage and may provide insight on the evolution of the antigen-specific response of that lineage. Therefore, we focused here on the high-confidence identification of CDRH3 peptides and of the corresponding clonotypes. As shown later, unique peptides derived from non-CDRH3 regions of Igs (i.e., from peptides containing all or portions of framework 1–3, CDR1, or CDR2 sequences) and corresponding to a single V gene in the database make only a minor contribution to the determination of the repertoire in immunized rabbits (contributing to <15% of the total identified clonotypes). Once the CDRH3s have been identified, the full-length sequences of corresponding VH genes are determined from the NextGen VH DNA database.
Fig. 2.
Immunoglobulin heavy chain variable region (VH gene) and circulating antibody repertoire characteristics. (A) Wu-Kabat variability plot representing the variability of the VH genes is shown on a residue-by-residue basis; (B) bar graphs representing VH germ-line family use (Left) and JH germ-line family use (Right) in the VH DNA repertoire determined by 454 sequencing of cDNA from CD138+ bone marrow plasma cells (n = 4729 VH genes, blue bars), peripheral B cells (n = 2788 VH genes, red bars), or from antibody proteins identified by proteomic analysis of the serum affinity purified IgGs (n = 334, green bars) for the CCH rabbit.
To generate antibodies, proteomically identified VH sequences must be paired with the VL genes. We therefore synthesized VH domain DNAs and used these to construct phage displayed scFv libraries comprising the VL cDNA repertoire. Functional scFv antibodies were isolated after two to three rounds of phage panning. Putative pairings of VH and VL genes were then expressed recombinantly as full-size IgGs in mammalian cells and isolated to characterize their antigen affinity and functionality.
V Gene Repertoire Analysis.
Preliminary studies in mice revealed that the methodology described in Fig. 1 was able to successfully identify abundant, antigen-specific IgGs in the serum of immunized mice. However, the small amount of serum that is obtained from mice at killing (0.8–1.5 mL) resulted in low counts of V-gene informative peptides and poor MS identification statistics. Larger amounts of serum can easily be obtained from humans, as well as other comparatively larger animals such as rabbits, an animal model that has been used extensively for immunological studies for nearly a century. For the present study, a New Zealand white rabbit (Oryctolagus cuniculus) was immunized with Concholepas concholepas hemocyanin (CCH) in complete Freund’s adjuvant (CFA), boosted with antigen in incomplete FA and killed 1 wk after the final boost (CCH rabbit). Additionally, to further validate our approach we performed serum IgG deconvolution on an unimmunized rabbit that, surprisingly, was found to exhibit a titer toward BSA (BSA rabbit; this observation occurred fortuitously because BSA was used as the generic blocking agent in our ELISA protocol). Unlike the BSA rabbit, the animal immunized with CCH did not exhibit any titer toward BSA. We prepared RNA samples from total peripheral B cells (PBCs), total bone marrow cells, and CD138+ bone marrow plasma cells (BM-PCs) isolated by magnetic sorting. First-strand cDNAs were generated using an oligo(dT) primer, and double-stranded products were amplified via 5′ RACE (16) using primers complementary to rabbit IgG CH1 (SI Appendix, Table S1). DNA amplification by 5′ RACE was preferable to using published FR1 and J region-specific primers designed for combinatorial library construction (17) because existing 5′ primer sets have not been validated for quantitative V gene amplification. In contrast, 5′ RACE circumvents the need for V gene-specific primers and provides a more accurate representation of the repertoire by avoiding biases introduced by the selection of PCR primer sets. V gene cDNA was sequenced using Roche 454 GS FLX Titanium (SI Appendix, Table S2). Germ-line V and J use were determined (Fig. 2B) as previously described (18). To reduce the impact of sequencing errors, the VH and VL protein sequence databases were compiled using sequences that occurred at n ≥ 2 reads.
