Abstract
During cerebral cortex development, a series of projection neuron types is generated in a fixed temporal order. In Drosophila neuroblasts, the transcription factor hunchback encodes first-born identity within neural lineages. One of its mammalian homologs, Ikaros, was recently reported to play an equivalent role in retinal progenitor cells, raising the question as to whether Ikaros/Hunchback proteins could be general factors regulating the development of early-born fates throughout the nervous system. Ikaros is also expressed in progenitor cells of the mouse cerebral cortex, and this expression is highest during the early stages of neurogenesis and thereafter decreases over time. Transgenic mice with sustained Ikaros expression in cortical progenitor cells and neurons have developmental defects, including displaced progenitor cells within the cortical plate, increased early neural differentiation, and disrupted cortical lamination. Sustained Ikaros expression results in a prolonged period of generation of deep layer neurons into the stages when, normally, only late-born upper layer neurons are generated, as well as a delayed production of late-born neurons. Consequently, more early-born and fewer late-born neurons are present in the cortex of these mice at birth. This phenotype was observed in all parts of the cortex, including those with minimal structural defects, demonstrating that it is not secondary to abnormalities in cortical morphogenesis. These data suggest that Ikaros plays a similar role in regulating early temporal fates in the mammalian cerebral cortex as Ikaros/Hunchback proteins do in the Drosophila nerve cord.
Keywords: cell fate, stem cells, developmental timing
As in most parts of the nervous system, multipotent progenitor cells of the cerebral cortex generate different types of neurons in a fixed temporal sequence (1). Here, the pyramidal neurons of the six cortical layers are produced in an inside-out order: The subplate and layer 6 neurons are formed first, followed by layer 5, layer 4, and, eventually, layer 2/3. This timing of neurogenesis is encoded within individual progenitor cells and depends mainly on cell-intrinsic mechanisms, including a progressive restriction in progenitor cell competence, from an early multipotent stage to increasingly restricted states in which progenitor cells are competent to generate only the last-born neurons of the lineage (1–3). Recently, a number of postmitotic transcription factors expressed in different subclasses of cortical neurons and necessary for their correct differentiation have been identified (4–15). However, the factors controlling particular temporal competence states at the level of cortical progenitor cells still remain unknown.
The neuroblasts of the Drosophila embryonic CNS constitute the best-understood example of the molecular control of neuronal temporal specification. Here, a series of “temporal transcription factor proteins” is expressed sequentially in neuroblasts: Hunchback, Krüppel, Pdm, Castor, and Grainyhead (16, 17). Loss- and gain-of-function experiments have demonstrated that these factors are required and sufficient for specification of sequential temporal fates in several neuroblast lineages (16, 18–20). Intriguingly, it was recently reported that Ikaros, a mammalian homolog of hunchback, the temporal factor specifying the first-born fate in Drosophila, serves a similar role in rodent retinal progenitor cells (21). In addition, the Caenorhabditis elegans hunchback homolog, hbl-1, is a heterochronic gene specifying early temporal fates in seam cell lineages (22, 23). The above observations raise the important question as to whether the function of Ikaros/Hunchback proteins in regulating the temporal order of neurogenesis identified in fly neuroblasts and rodent retina is more widely conserved throughout the mammalian nervous system, including the cerebral cortex.
The mammalian Ikaros family of transcription factors encompasses the closely related Ikaros, Helios, Aiolos, and Eos, as well as the more divergent member Pegasus (24–27). They are characterized by two highly conserved sets of zinc finger motifs: an N-terminal set of typically four DNA-binding zinc fingers and a C-terminal set of two zinc fingers involved in protein dimerization (24). Because of their high similarity, including binding to the same consensus sequence, the different family members act redundantly in many cases but can also have divergent roles (28–30). Ikaros, Helios, and Aiolos have primarily been studied in the hematopoietic system, where they are important regulators of cell fate determination and act as tumor suppressor genes (31, 32). Ikaros family members have also been found in the nervous system: Eos is expressed throughout the nervous system (33), and Ikaros and Helios are expressed in some striatal neurons (34, 35).
Here, we investigate a potentially conserved role for Ikaros/Hunchback proteins in controlling the temporal order of neurogenesis in the mammalian cerebral cortex. We find that one of the family members, Ikaros, is expressed preferentially in early cortical progenitor cells and is sufficient to promote early, at the expense of late, cortical neuronal fates.
Results
Ikaros Expression in Cortical Progenitor Cells Decreases over Time.
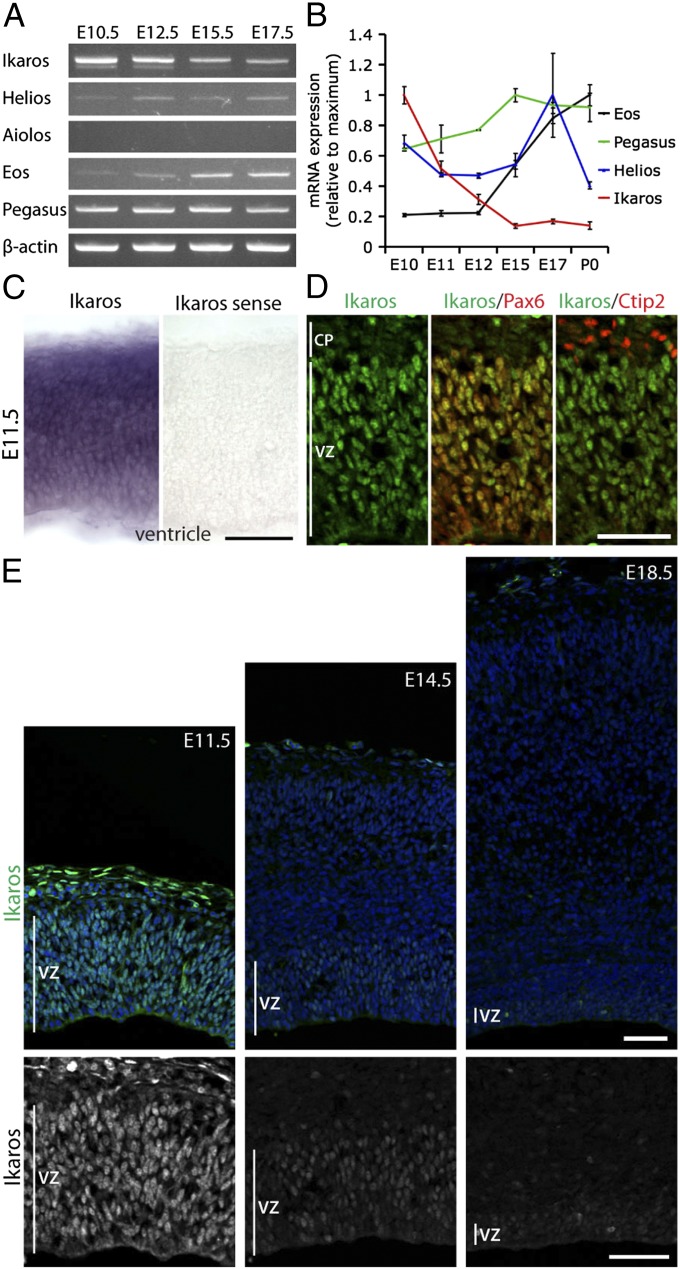
RT-PCR and quantitative RT-PCR (qRT-PCR) showed that four of the five Ikaros family members are expressed in the developing cortex in distinct but overlapping temporal patterns: Ikaros, Helios, Eos, and Pegasus (Fig. 1 A and B). Of the four genes, Ikaros had the most promising expression pattern for a candidate gene regulating early temporal competence. Its expression was high at early stages and decreased by over 80% from embryonic day (E) 10.5 to E15.5 (Fig. 1B). In situ hybridization confirmed that Ikaros mRNA was expressed throughout the cerebral cortex at E11.5 (Fig. 1C).
Fig. 1.
Ikaros is expressed in cortical progenitor cells at high levels early, decreasing over developmental time. (A) Semiquantitative RT-PCR for Ikaros family members in cortex of different developmental stages. Ikaros, Helios, Eos, and Pegasus are expressed in the developing cortex, but Aiolos is not. (B) Real-time qRT-PCR for Ikaros, Helios, Eos, and Pegasus in cortex from E10.5 to P0 shows that Ikaros mRNA levels decrease over developmental time in the cortex. Expression was normalized to the average expression of six housekeeping genes (GAPDH, β-actin, TBP, UBC, YWHAZ, and SDHA). For each gene, values are shown relative to the time point with the highest expression level and represent the mean normalized expression ± SEM (n = 3 independent RNA extractions). (C) In situ hybridization for Ikaros in E11.5 mouse cortex shows specific signal throughout the cortical wall that was not detected with a sense probe. (D and E) Immunostaining for Ikaros (green) in mouse cortical sections at different stages of development, as indicated. (D) At the beginning of neurogenesis, at E11.5, Ikaros is expressed in virtually all Pax6+ cortical progenitor cells of the VZ but not in neurons of the cortical plate (CP), as identified by Ctip2 staining. (E) At mid- (E14.5) and late (E18.5) neurogenic stages, Ikaros is still detected in cortical progenitor cells in the VZ but at much lower levels than at E11.5. All sections in E were processed in parallel from dissection through to imaging. (Scale bars: 50 μm.)
To determine Ikaros protein expression at cellular resolution, we used an antibody previously shown to be specific to Ikaros (26). In contrast to the ubiquitous mRNA, Ikaros protein was preferentially expressed in Pax6-expressing (Pax6+) radial glial progenitor cells in the ventricular zone (VZ), and not in neurons of the cortical plate at E11.5 (Fig. 1D). Also at mid- and late neurogenic stages (E14.5 and E18.5), Ikaros protein was present in Pax6+ VZ progenitor cells but at much lower levels than at E11.5 (Fig. 1E). A similar expression pattern was observed in rat cortex using two different Ikaros antibodies (Fig. S1C). This is similar to Hunchback/Ikaros protein expression in Drosophila neuroblasts and the rodent retina, and it is consistent with a potential role for Ikaros in controlling early temporal competence in cortical progenitor cells.
