Abstract
Photorespiratory carbon flux reaches up to a third of photosynthetic flux, thus contributes massively to the global carbon cycle. The pathway recycles glycolate-2-phosphate, the most abundant byproduct of RubisCO reactions. This oxygenation reaction of RubisCO and subsequent photorespiration significantly limit the biomass gains of many crop plants. Although photorespiration is a compartmentalized process with enzymatic reactions in the chloroplast, the peroxisomes, the mitochondria, and the cytosol, no transporter required for the core photorespiratory cycle has been identified at the molecular level to date. Using transcript coexpression analyses, we identified Plastidal glycolate glycerate translocator 1 (PLGG1) as a candidate core photorespiratory transporter. Related genes are encoded in the genomes of archaea, bacteria, fungi, and all Archaeplastida and have previously been associated with a function in programmed cell-death. A mutant deficient in PLGG1 shows WT-like growth only in an elevated carbon dioxide atmosphere. The mutant accumulates glycolate and glycerate, leading to the hypothesis that PLGG1 is a glycolate/glycerate transporter. This hypothesis was tested and supported by in vivo and in vitro transport assays and 18O2-metabolic flux profiling. Our results indicate that PLGG1 is the chloroplastidic glycolate/glycerate transporter, which is required for the function of the photorespiratory cycle. Identification of the PLGG1 transport function will facilitate unraveling the role of similar proteins in bacteria, archaea, and fungi in the future.
Keywords: metabolite transport, photosynthesis
Carbon flux through photorespiration is second in magnitude only to photosynthesis, and thus this metabolic pathway constitutes a major component of the global carbon cycle. Photorespiration is essential because the enzyme RubisCO, which assimilates CO2 from the atmosphere into biomass, also catalyzes a futile reaction, the oxygenation of the CO2 acceptor ribulose 1,5-bisphosphate (RuBP). This latter reaction leads to the formation of the toxic metabolite 2-phosphoglycolate (2-PG), which is detoxified and recycled to RuBP by a complex metabolic pathway called photorespiration. Photorespiration became an essential requirement for photosynthesis after the carbon composition of the atmosphere changed to an oxygen-rich atmosphere, as a consequence of oxygenic photosynthesis by cyanobacteria, algae, and plants. Large gains in photosynthetic efficiency can be achieved if photorespiration is suppressed by enriching CO2 in the vicinity of RubisCO. For example, plants carrying a metabolic bypass for photorespiration indeed produce more biomass (1, 2), providing a promising approach for increasing the productivity of some the most important crop plants, such as rice (Oryza sativa) or wheat (Triticum aestivum) that have to cope with high rates of photorespiration.
In plants, the photorespiratory cycle is a highly compartmentalized process with enzymatic reactions in chloroplasts, peroxisomes, and mitochondria as well as in the cytosol. In the chloroplast stroma (2-PG) resulting from the RubisCO oxygenation reaction is dephosphorylated to glycolate by 2-phosphoglycolate phosphatase (PGLP). Glycolate is exported from the chloroplasts to the peroxisomes, where it is oxidized to glyoxylate by glycolate oxidase (GOX) and transaminated to glycine by Ser:glyoxylate and Glu:glyoxylate aminotransferase (SGT and GGT, respectively). Glycine leaves the peroxisomes and enters the mitochondria, where two molecules of glycine are deaminated and decarboxylated by the glycine decarboxylase complex (GDC) and serine hydroxymethyltransferase (SHMT) to form one molecule each of serine, ammonia, and carbon dioxide. Serine is exported from the mitochondria to the peroxisomes, where it is predominantly converted to glycerate by SGT and hydroxypyruvate reductase (HPR). Glycerate leaves the peroxisomes and is taken up into the chloroplast, where it is phosphorylated by glycerate kinase (GLYK) to yield 3-phosphoglycerate (3-PGA). In essence, one of the four carbon atoms contained in two molecules of 2-PG entering the pathway is lost as CO2, whereas three are shuttled back into the Calvin-Benson cycle. Thus, photorespiration constitutes a metabolic repair cycle that is required to detoxify 2-PG, at the expense of energy and loss of carbon dioxide and ammonia. Most of the components of the photorespiratory cycle have been identified by forward genetic analyses starting in the 1980s. For example the first enzyme of the photorespiratory cycle PGLP (3) or the mitochondrial SHMT (4, 5) have been discovered by this approach. The hallmark of photorespiratory mutants is reduced or no growth under ambient air, but normal WT-like growth under CO2-enriched air. Only peroxisomal HPR mutants show no typical photorespiratory growth limitation in ambient air due to an alternative cytosolic pathway that suppresses the effect of the mutation (6). Photorespiratory mutants additionally contain elevated pools of photorespiratory metabolites.
The photorespiratory pathway intermediates have to be transported across multiple cellular membranes at high flux rates, which is facilitated by metabolite transporters residing in the organellar membranes. In total, two molecules of glycolate and one molecule of glycerate cross the chloroplast envelope and the peroxisomal membrane in one turn of the photorespiratory cycle. Two molecules of glycine and one molecule of serine cross the peroxisomal membrane and the mitochondrial envelope. These six transport steps in the carbon cycle of photorespiration together with the transport steps required for the nitrogen cycle result in 18 postulated transport processes (7). Although all soluble enzymes of the photorespiratory cycle are identified on the molecular level of the transporters, only the chloroplastidic DiT1 and DiT2 that are involved in nitrogen recycling during photorespiration are identified on the molecular level (8–11). Transporters required for the photorespiratory carbon cycle are still unknown, which represents a major gap in the knowledge about this important pathway.
Although most enzymes required for photorespiration have been identified by forward genetics, only one transporter associated with photorespiration has been found by this approach to date (8, 10). An alternative means for the identification of candidate genes is transcript coexpression analysis, which was developed by Eisen et al. (12) using microarray data for yeast. Coexpression analysis is based on the assumption that genes that function in the same pathway tend to display similar expression patterns. Hence, unknown genes that are coregulated with genes of a particular metabolic pathway are hypothesized to be involved in the same biological process. By this method, a wide range of genes were functionally characterized in yeast (13) or humans (14). In Arabidopsis thaliana, a coexpression analysis in combination with reverse genetics has been a successful strategy to find genes involved in, e.g., flavonoid metabolism (15), cellulose (16) and lignin biosynthesis (17, 18), and aliphatic glucosinolate biosynthesis (19).