Consistent with earlier reports of limited germ-line V gene diversity in rabbits (19, 20), we found that 89% of the VH genes in the IgG repertoire (CCH rabbit) were derived from only two germ-line V genes (1S40 and 1S45), and an overwhelming 75% contained the IGHJ4 segment. V gene and J gene use was highly similar in BM-PCs and PBCs. In the BSA rabbit, 86% of the VH sequences were derived from 1S44, 1S45, and 1S40, whereas 55% contained the IGHJ4 and 28% contained the IGHJ2 J-segment (SI Appendix, Fig. S1). CDRH3 lengths in the class-switched IgG repertoire of an immunized rabbit were longer than in immunized mice (SI Appendix, Fig. S2) (21). The rabbit class-switched V genes displayed a bimodal distribution of amino acid substitutions relative to the germline, with one peak centered at 9-aa substitutions, slightly higher than in the mouse, and a second peak at 24- to 25-aa substitutions (SI Appendix, Fig. S3). Because gene conversion is an important mechanism for Ig diversification in the rabbit (22), it cannot be ascertained whether the highly mutated V gene population resulted from activation-induced deaminase (AID) mutagenesis or from recombination events with V pseudogenes.
MS Proteomic Detection of Ig-Identifying Peptides.
IgG antibodies from a 2.5 mL rabbit serum sample were purified by Protein A affinity chromatography. F(ab)2 fragments were prepared by pepsin digestion and purified by affinity chromatography on immobilized CCH or BSA yielding ∼0.7 and 0.2 mg of high-purity F(ab)2 protein, respectively (>95% pure as determined by SDS/PAGE; typical results shown in SI Appendix, Fig. S4). The protein in the flow-through and wash fractions was also collected for LC-MS/MS analysis. The individual fractions were denatured, reduced, and alkylated with iodoacetamide to modify Cys residues before digestion with trypsin (SI Appendix, SI Materials and Methods). In silico analysis of the V gene database showed that digestion with trypsin should generate peptide fragments with enough coverage of the CDRH3 region and of lengths appropriate for MS detection to uniquely identify 91.4% of the putative antibody clones (SI Appendix, Fig. S5). Cleavage by chymotrypsin added less than 3% in observable VH clone coverage, and a similar effect was calculated for other cleavage-specific proteases. This slight potential gain in coverage was diminished when potential missed cleavages were considered.
Peptides were analyzed by nanospray LC-MS/MS, and collected spectra were searched by Sequest against an in-house NextGen rabbit VH+VL sequence database (containing sequences with n ≥ 2 reads) concatenated with the rabbit full protein-coding sequence database (OryCun2) and MaxQuant contaminants database (23, 24). Postsearch processing by the Percolator algorithm (25) generated a dataset of peptide-spectrum matches (PSMs) with an expected false-discovery rate <1%. False identifications were further controlled at the peptide level by accepting only those peptide identifications for which all PSMs exhibited an average deviation from the expected peptide mass of ≤1.5 ppm. Spectra were manually checked for consistency with the identified sequences, including the presence of modifications (static carbamidomethyl modification of Cys residues and dynamic oxidation of methionine to methionine-sulfoxide) and signature motifs such as the IGHJ-derived sequence, which gave a characteristic spectral pattern (Table 1).
Table 1.
