Abstract
The TrkC neurotrophin receptor belongs to the functional dependence receptor family, members of which share the ability to induce apoptosis in the absence of their ligands. Such a trait has been hypothesized to confer tumor-suppressor activity. Indeed, cells that express these receptors are thought to be dependent on ligand availability for their survival, a mechanism that inhibits uncontrolled tumor cell proliferation and migration. TrkC is a classic tyrosine kinase receptor and therefore generally considered to be a proto-oncogene. We show here that TrkC expression is down-regulated in a large fraction of human colorectal cancers, mainly through promoter methylation. Moreover, we show that TrkC silencing by promoter methylation is a selective advantage for colorectal cell lines to limit tumor cell death. Furthermore, reestablished TrkC expression in colorectal cancer cell lines is associated with tumor cell death and inhibition of in vitro characteristics of cell transformation, as well as in vivo tumor growth. Finally, we provide evidence that a mutation of TrkC detected in a sporadic cancer is a loss-of-proapoptotic function mutation. Together, these data support the conclusion that TrkC is a colorectal cancer tumor suppressor.
Keywords: neurotrophin-3, caspase-3, genetic, epigenetic
The Trk tyrosine kinase receptors and their ligands, the neurotrophins, have been studied extensively for their role in nervous system development. However, TrkA was originally cloned as an oncogene from colon carcinoma tumors in which the TrkA kinase domain was fused to the tropomyosin gene in the extracellular domain (1). This discovery motivated a great number of studies, which showed that neurotrophins (NGF, BDNF, and NT-4/5, NT-3) and their respective Trk receptors (TrkA, TrkB, and TrkC), are all involved in various malignancies (for review, see ref. 2). The initial (and still generally accepted) view is that Trks, like other tyrosine kinase receptors, are oncogenic receptors, and therefore pan-Trk kinase inhibitors are currently being tested in clinical trials (3–5). Somewhat surprisingly, however, it has turned out that, at least in tumors such as neuroblastoma and medulloblastoma, TrkA, TrkB, and TrkC behave very differently, despite their close homology. TrkA and TrkC expression is associated with a good prognosis, whereas TrkB is expressed in very aggressive tumors (for review; see ref. 2). The fact that the high expression of a tyrosine kinase receptor known to activate prooncogenic pathways (like the MAPK and PI3K-AKT pathways) is associated with a better outcome is counter intuitive, and suggests the possibility that TrkA and TrkC, rather than functioning solely as oncogenes, may also, in at least some cases, act as tumor suppressors. Although this notion may be ostensibly paradoxical, two recent independent studies have lent support to it, by demonstrating that both TrkA and TrkC, but not TrkB, act as dependence receptors (6, 7).
Dependence receptors, which also include DCC (Deleted in Colorectal Carcinoma), UNC5H, Patched, Neogenin, and the RET, EPHA4, IR, IGF1R, and Alk tyrosine kinase receptors (8–14), share the functional property of inducing cell death when disengaged from their ligands, but suppressing their proapoptotic activity when bound by their respective trophic ligands. These receptors thus create cellular states of dependence on their respective ligands (15, 16). The molecular mechanisms used by these unbound receptors to trigger apoptosis are in large part unknown (15, 16), but it has been hypothesized that this characteristic acts as a means of eliminating tumor cells that would otherwise proliferate in settings of ligand unavailability, such as exist for invasive or metastatic neoplasms. The proapoptotic activity of dependence receptors has thus been proposed to confer a tumor suppressor activity that is in turn suppressed by the presence of trophic ligands. This was formally demonstrated for the prototype dependence receptors DCC and UNC5H3/C, which bind the ligand netrin-1 (16). Both DCC and UNC5C have been suggested to be colon cancer tumor suppressors because their expression is lost or markedly decreased in the vast majority of colorectal cancers (17–20). Moreover, inactivation of UNC5C or specific inactivation of DCC’s proapoptotic activity in mice was shown to promote intestinal tumor progression (20, 21). Moreover, it was recently shown that missense mutations in UNC5C are associated with risk of familial colorectal cancer (22). As expected for a receptor that triggers apoptosis in settings of ligand limitation, tumor survival may be achieved not only by downregulating the proapoptotic receptor but also by autocrine production of the associated ligand. In support of this prediction, it was shown that in several cancers, such as metastatic breast cancer, lung cancer, and neuroblastoma, netrin-1 is produced in an autocrine manner to block netrin-1 receptor-induced apoptosis (23–25).
Interestingly, NT-3, the ligand for TrkC, has also been shown to be up-regulated in neuroblastoma, potentially inhibiting TrkC-induced apoptosis (26). Therefore, we tested the hypothesis that TrkC functions as a suppressor of colorectal malignancies, in an analogous fashion to the netrin-1 receptors. We demonstrate here that expression of the TrkC gene is down-regulated in colorectal cancers, primarily because of tumor-associated specific promoter methylation. We also show that, in colorectal cancer cell lines, re-expression of TrkC is associated with cancer cell death and loss of traits of cell transformation in vitro. We also show that TrkC expression constrains tumor growth in an avian model. Altogether, our data support the conclusion that TrkC is a colorectal cancer tumor suppressor.
Results
Expression of TrkC Is Down-Regulated in Human Colorectal Cancers.