In this work we have used coexpression analysis in combination with reverse genetics to identify a transporter involved in the photorespiratory cycle. Coexpression analysis revealed that the genes encoding photorespiratory enzymes are coexpressed with each other and with the candidate transporter Plastidal glycolate glycerate translocator 1 (PLGG1), which was previously identified in proteomics of chloroplasts (20) and was assumed to be involved in programmed cell death (21, 22). By analyzing an A. thaliana transferred DNA (T-DNA) knockout mutant deficient in PLGG1 (plgg1-1), we could demonstrate that the mutant has a strong photorespiratory phenotype and is no longer able to transport glycolate and glycerate across the chloroplast envelope. The chloroplastidic glycolate/glycerate transporter PLGG1 thus defines a unique class of metabolite transporters that is present in Archaeplastida, fungi, bacteria, and archaea.
Results
PLGG1 Is Coexpressed with Known Photorespiratory Enzymes.
To identify transporters involved in the photorespiratory pathway, a coexpression analysis with 11 known photorespiratory pathway enzymes was conducted by using publicly available coexpression databases [AtGenExpress and NASC Array, CSB.DB (http://csbdb.mpimp-golm.mpg.de)]. Photorespiratory genes were significantly coexpressed with ∼100–150 genes, depending on the coexpression matrix used. These included genes involved in photorespiration, photosynthesis, and chloroplast function (23). The Spearman and Pearson coexpression coefficients for 10 photorespiratory genes were >0.9 for 73 of 98 tested cases (Table 1). PGLP is coexpressed with all genes involved in the pathway, with the exception of GLYK (Table 1). For some of the enzymes, several isoforms of the enzymes exist. In these cases, only one distinct isoform was found to be coexpressed with other photorespiratory genes. For example, of the five isoforms encoding enzymes for glycolate oxidation, only AtGOX1 was strongly correlated (AtGOX2 was indistinguishable because of the probe on the ATH1 chip) (Table 1). Likewise, only GGT1 (but not GGT2), only one isoform each of the subunits of GDC, and only the SHM1 isoform were coexpressed in the context of photorespiration (Table 1). Only GLYK was not coexpressed with the other genes in the pathway (Table1).
Table 1.
Coexpression Spearman coefficients for genes involved in photorespiration and for the candidate gene PLGG1
| Enzyme | AGI | Abbrev. | PGLP1 | GOX1 | AGT1 | GGT1 | GLDP1 | GLDH1 | GLDH3 | GLDT1 | SHM1 | HPR1 | GlyK | PLGG1 |
| 2-PG phosphatase | At5g36790 | PGLP1 | 1.00 | — | — | — | — | — | — | — | — | — | — | — |
| GOX | At3g14420 | GOX1 | 0.93 | 1.00 | — | — | — | — | — | — | — | — | — | — |
| SGT | At2g13360 | AGT1 | 0.91 | 0.95 | 1.00 | — | — | — | — | — | — | — | — | — |
| GGT | At1g23310 | GGT1 | 0.92 | 0.96 | 0.96 | 1.00 | — | — | — | — | — | — | — | — |
| GDC P-protein | At4g33010 | GLDP1 | 0.93 | 0.95 | 0.93 | 0.94 | 1.00 | — | — | — | — | — | — | — |
| GDC H-protein | At2g35370 | GLDH1 | 0.93 | 0.92 | <0.9 | <0.9 | <0.9 | 1.00 | — | — | — | — | — | — |
| GDC H-protein | At1g32470 | GLDH3 | 0.91 | <0.9 | <0.9 | <0.9 | <0.9 | 0.94 | 1.00 | — | — | — | — | — |
| GDC T-protein | At1g11860 | GLDT1 | 0.92 | 0.92 | <0.9 | <0.9 | <0.9 | 0.92 | 0.95 | 1.00 | — | — | — | — |
| SHMT | At4g37930 | SHM1 | 0.93 | 0.96 | 0.94 | 0.96 | 0.95 | 0.91 | <0.9 | 1.00 | 1.00 | — | — | — |
| HPRs | At1g68010 | HPR1 | 0.93 | 0.97 | 0.96 | 0.97 | 0.94 | <0.9 | <0.9 | 0.91 | 0.96 | 1.00 | — | — |
| GLYK | At1g80380 | GlyK | <0.9 | <0.9 | <0.9 | <0.9 | <0.9 | <0.9 | <0.9 | <0.9 | <0.9 | <0.9 | 1.00 | — |
| PLGG1 | At1g32080 | PLGG1 | 0.93 | 0.9 | 0.9 | 0.9 | <0.9 | 0.93 | 0.93 | 0.93 | 0.91 | 0.93 | <0.9 | 1.00 |
Only the isoform with the best correlation coefficient is shown; correlations with P < 0.1 are in boldface type. Abbrev., abbreviation; —, reciprocal coexpression value.
To identify unknown transporters in the photorespiratory cycle, proteins of unknown function with a coexpression coefficient of at least 0.9 with the majority of photorespiratory genes were tested for the presence of predicted membrane spanning helices. One protein of unknown function previously identified by proteomics and named PLGG1 (20) was both coexpressed with genes involved in photorespiration and had 12 predicted membrane spanning helices [ARAMEMNON (24)]. We hypothesized that PLGG1 is a transporter involved in photorespiration.
plgg1-1 Insertion Mutant Exhibits a Photorespiratory Phenotype.