In contrast to what has been reported from Drosophila and the rodent retina, we did not detect Ikaros in early-born neurons in the cortex. Interestingly, however, we did find that another family member, Helios, is expressed specifically in layer 6 cortical neurons (the first-born neurons) in a mediolateral gradient (Fig. S1D). Because a true regulator of progenitor cell competence should primarily act in progenitor cells, and because Drosophila Hunchback induces early temporal fates when misexpressed in neuroblasts but not postmitotically in neurons (36), we focused our studies on Ikaros because of its expression in early progenitor cells.
Ikaros-Expressing Cortical Progenitor Cells Give Rise to Projection Neurons of All Laminar Fates.
If Ikaros is a temporal factor specifying early competence in cortical progenitor cells, the prediction would be that Ikaros-expressing progenitor cells are multipotent and give rise to neurons of all laminar types. To test this hypothesis and to study the neuronal output from Ikaros-expressing progenitor cells, we took advantage of a BAC transgenic mouse line expressing Cre recombinase under the control of the Ikaros promoter (37). On crossing this line to the R26-YFP reporter line (38), Ikaros-Cre–dependent YFP expression was observed in the cortex. Recombination rates from this BAC-Ikaros-Cre are relatively low, and clones of YFP+ progenitor cells with radial glial morphology and their progeny were distinguishable in the cortex at early developmental stages (Fig. 2 A and B).
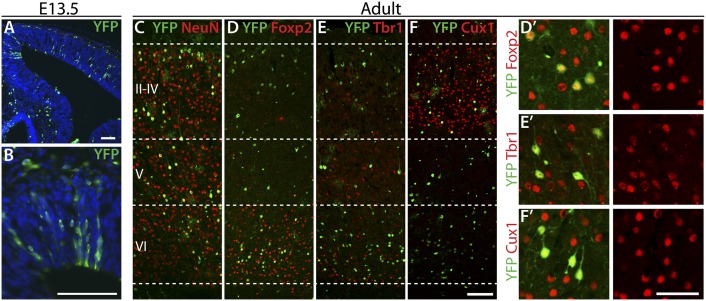
Fig. 2.
Ikaros-expressing progenitor cells generate projection neurons of all laminar fates. (A and B) BAC-Ikaros-Cre transgenic mouse crossed to the R26-YFP reporter mouse was used for lineage analysis of neurons produced from Ikaros-expressing progenitor cells. A low recombination rate gives rise to distinguishable clones of progenitor cells and their progeny at E13.5. (C–F) In the adult brain, progeny of Ikaros-Cre–expressing progenitor cells (YFP+) were mainly neurons (NeuN+) and found throughout the layers of the cortex. (D and E) YFP+ progeny in deep cortical layers frequently expressed Foxp2 and Tbr1, two transcription factors expressed in layer 6 projection neurons. (F) YFP+ neurons in the upper layers were often positive for Cux1, a marker of cortical projection neurons in layers 2–3. (Scale bars: A–F, 100 μm; D′–F′, 50 μm.)
The fates of neurons produced from Ikaros-Cre–expressing progenitor cells were examined in the adult brain. Consistent with our predictions, YFP+ neurons were dispersed throughout all cortical layers (Fig. 2C). The YFP+ neurons within layer 6 frequently expressed the layer 6-specific transcription factors Foxp2 and Tbr1 (Fig. 2 D and E), and YFP+ neurons located in the upper layers expressed the upper layer-specific transcription factor Cux1 (Fig. 2F), indicating that the early Ikaros-expressing progenitor cells generate neurons of all laminar fates.
Increased Ikaros Expression Alters Cortical Development.
To determine if Ikaros is required for specification of early temporal fates in cortical progenitor cells, we analyzed cortical development in a mutant mouse lacking Ikaros C-terminal exon 7, containing the dimerization domain. In the hematopoietic system, this mouse was reported to constitute an Ikaros null (30). These mice had no obvious cortical phenotype, in terms of overall morphology, cortical growth, or the proportions of early- and late-born neurons present in the cortex at birth (Fig. S2). However, redundancy between Ikaros proteins is well described in other systems (28–30) and could be masking a phenotype in the cortex, considering the cortical expression of four homologous family members.
As an alternative approach to assess a role for Ikaros in regulating temporal fates in the cortex, we created a mouse with sustained Ikaros expression in the developing cortex. If Ikaros confers early temporal competence to progenitor cells, this mouse should then display a prolonged and increased production of early-born cortical neurons. To this end, we generated transgenic mice expressing Ikaros from the D6 enhancer. This 6-kb-long genomic fragment from the promoter region of the mouse Dach1 gene mediates specific cortical and hippocampal expression in progenitor cells and neurons starting at E10 and lasting throughout development (39, 40). For easier visualization of transgene expression, an internal ribosome entry site (IRES)-GFP sequence was also included (Fig. 3A). To control for the possibility that any observed phenotype could be due to abnormally high levels of Ikaros or GFP interacting nonspecifically with and disrupting the function of other proteins or complexes, we also generated transgenic mice expressing a DNA-binding deficient form of Ikaros, Ikaros159A. The Ikaros159A has a point mutation causing an asparagine-to-alanine substitution in the second DNA-binding zinc finger, which disrupts its ability to bind DNA (41, 42) (Fig. 3B).
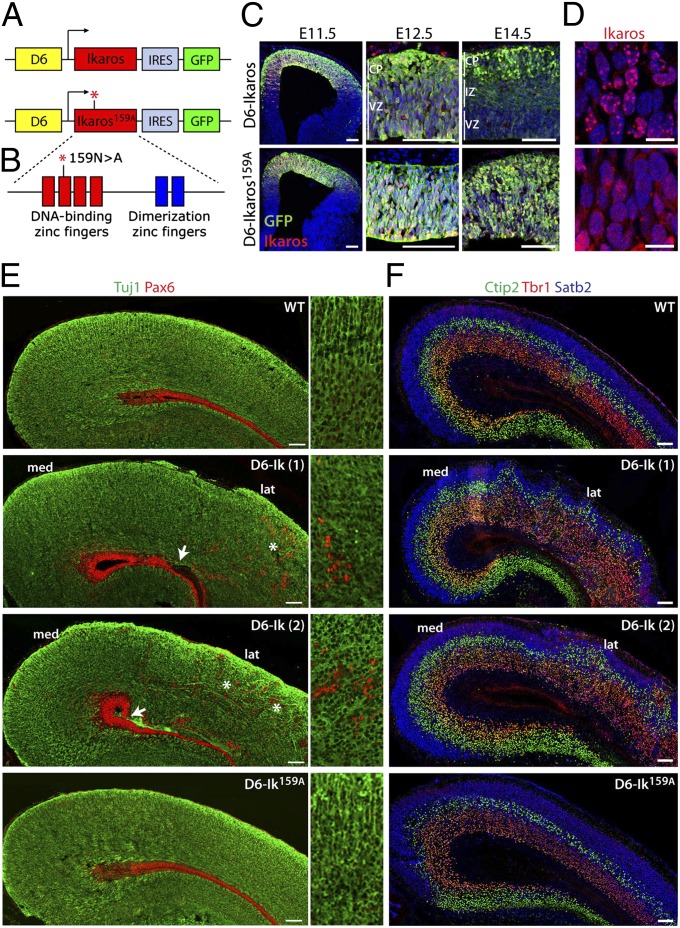
Fig. 3.
Increased Ikaros expression in the cortex affects cortical architecture and is dependent on Ikaros DNA binding ability. (A and B) Constructs used for generation of D6-Ikaros and D6-Ikaros159A transgenic mice. (A) Ikaros and Ikaros159A followed by IRES-GFP were cloned downstream of the D6 enhancer sequence, which drives expression specifically in the cortex and hippocampus. (B) Ikaros159A protein has an asparagine-to-alanine point mutation in the second DNA-binding zinc finger, which disrupts its ability to bind DNA (41). (C) Sections of D6-Ikaros (Upper) and D6-Ikaros159A (Lower) cortex at different developmental stages stained for GFP and Ikaros. Both transgenes are expressed specifically in the cortex. GFP and Ikaros expression from the D6-Ikaros transgene, but not from the D6-Ikaros159A transgene, is down-regulated in the VZ and IZ starting at E12.5. (D) In the D6-Ikaros cortex, Ikaros protein is localized to pericentromeric foci (Upper), whereas in D6-Ikaros159A, the Ikaros159A protein is more diffusely localized throughout the nucleus (Lower). (E) Sections of WT, two lines of D6-Ikaros, and one line of D6-Ikaros159A P0 cortex immunostained for neurons (Tuj1, green) and progenitor cells (Pax6, red). In WT and D6-Ikaros159A cortex, the large majority of Pax6+ progenitor cells are confined to the VZ. In D6-Ikaros brains, ectopic Pax6+ cells were also found scattered throughout the cortical wall (asterisks and Insets). In severely affected D6-Ikaros brains, the integrity of the VZ was also disrupted (arrows). This phenotype was more severe in the lateral cortex (lat), whereas the most medial cortex (med) and hippocampus displayed a grossly normal structure. (F) Sections of WT, two lines of D6-Ikaros, and one line of D6-Ikaros159A P0 cortex stained for transcription factors expressed in neurons of specific cortical layers: Tbr1 (layer 6 neurons, red), Ctip2 (layer 5 neurons, green), and Satb2 (layer 2–4 neurons, blue). D6-Ikaros, but not D6-Ikaros159A, displays a disorganized laminar structure and an increased width of layer 6. (Scale bars: C and E–F, 100 μm; D, 10 μm.)