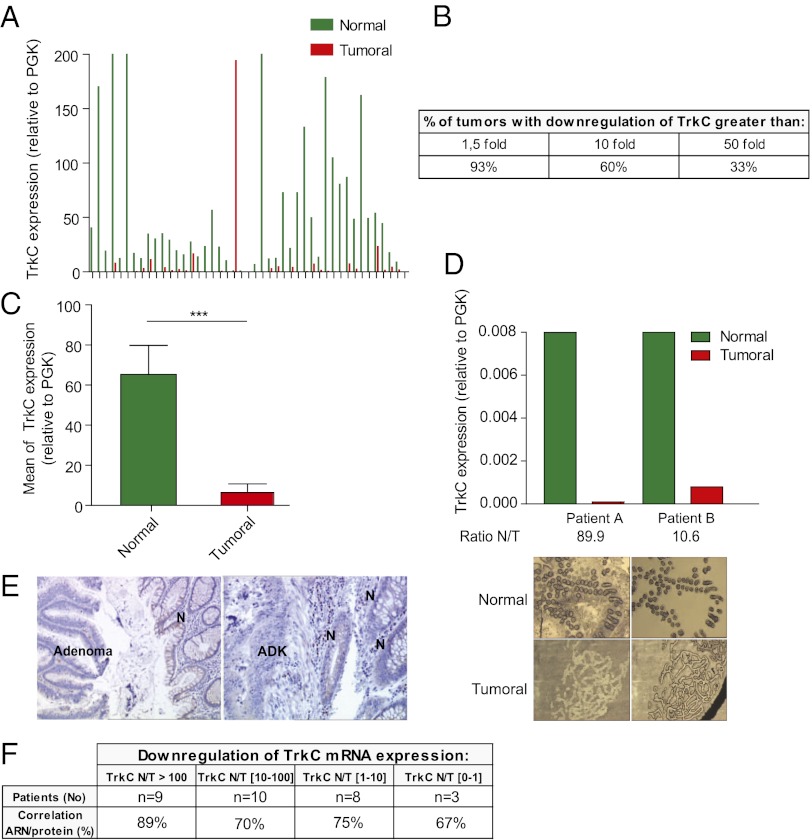
TrkC has been shown to be expressed by epithelial cells (27). We therefore assessed TrkC gene expression by quantitative real-time reverse transcriptase PCR (Q-RT-PCR) in a panel of 45 colorectal cancers, and compared the results to those from corresponding matched normal tissues. As shown in Fig. 1A, although expression of TrkC was variable but relatively high in most normal tissues, a markedly decreased expression was observed in matched tumors. A 10-fold decrease of TrkC expression was observed in over 60% of the tested tumors, and one-third of the tumors showed a 50-fold decrease in expression (Fig. 1B). Mean TrkC expression was more than 10-fold lower in normal tissues than in the corresponding neoplasms (Fig. 1C, P < 0.001). This marked TrkC decrease was found to be independent of the stage of the pathology, as it was similarly detected in stage I, II, III, and IV colorectal cancers (Fig. S1A). Furthermore, TrkC expression was reduced in tumors not only in comparison with adjacent normal tissue, but also in comparison with normal tissues from age-matched healthy patients (Fig. S1B).
Fig. 1.
TrkC expression is lost in colorectal tumors. (A–C) Quantitative real-time RT-PCR was performed using total RNA extracted from normal (N) and tumoral (T) tissues with specific human TrkC primers. PGK showing the less variability in their expression between normal and colorectal tumoral tissues, as described previously (20), was used as housekeeping gene. (A) The expression levels in 45 colorectal tumors (Tumoral) and corresponding normal tissue (Normal) are given as a ratio between TrkC and PGK, the internal control. (B) Table indicating the percentage of patients showing a loss of TrkC expression in tumor compared with normal tissue. (C) Mean TrkC expression in tumoral tissues versus normal tissues is presented. ***P < 0.001, two-sided Mann–Whitney test, the two means being compared. (D) Laser capture microdissection (LCM) was performed on 8 pairs of tumor/normal tissues (two representative pairs being presented here), and TrkC expression was determined as in A. (Upper) The expression levels in 2 colorectal tumors (T) and corresponding normal tissue (N) are given as a ratio between TrkC and PGK. (Lower) Typical microscopic visualization of LCM on colon section from human normal (N) or tumoral biopsies (T). Sections are shown before LCM (Left) and after LCM (Right), as the captured material is confirmed under microscopic visualization before processing for RNA extraction. (E) Immunostaining of TrkC protein was performed on tissue sections isolated from biopsies of two different patients and counterstained with Mayer’s hematoxylin. N, normal tissue; ADK, adenocarcinoma. (F) Table showing the correlation between TrkC mRNA expression and TrkC protein expression determined by immunostaning in pair normal/tumor for 30 patients in the panel of 45 patients.
To eliminate any potential bias due to heterogeneity in tumor samples that may exceed that in normal tissue samples, epithelial cells from tumors and normal tissue were laser microdissected and assessed for TrkC expression by Q-RT-PCR. In the 8 pairs of samples assessed, a striking decrease in TrkC expression was detected in tumor cells (Fig. 1D and Fig. S1C). The loss of TrkC in neoplastic tissue observed at the RNA level was confirmed at the protein level by TrkC immunohistochemistry on biopsy sections from 30 patients (Fig. 1 E and F).
Whereas TrkC level was observed to be decreased markedly in colon cancer samples, we failed to detect a gain of NT-3 expression in these samples (Fig. S1D). This finding argues that, in contrast to neuroblastoma cells, which typically select a gain of NT-3 to survive ligand limitation (26) in colon cancer, TrkC loss is preferentially selected.
TrkC expression was next analyzed in a panel of colorectal cancer cell lines. As shown in Fig. S1E, most colorectal cancer cell lines screened were either negative for TrkC or showed only a modest level of expression compared with the neuroblastoma cell lines IMR32 and CLB-Ge2, in which TrkC and its ligand NT-3 were shown to be expressed (26). In this study, we used colorectal cancer HCT116 and HCT8 cells as a model. We confirmed that, in HCT116 and HCT8 cells, TrkC was not detected by immunohistochemistry (Fig. S1F).