To test the hypothesis that PLGG1 is involved in photorespiration, we isolated an A. thaliana T-DNA insertional mutant. The plgg1-1 mutant carries the T-DNA insertion in the first intron of the respective gene At1g32080 (Fig. S1). PLGG1 cDNA can only be detected in WT, but not in plgg1-1 mutant, plants (Fig. S1). Under ambient CO2 conditions (380 ppm) the plgg1-1 mutant developed yellow and bleached lesions on the leaf lamina, but not along the veins (Fig. 1A). This phenotype was suppressed under high CO2 (3,000 ppm) conditions (Fig. 1B). The phenotype was complemented when PLGG1 was expressed under its own promoter in plgg1-1 plants (Fig. 1C). We conclude that plgg1-1 displays a bleached leaf phenotype in ambient CO2 that can be suppressed by elevated CO2, which is consistent with a function of PLGG1 in photorespiration.
Fig. 1.
Photorespiratory phenotype of plgg1-1 plants. (A, C, and D) plgg1-1 (A), WT (C), and comp (D) plants grown in high CO2 for 4 wk and shifted to ambient CO2 for 1 wk. (B) PLGG1-1 plant grown in high CO2 for 5 wk.
Photorespiratory Metabolites Accumulate in plgg1-1 Plants.
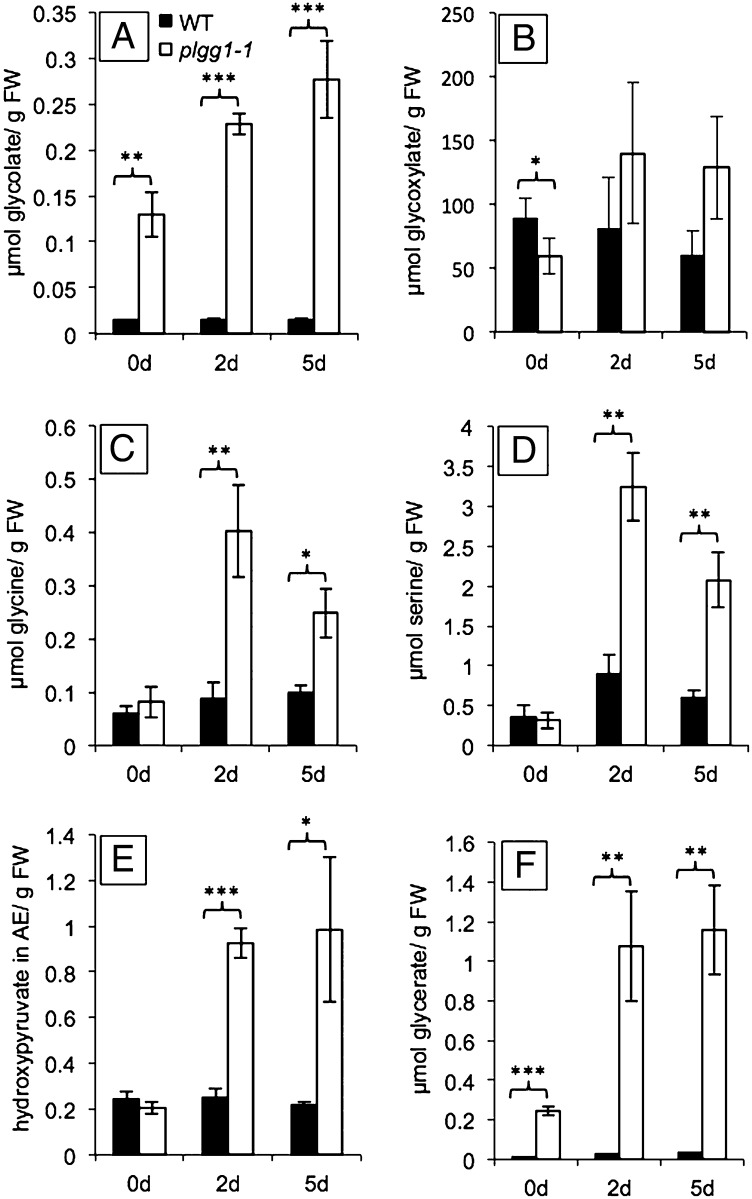
To determine the position of PLGG1 in the photorespiratory pathway, we analyzed metabolites in WT and plgg1-1 plants under high and ambient CO2 conditions. The chosen time points represent metabolite levels under: (i) high CO2 (0 d), conditions under which the rate of photosynthesis was nearly identical in WT and mutant (14.56 ± 0.42 and 13.87 ± 0.23 µmol of CO2 per m2⋅s−1, respectively; Table S1); (ii) after shift to ambient CO2 conditions of plants with no or very weak visible phenotype but already decreased photosynthetic capacity in the mutant (2 d; 6.7 ± 0.76 µmol of CO2 per m2⋅s−1); and (iii) after a pronounced visible phenotype and low rates of photosynthesis were observed in the mutant (5 d; 4.62 ± 0.47 µmol of CO2 per m2⋅s−1). The largest and most significant differences between plgg1-1 mutant plants and WT were found for glycolate, glycine, serine, hydroxypyruvate, and glycerate, which are all photorespiratory intermediates. Already in CO2-enriched air (0 d), glycolate and glycerate were significantly elevated in the mutant compared with WT (Fig. 2 A and F). This accumulation escalated when plants were shifted from elevated CO2 to ambient air. Glycine, serine, and hydroxypyruvate did not accumulate in mutant plants under high CO2, but when shifted to ambient air (Fig. 2 C–E). Steady-state glyoxylate levels in mutant plants did not differ significantly from those in WT. After shift from high CO2 to ambient CO2 conditions, only serine levels changed in WT plants (Fig. 2C).
Fig. 2.
Accumulation of photorespiratory metabolites in WT and plgg1-1 plants. WT and plgg1-1 plants were grown in 3,000-ppm CO2 conditions for 4 wk and then shifted to ambient air. Steady-state metabolite levels were measured for plants kept at high CO2 and after 2- and 5-d shift to ambient air. All values are measured in micromole per gram of fresh weight, except hydroxypyruvate, which is shown as arbitrary units. (A) Glycolate. (B) Glyoxylate. (C) Glycine. (D) Serine. (E) Hydroxypyruvate. (F) Glycerate. Error bars indicate SD; n = 3.