Two stable lines of D6-Ikaros and four of D6-Ikaros159A were obtained, all of which were viable and fertile. GFP fluorescence was only detected in the cortex, confirming cortex-specific expression and absence of position effects on transgene expression (Fig. 3C). Ikaros immunostaining revealed that overexpressed WT Ikaros was localized to typical pericentromeric foci (43), whereas Ikaros159A had a more diffuse expression throughout the nucleus, in agreement with the inability of Ikaros159A to bind DNA (Fig. 3D). In the D6-Ikaros lines, transgene expression was ubiquitous throughout cortex at early stages (E11.5), with transgene expression of both GFP and Ikaros gradually reducing in expression in the VZ and intermediate zone (IZ) between E12.5 and E17.5 (Fig. 3C and Fig. S3). This reduction in transgene expression over time was dependent on expressing a functional Ikaros with intact DNA-binding ability, because it was observed in the two separate D6-Ikaros lines but not in the D6-Ikaros159A lines (Fig. 3C).
Examination of gross cortical anatomy at birth showed that overexpression of WT Ikaros protein resulted in a disrupted cortical structure. In WT brains, Pax6+ progenitor cells are almost exclusively found in the VZ; however, in the two independent D6-Ikaros lines, Pax6+ cells were also found scattered through the cortical plate and IZ (Fig. 3E). Costaining with the proliferation marker Ki67, the mitotic marker phosphohistone H3 (pH3), and BrdU incorporation confirmed that the ectopic Pax6+ cells were proliferating progenitor cells and not neurons misexpressing Pax6 (Fig. S4). In the most severely affected brains, the integrity of the VZ was sometimes disrupted, as seen by a lack of Pax6+ cells lining the ventricle (Fig. 3E). Together, this suggests a misplacement and delamination of progenitor cells from the VZ, potentially due to defects in neuroepithelial polarity or integrity. Ikaros overexpression also resulted in a disorganized cortical plate, as seen by disruption of the organization of axonal fibers labeled with Tuj1 (Fig. 3E). Additionally, immunostaining for neurons of the different laminar fates showed that even though the D6-Ikaros cortex had a clear separation of deep and upper layer neurons, the layers were not as uniformly linear as in WT brains (Fig. 3F). All structural defects observed were much more pronounced in the lateral cortex, whereas the most medial part and the hippocampus had a grossly normal laminar organization. In contrast, cortical structure in mice overexpressing the DNA-binding deficient Ikaros protein (Ikaros159A) was indistinguishable from that of WT littermates (Fig. 3 E and F).
Cortical Progenitor Cell Populations in the Ikaros Transgenic Cortex.
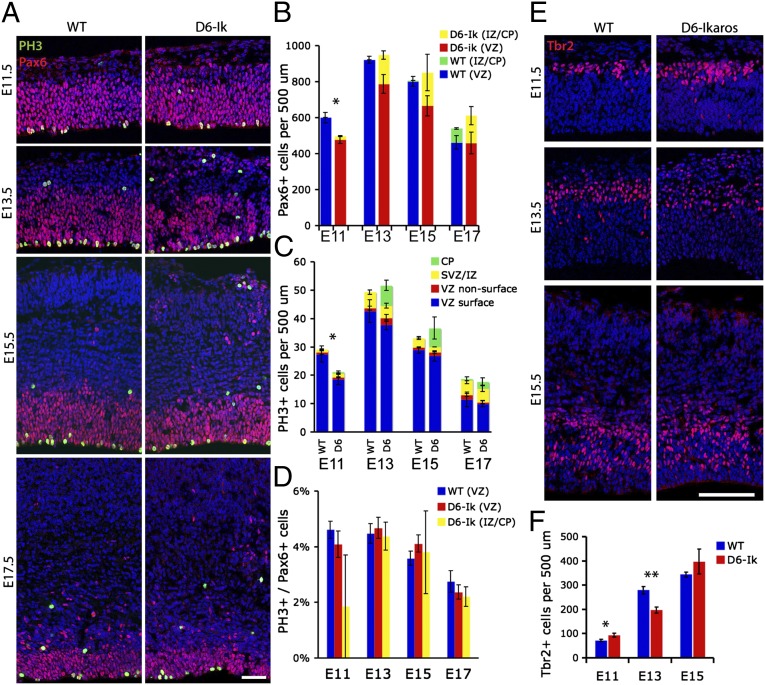
To understand better when and how the disorganization of progenitor cells in the D6-Ikaros cortex is first manifest, we examined Pax6+ and mitotic (pH3+) cells from the beginning (E11.5) to the end (E17.5) of the neurogenic period (Fig. 4 A–D). At E11.5, the first ectopic Pax6+ cells were observed in the early cortical plate/preplate. At this stage, they were relatively few; however, between E13.5 and E17.5, they were present in more substantial numbers. At E11.5, we also observed a small but significant reduction in total numbers of Pax6+ cells in the D6-Ikaros cortex; however, after this stage, there was no significant difference in total numbers of Pax6+ cells (Fig. 4B). Consistent with this, we also observed a displacement of mitotic (pH3+) cells to the cortical plate in D6-Ikaros mice, as well as decreased numbers of pH3+ cells at E11.5 but no significant difference in numbers from E13.5–E17.5 (Fig. 4C). The mitotic index of Pax6+ cells (percentage of Pax6+ cells in mitosis) was unaltered in D6-Ikaros compared with WT cortex, and also between Pax6+ cells in the VZ and ectopic Pax6+ cells, over the whole neurogenic period (Fig. 4D). This indicates that proliferative activity and cell cycle length are not affected by overexpression of Ikaros or by the displacement of progenitor cells. Examination of Tbr2+ intermediate/basal progenitor cells (IPCs) showed increased numbers of IPCs in D6-Ikaros at E11.5, followed by decreased numbers at E13.5 and normal numbers at E15.5 (Fig. 4 E and F). These data suggest that there is an early but transient increased production of IPCs at the expense of maintenance of radial glial/VZ cells, which is no longer present at E13.5.
Fig. 4.
Ectopic progenitor cells and proliferation in the Ikaros-overexpressing cortex. (A) Immunostaining for Pax6+ radial glial progenitor cells and the mitotic marker pH3 in WT and D6-Ikaros cortex shows that ectopic Pax6 progenitor cells are present in the cortical plate (CP) and IZ throughout developmental stages E11.5–E17.5 in the D6-Ikaros cortex. (B) Total number of Pax6+ progenitor cells (including both VZ-bound and ectopic progenitors) was significantly lower in D6-Ikaros at E11.5 but normal from E13.5–E17.5 (n = 3–4 brains). (C) More mitotic (ph3+) cells are found in the cortical plate/IZ of D6-Ikaros compared with WT littermates throughout the stages. The total number of mitotic cells is decreased in D6-Ikaros at E11.5 but unaffected at later stages (n = 3–4 brains). SVZ, subventricular zone. (D) Mitotic index of Pax6+ progenitor cells is not affected in D6-Ikaros cortex (n = 3–4 brains). (E and F) Numbers of Tbr2+ IPCs in D6-Ikaros compared with WT cortex were increased at E11.5, decreased at E13.5, and indistinguishable at E15.5 (n = 3 brains; *P < 0.05; **P < 0.01). (Scale bars: A, 50 μm; E, 100 μm.)
Quantifications of cortical thickness and the width of the Tuj1+ neuronal layer (cortical plate and IZ) over developmental stages E11.5 to postnatal day (P) 0 showed that even though overall cortical growth was relatively normal in D6-Ikaros mice compared with WT littermates, there was a significant increase in neurons from E11.5 to E13.5 (Fig. S5). This effect was, however, transient: the width of the Tuj1+ layer reverted to WT levels by E15.5. Together with the transient early decrease in Pax6+ cells and increase in Tbr2+ cells, this indicates that overexpression of Ikaros pushes the cells toward differentiation and neurogenesis at early developmental stages, consistent with Ikaros’ previously reported role as a tumor suppressor and prodifferentiation factor (35, 44, 45). The reversal of this phenotype at E13.5–E15.5 correlated with the down-regulation of the transgene in the VZ and may reflect a later correction of cortical neuronal output or increased cell death.
The disorganized structure and effects on neurogenesis, as well as all other aspects of the phenotype described here, were essentially identical between the two independent D6-Ikaros lines. In contrast, no effect on either cortical architecture or cortical growth and neurogenesis was observed in D6-Ikaros159A (Fig. S6 A–D). This confirms that the phenotypes observed in D6-Ikaros are not due to insertional mutagenesis or nonspecific interactions but are dependent on Ikaros’ DNA-binding activity. In certain cases, DNA-binding deficient Ikaros proteins have been reported to act as dominant negatives (DNs), through formation of nonfunctional dimers with WT Ikaros family proteins (46–48). To determine the ability of Ikaros159A to act as a DN in cortex, we generated double transgenic D6-Ikaros/D6-Ikaros159A embryos to examine whether Ikaros159A could rescue the cortical defects caused by overexpression of WT Ikaros. The phenotype of double transgenic embryos was indistinguishable from single transgenic D6-Ikaros embryos, both in terms of ectopic progenitor cells and increased early neurogenesis (Fig. S7 A–J). Ikaros159A also failed to affect pericentromeric localization (i.e., DNA binding) of WT overexpressed Ikaros (Fig. S7 K–M). Together, this shows that Ikaros159A is nonfunctional, does not act as a DN in the cortex, and is therefore a useful control for D6-Ikaros.
Sustained Ikaros Expression Results in More Deep Layer Neurons and Fewer Upper Layer Neurons at E17.5.