Taken together, these data support the view that, although TrkC is expressed in normal colon epithelium, TrkC is silenced in a large fraction of colon cancer samples and cell lines.
TrkC Expression Is Inhibited via Tumor-Associated TrkC Promoter Methylation.
In an effort to delineate the mechanisms that may underlie TrkC down-regulation in colon cancer cells, we first analyzed whether loss of heterozygosity (LOH) may occur at the TrkC locus. Using a restricted panel of colorectal cancers and matched normal tissues, and a series of microsatellite markers within the TrkC gene, we failed to observe any significant occurrence of LOH, supporting the view that TrkC down-regulation results from an alternative mechanism (Fig. S2 A and B). Therefore, we transfected a TrkC promoter–luciferase reporter construct into HCT116 and HT29 TrkC-negative cells and into MDA-MB-436 TrkC-positive cells, and compared luciferase expression. As shown in Fig. S2C, the exogenous TrkC promoter was activated in both cell lines, supporting the view that TrkC silencing is not due to indirect modulation of regulators of TrkC transcription, but rather to a direct silencing of the endogenous TrkC promoter.
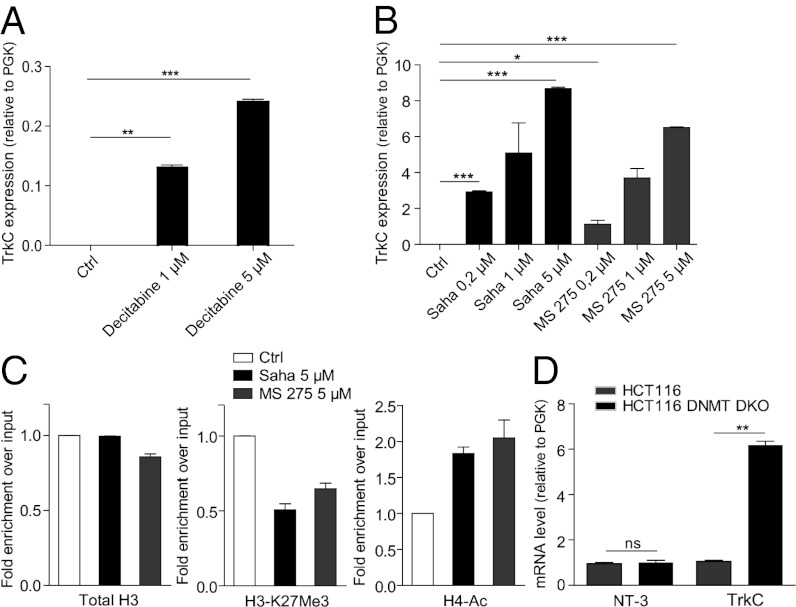
We thus analyzed whether TrkC expression in colorectal cancer cell lines is restored in the presence of a DNA methylation inhibitor. As shown in Fig. 2A and Fig. S2D, treatment with 5-aza-2’-deoxycytidine (Decitabine) of HCT116 and HCT8 cells, respectively, was associated with restoration of TrkC expression. Similar results were obtained when HCT116 or HCT8 cells were treated with inhibitors of histone deacetylases (HDACi), such as Saha or MS275 (Fig. 2B and Fig. S2E). HCT116 cells treated with Saha or MS275 accumulated acetylated H4 on the TrkC promoter, as revealed by chromatin immunoprecipitation (ChIP) using an H4 pan-acetylated antibody (Fig. 2C). Because H4 acetylation is generally associated with activation of transcription (28), this observation supports the view that HDACs repress TrkC expression in HCT116 cells. In support of this view, a decrease in the level of the repressive H3K27Me3 epitope (29) was observed at the TrkC promoter upon treatment with Saha and MS275, in association with the induction of its transcription (Fig. 2C).
Fig. 2.
TrkC is re-expressed in HCT116 colorectal cancer cell line following epi-drugs treatment. (A and B) HCT116 cells were treated for 72 h with decitabine (A) or for 24 h with Saha or MS 275 (B) at the indicated concentrations. TrkC expression was measured by Q-RT-PCR, using PGK as internal control. (C) Chomatin immunoprecipitation performed on HCT116 cells treated or not with 5 μM Saha or 5 μM MS275 using the following antibodies: total H3, H3-K27Me3 (mark of transcription repression) or H4-pan-acetyl (mark of transcription activation). The experiments were done with one primers couple. Graphs show relative enrichment over input. Enrichment for control cells is set at 1 in each individual experiment. Data are the mean ± SEM of at least two independent experiments. (D) The level of TrkC and NT-3 mRNA was measured in wild-type (WT) and HCT116 cells invalidated for DNMT1 and -3B (DNMT DKO) by Q-RT-PCR.
To exclude the possibility that these data were due to drug toxicity, we analyzed TrkC expression in HCT116 cells null for both DNMT1 and DNMT3B, two important DNA methyltransferases (30). As shown in Fig. 2D, TrkC expression was increased in HCT116 DKO DNMT1 and DNMT3b cells compared with wild-type HCT116. Taken together, these data indicate that TrkC expression is repressed by methylation and HDAC-driven deacetylation in HCT116 colorectal cancer cells.