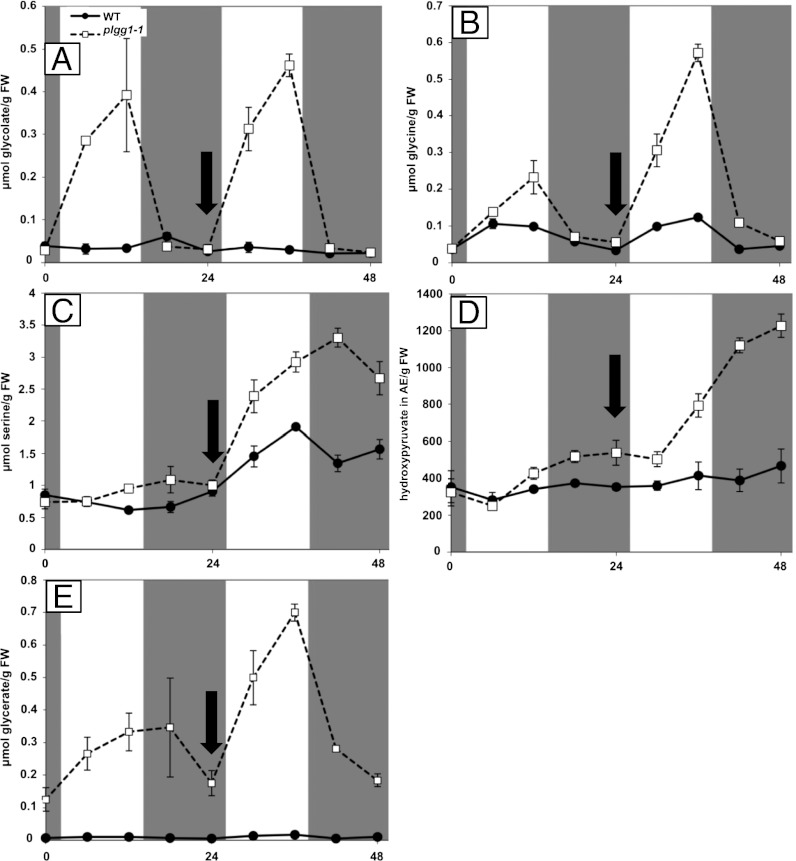
Because photorespiration occurs only during the day when RubisCO is active, we expected daytime-dependent metabolite accumulation patterns in the mutant. During the night in elevated CO2, in WT and plgg1-1 plants, photorespiratory metabolite levels with the exception of glycerate did not differ strongly (Fig. 3). During the day in elevated CO2, only glycerate and glycolate accumulated in a time-dependent manner (Fig. 3 A and E). After the shift to ambient air, all five metabolites showed a light-dependent accumulation that became more pronounced throughout the light period. During the night in ambient air, glycolate and glycine levels in plgg1-1 plants dropped to WT levels. Serine and hydroxypyruvate levels did not mirror the values during the night in CO2-enriched air, but stayed elevated (Fig. 3 C and D). The metabolite profile of the plgg1-1 mutant is consistent with a role of PLGG1 in photorespiration. The pronounced accumulation of glycolate and glycerate and the localization of this transporter at the chloroplast envelope membrane (ref. 25; Fig. S2) point to PLGG1 as a glycolate/glycerate transporter.
Fig. 3.
Time course of photorespiratory metabolite accumulation in WT and plgg1-1 plants. WT and plgg1-1 plants were grown in 3,000-ppm CO2 conditions for 4 wk and shifted to ambient air (300 ppm, at 24 h; indicated by black arrow). All values are measured in micromole per gram of fresh weight, except hydroxypyruvate, which is shown as arbitrary units. (A) Glycolate. (B) Glycine. (C) Serine. (D) Hydroxypyruvate. (E) Glycerate. Error bars indicate SD; n = 3.
Glycerate- and Light-Dependent O2 Evolution in Intact Chloroplasts.
Transport experiments with isolated chloroplasts showed that a single transporter transports glycolate and glycerate across the envelope (26, 27). Therefore, we expressed PLGG1 heterologously and measured active 14C-glycerate uptake in an in vitro uptake system. A Michaelis–Menten-type saturation kinetics of glycerate uptake was observed when liposomes were preloaded with glycerate or glycolate. However, a high background signal was detected in the liposome system due to high rates of unspecific diffusion of glycerate in the liposome system (Fig. S3). Therefore, to corroborate the result of the liposome uptake assays, we used an in vivo system that had been used successfully (26) to test whether the plgg1-1 mutant was affected in the transport of glycerate. Chloroplasts show light-dependent oxygen evolution in the presence of glycerate (26) because imported glycerate is converted by GLYK to 3-PGA, which is further reduced to triose phosphate. The reduction consumes NADPH, and the photosynthetic regeneration of NADPH produces oxygen. Hence, feeding of glycerate to isolated chloroplasts drives oxygen evolution (for a detailed scheme, see Fig. S4).
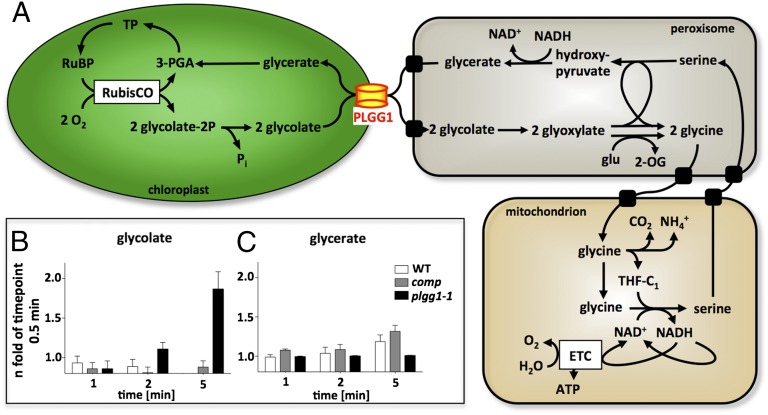
To test whether plgg1-1 chloroplasts were physiologically intact and transport-competent, 3-PGA–dependent oxygen evolution was measured. The rates of 3-PGA–dependent oxygen evolution observed for WT and complemented plgg1-1 mutant (comp) chloroplasts (20.32 µmol per mg of Chl per h) did not differ significantly from the rates observed for plgg1-1 chloroplasts (Table 2). With WT and comp chloroplasts, O2 evolution rates of 22.54 and 18.77 µmol per mg of Chl per h, respectively, were observed with 1 mM glycerate. The rates observed with plgg1-1 chloroplasts did not exceed the background signal and differed significantly (P < 0.0001) from the WT rates. Hence, chloroplasts of the plgg1-1 mutant do not display glycerate-dependent oxygen evolution, which supports PLGG1’s function as a glycerate transporter.