Highest count iCDRH3 peptides from the CCH rabbit detected in the affinity chromatography elution fraction
| Rank/name | iCDRH3 MS sequence | Spectral counts (%) | Full CDRH3 transcript sequence (% oxidation products in parentheses) | Fractions where peptide was detected | No. somatic variants (from VH gene repertoire) | V gene origin |
| 1* | NVAGYLCAPAFNFR | 115 (8.00) | ARNVAGYLCAPAFNFRSPGTLVTVSSGQPK | E | 1 | BM-PC |
| 2* | NFKLWGPGTLVTVSSGQPK | 88 (6.11) | ARNFKLWGPGTLVTVSSGQPK | E | 3 | BM-PC, PBC |
| 3* | MDSHSDGFDPWGPGTLVSVSSGQPK | 82 (5.70) | ARMDSHSDGFDPWGPGTLVSVSSGQPK (51%) | E | 7 | PBC |
| 4 | FTISSDNAQNTVDLK | 64 (4.45) | AREGYGGYVGYMGLWGPGTLVTVSSGQPK | E+W+F | 3 | BM-PC, PBC |
| 5* | VCGMDLWGPGTLVTVSSGQPK | 50 (3.48) | ARNVYGASRVCGMDLWGPGTLVTVSSGQPK (43%) | E | 2 | BM-PC |
| 6* | NPGGTSNLWGPGTLVTVSSGQPK | 47 (3.27) | ARNPGGTSNLWGPGTLVTVSSGQPK | E | 1 | BM-PC |
| 7* | KFNLWGPGTLVTVSSGQPK | 44 (3.05) | ARDADDYRKFNLWGPGTLVTVSSGQPK | E | 1 | PBC |
| 8 | AFNLWGPGTLVTVSSGQPK | 43 (3.00) | ARDVGYGNDNYRAFNLWGPGTLVTVSSGQPK | E+W+F | 1 | PBC |
| 9* | SPSSGSSNLWGPGTLVTVSSGQPK | 39 (2.71) | ARSPSSGSSNLWGPGTLVTVSSGQPK | E | 2 | BM-PC |
| 10 | NSGSASNLWGPGTLVTVSSGQPK | 31 (2.16) | ARNSGSASNLWGPGTLVTVSSGQPK | E | 2 | BM-PC |
| 11 | GMDLWGPGTLVTVSSGQPK | 29 (2.02) | AREDTYGDANTDYLYRGMDLWGPGTLVTVSSGQPK | E+W | 3 | BM-PC, PBC |
| 12 | NAGTASNLWGPGTLVTVSSGQPK | 28 (1.95) | ARNAGTASNLWGPGTLVTVSSGQPK | E | 1 | BM-PC |
| 13 | GLTAADTATYFCAR | 28 (1.95) | ARDGIDGNGYNDLNLWGPGTLVTVSSGQPK | E+F | 1 | BM-PC |
| 14 | ELTGNGIYALK | 27 (1.88) | ARELTGNGIYALKLGGPGTLVTVSSGQPK | E | 1 | BM-PC |
| 15 | TSSTTVPLQMTSLTAADTATYFCGR | 27 (1.88) | GRGYTDGMDLGGPGTLVTVSSGQPK | E | 1 | BM-PC |
iCDRH3 peptide sequences, counts of affiliated mass spectra, and their frequencies relative to all spectral counts in the eluent (in parentheses), corresponding full-length CDRH3 sequences, and numbers of somatic variants deduced from the VH DNA sequence database. BM-PC, VH genes from the bone marrow PCs; PBC, VH genes from peripheral B-cells. Percentages of spectral counts corresponding to oxidation products (oxidized L-Met) are shown in parentheses in the full CDRH3 sequences column. iCDRH3 peptides detected in the affinity chromatography eluent are marked as “E,” in the wash buffer as “W,” or in the flow-through as “F.”
*VH synthesized for phage panning and binding validation.
Peptides identified by MS analysis were classified according to their cooccurrence with CDRH3 sequences in the V gene DNA sequence database. Peptides that uniquely identified specific CDRH3s were defined as informative of CDRH3 (iCDRH3) peptides; in contrast, peptides mapping to multiple CDRH3 sequences were defined as noninformative (niCDRH3). iCDRH3 peptides were considered antigen-specific if the frequency of spectral counts in the affinity chromatography elution fraction was 10-fold greater than in the combined wash and flow-through fractions. More than three-quarters of all peptides containing portions of a CDRH3 sequence were found to correspond to iCDRH3s (SI Appendix, Fig. S6) (75% and 85% of CDRH3-derived peptides in CCH and BSA rabbits, respectively). A small fraction of upstream peptides (i.e., from CDR1 or CDR2) mapping to nondegenerate V gene sequences (all with the same CDRH3) in the database were also identified, providing additional coverage and identification of antigen-specific clonotypes. By including the information deduced from these upstream iCDRH3 peptides, the number of antigen-specific iCDRH3s in CCH rabbit serum increased from 29 to 34. An additional 25 peptides defining 83 full-length V-gene sequences were detected in the elution fraction but were overwhelmingly present in the wash and flow-through fractions (elution frequency:flowthrough + wash frequency ratio <10), indicative of very weakly binding/low-specificity and very low abundance antibodies (Fig. 3A; also SI Appendix, Fig. S7 for the BSA-specific iCDRH3 peptides). SI Appendix, Fig. S8 documents the high reproducibility among technical replicates. V-family and J-family gene use of full VH sequences corresponding to identified iCDRH3 peptides were found to be consistent with the germ-line gene use data obtained from 454 sequencing (Fig. 2B) and BSA rabbits (SI Appendix, Fig. S1; also SI Appendix, Fig. S9 for CDRH3 statistics).