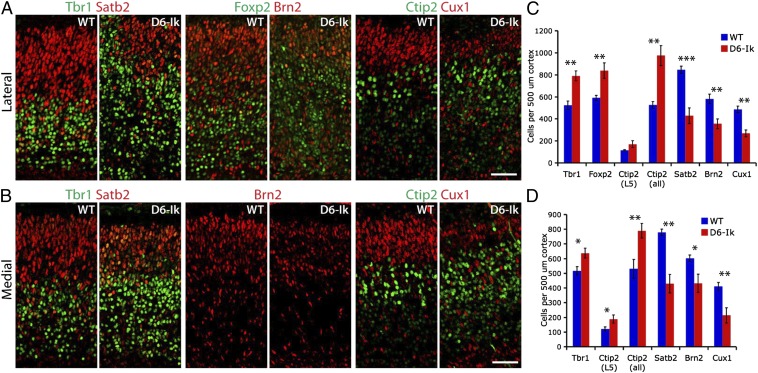
To examine the consequences of sustained Ikaros expression on temporal fates in the cortex, we quantified early- and late-born neurons at E17.5, a stage when all deep layer neurons and most upper layer neurons have been generated in the WT cortex (Fig. 5). Because the disorganized structure of the D6-Ikaros cortex could potentially affect distribution of cell types, quantifications were made both for the lateral, more affected cortex and for the most medial part of the cortex, which displays milder structural defects. In D6-Ikaros, we found a significant increase in Tbr1+ and Foxp2+ cells, two transcription factors expressed specifically in deep layer 6 cells, the earliest born cortical neurons, in both lateral and medial cortex (n = 4; Fig. 5). We also found a significant increase in Ctip2+ cells, a transcription factor specifically expressed in all deep layer neurons (layers 5–6) at this stage (n = 4; Fig. 5). When quantifying only the strong Ctip2+ cells in layer 5, a significant increase was detected in the medial cortex and a trend of more layer 5 neurons was observed in the lateral cortex (n = 4; Fig. 5). Accompanying the increase in deep layer neurons, the D6-Ikaros cortex had a significant decrease in neurons expressing three different upper layer-specific transcription factors, Satb2 (layers 2–4), Brn2 (layers 2–3 and 5), and Cux1 (layers 2–3), compared with WT littermates (n = 4; Fig. 5) in both the lateral and most medial parts of cortex. In contrast, no change in the numbers of early- or late-born neurons was detected in the D6-Ikaros159A cortex (Fig. S6 E and F).
Fig. 5.
More deep layer neurons and fewer upper layer neurons following sustained Ikaros expression in the cortex. (A and B) Immunostaining for deep layer-specific transcription factors Tbr1 (layer 6), Foxp2 (layer 6), and Ctip2 (weak in layer 6, strong in layer 5), and late-born, upper layer-specific transcription factors Satb2 (layers 2–4), Brn2 (layers 2–5), and Cux1 (layers 2–3) in E17.5 WT and D6-Ikaros lateral (A) and medial (B) cortex. (C and D) Significantly more early-born, deep layer neurons and significantly fewer late-born, upper layer neurons were observed in D6-Ikaros than in WT littermates, both in the lateral (C) and medial (D) parts of the cortex. Values represent mean ± SEM (n = 4 brains). *P < 0.05; **P < 0.01; ***P < 0.001. (Scale bars: 50 μm.)
Sustained Ikaros Expression Leads to an Early and Sustained Increase in Deep Layer Neuron Production.
The observed increase in deep layer neurons and decrease in upper layer neurons in the D6-Ikaros cortex at E17.5 is what we would expect if Ikaros were encoding early temporal competence in progenitor cells. Alternatively, the increased numbers of deep layer neurons present at E17.5 could be a consequence of the increased neuron production between E11 and E13, if all the extra neurons produced were deep layer neurons. To distinguish between these possibilities, we examined the effect of sustained Ikaros expression on the numbers of deep layer (Tbr1+ and Ctip2+) and upper layer (Satb2+) neurons at 2-d intervals from the beginning (E11.5) to the end (P0) of neurogenesis (Fig. 6).
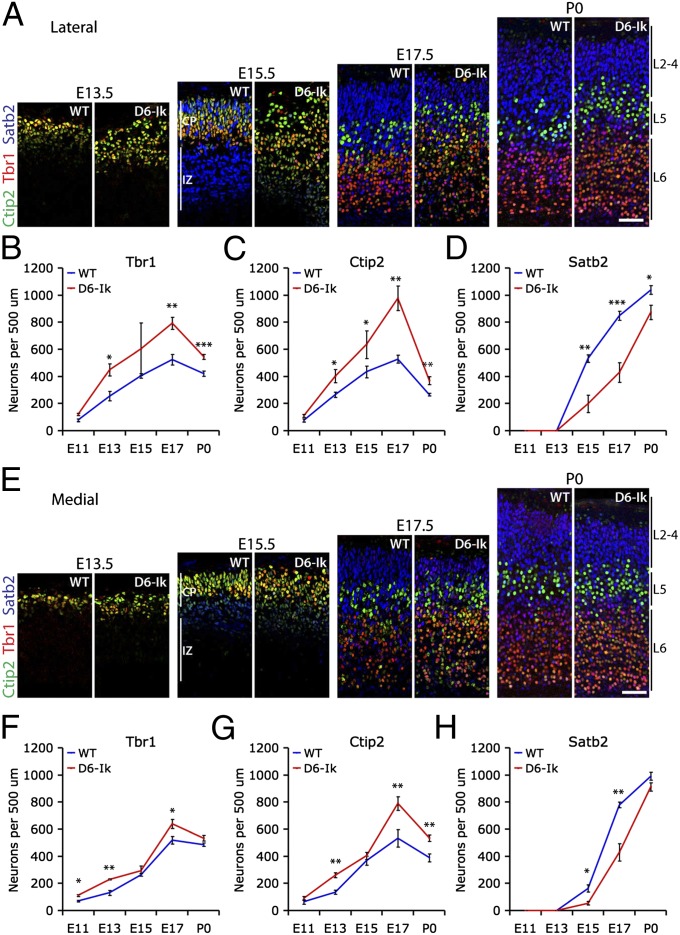
Fig. 6.
Early and sustained increase in deep layer genesis and delayed upper layer genesis following sustained Ikaros expression in the cortex. (A–D) Immunostaining and quantifications of Tbr1 (layer 6, red), Ctip2 (weak in layer 6 and strong in layer 5, green), and Satb2 (layers 2–4; blue) in WT and D6-Ikaros lateral cortex at different stages of cortical development as indicated. (E–H) Immunostaining and quantifications of the same markers in WT and D6-Ikaros medial cortex. Increased numbers of Tbr1+ and Ctip2+ deep layer neurons were observed in D6-Ikaros compared with WT littermates throughout development both laterally and medially, as well as a delayed appearance and decreased numbers of Satb2+ upper layer neurons throughout development. In B–D and F–H, values represent mean ± SEM (n = 3–7 brains). *P < 0.05; **P < 0.01; ***P < 0.001. (Scale bars: 50 μm.)
Consistent with the overall increase in neurons observed in D6-Ikaros at early stages, more deep layer neurons (Tbr1 and Ctip2) were present in the D6-Ikaros cortex compared with WT at E11.5–E13.5 both laterally and medially (Fig. 6). The number of deep layer neurons per unit width of cortex increased in both WT and D6-Ikaros cortex until E17.5. At all stages, the Ikaros-overexpressing cortex had more deep layer neurons than the cortex of WT littermates. Importantly, the total increase in deep layer neurons was larger at E17.5 than at E12.5 or E13.5, particularly if also considering the larger surface area of the cortex at E17.5. Therefore, the early increase in neurogenesis is unable to account for all the extra deep layer neurons present at E17.5. These results therefore suggest a sustained increased production of deep layer neurons in the Ikaros-overexpressing cortex.
We also observed a decrease in upper layer neurons after E17.5 (Fig. 6). In the WT cortex, this likely reflects a dilution of neurons due to the expansion of the cortical area by the addition of other cell types, such as astrocytes and interneurons, as well as to the growth of dendrites and axons. Interestingly, however, the decrease in deep layer neurons was more pronounced in D6-Ikaros than in WT, resulting in a partial normalization and correction of deep layer neuron numbers toward WT levels. A likely mechanism for this is apoptosis, which was increased in D6-Ikaros compared with WT cortex at late stages of development (Fig. S8).
Sustained Ikaros Expression Results in the Delayed Appearance of Upper Layer Neurons.
Accompanying the increase in deep layer neurons, the D6-Ikaros cortex displayed a delayed appearance of upper layer (Satb2+) neurons. In WT brains, the first Satb2+ neurons were observed at E15.5. At this stage, many of the immature neurons in the IZ migrating out toward the cortical plate were Satb2+, and the first Satb2 cells were also observed in the cortical plate (Fig. 6 A and E). In D6-Ikaros brains of the same stage, the number of Satb2+ neurons in the cortical plate was considerably lower, and most of the immature migrating neurons of the IZ were Satb2− or expressed much weaker levels of Satb2 (Fig. 6 A and E; n = 3). Two days later at E17.5, upper layer neuron production was still lagging behind in D6-Ikaros, with only half the number of Satb2 neurons compared with those in WT littermates (Fig. 6; n = 4). At P0, the difference was smaller but there were still significantly fewer Satb2+ neurons in the Ikaros-overexpressing cortex, most notably in the lateral cortex (Fig. 6; n = 6). This delayed onset of upper layer neurogenesis, coupled with the later continued increase in upper layer genesis until P0, indicates that the decrease in upper layer neurons cannot be explained by a depletion of radial glial cells or IPCs, consistent with the normal numbers of Pax6+ and Tbr2+ cells observed in the Ikaros-overexpressing cortex at late neurogenic stages (Fig. 4). These results therefore suggest that sustained Ikaros levels extend the window of deep layer neurogenesis at the expense of upper layer neuron production.