Table 2.
3-PGA–dependent and glycerate-dependent O2 evolution in isolated intact WT, plgg1-1, and comp chloroplasts
| Chloroplast | Glycerate | 3-PGA |
| WT | 22.54 (±1.11) | 22.12 (±3.58) |
| plgg1-1 | 1.61 (±1.72) | 19.91 (±1.72) |
| comp | 18.77 (±0.66) | 20.32 (±2.22) |
Values are in µmol per mg of Chl per h ± SD. n = 3.
Glycolate and Glycerate Flux Are Impaired in plgg1-1 Plants.
PLGG1’s function as a glycolate transporter was further tested by [18O]oxygen flux analysis. Transfer of label from glycolate to downstream metabolites would be impaired in plgg1-1 if PLGG1 were involved in catalyzing the efflux of glycolate from the chloroplast. To follow photorespiratory flux in plgg1-1 in relation to WT plants, we incubated plants in an [18O]oxygen atmosphere and followed the incorporation of the label into metabolites, a method that was used to identify metabolites and the kinetics of the photorespiratory cycle (28). The label was incorporated into glycolate within 5 s and into glycerate after 3 min (28). Therefore, we chose time points of 30 s and 1, 2, and 5 min for a kinetic analysis. All values were expressed relative to the initial label (30 s). For glycolate, the values observed with WT and comp plants were similar to those obtained by Berry et al. (ref. 28; Fig. 4). After 30-s exposure to 18O, the incorporation reached a plateau, and no further increase in label incorporation was detectable after 1 min (0.93-fold for WT and 0.86-fold for comp), 2 min (0.88-fold for WT and 0.81-fold for comp), and 5 min (0.88-fold for WT and 0.88-fold for comp), respectively, indicating that a steady state was achieved. For plgg1-1 plants, no increase in label incorporation into glycolate was obtained after 1 min compared with the value measured after 30 s. In contrast to WT, the label in glycolate continued to accumulate after 2 min (1.11-fold) and 5 min (1.86-fold) in the mutant, which indicates a slower removal of label from the glycolate pool. This finding was consistent with a reduced export of glycolate from the chloroplast. In WT and comp plants, the 18O2 label incorporation in glycerate increased slowly. This result was not the case for the mutant plants. Here, no increase in label incorporation into glycerate could be observed, with continuous low values for all time points. Again, these data indicate that the transfer of label from the glycolate to the glycerate pool is impaired, which can be explained by reduced export of glycolate from the chloroplast. Thus, plgg1-1 is impaired in the transport of both glycolate and glycerate.
Fig. 4.
Revised model of the photorespiratory cycle and 18O2 incorporation in glycolate and glycerate; (A) The photorespiratory cycle including the chloroplastidic glycolate/glycerate transporter PLGG1. (B and C) 18O2 incorporation into glycolate (B) and glycerate (C) was determined in WT, complemented mutants (comp), and plgg1-1 mutant plants. Plants were grown in ambient air for 4 wk and incubated in 18O2 air for 0.5, 1, 2, and 5 min, and metabolite levels were analyzed by using GC/MS. Metabolite levels were normalized to the relative abundance after 0.5 min to calculate the enrichment of 18O2 incorporation in glycolate and glycerate. ETC, electron transport chain; 2-OG, 2-oxoglutarate; TP, triose phosphate. n = 3.
Discussion
Photorespiration is a highly compartmentalized pathway that requires multiple transmembrane transport steps across organellar membranes to enable the high flux of metabolites in this process. Despite their importance, no transporter involved in the carbon cycle of photorespiration has been identified to date. Here we report the identification of the chloroplastidic glycolate/glycerate transporter PLGG1 as a transporter of the photorespiratory carbon cycle.
Coexpression analysis showed that PLGG1 is coregulated with photorespiratory enzymes (Table 1), and an A. thaliana T-DNA insertional mutant deficient in plgg1-1 (Fig. S1) is only able to grow WT-like and exhibit WT-like rates of photosynthesis in elevated CO2 (Fig. 1 A and B and Table S1). In previous studies, this CO2-dependent phenotype was also observed for other photorespiratory mutants (4, 29–31). Those plants grew normally in CO2-enriched air but had reduced growth or were even inviable under ambient CO2. Compared with other photorespiratory mutants, the phenotype observed for plgg1-1 plants is relatively mild because plants can still grow—albeit more slowly and with visible symptoms (Fig. 1)—in ambient air, which is likely the reason it was not identified in previous photorespiratory mutant screens. Glycolate, a small organic acid, is probably able to diffuse out of chloroplasts, as do other small organic acids (32), once it accumulates to high levels (Fig. 2). In the absence of high photorespiratory flux during nighttime, the glycerate and glycolate pools approach WT levels (Fig. 3), which likely alleviated the photorespiratory symptoms in the plgg1-1 mutant. A shift in nighttime metabolism at least partially metabolized extraplastidial glycerate, likely toward serine synthesis, which explains why, during the night in ambient air, serine and hydroxypyruvate levels continued to rise, whereas glycerate levels fell but did not reach WT levels. Accumulation of the photorespiratory metabolites glycine, serine, and hydroxypyruvate is likely driven by feedback through cytoplasmatic and peroxisomal glycerate accumulation and was previously observed in other photorespiratory mutants with elevated glycerate levels (6, 30).