Fig. 3.
Identified iCDRH3 peptides from affinity chromatography and alignment of corresponding CDRH3s. (A) Histogram showing frequencies of identified informative peptides corresponding to a unique clonotype (V genes with same VH, JH CDRH3 sequence and 80% homology in the CDRH3) in the antigen-affinity chromatography elution, flow-through, and wash fractions. (Inset) Magnified histogram of the top 15 highest count unique peptides detected in the antigen-affinity chromatography elution, flow-through, and wash fractions. Peptide IDs are ranked by relative abundance in elution. Identified peptides in the affinity chromatography elution fraction that are found overwhelmingly in the flow-through and wash buffer fractions likely correspond to antibodies that bind antigen very weakly or nonspecifically. (B) Pairwise alignment of CCH-immunized rabbit CDRH3s in the antigen-specific serum IgG repertoire and observed exclusively in the affinity chromatography elution. The dendrogram shows hierarchical clustering (based on pairwise sequence alignments at the amino acid level) of CDRH3 sequences detected in the elution at >10-fold higher number of counts relative to the affinity chromatography wash and flow-through. (Numbered sequences represent VH synthesized for binding validation.)
In total, our analysis of the CCH rabbit revealed that the antigen-specific polyclonal response is composed of ∼34 IgG antibodies that are classified into 30 different clonotypes [same V and J, same CDRH3 length, 80% aa homology (14, 15)]; 4 of the 34 antibodies that constitute the majority of the antigen-specific IgG repertoire in this animal differed by only 1–3 aa and therefore corresponded to clonally related antibodies (Fig. 3B; also SI Appendix, Fig. S10 for BSA). The top 12 iCDRH3 peptides accounted for ∼60% of all of the antigen-specific peptide counts, suggesting that the serum response was dominated by yet a smaller set of antibodies (sequence logo shown in SI Appendix, Fig. S11). The BSA rabbit data revealed a more restricted response comprising 18 distinct antibodies, as shown in SI Appendix, Fig. S10 and Table S3.
Comparison of the composition of the serum IgG response with the V gene repertoires obtained by NextGen sequencing of B cells in different compartments can provide useful insights on the dynamics of the humoral response. For example, in the CCH rabbit, 18 of 34 of the VH sequences encoding the serum antibody repertoire correspond to BM-PC sequences in the sequencing database (database comprising sequences from BM and PBCs as detailed in SI Appendix, Table S2). The remainder (16 of 34) of the identified serum antibody repertoire map to PBC sequences in the database. Thus, 7 d after boost immunization nearly half of the serum antibodies seem to be expressed predominantly by plasma cells that had migrated into the bone marrow.
Construction and Characterization of Serum MAbs.
To evaluate whether the identified VH genes encoded proteins that recognize the antigen, it was first necessary to identify the VL domains to which they pair. The in vivo VH:VL pairing problem cannot be universally solved (i.e., for all identified VH sequences) by proteomic approaches for two reasons: (i) the abundances of VH and VL chains do not correlate because an excess of VL chains are secreted in the serum, and VH chains can pair with more than one VL; and (ii) because of the lower sequence complexity of VL chains relative to VH, higher proportions of VL peptides share partial sequence identity, resulting in increased ambiguity in PSMs and peptide-sequence mappings. Hence, the proteomic problem of distinguishing false-positive identifications increases significantly, and the confidence in the analysis is weak.