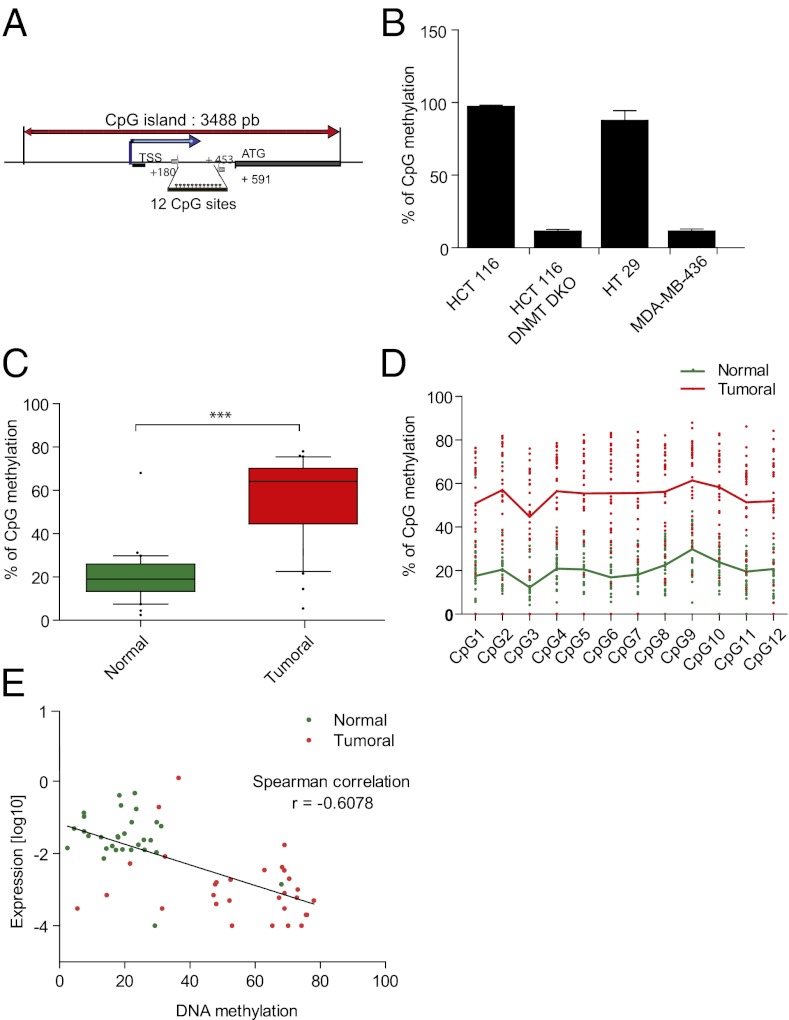
We then investigated whether methylation of the TrkC promoter occurs in colorectal cancer. This was assessed in a panel of 30 colorectal tumors and compared with adjacent normal tissues. Putative CpG islands found in the TrkC promoter (Fig. 3A) were investigated for methylation by bisulfite sequencing. Following PCR amplification of the TrkC promoter (Fig. 3A), pyrosequencing was used to determine the bisulfite-converted sequence of 12 CpG sites in this amplicon. The ratio of C-to-T at individual sites was determined quantitatively, based on the amount of C and T incorporation during the sequence extension (Fig. S3). In HCT116 and HT29 colon cancer cell lines lacking TrkC, the 12 CpG sites displayed a methylation rate of 100%, whereas they were not methylated in TrkC-positive cells (HCT116 DNMTs DKO and MDA-MB-436) (Fig. 3B). We then analyzed TrkC promoter methylation in matched normal and neoplastic colorectal tissues. The frequency of methylation was highly significantly increased in the tumor samples, compared with the normal samples (Fig. 3C). We verified that the methylation was observed homogeneously on each of the 12 CpG sites analyzed (Fig. 3D). We observed a statistically significant inverse correlation between TrkC expression and the intensity of methylation (Fig. 3E). Thus, TrkC expression is lost or strongly decreased in colorectal tumors through specific tumor-associated promoter methylation.
Fig. 3.
TrkC promoter is hyper-methylated in tumoral colorectal tissue. (A) A schematic representation of TrkC promoter is shown. The main CpG island, the transcription start site (TSS), and translation starting codon (ATG) are represented. The primers used for the pyrosequencing of the 12 CpG sites indicated were located in (+180;+205) and (+453;+476). (B) Analysis by pyrosequencing of the TrkC promoter methylation in TrkC-negative cells (HCT116 WT, HT29) and in TrkC-positive cells (HCT116 DNMT DKO colorectal cell lines and MDA-MB-436 breast cell line). (C) Analysis by pyrosequencing of the TrkC promoter methylation in matched normal and tumoral colorectal tissues from 30 patients in the panel of 45 patients. Statistical analysis has been performed using two-sided Mann–Witney test. ***P < 0.001. (D) Pyrosequencing of the TrkC promoter methylation in matched normal and tumoral colorectal tissues from a single patient. A representative diagram showing the methylation level on each of the 12 CpG sites analyzed is shown. (E) Inverse correlation between the decrease of TrkC mRNA level and DNA methylation of the TrkC promoter.
TrkC Expression Inhibits Colorectal Tumor Growth by Triggering Tumor Cell Apoptosis.