Glycerate and glycolate are transported across the chloroplast envelope in the photorespiratory cycle by the same transport protein (26). The localization of PLGG1 in the chloroplast envelope (Fig. S3), together with the metabolite accumulation patterns (Figs. 2 and 3), identify PLGG1 as the chloroplastidic glycolate/glycerate transporter that was characterized biochemically the 1980s and 1990s in McCarty’s laboratory (26, 27, 33–35). It was shown that the transport rates observed with isolated chloroplasts are sufficient to cope with the high photorespiratory carbon flux, that the transporter exhibits a proton/substrate symport activity, and that glycolate and glycerate are transported through the same transport protein.
To verify that PLGG1 is the glycolate/glycerate transporter, we expressed PLGG1 heterologously and measured 14C-glycerate/glycolate counterexchange activity in a reconstituted liposome assay. Although a saturable and preloading-dependent uptake kinetics could be observed, high rates of unspecific diffusion were also detected (Fig. S3). This high rate of diffusion is typical for small organic acids in artificial membrane systems and was detected before (27). We therefore next used an in vivo approach that is more suitable to verify glycolate/glycerate transport. To this end, we used an in vivo approach with isolated chloroplasts that was developed by Howitz and McCarty (26) and 18O2 flux analysis in plgg1-1 plants. WT chloroplasts evolved oxygen when provided with 3-PGA or glycerate, proving that they were transport-competent and biochemically active. PLGG1-1 chloroplasts were also transport-competent and biochemically active because they were capable of evolving oxygen when supplied with 3-PGA (Table 2). They were only unable to transport glycerate because no glycerate-dependent oxygen evolution was observed with plgg1-1 chloroplasts (Table 2). Flux analysis further supported PLGG1’s role as the glycolate/glycerate transporter because the 18O2 label accumulation in glycolate increased in the plgg1-1 plants over time, indicating that glycolate is trapped in the chloroplast and is not accessible to enzymes that process glycolate outside of the chloroplast (Fig. 3). In contrast, the label accumulation in glycolate in WT plants already reached a plateau after 30 s and did not increase, indicating that glycolate was processed outside of the chloroplast and could thus proceed in the photorespiratory cycle. In WT plants, label incorporation into glycerate increased over time because it takes a few minutes until turnover of the majority of the glycerate pool is achieved (28). In plgg1-1 plants, the transfer of the label from glycolate to other metabolites is mostly blocked, and thus no increase in label in the glycerate pool is detectable.
Two recent publications hypothesized that PLGG1 is involved in chloroplast development or functions against cell death (21, 22), but in neither publication was the molecular function of PLGG1 identified. The conclusions were drawn from the visible observations that true leaves develop chlorotic regions in which chloroplasts are destroyed. We demonstrate that elevated CO2 alleviates the symptoms in all developmental stages (Fig. 1 and Fig. S5), that the accumulation of photorespiratory metabolites predates the occurrence of visible symptoms (Fig. 2), and that PLGG1 is the chloroplast glycerate/glycolate carrier (Fig. 4 and Table 2). Because the phenotype can be suppressed with high CO2 (Fig. 1) and biochemically active chloroplasts can be isolated from plgg1-1 mutant plants grown in high CO2 (Table 2), we posit that the visible symptoms are due to the accumulation of toxic concentrations of glycolate and glycerate. Indeed, photorespiratory intermediates, including glycerate, can be toxic to chloroplasts at high concentrations (36), explaining chloroplast and cell disruption in the plgg1-1 mutant. Thus, the chloroplastidic glycolate/glycerate transporter PLGG1 defines a unique class of metabolite transporters, which is present in Archaeplastida, fungi, bacteria, and archaea (Fig. S6).
Materials and Methods
Plant Growth and Conditions.
A. thaliana ecotype Columbia (Col-0) was used as WT reference. The SALK line SALK_053469 (plgg1-1) was obtained from the NottinghamArabidopsis Stock Centre (37). Complemented plgg1-1 mutant plants (comp) were used as control for the SALK line. Unless stated otherwise, plants were grown in normal air (380 ppm CO2) and in air with elevated CO2 (3,000 ppm; high CO2) at a 12-h light/12-h dark cycle (22/18 °C) in growth chambers (150 µmol⋅m−2⋅s−1 light intensity) on soil (mixture of 1/4 Floraton and 3/4 Arabidopsis root substrate).
Isolation of the T-DNA Insertion Line.
PCR-based screening was used to isolate a homozygous T-DNA insertion line for Plgg1. Primers P1, P2, and P3 were used for the genomic DNA screening (for primer sequences, see Table S2). P1 and P2 for were used for amplification of the WT gene, and P1 and P3 were used for the T-DNA/gene junction. The effect of the T-DNA insertion on the amount of PLGG1 transcript amounts was tested by using quantitative PCR with cDNA of WT and plgg1-1 plants as template and primers P4 and P5 for amplification. As a positive control ACTIN7 (AtACT7, At5g09810) was amplified by using P6 and P7.
Statistical Analysis.
Curve fits and Student t tests were performed with PRISM 5.0a (GraphPad). Results were called extremely significant if P was <0.0001 and very significant if P was <0.001 and is indicated by three and two asterisks, respectively.
β-Glucuronidase Expression and Establishment of a Complementation Line.
To assess the expression profile of the PLGG1 gene, a 3.5-kb gDNA fragment upstream of the ATG start site including the first nine bases of the PLGG1 gene was amplified by using P8/P9 and cloned into vector pCAMBIA3301 for a C-terminal β-glucuronidase (GUS) fusion. For complementation analysis, a 9-kb gDNA fragment, including 3.5 kb upstream of the ATG-start site, the full genomic PLGG1 sequence, and a 0.5-kb 3′-UTR was amplified by using P10/P11 and cloned into vector pCAMBIA3301. A. thaliana plants were stably transformed by using the floral dip method (38). GUS staining of 2-wk-old seedlings was performed by using the method described in ref. 39.