We therefore addressed the VH:VL pairing problem by synthesizing select VH genes and then performing two to three rounds of phage panning of scFv libraries constructed with amplified VL cDNA (i.e., a VL chain shuffled library for each selected VH). Phage panning of a fixed VH with a library of VL genes is an established method for identifying functional pairs that bind antigen with high affinity (26). The VH genes corresponding to seven of the most abundant proteomically identified iCDRH3s were synthesized by automated DNA synthesis (18). In instances where more than one full-length VH gene in the database corresponded to an iCDRH3 (owing to additional somatic mutations within the V gene), the most common somatic variant was selected for synthesis. The libraries were confirmed to be of sufficient size to cover the entire VL gene repertoire deduced from DNA sequencing, as estimated by rarefaction analysis (SI Appendix, Fig. S12).
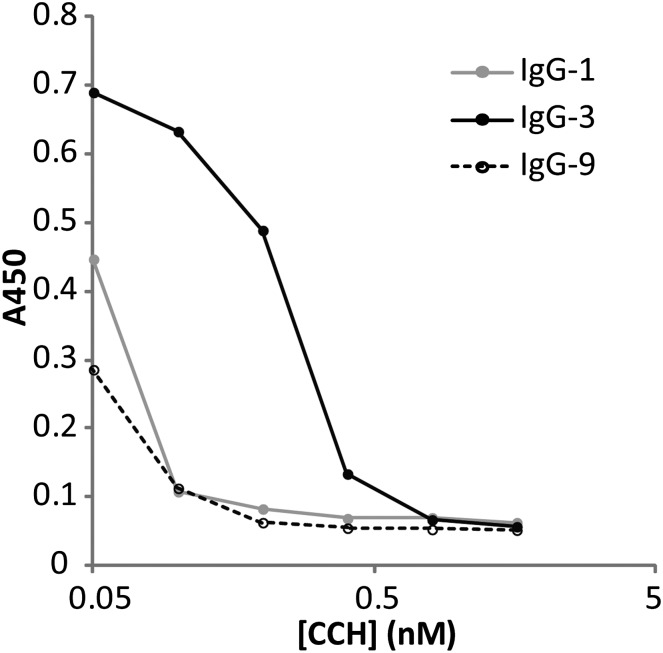
For the antigen-specific full-length scFvs isolated by panning (SI Appendix, Table S4), DNA sequencing revealed that the synthetic VH genes paired with one or, in the case of VH gene 6, two clonally related VL domains (SI Appendix, Fig. S13). The VH and VL genes were inserted into vectors encoding rabbit H and L chains, respectively, and cotransfected into HEK293 cells to produce the respective IgGs. The recombinant antibodies were found to display subnanomolar KD for the antigen by competitive ELISA (Fig. 4) and shown to be effective for immunoprecipitation of CCH from mock mixtures (SI Appendix, Fig. S14). Certain proteomically identified VH genes (2, 5, 7) did not yield specific binders by phage panning. This is not surprising, because it is well established that recombinant rabbit antibody fragments are particularly difficult to express in bacteria (27). Panning phage display of libraries using synthetic VH genes from the BSA library also showed significant enrichment after two rounds.
Fig. 4.
Competitive ELISA of full-length IgG containing the VH genes corresponding to selected abundant iCDRH3s (Table 1) identified in the CCH immunized rabbit. Antibodies identified using the proposed methodology (Fig. 1) were shown to display subnanomolar affinities. The expression yield of the rabbit IgG-6 antibody in HEK-293 cells was too low for accurate quantitative affinity measurements.
Discussion
We report a general strategy for the molecular deconvolution of the monoclonal antibodies that constitute the polyclonal response to antigen. We found that the serum IgG response in an immunized rabbit is oligoclonal, comprising 34 individual antibodies with high antigen selectivity that grouped into 30 distinct clonotypes of antigen-specific antibodies likely to have been derived from different progenitor B cells (SI Appendix, Fig. S15). An unimmunized animal that fortuitously had a titer to BSA contained 18 antigen-specific antibodies (12 clonotypes).