After completion of upper layer genesis, cortical progenitor cells switch to astrogliogenesis. This switch was, however, not noticeably affected in the D6-Ikaros cortex, where the astrocytic protein GFAP turned on normally in radial glial cells around P0 (Fig. S9A), and the number of S-100β–expressing astrocytes was not different from that of WT littermates (Fig. S9 B and C). This could imply that the molecular mechanisms regulating the switch from neurogenesis to gliogenesis are independent from, and overlying, the temporal specification of neuronal subtypes in the cortex.
Sustained Ikaros Expression Prolongs the Period of Production of Deep Layer Neurons.
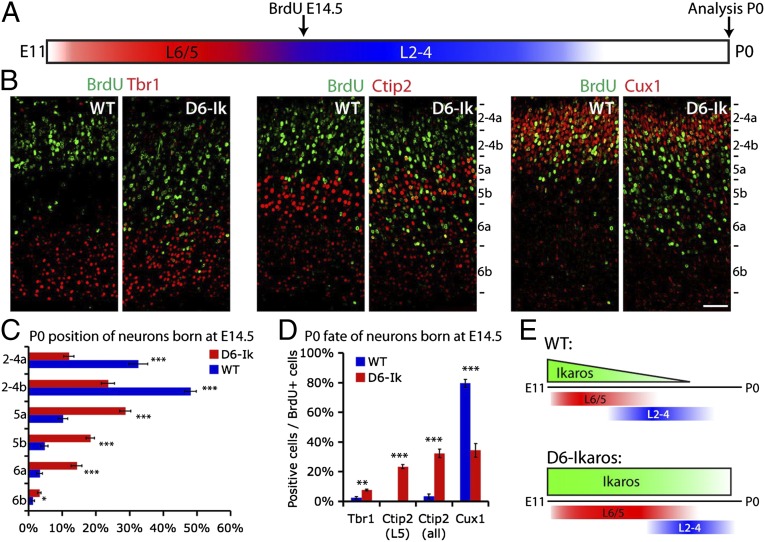
To test directly if sustained Ikaros expression can extend the period during which deep layer neurons are generated, we used BrdU birth-dating to determine fates of neurons in mice born at E14.5. At this stage, deep layer neurogenesis is essentially completed in WT cortex and the majority of neurons produced are upper layer neurons (49). BrdU was administered to pregnant mice at E14.5, and laminar fates of BrdU-labeled cells were examined at P0 (Fig. 7A). Comparing the position of strongly BrdU-labeled neurons within the cortical plate, an obvious difference was detected between WT and D6-Ikaros brains. In WT cortex, most BrdU+ cells were located in the upper layers as expected, whereas in D6-Ikaros cortex, many BrdU-labeled cells were also located at deeper positions (Fig. 7 B and C; n = 4).
Fig. 7.
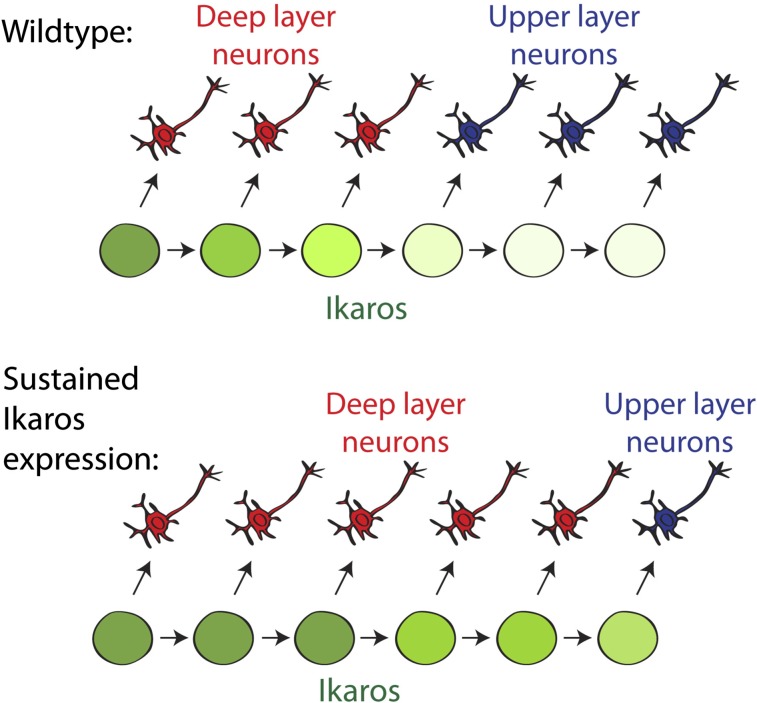
BrdU birth-dating shows an extended period of deep layer neuron production following sustained Ikaros expression in the cortex. (A) Birth-dating experimental design. BrdU was administered to pregnant females at E14.5 and incorporated by cycling progenitor cells. Laminar positions and neuronal fates of strongly BrdU+ neurons born at E14.5 were analyzed at P0. (B and C) Cells born at E14.5 in the WT cortex mainly populate the upper layers (Cux1+), and very few are found in the deeper layers (Tbr1+ and Ctip2+). Cells born at E14.5 in the D6-Ikaros cortex are found at deeper positions, within the Tbr1+ and Ctip2+ layers, and also in Cux1+ upper layers. (Scale bar: B, 50 μm.) (D) In the WT cortex, fewer than 5% of neurons born at E14.5 express deep layer markers Tbr1 or Ctip2, and most (80%) express upper layer marker Cux1. In D6-Ikaros, the proportion of deep layer neurons produced at E14.5 was greatly increased (10-fold for Ctip2) and the proportion of upper layer neurons was less than half of the WT level. In C and D, values represent mean ± SEM (n = 4 brains). *P < 0.05; **P < 0.01; ***P < 0.001. (E) Schematic drawing summarizes the D6-Ikaros developmental timing phenotype. In WT cortex, Ikaros levels decrease in progenitor cells over time. Sustained Ikaros expression beyond its normal period leads to prolonged and increased production of deep layer neurons coupled to delayed production of upper layer neurons.
The BrdU+ cells found at deeper positions could reflect a prolonged production of deep layer neurons in D6-Ikaros or, alternatively, could indicate a defect in neuron migration. To distinguish between these two possibilities, the identity of BrdU-labeled cells was confirmed by costaining for the deep layer-specific transcription factors Tbr1 and Ctip2 and the upper layer-specific transcription factor Cux1 (Fig. 7D). In the WT cortex, as expected, most neurons born at E14.5 were upper layer, Cux1+ neurons (80%) and very few were Tbr1+ or Ctip2+ deep layer neurons (<5%). In the D6-Ikaros cortex, the proportion of deep layer neurons produced at E14.5 was greatly increased (from 3% to 32% for Ctip2 and from 3% to 8% for Tbr1; n = 4). In addition, the proportion of upper layer Cux1+ neurons was reduced to lower than half of the WT level (from 80% to 34%; n = 4). Together, this confirms that the D6-Ikaros cortex has a prolonged production of deep layer neurons into the stages when only upper layer neurons are generated normally. This prolonged production is especially remarkable because increased numbers of deep layer neurons are already present due to the early increase in differentiation.
To study how Ikaros performs these functions, we compared whole-genome mRNA expression between the D6-Ikaros and control WT cortex at E11.5 (Datasets S1 and S2). This stage was chosen because it would avoid confounding effects on gene expression of the marked changes in neuronal cell type that are found in the D6-Ikaros cortex at later stages. At E11.5, a set of 149 genes was found with increased expression in the D6-Ikaros cortex (P < 0.05), together with a set of 174 down-regulated genes (P < 0.05). In contrast, no changes in mRNA expression (except Ikaros and GFP itself) were detected in the D6-Ikaros159A cortex compared with that of WT littermates.
The sets of genes with altered expression are functionally heterogeneous. We observed up-regulation of many neuronal genes, including a glutamate receptor subunit, the neuron-specific RNA binding protein HuD/Elavl4, and a voltage-gated T-type calcium channel (Cacna1h). In addition, positive regulators of neurogenesis (Sox4, Sox6, and Sox21) and differentiation/cell cycle exit genes (Cdkn1c, Gadd45a, and Gadd45g) were up-regulated. This was accompanied by down-regulation of positive regulators of the cell cycle, cyclins D1 and D2. Together, these changes in gene expression are consistent with the early increase in neuronal differentiation observed at this stage. In addition, Vangl2/Strabismus, a component of the planar cell polarity pathway that regulates cell polarity, was significantly down-regulated in the Ikaros transgenic cortex. Previous reports have shown that loss of Vangl2 function results in neural tube defects, and Vangl2 mutant mice have alterations in spindle orientation in cortical progenitor cells (50), which could potentially contribute to the misplacement of progenitor cells observed in D6-Ikaros cortex.
Reintroduction of Ikaros in Late Cortical Progenitor Cells and Later Born Neurons Is Not Sufficient to Generate Additional Early-Born Neurons.
In Drosophila, even though sustained hunchback expression in neuroblasts extends generation of early-born neurons, reintroduction of hunchback into later neuroblasts fails to induce a fate change to early-born neurons, which was attributed to a loss of competence to respond to Hunchback (36, 51). In contrast, retinal progenitor cells are competent to generate low numbers of early-born neurons on reintroduction of Ikaros at late stages (21).