Metabolite Extraction and Gas Chromatography–Time-of-Flight Mass Spectroscopy Analysis.
Methanolic extraction of leaf material was performed according to the method described by Fiehn et al. (40), and gas chromatography–time-of-flight mass spectroscopy (GC/MS) was performed according to Lee and Fiehn (41). Analysis of metabolites was performed by GC/MS (Agilent Technologies 5973). Results were analyzed by using the MassLynx software package supplied with the instrument (Waters). As an internal standard ribitol was added and relative metabolite levels were determined from the ratio of the area of each metabolite and the corresponding ribitol area. A detailed description of the methods is available in SI Materials and Methods.
Transient Expression of GFP Fusions in Tobacco Protoplast.
For localization studies a C-terminal GFP fusion construct was cloned by using Gateway vectors to insert PLGG1 into pMDC83 (42) via pDONR207 (Invitrogen; Gateway). For amplification primers P12/P13 were used. Vectors were transformed into Nicotiana benthamiana leaves via infection with Agrobacterium tumefaciens strain GV3101 (43, 44); protoplasts were isolated after 3 d, and localization was visualized by using a Zeiss laser scanning microscope 510 Meta as described in detail in Breuers et al. (25).
Chloroplast Isolation and Glycerate-Dependent Oxygen Evolution.
Chloroplasts were isolated according to the method described by Aronsson and Jarvis (45) using a two-step Percoll gradient, with the modification that plants were grown on soil for 3 wk in high CO2. Intactness of chloroplasts was determined by using the Hill reaction (46). The 3-PGA and glycerate-dependent oxygen evolution was measured according to the method described by Howitz and McCarty (26).
18O2 Feeding.
A. thaliana WT and plgg1-1 plants were fed with 18O2 according to the method described in ref. 28. Plants were grown on soil for 4 wk in ambient air in 8-h light/16-h dark cycle (20/16 °C). A single plant in a pot was placed in a 1-L plastic bag with a plastic seal (Paclan GmbH). Air was removed by vacuum and replaced by air-mixture of 0.03% CO2, 78.97% N2, and 21% 18O2. After 0.5, 1, 2, and 5 min, plants were freeze-quenched within <2 s in liquid nitrogen and processed for metabolite analysis. Metabolites were extracted by the method optimized for photorespiratory metabolites. A detailed description of the methods is available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank D. Weits and J. van Dongen for providing and helping with the gas mixer equipment.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. W.B.F. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215142110/-/DCSupplemental.
References
- 1.Kebeish R, et al. Chloroplastic photorespiratory bypass increases photosynthesis and biomass production in Arabidopsis thaliana. Nat Biotechnol. 2007;25(5):593–599. doi: 10.1038/nbt1299. [DOI] [PubMed] [Google Scholar]
- 2.Maier A, et al. Transgenic Introduction of a glycolate oxidative cycle into A. thaliana chloroplasts leads to growth improvement. Front Plant Sci. 2012;3:38. doi: 10.3389/fpls.2012.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Somerville CR, Ogren WL. Phosphoglycolate phosphatase-deficient mutant of Arabidopsis. Nature. 1979;280(5725):833–836. [Google Scholar]
- 4.Somerville CR, Ogren WL. Photorespiration-deficient mutants of Arabidopsis thaliana lacking mitochondrial serine transhydroxymethylase activity. Plant Physiol. 1981;67(4):666–671. doi: 10.1104/pp.67.4.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voll LM, et al. The photorespiratory Arabidopsis shm1 mutant is deficient in SHM1. Plant Physiol. 2006;140(1):59–66. doi: 10.1104/pp.105.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Timm S, et al. A cytosolic pathway for the conversion of hydroxypyruvate to glycerate during photorespiration in Arabidopsis. Plant Cell. 2008;20(10):2848–2859. doi: 10.1105/tpc.108.062265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reumann S, Weber APM. Plant peroxisomes respire in the light: Some gaps of the photorespiratory C-2 cycle have become filled - Others remain. Biochimica Et Biophysica Acta-Molecular Cell Research. 2006;1763(12):1496–1510. doi: 10.1016/j.bbamcr.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Somerville SC, Ogren WL. An Arabidopsis thaliana mutant defective in chloroplast dicarboxylate transport. Proc Natl Acad Sci USA. 1983;80(5):1290–1294. doi: 10.1073/pnas.80.5.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Somerville SC, Somerville CR. A mutant of Arabidopsis deficient in chloroplast dicarboxylate transport is missing an envelope protein. Plant Sci Lett. 1985;37(3):217–220. [Google Scholar]
- 10.Renné P, et al. The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2. Plant J. 2003;35(3):316–331. doi: 10.1046/j.1365-313x.2003.01806.x. [DOI] [PubMed] [Google Scholar]
- 11.Schneidereit J, Häusler RE, Fiene G, Kaiser WM, Weber APM. Antisense repression reveals a crucial role of the plastidic 2-oxoglutarate/malate translocator DiT1 at the interface between carbon and nitrogen metabolism. Plant J. 2006;45(2):206–224. doi: 10.1111/j.1365-313X.2005.02594.x. [DOI] [PubMed] [Google Scholar]
- 12.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu LF, et al. Large-scale prediction of Saccharomyces cerevisiae gene function using overlapping transcriptional clusters. Nat Genet. 2002;31(3):255–265. doi: 10.1038/ng906. [DOI] [PubMed] [Google Scholar]
- 14.Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004;14(6):1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yonekura-Sakakibara K, Tohge T, Niida R, Saito K. Identification of a flavonol 7-O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. J Biol Chem. 2007;282(20):14932–14941. doi: 10.1074/jbc.M611498200. [DOI] [PubMed] [Google Scholar]
- 16.Persson S, Wei HR, Milne J, Page GP, Somerville CR. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA. 2005;102(24):8633–8638. doi: 10.1073/pnas.0503392102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehlting J, et al. Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J. 2005;42(5):618–640. doi: 10.1111/j.1365-313X.2005.02403.x. [DOI] [PubMed] [Google Scholar]