The first step in the analysis pipeline involved the determination of the V gene repertoire using NextGen sequencing. In the rabbit we find that the class-switched repertoire is dominated by the use of two to three germ-line VH families and by two germ-line JH families [J4 and J2 in both CCH and BSA rabbits (Fig. 2B and SI Appendix, Fig. S1)]. The analysis of the V gene repertoire further revealed that a subpopulation of rabbit V genes contains a large number of amino acid substitutions relative to the germline, likely a consequence of the gene conversion processes that occur during V gene diversification in rabbits. The deconvolution of the serum IgGs was made possible by LC/MS-MS shotgun proteomic analysis using an individually derived B-cell V gene sequence database, combined with strict filtering criteria for the confident identification of peptide sequences, including the use of a high mass accuracy filter (≤1.5 ppm). Although we have provided evidence that our analysis correctly captures the key features of the serological IgG repertoire, we do acknowledge that as a method that relies on LC-MS, it is subject to the experimental constraints of shotgun proteomic analyses (28).
Collectively, the identification of the repertoire of antigen-specific antibodies in serum leads to several interesting observations. First, the serum response in these animals seemed to be oligoclonal (i.e., neither is it highly diverse nor does it comprise only of a handful different antibodies). Second, VH use in the circulating antibodies was entirely consistent with the VH gene repertoire in the animal as determined by NextGen sequencing, highlighting the significance of the cellular repertoire in shaping humoral immunity (Fig. 2B and SI Appendix, Fig. S1). Third, it is interesting that most of the antigen-specific VH clonotypes identified proteomically correspond to a single V gene or at most to only a few somatic variants. However, this may not always be the case, especially when antibodies are generated in response to persistent or recurrent infections (29–32). In those instances, detection of peptides from CDR1 and CDR2 might be used to identify the dominant somatic variant(s) in serum. Fourth, as expected, many of the proteomically identified CDRH3s corresponded to VH genes isolated from BM-PCs. Approximately half of the iCDRH3s, however, were found to map only to PBCs and may be derived from recently activated plasmablasts in transit to the bone marrow. We note that because the formation of plasmablasts in the course of B-cell expansion is a consequence of asymmetric division that should also give rise to B memory cells, highly exhaustive DNA sequencing of the peripheral B memory V gene repertoire should be sufficient to yield all of the Ig sequences found in circulation at steady state (33). Fifth, the CCH rabbit data revealed that two of the top ranked iCDRH3s in terms of spectral counts display evidence of oxidative modifications, as shown in Table 1. Although it is known that amino acid oxidation can occur during sample preparation, it is tempting to speculate that the oxidative modification of certain circulating IgGs may have resulted from in vivo posttranslational modifications rather than processing artifacts, because plasma cells experience high levels of oxidative stress (34), and the t1/2 of circulating Igs in serum is >1 wk, resulting in extensive exposure of antibodies to oxidizing conditions. L-Methionine oxidation has also been observed repeatedly in recombinantly expressed therapeutic antibodies from CHO cells (35, 36). Additional studies will be needed to determine the extent of L-Methionine oxidation during sample processing and the frequency of in vivo modification.
At present the molecular deconvolution of antigen-specific serum antibodies requires a sample size of ∼3–5 mL of whole blood, an amount that is easily obtainable from most laboratory animals down to rats and also from humans. With further advances in the sensitivity of MS, it may prove possible to also analyze the Ig serum composition from a single mouse, including genetically homogenous transgenic animals displaying well-characterized defects in B-cell development. Importantly, the approach we have developed may be extended to the analysis of the serological response in humans after vaccination or related to pathologic states.
Materials and Methods
All materials used in this study, including vendor source are provided in SI Appendix, SI Materials and Methods.
Rabbit immunization, serum IgG isolation and sample preparation for LC-MS/MS measurements, proteomics data analysis, and all related computational analyses are described in SI Appendix, SI Materials and Methods.