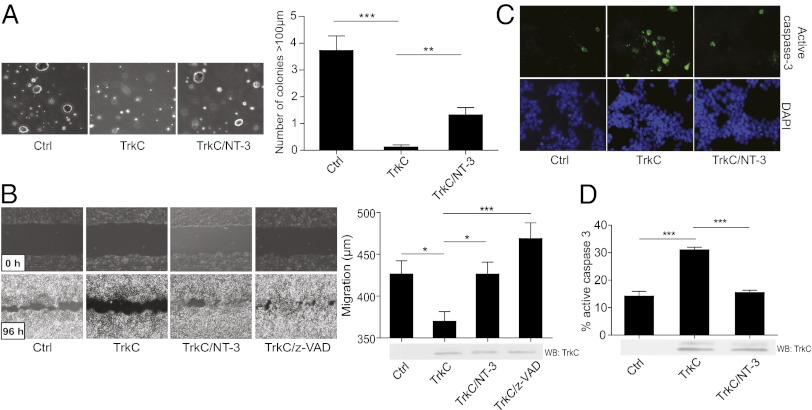
The very frequent loss or reduction of TrkC expression in colorectal cancer suggests that the presence of TrkC acts as a constraint on colorectal tumor development. Therefore, we analyzed whether TrkC inhibits in vitro the phenotypic hallmarks of malignant transformation: anchorage-independent growth and the ability to migrate (31). HCT116 cells were transiently transfected with a TrkC-expressing construct and allowed to grow in soft agar. Expression of TrkC receptor in HCT116 cells prominently inhibited growth in soft agar (Fig. 4A and Fig. S4A). Furthermore, by following the closure of a scratch performed in a HCT116 monolayer (Fig. 4B), or their migration through a transwell device (Fig. S4B), we observed that TrkC expression inhibited the migratory capacity of HCT116 cells. This observation is somewhat surprising, because one might expect forced expression of a tyrosine kinase receptor to promote anchorage-independent growth and migration. Concurrently, when the same anchorage-independent growth and migration assays were performed in the presence of the TrkC ligand NT-3, the TrkC effect was antagonized (Fig. 4 A and B and Fig. S4B). Together, these data support the view that unliganded TrkC constrains colorectal cancer cell growth and migration, supporting the hypothesis that TrkC functions as a dependence receptor that is frequently down-regulated in colorectal malignancies.
Fig. 4.
Re-expression of TrkC limits the hallmarks of colorectal cancer cells transformation via apoptosis induction. (A) Control, TrkC, or both TrkC and NT-3 overexpressing HCT116 cells were grown in soft agar for two weeks. The number of colonies was counted in five random fields and the average number per field was calculated. Data represent mean ± SEM *P < 0.05, ***P < 0.001, two-sided Mann–Whitney test, compared with control. Photographs of representative colonies for each condition are shown. (B) Scratch assay was performed on HCT116 cells expressing control vector and TrkC with or without NT-3 and z-VAD-fmk addition. The scratch open area was measured at 0 h and 96 h and the migration distance was determined. Representative photographs are shown. (C and D) Control, TrkC or both TrkC and NT-3 overexpressing HCT116 cells were immunolabeled for active caspase-3. Representative photographs are shown in C. The quantification and the corresponding Western blot controlling TrkC expression level are shown in D. ***P < 0.001, two-sided Mann–Whitney test.
We next determined whether this in vitro tumor suppressive effect is related to the ability of TrkC to behave as a dependence receptor. As noted above, we observed that upon scratch or transwell assays, the inhibitory effect of enforced expression of TrkC was completely inhibited by addition of the general apoptosis inhibitor zVAD-fmk (Fig. 4B and Fig. S4B). TrkC was then expressed in HCT116 cells, and cell death was determined by following caspase-3 activation, either by measurement of caspase-3 substrate cleavage (Fig. S4A) or by cleaved caspase-3 immunostaining (Fig. 4 C and D). As shown in Fig. 4 C and D and Fig. S4C, enforced TrkC expression efficiently triggered HCT116 cell apoptosis, and the addition of NT-3 was sufficient to block TrkC-induced apoptosis. Similar results were obtained with the HCT8 TrkC-negative colon cancer cell line (Fig. S4 D and E). Moreover, we observed that enforced expression of the TrkC domain that is necessary and sufficient to induce apoptosis, the so-called TrkC Killer Fragment (TrkC KF) (6), is sufficient to inhibit anchorage-independent growth to an extent similar to that of TrkC (Fig. S4C). Together, these data support the view that TrkC is a colon cancer tumor suppressor due to its dependence receptor activity.