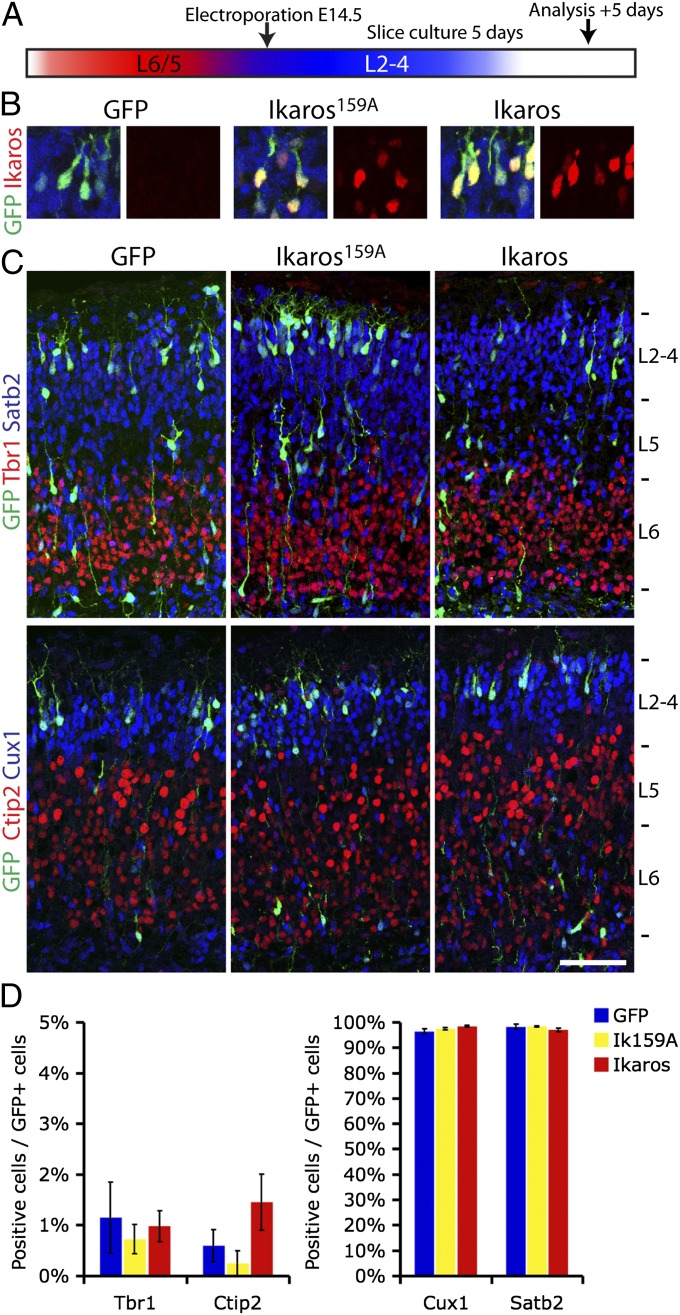
To investigate this in the cortex, we electroporated an Ikaros-ires-GFP construct into WT E14.5 ventricular progenitor cells, as well as two control constructs expressing the inactive Ikaros form, Ikaros159A, or ires-GFP only (Fig. 8A). After culturing cortical slices for 5 d ex vivo, Ikaros protein expression in electroporated cells was confirmed (Fig. 8B) and the identity of GFP+ neuronal progeny in the cortical plate was determined by costaining for deep and upper layer markers. In all conditions, over 95% of neurons were positive for the upper layer markers Cux1 and Satb2 and less than 2% were positive for the deep layer markers Tbr1 and Ctip2, with no significant differences between Ikaros and GFP or Ikaros159A (Fig. 8 C–E; n = 3 brains, 100–300 cells per brain counted). Therefore, reintroduction of Ikaros in late cortical progenitor cells that have already switched to producing upper layer neurons is not sufficient to induce earlier born type neurons. This suggests that late cortical progenitor cells lose competence to respond to Ikaros, similar to what has been observed in Drosophila. Furthermore, expression of Ikaros in late-born postmitotic neurons does not respecify upper layer identities, arguing that the altered cell fates observed in the D6-Ikaros cortex are due to sustained expression of Ikaros specifically in progenitor cells.
Fig. 8.
Reintroduction of Ikaros in late progenitor cells is not sufficient to induce heterochronically early fates. (A) Electroporation experimental design. Ikaros-ires-GFP, inactive Ikaros159A-IRES-GFP, and GFP only were electroporated into VZ progenitors of the lateral ventricle at E14.5. Brains were cultured as slices for 5 d before fates of GFP+ neurons produced from these progenitor cells were analyzed by immunohistochemistry. (B) Ikaros-IRES-GFP and Ikaros159A-IRES-GFP constructs, but not the IRES-GFP control construct, express Ikaros in GFP-positive neurons after 5 d of culture. (C and D) No differences in fates of neurons born from Ikaros and control electroporated progenitor cells were observed. In all conditions, less than 2% of neurons were expressing deep layer markers Tbr1 and Ctip2 and most neurons (>95%) were upper layer neurons expressing Cux1 and Satb2. In C, values represent mean ± SEM (n = 3 brains; 100–300 cells per brain counted). (Scale bar: C, 100 μm.)
Discussion
We report here that Ikaros is expressed in neural progenitor cells of the mammalian cerebral cortex at high levels during early developmental stages, decreasing over time. Transgenic mice in which Ikaros expression in the developing cortex was increased and sustained beyond its normal period displayed a striking phenotype of three main components: (i) a transient increase in neuronal differentiation at early stages, (ii) a disrupted cortical structure with ectopic Pax6+ progenitor cells, and (iii) a prolonged and increased production of deep layer neurons coupled to a delayed onset of upper layer genesis. The latter phenotype resulted in a cortex containing more deep layer neurons and fewer upper layer neurons.
Importantly, the altered proportions of deep and upper layer neurons could not be attributed to alterations in progenitor cell numbers or proliferation, or to the early increase in neurogenesis. In fact, previous studies have shown that early excess neuronal differentiation results in a shortened period of deep layer genesis and a precocious switch to upper layer genesis, keeping the numbers of early-born neurons at normal levels (52). Given that at early stages of cortical development, the D6-Ikaros cortex contains more deep layer neurons than in littermate controls due to the increased early differentiation, it would be predicted that this would repress further genesis of deep layer neurons by negative feedback from postmitotic neurons, as occurs in the retina (53, 54) and as has been proposed to operate in the cortex (55). In contrast, our birth-dating results identified an extended period of deep layer neuron production in the D6-Ikaros cortex compared with that of littermate controls.
It is formally possible that Ikaros indirectly promotes early fates through the disorganization of cortical structure or the displacement of progenitor cells if misplaced progenitor cells or ectopic neurogenesis positions result in biased production of neurons of deep layer fates. However, no such effect has been previously reported in mice with similarly disrupted structure and displaced cortical progenitor cells due to mutations in other genes (56–58). Furthermore, we also found increased deep layer neurons coupled to decreased upper layer neurons in the most medial part of cortex, in which there were minimal anatomical defects in lamination and cortical structure. Our results therefore support a model in which Ikaros directly promotes early-born fates at the expense of late-born fates in the cortex.
The effect of Ikaros in promoting early-born cortical fates could, in principle, be exerted either in progenitor cells or in neurons, because D6 drives expression in both cell types. However, the expression of Ikaros in late-born neurons when introduced by electroporation of cortical progenitor cells at E14.5 did not result in changes in layer identities, even though neurons born at the same stage in the D6-Ikaros cortex did change fates toward early-born identities (as shown by BrdU birth-dating). Therefore, acute introduction of Ikaros to progenitor cells or neurons after E14.5 is not sufficient, on its own, to specify deep neuronal fates, acting within either progenitor cells or neurons.
It is likely, therefore, that the sustained expression of Ikaros in progenitor cells from early stages underlies the increase in early-born and/or deep layer neuronal fates in the D6-Ikaros cortex, although we currently cannot exclude that neuronal Ikaros expression also contributes to the phenotype. Our results favor a model in which Ikaros regulates early temporal fates or competence in cortical progenitor cells, similar to the function of the Ikaros homolog, Hunchback, in Drosophila. Such a mode of action would be in contrast to other transcription factors that control neuronal identity in the cortex, including Satb2, Fezf2, Ctip2, Tbr1, and Sox5, which are expressed postmitotically in neurons of specific laminar fates (4–13). Such postmitotic temporal factors are likely to act downstream of progenitor cell temporal factors and to function by directing correct differentiation programs of particular neuronal types (59). They may also fine-tune temporal fates by mutual repression of transcription factors in alternative layers (60).
The sustained Ikaros expression in D6-Ikaros led to a delayed onset of upper layer neuron production but did not completely prevent upper layer genesis. This could suggest that Ikaros is not instructive, but permissive, for early fates. However, the Ikaros transgene was down-regulated in progenitor cells beginning from E12.5. It is therefore possible that a stronger effect on extending deep layer genesis at the expense of upper layer genesis would be observed if this down-regulation could be avoided. An intriguing possibility is that the transgene down-regulation may reflect a normal repression of Ikaros to allow for progression to later temporal states. Transgene down-regulation was dependent on having a functionally intact Ikaros (it did not occur in lines expressing a point mutant DNA-binding deficient Ikaros), and it may therefore reflect an autoregulatory feedback mechanism, which is something that has been reported for Hunchback in flies (61). For such a mechanism to act on the transgene, it cannot represent transcriptional repression, because the transgene is not transcribed from the Ikaros promoter, or protein degradation, because GFP expressed from IRES was also down-regulated. The down-regulation therefore likely occurs at the mRNA level. An interesting possibility is that this repression is micro-RNA (miRNA)-mediated, because the C. elegans homolog hbl-1 is repressed by miRNAs to allow for progression to later temporal states (22, 23). In addition, miRNAs were recently shown to be required for temporal progression of retinal progenitor cell competence, which could potentially occur through repression of Ikaros (62).