Validation and characterization of proteomically identified serum antibodies including synthetic gene synthesis, recombinant IgG cloning, and ELISA methodologies are provided in detail in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Bob Glass for assistance with rabbit immunization and bone marrow isolation, Dr. Sai Reddy for help with flow cytometry, Dr. J. Borrok for initial experiments, Dr. Scott Hunicke-Smith for assistance with Next-Gen DNA sequencing, Constantine Chrysostomou for assistance in bioinformatics analysis, Chhaya Das for recombinant IgG expression, Dr. Greg Ippolito for reading the manuscript, Prof. Itai Benhar for important input and comments on the manuscript, and Prof. Brent L. Iverson for useful discussions. Funding for this work was provided by the Clayton Foundation (G.G.), Welch Foundation Grant F1515 (to E. M. Marcotte), Defense Advanced Research Projects Agency (G.G. and A.D.E.), and National Institutes of Health (NIH) Grants 5 RC1DA028779 (to G.G. via a subcontract from University of Chicago) and GM 076536 (to E. M. Marcotte). J.J.L. was supported by a postdoctoral fellowship by Cancer Prevention and Research Institute of Texas. The Linear Trap Quadrupole (LTQ) Orbitrap Velos MS was purchased with generous support by the NIH Western Research Center of Excellent in Biodefense (NIH Grant 5U54AI057156) and the Texas Institute for Drug and Diagnostics Development (TI-3D).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213737110/-/DCSupplemental.
References
- 1.Browning CH. Emil Behring and Paul Ehrlich: Their contributions to science. Nature. 1955;175(4457):570–575. doi: 10.1038/175570a0. [DOI] [PubMed] [Google Scholar]
- 2.Kantha SS. A centennial review; the 1890 tetanus antitoxin paper of von Behring and Kitasato and the related developments. Keio J Med. 1991;40(1):35–39. doi: 10.2302/kjm.40.35. [DOI] [PubMed] [Google Scholar]
- 3.Radbruch A, et al. Competence and competition: The challenge of becoming a long-lived plasma cell. Nat Rev Immunol. 2006;6(10):741–750. doi: 10.1038/nri1886. [DOI] [PubMed] [Google Scholar]
- 4.Scheid JF, et al. A method for identification of HIV gp140 binding memory B cells in human blood. J Immunol Methods. 2009;343(2):65–67. doi: 10.1016/j.jim.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453(7195):667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dekker LJ, et al. An antibody-based biomarker discovery method by mass spectrometry sequencing of complementarity determining regions. Anal Bioanal Chem. 2011;399(3):1081–1091. doi: 10.1007/s00216-010-4361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Costa D, et al. Sequencing and quantifying IgG fragments and antigen-binding regions by mass spectrometry. J Proteome Res. 2010;9(6):2937–2945. doi: 10.1021/pr901114w. [DOI] [PubMed] [Google Scholar]
- 8.Bandeira N, Pham V, Pevzner P, Arnott D, Lill JR. Automated de novo protein sequencing of monoclonal antibodies. Nat Biotechnol. 2008;26(12):1336–1338. doi: 10.1038/nbt1208-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung WC, et al. A proteomics approach for the identification and cloning of monoclonal antibodies from serum. Nat Biotechnol. 2012;30(5):447–452. doi: 10.1038/nbt.2167. [DOI] [PubMed] [Google Scholar]
- 10.Reddy S, et al. Rapid Isolation of Monoclonal Antibodies from Animals. Patent 20110312505 (Filed May 17, 2011)
- 11.Lindop R, et al. Molecular signature of a public clonotypic autoantibody in primary Sjögren’s syndrome: A “forbidden” clone in systemic autoimmunity. Arthritis Rheum. 2011;63(11):3477–3486. doi: 10.1002/art.30566. [DOI] [PubMed] [Google Scholar]
- 12.Sato S, et al. Proteomics-directed cloning of circulating antiviral human monoclonal antibodies. Nat Biotechnol. 2012;30(11):1039–1043. doi: 10.1038/nbt.2406. [DOI] [PubMed] [Google Scholar]
- 13.Murphy K, Travers P, Walport M, editors. Janeway’s Immunobiology. New York: Garland Science; 2007. [Google Scholar]