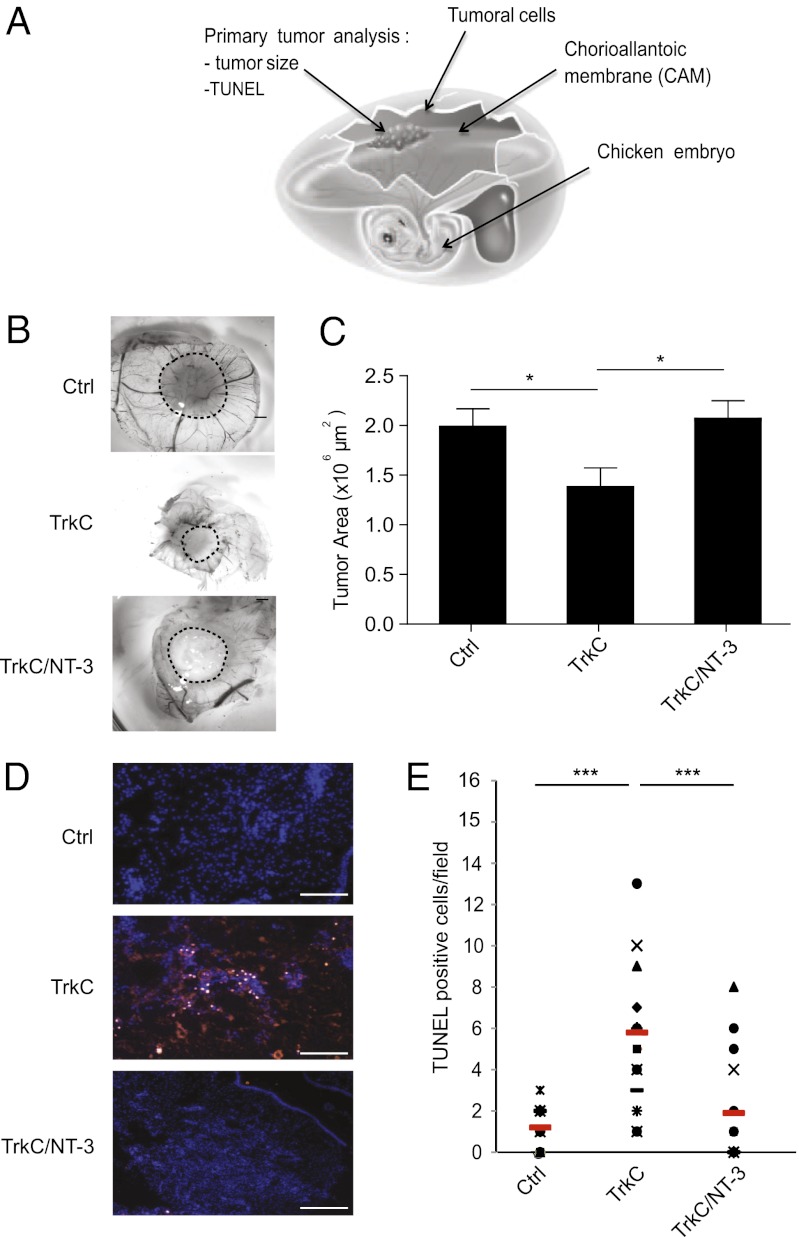
To analyze the tumor suppressive activity of TrkC in vivo, HCT116 cells with or without enforced TrkC expression were xenografted to the chorioallantoic membrane (CAM) of 10-d-old chicken embryos (Fig. 5A). The CAM of chicken embryos is a well described model to study primary tumor growth and metastasis (25, 26, 32). Seventeen-day-old chicken embryos were analyzed for primary tumor size. As shown in Fig. 5 B and C, tumors derived from the TrkC-transfected cells were significantly smaller than tumors derived from mock-transfected cells. TUNEL staining of the tumors revealed that the inhibition of tumor growth triggered by unliganded TrkC was associated with its proapoptotic activity, because an increased number of apoptotic cells was detected in tumors derived from TrkC expressing cells compared with control tumors (Fig. 5 D and E). Conversely, the presence of NT-3 completely suppressed the inhibitory activity of TrkC, supporting the view that TrkC functions as a colon cancer tumor suppressor in vivo by virtue of its dependence receptor effect.
Fig. 5.
TrkC expression induces tumor growth inhibition in vivo. (A) Schematic representation of the experimental chick model. HCT116 cells, transiently transfected with various plasmids, were grafted in CAM at day 10. Tumors were harvested on day 17, measured, and sectioned for TUNEL staining. n = 10–15 for each condition. (B) Representative images of HCT116 primary tumors formed. HCT116 cells were either transfected with empty vector (Ctrl), TrkC and treated with NT-3 (10 ng/mL) or not, before graft. (Scale bar: 2 μm.) (C) Quantitative analysis showing the size of the respective primary tumors described in B; *P < 0.05, two-sided Mann–Whitney test. (D) Representative images of TUNEL-positive cells in sections performed on the respective primary tumors described in B. (Scale bar: 100 μm.) (E) Quantification of the TUNEL-positive cells described in D. The red bars indicate the respective mean of the various measures performed for each sample. ***P < 0.001, two-sided Mann–Whitney test.
TrkC Gene Is Mutated in Human Sporadic Cancer.
The view that TrkC expression down-regulation is selected in most sporadic colorectal cancers, and that this down-regulation is associated with a reduction in tumor cell death, suggests the possibility that some sporadic cancers may exhibit specific loss-of-proapoptotic function mutations rather than overall down-regulation of TrkC. We therefore searched for putative TrkC gene mutations in the panel of tumors submitted to deep-sequencing and included in the Cosmic database (www.sanger.ac.uk/perl/genetics/CGP/cosmic?action=gene&ln=NTRK3) or described in ref. 33. As shown in Fig. S5A, somatic missense mutations of TrkC have been detected in gastrointestinal tract cancers. Of interest, several missense mutations were detected within the killer fragment of TrkC (amino acids 495–641), which also encompasses part of the TrkC tyrosine kinase domain (amino acids 538–839). Therefore, we determined whether these mutations were gain-of-function mutations constitutively inducing kinase activation (as classically observed for other tyrosine kinase receptors) or whether they represented instead loss-of-proapoptotic function mutations, as expected based on the dependence receptor paradigm. The various mutations shown in Fig. S5A were therefore introduced into a TrkC-expressing construct, focusing on the TrkC E543D and D584E mutations, and these were expressed in HCT116 cells and HCT8 cells (Fig. S5B). In both HCT116 (Fig. S5 B and C) and HCT8 (Fig. S5D) cells, the E543D mutation did not affect TrkC proapoptotic activity; however, the expression of the TrkC E543D mutant constitutively induced Erk-1/2 activation, compared with wild-type TrkC (Fig. S5E). Thus, as expected for a tyrosine kinase receptor acting as a classical oncogene, the sporadic mutation E543D is most likely to be a gain-of-function mutation. In contrast, however, the D584E of TrkC did not affect Erk-1/2 activity (Fig. S5E) but completely abrogated TrkC proapoptotic activity (Fig. S5 B–D). Furthermore, one of the detected mutations (TrkC E556*) is a nonsense mutation that truncated TrkC aminoterminally to most of its proapoptotic domain; in addition, we failed to detect any TrkC protein production when expressing D565H and D609V mutants. These data demonstrate that at least two different classes of tumor-associated TrkC mutants may be distinguished: those that result in a gain-of-oncogenic function, and those that result in a loss-of-proapoptotic-function, destroying the tumor-suppressive effect of TrkC.