Even though Ikaros is sufficient for promoting early temporal fates in the cortex, no phenotype was observed in the cortex of Ikaros mutant mice. This is potentially due to a well-described redundancy between the four Ikaros family proteins expressed in the cortex (28, 30). Additionally, the truncated Ikaros protein produced in these mice still has an intact DNA-binding domain and may retain some function in neural progenitor cells, even though it was reported as nonfunctional in the thymus (30). Varying degrees of redundancy between tissues likely affect phenotypic severity, which could explain why a mild defect was observed in the retina [30% reduction in early-born neurons (21)] but not in the cortex. To confirm the requirement for Ikaros proteins in developmental timing in the cortex, appropriate loss-of-function models, including compound conditional KOs for several family members, will be essential. We predict that such mice will have a loss or reduction of early-born neurons in the cortex, and possibly also in other parts of the nervous system.
In Drosophila neuroblasts, Hunchback acts both as a temporal identity factor and to maintain early competence. Overexpression of Hunchback from early stages leads to continued overproduction of early neurons, and down-regulation of Hunchback is required to initiate the progressive restriction and loss of competence to respond to the hunchback-Krüppel-pdm-castor series. When reexpressed into neuroblasts after its down-regulation, Hunchback can induce ectopic first-born neurons, although this ability is lost after five divisions (18, 36, 51). We found evidence here for a similar function of Ikaros in the cerebral cortex: Cortical progenitor cells with sustained expression of Ikaros displayed an extended period of production of early-born neurons, but reintroduction of Ikaros into later stage progenitor cells after the developmental down-regulation of Ikaros did not result in the renewed production of deep layer neuron types.
In conclusion, the role proposed here for Ikaros in cortical development supports conservation of Hunchback/Ikaros function in several parts of the vertebrate nervous system, arguing for an integral role for Ikaros in the core timing mechanism. Future studies of the mechanisms, targets, and regulation of Ikaros/Hunchback proteins in different parts of the nervous system and comparisons between them therefore can potentially provide fundamental insights into the control of the temporal order of neurogenesis.
Materials and Methods
Animals.
All animal work was carried out in accordance with the Canadian Council on Animal Care and the IRCM Animal Care Committee, and UK local and national (Home Office) ethical and legal regulations. For birth-dating studies, 40 mg/kg BrdU (b5002; Sigma–Aldrich) was administered i.p. to pregnant dams. WT mice for expression studies were MF1 × MF1, rats were Sprague–Dawley (Charles River Laboratories), and Ikaros mutant mice (30) were on the CBA/CaH background. For generation of D6-Ikaros and D6-Ikaros159A mice, Ikaros-IRES-GFP and Ikaros159A-IRES-GFP sequences were PCR-amplified from retroviral vectors (41), and cloned into the Not1 site of the D6 vector (39). Transgenic mice were generated by pronuclear injection of Sal1 linearized vector in CBA × C57BL6 F1 zygotes, and transgenic lines were established in the same genetic background. Transgenic embryos and pups were genotyped by their GFP expression in the cortex, which was visible through the skull until P3. Two independent lines of D6-Ikaros and four of D6-Ikaros159A were used.
RT-PCR and qRT-PCR.
RNA was extracted using the RNeasy kit (Qiagen), and cDNA was prepared using SuperScriptIII (Invitrogen) and random hexamers. For semiquantitative RT-PCR, 5 ng of cDNA was used per reaction at 28 cycles for Ikaros family genes and 22 cycles for actin. Quantitative PCR was carried out using SYBR Green JumpStart Taq ReadyMix (Sigma) on a StepOnePlus Real-Time PCR System (Applied Biosystems) with 5 ng of cDNA per reaction and standard cycling conditions. The relative abundance of each gene was normalized to the average abundance of six housekeeping genes (Gapdh, β-actin, Tbp, Ubc, Ywhaz, and Sdha) to obtain the relative expression level. Three independent experiments and two technical replicates per sample were used. Statistical comparisons were done using the Student t test (two-sample assuming equal variance). Sequences of intron spanning primers, compared against the mouse genome and transcriptome using BLAST (63) to ensure specificity, are shown in Tables S1 and S2.
In Situ Hybridization, Immunofluorescence, and Data Analysis.
Whole-mount in situ hybridization was carried out on mouse embryo heads as described (64) using probes of antisense and sense Ikaros full-length coding sequence (1.8 kb). Heads were then cryosectioned at 25 μm and imaged in Volocity (PerkinElmer).
Fixation, cryosectioning, and immunohistochemistry were carried out as described (52), with minor modifications. For Ikaros immunostaining, fixation time was reduced to less than 3 h and freshly sectioned tissue was always used for staining. To quench the GFP signal in D6-Ikaros brains, sections were heated to 100 °C in 0.01 M sodium citrate. Primary antibodies used were rat anti-BrdU (ab6326; Abcam), mouse anti-BrdU (B8434; Sigma–Aldrich), goat anti-Brn2 (sc-6029; Santa Cruz Biotechnology), rat anti-CTIP2 (ab18465; Abcam), rabbit anti-Cux1 (sc-13024; Santa Cruz Biotechnology), rabbit anticleaved caspase-3 (96612; Cell Signaling Technology), rabbit anti-Foxp2 (ab16046; Abcam), rabbit anti-Ikaros (26), mouse anti-Ikaros 4E9 (65), rabbit anti-Helios (26), mouse anti-NeuN (ab13938; Abcam), mouse anti-Ki67 (550609; Becton Dickinson), rabbit anti-Pax6 (PRB-278P; Covance), mouse anti-Pax6 (Developmental Studies Hybridoma Bank), rat anti-pH3 (ab10543; Abcam), mouse anti-SATB2 (ab51502; Abcam), rabbit anti-TBR1 (ab31940; Abcam), and mouse anti-Tuj1 (MMS-435P; Covance). Secondary antibodies were Alexa Fluors 488, 546, and 647 (Invitrogen).
Counts of cells expressing specific proteins were carried out on 20× confocal images (Zeiss 510 Meta, Olympus FV1000, or Leica SP5) using the ImageJ (National Institutes of Health) plug-in ITCN or in the case of cells double-positive for BrdU and cell type-specific markers, using Volocity (PerkinElmer). The exact field of pictures taken was chosen in the DAPI channel, so that no biased sampling errors were made toward areas with more or less neurons of the different subtypes. For all quantifications, at least three transgenic/mutant mice and three WT littermates were used, with at least two sections per mouse. Statistical comparisons were done using the Student t test (two-sample assuming equal variance).
Electroporation and Slice Culture.
Ikaros and Ikaros159A were cloned into the pCIG2-ires-GFP gateway vector (gift from P. Barnes and F. Polleux, University of North Carolina, Chapel Hill). The constructs were injected into the lateral ventricle of E14.5 heads and electroporated at 60 V with five pulses of 50 ms with a 1-s interval. Vibratome slices of forebrain (250 μm) were cultured as described (66), followed by fixation, cryosectioning, and immunohistochemistry as described above.
Gene Expression Profiling.
Three pools of WT embryos and three pools of transgenic embryos were used for each study, and each pool consisted of the cortices from three to four embryos. RNA was extracted using TRIzol, amplified and hybridized to Illumina beadchip Mouse WG6 v3 arrays (Cambridge Genomic Services, Department of Pathology, University of Cambridge). Data were analyzed using the Illumina R packages lumi and limma, with a false discovery rate of 5%.
Supplementary Material
Acknowledgments
We thank Katia Georgopoulos for the Ikaros KO mice and Ikaros antibody, Stephen Smale for Ikaros and Helios antibodies, Brad Cobb for Ikaros and Ikaros159A constructs, and Stefan Krauss for the D6 construct. J.M.A. was supported by the Cambridge/Wellcome Trust 4-y doctoral program in developmental biology. Research in F.J.L.’s group benefits from core support to the Gurdon Institute from the Wellcome Trust and Cancer Research UK.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 2710 (volume 110, number 8).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215707110/-/DCSupplemental.