- 18.Alejandro S, et al. AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr Biol. 2012;22(13):1207–1212. doi: 10.1016/j.cub.2012.04.064. [DOI] [PubMed] [Google Scholar]
- 19.Hansen BG, Kliebenstein DJ, Halkier BA. Identification of a flavin-monooxygenase as the S-oxygenating enzyme in aliphatic glucosinolate biosynthesis in Arabidopsis. Plant J. 2007;50(5):902–910. doi: 10.1111/j.1365-313X.2007.03101.x. [DOI] [PubMed] [Google Scholar]
- 20.Bräutigam A, Hoffmann-Benning S, Weber AP. Comparative proteomics of chloroplast envelopes from C3 and C4 plants reveals specific adaptations of the plastid envelope to C4 photosynthesis and candidate proteins required for maintaining C4 metabolite fluxes. Plant Physiol. 2008;148(1):568–579. doi: 10.1104/pp.108.121012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamaguchi M, et al. Loss of the plastid envelope protein AtLrgB causes spontaneous chlorotic cell death in Arabidopsis thaliana. Plant Cell Physiol. 2012;53(1):125–134. doi: 10.1093/pcp/pcr180. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, et al. A chloroplast envelope membrane protein containing a putative LrgB domain related to the control of bacterial death and lysis is required for chloroplast development in Arabidopsis thaliana. New Phytol. 2012;193(1):81–95. doi: 10.1111/j.1469-8137.2011.03867.x. [DOI] [PubMed] [Google Scholar]
- 23.Horan K, et al. Annotating genes of known and unknown function by large-scale coexpression analysis. Plant Physiol. 2008;147(1):41–57. doi: 10.1104/pp.108.117366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwacke R, et al. ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 2003;131(1):16–26. doi: 10.1104/pp.011577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breuers FKH, et al. Dynamic remodeling of the plastid envelope membranes: A tool for chloroplast envelope in vivo localizations. Front Plant Sci. 2012;3:7. doi: 10.3389/fpls.2012.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howitz KT, McCarty RE. d-Glycerate transport by the pea chloroplast glycolate carrier: Studies on [1-C]d-glycerate uptake and d-glycerate dependent O(2) evolution. Plant Physiol. 1986;80(2):390–395. doi: 10.1104/pp.80.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howitz KT, McCarty RE. Solubilization, partial purification, and reconstitution of the glycolate/glycerate transporter from chloroplast inner envelope membranes. Plant Physiol. 1991;96(4):1060–1069. doi: 10.1104/pp.96.4.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berry JA, Osmond CB, Lorimer GH. Fixation of 1802 during photorespiration. Plant Physiol. 1978;62:954–967. doi: 10.1104/pp.62.6.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Somerville CR, Ogren WL. Inhibition of photosynthesis in Arabidopsis mutants lacking leaf glutamate synthase activity. Nature. 1980;286(5770):257–259. [Google Scholar]
- 30.Boldt R, et al. D-GLYCERATE 3-KINASE, the last unknown enzyme in the photorespiratory cycle in Arabidopsis, belongs to a novel kinase family. Plant Cell. 2005;17(8):2413–2420. doi: 10.1105/tpc.105.033993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Igarashi D, et al. Identification of photorespiratory glutamate:Glyoxylate aminotransferase (GGAT) gene in Arabidopsis. Plant J. 2003;33(6):975–987. doi: 10.1046/j.1365-313x.2003.01688.x. [DOI] [PubMed] [Google Scholar]
- 32.Benning C. Evidence supporting a model of voltage-dependent uptake of auxin into cucurbita vesicles. Planta. 1986;169(2):228–237. doi: 10.1007/BF00392319. [DOI] [PubMed] [Google Scholar]
- 33.Howitz KT, McCarty RE. Kinetic characteristics of the chloroplast envelope glycolate transporter. Biochemistry. 1985;24(11):2645–2652. [Google Scholar]
- 34.Howitz KT, McCarty RE. Substrate-specificity of the pea chloroplast glycolate transporter. Biochemistry. 1985;24(14):3645–3650. [Google Scholar]
- 35.Young XK, McCarty RE. Assay of proton-coupled glycolate and D-glycerate transport into chloroplast inner envelope membrane vesicles by stopped-flow fluorescence. Plant Physiol. 1993;101(3):793–799. doi: 10.1104/pp.101.3.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Enser U, Heber U. Metabolic regulation by pH gradients. Inhibition of photosynthesis by indirect proton transfer across the chloroplast envelope. Biochim Biophys Acta. 1980;592(3):577–591. doi: 10.1016/0005-2728(80)90102-4. [DOI] [PubMed] [Google Scholar]
- 37.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301(5633):653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 38.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 39.Lagarde D, et al. Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J. 1996;9(2):195–203. doi: 10.1046/j.1365-313x.1996.09020195.x. [DOI] [PubMed] [Google Scholar]
- 40.Fiehn O, et al. Metabolite profiling for plant functional genomics. Nat Biotechnol. 2001;19(2):173–173. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- 41.Lee DY, Fiehn O. High quality metabolomic data for Chlamydomonas reinhardtii. Plant Methods. 2008;4:7. doi: 10.1186/1746-4811-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133(2):462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koncz C, Schell J. The promoter of Tl-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of agrobacterium binary vector. Mol Gen Genet. 1986;204(3):383–396. [Google Scholar]
- 44.Bendahmane A, Kanyuka K, Baulcombe DC. The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell. 1999;11(5):781–792. doi: 10.1105/tpc.11.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aronsson H, Jarvis P. A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 2002;529(2-3):215–220. doi: 10.1016/s0014-5793(02)03342-2. [DOI] [PubMed] [Google Scholar]
- 46.Bregman A. Laboratory Investigations in Cell and Molecular Biology. New York: John Wiley & Sons; 1990. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.