Discussion
Here we have presented evidence that TrkC is a colorectal cancer tumor suppressor, whose function is abrogated or strongly down-regulated in the vast majority of colorectal tumors. In support of the hypothesis that TrkC functions as a dependence receptor, we show that a reduction in TrkC expression allows survival in settings of NT-3 limitation, which may be a selective advantage for colorectal cancer cells. In human colorectal tumors, this selective advantage is achieved primarily by promoter methylation.
According to the dependence receptor theory, an equivalent selective advantage to losing TrkC would occur through a gain of autocrine expression of NT-3. However, whereas it was shown that in other neoplasms such as neuroblastomas, NT-3 is part of an autocrine loop that blocks TrkC-induced apoptosis (26), this mechanism is absent or rare in colon cancer. Why TrkC is down-regulated in some tumors, such as colorectal cancer, whereas NT-3 expression is gained in others, remains to be investigated. Of interest, although the detected gain of NT-3 in some tumors could be interpreted as a classic oncogenic stimulation of a tyrosine kinase receptor, similarly to what is seen for many other oncogenic tyrosine kinase receptors (34, 35), the loss of TrkC expression in colorectal cancer clearly does not support the notion that TrkC functions solely as an oncogenic tyrosine kinase receptor. Instead, these findings support the view that TrkC is a tumor suppressor, and we present evidence both in vitro and in vivo that TrkC acts as a tumor suppressor via its dependence receptor function. Moreover, the expression of TrkC is a marker of favorable outcome in cancers of nervous tissue origin such as neuroblastoma (36, 37) and medulloblastoma (38, 39), in favor of the argument that in most cancers, TrkC is primarily a tumor suppressor, via its dependence receptor function, rather than a proto-oncogene via its tyrosine kinase activity. This does not hold true, however, for the translocation resulting in the ETV6 fusion to the intracellular domain of TrkC first reported in acute myeloid leukemia with t(12, 15)(p13;q25) (40) and later also observed in breast carcinoma (41), T-cell lymphoma (42), and congenital fibrosarcoma (43, 44), for which the balance between TrkC proapoptotic activity and prokinase activity is probably shifted toward the prokinase activity.
Although the data presented here support the hypothesis that TrkC triggers apoptosis of cancer cells or precancerous cells in settings of ligand limitation, it may be of interest to compare the effects of TrkA expression to those of TrkC expression. Although TrkA has recently been shown to be a dependence receptor, triggering apoptosis in the absence of ligand (7), and although preliminary data support the view that TrkA expression is also decreased in colorectal cancer, TrkA, in contrast to TrkC, has been reported to induce apoptosis not only following NGF removal, but also in at least some cancer cell lines when engaged by NGF. For example, NGF treatment of TrkA-expressing medulloblastoma cells induces cell death (45), and pheochromocytoma PC12 cells expressing TrkA were rendered susceptible to cell death following NGF-treatment (46). Similarly, the expression of TrkA in rat glioma cell line reduced its invasiveness in correlation with a higher rate of apoptosis (47), whereas TrkA-transfected U2OS osteosarcoma cells also underwent cell death (48). These results with TrkA and NGF raise the question of whether TrkC functions as a tumor suppressor only via its dependence receptor activity, or whether NT-3 contributes to this activity. To date, we have not observed NT-3 mediated TrkC-induced apoptosis. Moreover, the absence of significant decrease of NT-3 expression in colon cancer supports the notion that the tumor-suppressive activity of TrkC is explained by its dependence receptor function.
Bardelli et al. (33) reported the spectrum of mutations within the kinase domain of tyrosine kinase proteins detected in sporadic colorectal cancers, with the view that these mutations were gain-of-function, oncogenic mutations. Interestingly, TrkC was one of the most frequently mutated kinases identified (33). That report, together with the recent effort in deep-sequencing of human tumors, demonstrates that TrkC displays a large spectrum of missense mutations. The large spectrum of mutations covering the entire TrkC coding sequence (Fig. S5) is a further argument in support of a role of TrkC as a tumor suppressor. Moreover, here we distinguish two classes of tumor-associated TrkC mutants: those acting as oncogenic mutants (such as TrkC E543D), and those acting to abrogate the TrkC proapoptotic effect. It remains to be investigated whether or not each member of this latter class of mutants affects the generation of the proapoptotic TrkC killer fragment, the recruitment of proapoptotic proteins to this killer fragment, or the TrkC protein stability. Ongoing deep-sequencing should reveal novel mutations of TrkC in neoplasms, and further work should determine whether these mutations are oncogenic-gain-of-function mutations or, as shown above, they are loss-of-proapoptotic-function mutations. We present data demonstrating that the TrkC dependence receptor is a colon cancer tumor suppressor and, as such, plays a similar role to DCC and UNC5H, other dependence receptors whose expression is abrogated or markedly reduced in colorectal cancers (20, 22, 49). Similarly to what we observed for TrkC, promoter methylation is a mechanism that underlies colorectal cancer-associated down-regulation of DCC and UNC5H. This result is of particular interest in the light of the currently clinically tested inhibitors of methylation or of histone deacetylases that have an anticancer effect (for review, see ref. 50). Whether part of this efficiency is due to reactivation of DCC, UNC5H, or TrkC dependence receptor and associated apoptosis induction remains to be investigated.