References
- 1.Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: Lessons from the retina. Nat Rev Neurosci. 2001;2(2):109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- 2.McConnell SK, Kaznowski CE. Cell cycle dependence of laminar determination in developing neocortex. Science. 1991;254(5029):282–285. doi: 10.1126/science.254.5029.282. [DOI] [PubMed] [Google Scholar]
- 3.Frantz GD, McConnell SK. Restriction of late cerebral cortical progenitors to an upper-layer fate. Neuron. 1996;17(1):55–61. doi: 10.1016/s0896-6273(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 4.Arlotta P, et al. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45(2):207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 5.Alcamo EA, et al. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57(3):364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 6.Britanova O, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57(3):378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 7.Chen B, et al. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc Natl Acad Sci USA. 2008;105(32):11382–11387. doi: 10.1073/pnas.0804918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47(6):817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 9.Kwan KY, et al. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci USA. 2008;105(41):16021–16026. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai T, et al. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 2008;57(2):232–247. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 11.Bedogni F, et al. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc Natl Acad Sci USA. 2010;107(29):13129–13134. doi: 10.1073/pnas.1002285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han W, et al. TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc Natl Acad Sci USA. 2011;108(7):3041–3046. doi: 10.1073/pnas.1016723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKenna WL, et al. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J Neurosci. 2011;31(2):549–564. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci USA. 2005;102(47):17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen JG, Rasin MR, Kwan KY, Sestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc Natl Acad Sci USA. 2005;102(49):17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106(4):511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- 17.Brody T, Odenwald WF. Programmed transformations in neuroblast gene expression during Drosophila CNS lineage development. Dev Biol. 2000;226(1):34–44. doi: 10.1006/dbio.2000.9829. [DOI] [PubMed] [Google Scholar]
- 18.Grosskortenhaus R, Pearson BJ, Marusich A, Doe CQ. Regulation of temporal identity transitions in Drosophila neuroblasts. Dev Cell. 2005;8(2):193–202. doi: 10.1016/j.devcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 19.Novotny T, Eiselt R, Urban J. Hunchback is required for the specification of the early sublineage of neuroblast 7-3 in the Drosophila central nervous system. Development. 2002;129(4):1027–1036. doi: 10.1242/dev.129.4.1027. [DOI] [PubMed] [Google Scholar]
- 20.Grosskortenhaus R, Robinson KJ, Doe CQ. Pdm and Castor specify late-born motor neuron identity in the NB7-1 lineage. Genes Dev. 2006;20(18):2618–2627. doi: 10.1101/gad.1445306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elliott J, Jolicoeur C, Ramamurthy V, Cayouette M. Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron. 2008;60(1):26–39. doi: 10.1016/j.neuron.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 22.Lin SY, et al. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003;4(5):639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 23.Abrahante JE, et al. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4(5):625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 24.Molnár A, Georgopoulos K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol Cell Biol. 1994;14(12):8292–8303. doi: 10.1128/mcb.14.12.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan B, et al. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 1997;16(8):2004–2013. doi: 10.1093/emboj/16.8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hahm K, et al. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes Dev. 1998;12(6):782–796. doi: 10.1101/gad.12.6.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perdomo J, Holmes M, Chong B, Crossley M. Eos and pegasus, two members of the Ikaros family of proteins with distinct DNA binding activities. J Biol Chem. 2000;275(49):38347–38354. doi: 10.1074/jbc.M005457200. [DOI] [PubMed] [Google Scholar]
- 28.Cai Q, Dierich A, Oulad-Abdelghani M, Chan S, Kastner P. Helios deficiency has minimal impact on T cell development and function. J Immunol. 2009;183(4):2303–2311. doi: 10.4049/jimmunol.0901407. [DOI] [PubMed] [Google Scholar]
- 29.Georgopoulos K, et al. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79(1):143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- 30.Wang JH, et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5(6):537–549. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 31.Merkenschlager M. Ikaros in immune receptor signaling, lymphocyte differentiation, and function. FEBS Lett. 2010;584(24):4910–4914. doi: 10.1016/j.febslet.2010.09.042. [DOI] [PubMed] [Google Scholar]
- 32.Ng SY, Yoshida T, Georgopoulos K. Ikaros and chromatin regulation in early hematopoiesis. Curr Opin Immunol. 2007;19(2):116–122. doi: 10.1016/j.coi.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 33.Honma Y, et al. Eos: A novel member of the Ikaros gene family expressed predominantly in the developing nervous system. FEBS Lett. 1999;447(1):76–80. doi: 10.1016/s0014-5793(99)00265-3. [DOI] [PubMed] [Google Scholar]
- 34.Agoston DV, et al. Ikaros is expressed in developing striatal neurons and involved in enkephalinergic differentiation. J Neurochem. 2007;102(6):1805–1816. doi: 10.1111/j.1471-4159.2007.04653.x. [DOI] [PubMed] [Google Scholar]
- 35.Martín-Ibáñez R, et al. Ikaros-1 couples cell cycle arrest of late striatal precursors with neurogenesis of enkephalinergic neurons. J Comp Neurol. 2010;518(3):329–351. doi: 10.1002/cne.22215. [DOI] [PubMed] [Google Scholar]
- 36.Pearson BJ, Doe CQ. Regulation of neuroblast competence in Drosophila. Nature. 2003;425(6958):624–628. doi: 10.1038/nature01910. [DOI] [PubMed] [Google Scholar]
- 37.Tarchini B, Jolicoeur C, Cayouette M. In vivo evidence for unbiased ikaros retinal lineages using an ikaros-cre mouse line driving clonal recombination. Dev Dyn. 2012;241(12):1973–1985. doi: 10.1002/dvdy.23881. [DOI] [PubMed] [Google Scholar]
- 38.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Machon O, et al. Forebrain-specific promoter/enhancer D6 derived from the mouse Dach1 gene controls expression in neural stem cells. Neuroscience. 2002;112(4):951–966. doi: 10.1016/s0306-4522(02)00053-2. [DOI] [PubMed] [Google Scholar]
- 40.van den Bout CJ, Machon O, Røsok O, Backman M, Krauss S. The mouse enhancer element D6 directs Cre recombinase activity in the neocortex and the hippocampus. Mech Dev. 2002;110(1-2):179–182. doi: 10.1016/s0925-4773(01)00597-4. [DOI] [PubMed] [Google Scholar]
- 41.Cobb BS, et al. Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes Dev. 2000;14(17):2146–2160. doi: 10.1101/gad.816400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papathanasiou P, et al. Widespread failure of hematolymphoid differentiation caused by a recessive niche-filling allele of the Ikaros transcription factor. Immunity. 2003;19(1):131–144. doi: 10.1016/s1074-7613(03)00168-7. [DOI] [PubMed] [Google Scholar]
- 43.Brown KE, et al. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91(6):845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 44.Kathrein KL, Lorenz R, Innes AM, Griffiths E, Winandy S. Ikaros induces quiescence and T-cell differentiation in a leukemia cell line. Mol Cell Biol. 2005;25(5):1645–1654. doi: 10.1128/MCB.25.5.1645-1654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winandy S, Wu P, Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell. 1995;83(2):289–299. doi: 10.1016/0092-8674(95)90170-1. [DOI] [PubMed] [Google Scholar]
- 46.Ruiz A, Williams O, Brady HJ. The Ikaros splice isoform, Ikaros 6, immortalizes murine haematopoietic progenitor cells. Int J Cancer. 2008;123(6):1240–1245. doi: 10.1002/ijc.23706. [DOI] [PubMed] [Google Scholar]
- 47.Sun L, Liu A, Georgopoulos K. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J. 1996;15(19):5358–5369. [PMC free article] [PubMed] [Google Scholar]
- 48.Trinh LA, et al. Down-regulation of TDT transcription in CD4(+)CD8(+) thymocytes by Ikaros proteins in direct competition with an Ets activator. Genes Dev. 2001;15(14):1817–1832. doi: 10.1101/gad.905601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hevner RF, et al. Beyond laminar fate: Toward a molecular classification of cortical projection/pyramidal neurons. Dev Neurosci. 2003;25(2-4):139–151. doi: 10.1159/000072263. [DOI] [PubMed] [Google Scholar]
- 50.Lake BB, Sokol SY. Strabismus regulates asymmetric cell divisions and cell fate determination in the mouse brain. J Cell Biol. 2009;185(1):59–66. doi: 10.1083/jcb.200807073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cleary MD, Doe CQ. Regulation of neuroblast competence: Multiple temporal identity factors specify distinct neuronal fates within a single early competence window. Genes Dev. 2006;20(4):429–434. doi: 10.1101/gad.1382206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pereira JD, et al. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci USA. 2010;107(36):15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belliveau MJ, Cepko CL. Extrinsic and intrinsic factors control the genesis of amacrine and cone cells in the rat retina. Development. 1999;126(3):555–566. doi: 10.1242/dev.126.3.555. [DOI] [PubMed] [Google Scholar]
- 54.Waid DK, McLoon SC. Ganglion cells influence the fate of dividing retinal cells in culture. Development. 1998;125(6):1059–1066. doi: 10.1242/dev.125.6.1059. [DOI] [PubMed] [Google Scholar]
- 55.Desai AR, McConnell SK. Progressive restriction in fate potential by neural progenitors during cerebral cortical development. Development. 2000;127(13):2863–2872. doi: 10.1242/dev.127.13.2863. [DOI] [PubMed] [Google Scholar]
- 56.Cappello S, et al. The Rho-GTPase cdc42 regulates neural progenitor fate at the apical surface. Nat Neurosci. 2006;9(9):1099–1107. doi: 10.1038/nn1744. [DOI] [PubMed] [Google Scholar]
- 57.Konno D, et al. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat Cell Biol. 2008;10(1):93–101. doi: 10.1038/ncb1673. [DOI] [PubMed] [Google Scholar]
- 58.Imai F, et al. Inactivation of aPKClambda results in the loss of adherens junctions in neuroepithelial cells without affecting neurogenesis in mouse neocortex. Development. 2006;133(9):1735–1744. doi: 10.1242/dev.02330. [DOI] [PubMed] [Google Scholar]
- 59.Jacob J, Maurange C, Gould AP. Temporal control of neuronal diversity: Common regulatory principles in insects and vertebrates? Development. 2008;135(21):3481–3489. doi: 10.1242/dev.016931. [DOI] [PubMed] [Google Scholar]
- 60.Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Curr Opin Neurobiol. 2008;18(1):28–35. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kohwi M, Hiebert LS, Doe CQ. The pipsqueak-domain proteins Distal antenna and Distal antenna-related restrict Hunchback neuroblast expression and early-born neuronal identity. Development. 2011;138(9):1727–1735. doi: 10.1242/dev.061499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Georgi SA, Reh TA. Dicer is required for the transition from early to late progenitor state in the developing mouse retina. J Neurosci. 2010;30(11):4048–4061. doi: 10.1523/JNEUROSCI.4982-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 64.Kondo T, Zákány J, Duboule D. Control of colinearity in AbdB genes of the mouse HoxD complex. Mol Cell. 1998;1(2):289–300. doi: 10.1016/s1097-2765(00)80029-5. [DOI] [PubMed] [Google Scholar]
- 65.Kim J, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10(3):345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 66.Polleux F, Ghosh A. The slice overlay assay: A versatile tool to study the influence of extracellular signals on neuronal development. Sci STKE. 2002;2002(136):pl9. doi: 10.1126/stke.2002.136.pl9. [DOI] [PubMed] [Google Scholar]