- 14.Poulsen TR, Jensen A, Haurum JS, Andersen PS. Limits for antibody affinity maturation and repertoire diversification in hypervaccinated humans. J Immunol. 2011;187(8):4229–4235. doi: 10.4049/jimmunol.1000928. [DOI] [PubMed] [Google Scholar]
- 15.Moody MA, et al. H3N2 influenza infection elicits more cross-reactive and less clonally expanded anti-hemagglutinin antibodies than influenza vaccination. PLoS ONE. 2011;6(10):e25797. doi: 10.1371/journal.pone.0025797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rader C, et al. The rabbit antibody repertoire as a novel source for the generation of therapeutic human antibodies. J Biol Chem. 2000;275(18):13668–13676. doi: 10.1074/jbc.275.18.13668. [DOI] [PubMed] [Google Scholar]
- 18.Reddy ST, et al. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat Biotechnol. 2010;28(9):965–969. doi: 10.1038/nbt.1673. [DOI] [PubMed] [Google Scholar]
- 19.Knight KL. Restricted VH gene usage and generation of antibody diversity in rabbit. Annu Rev Immunol. 1992;10:593–616. doi: 10.1146/annurev.iy.10.040192.003113. [DOI] [PubMed] [Google Scholar]
- 20.Knight KL, Becker RS. Molecular basis of the allelic inheritance of rabbit immunoglobulin VH allotypes: Implications for the generation of antibody diversity. Cell. 1990;60(6):963–970. doi: 10.1016/0092-8674(90)90344-e. [DOI] [PubMed] [Google Scholar]
- 21.Wu TT, Johnson G, Kabat EA. Length distribution of CDRH3 in antibodies. Proteins. 1993;16(1):1–7. doi: 10.1002/prot.340160102. [DOI] [PubMed] [Google Scholar]
- 22.Becker RS, Knight KL. Somatic diversification of immunoglobulin heavy chain VDJ genes: Evidence for somatic gene conversion in rabbits. Cell. 1990;63(5):987–997. doi: 10.1016/0092-8674(90)90502-6. [DOI] [PubMed] [Google Scholar]
- 23.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass-spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 24.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 25.Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4(11):923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 26.Marks JD, et al. By-passing immunization: Building high affinity human antibodies by chain shuffling. Biotechnology (N Y) 1992;10(7):779–783. doi: 10.1038/nbt0792-779. [DOI] [PubMed] [Google Scholar]
- 27.Popkov M, et al. Rabbit immune repertoires as sources for therapeutic monoclonal antibodies: The impact of kappa allotype-correlated variation in cysteine content on antibody libraries selected by phage display. J Mol Biol. 2003;325(2):325–335. doi: 10.1016/s0022-2836(02)01232-9. [DOI] [PubMed] [Google Scholar]
- 28.Mallick P, Kuster B. Proteomics: A pragmatic perspective. Nat Biotechnol. 2010;28(7):695–709. doi: 10.1038/nbt.1658. [DOI] [PubMed] [Google Scholar]
- 29.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333(6044):850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 30.Wu X, et al. NISC Comparative Sequencing Program Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333(6049):1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333(6049):1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker LM, et al. Protocol G Principal Investigators Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477(7365):466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barnett BE, et al. Asymmetric B cell division in the germinal center reaction. Science. 2012;335(6066):342–344. doi: 10.1126/science.1213495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slifka MK, Antia R, Whitmire JK, Ahmed R. Humoral immunity due to long-lived plasma cells. Immunity. 1998;8(3):363–372. doi: 10.1016/s1074-7613(00)80541-5. [DOI] [PubMed] [Google Scholar]
- 35.Boyd D, Kaschak T, Yan B. HIC resolution of an IgG1 with an oxidized Trp in a complementarity determining region. J Chromatogr B Analyt Technol Biomed Life Sci. 2011;879(13-14):955–960. doi: 10.1016/j.jchromb.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Houde D, Peng Y, Berkowitz SA, Engen JR. Post-translational modifications differentially affect IgG1 conformation and receptor binding. Mol Cell Proteomics. 2010;9(8):1716–1728. doi: 10.1074/mcp.M900540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.