Experimental Procedures
A full description of the experimental procedures is given in SI Experimental Procedures.
Analysis of Colorectal Samples.
This study is based on 90 colorectal samples from a cohort of 45 patients. To assay TrkC expression in human colorectal samples, total RNA was extracted from biopsies of patients undergoing surgery for colorectal cancer. Q-RT-PCR was performed using standard procedures and primers as described in SI Experimental Procedures. Immunostaining of patient biopsies sections was performed with an anti-TrkC antibody. Sections were counterstained with a Mayer’s Hematoxylin coloration.
Cell Lines, Treatment, Plasmids, TrkC Site-Directed Mutagenesis, and Transfection Procedure.
Colorectal cancer cell lines HCT116, SW480, HT29, V9P, Caco-2, Colo320, HCT-15, SW48, SW620, SW116, SW480, ISP1, LS1034, SW837, and HCT8 have been described. The HCT116 double knockout for DNMT1 and DNMT3b (DKO) have been described and were kindly provided by B. Vogelstein (Ludwig Center at Johns Hopkins, Baltimore, MD) (30). The full-length TrkC expressing plasmid was described (6). Mutations were generated on the human TrkC gene using the QuikChange Site-Directed Mutagenesis kit (Stratagene). The plasmid constructs were transfected using JetPrime transfectant (PolyPlus) following manufacturer’s instructions.
Supplementary Material
Acknowledgments
We thank Prof. P. J. Donovan, Dr. H. Fong, Prof. P. H. Sorensen, and Prof. Y. A. Barde for materials; R. Dante, M. Grandin, G. Devailly, E. Dardenne, and B. Gras for important advice; H. Bilak and Prof D. E. Bredesen for editing the manuscript; C. Guix, A. S. Campos, P. A. Bissey, C. Cuenin, B. Boucher, and G. Grelier for technical help; and C. Rey in the ProfileXpert platform for the laser microdissection. This work was supported by institutional grants from Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, Centre Léon Bérard, and University of Lyon; and grants from the Ligue Contre le Cancer, Fondation pour la Recherche Médicale, Institut National du Cancer, l'Agence Nationale de la Recherche, Association pour la Recherche contre le Cancer, Cancéropole Rhone Alpes Auvergne, and the European Research Council.
Footnotes
The authors declare no conflict of interest.
†This Direct Submission article had a prearranged editor.
See Commentary on page 2697.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212333110/-/DCSupplemental.
References
- 1.Martin-Zanca D, Hughes SH, Barbacid M. A human oncogene formed by the fusion of truncated tropomyosin and protein tyrosine kinase sequences. Nature. 1986;319(6056):743–748. doi: 10.1038/319743a0. [DOI] [PubMed] [Google Scholar]
- 2.Nakagawara A. Trk receptor tyrosine kinases: A bridge between cancer and neural development. Cancer Lett. 2001;169(2):107–114. doi: 10.1016/s0304-3835(01)00530-4. [DOI] [PubMed] [Google Scholar]
- 3.Marshall JL, et al. Phase I trial of orally administered CEP-701, a novel neurotrophin receptor-linked tyrosine kinase inhibitor. Invest New Drugs. 2005;23(1):31–37. doi: 10.1023/B:DRUG.0000047103.64335.b0. [DOI] [PubMed] [Google Scholar]
- 4.Knapper S, et al. A phase 2 trial of the FLT3 inhibitor lestaurtinib (CEP701) as first-line treatment for older patients with acute myeloid leukemia not considered fit for intensive chemotherapy. Blood. 2006;108(10):3262–3270. doi: 10.1182/blood-2006-04-015560. [DOI] [PubMed] [Google Scholar]
- 5.Chan E, et al. A phase I trial of CEP-701 + gemcitabine in patients with advanced adenocarcinoma of the pancreas. Invest New Drugs. 2008;26(3):241–247. doi: 10.1007/s10637-008-9118-3. [DOI] [PubMed] [Google Scholar]
- 6.Tauszig-Delamasure S, et al. The TrkC receptor induces apoptosis when the dependence receptor notion meets the neurotrophin paradigm. Proc Natl Acad Sci USA. 2007;104(33):13361–13366. doi: 10.1073/pnas.0701243104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nikoletopoulou V, et al. Neurotrophin receptors TrkA and TrkC cause neuronal death whereas TrkB does not. Nature. 2010;467(7311):59–63. doi: 10.1038/nature09336. [DOI] [PubMed] [Google Scholar]
- 8.Mehlen P, et al. The DCC gene product induces apoptosis by a mechanism requiring receptor proteolysis. Nature. 1998;395(6704):801–804. doi: 10.1038/27441. [DOI] [PubMed] [Google Scholar]
- 9.Matsunaga E, et al. RGM and its receptor neogenin regulate neuronal survival. Nat Cell Biol. 2004;6(8):749–755. doi: 10.1038/ncb1157. [DOI] [PubMed] [Google Scholar]
- 10.Thibert C, et al. Inhibition of neuroepithelial patched-induced apoptosis by sonic hedgehog. Science. 2003;301(5634):843–846. doi: 10.1126/science.1085405. [DOI] [PubMed] [Google Scholar]
- 11.Bordeaux MC, et al. The RET proto-oncogene induces apoptosis: A novel mechanism for Hirschsprung disease. EMBO J. 2000;19(15):4056–4063. doi: 10.1093/emboj/19.15.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furne C, et al. EphrinB3 is an anti-apoptotic ligand that inhibits the dependence receptor functions of EphA4 receptors during adult neurogenesis. Biochim Biophys Acta. 2009;1793(2):231–238. doi: 10.1016/j.bbamcr.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boucher J, et al. A kinase-independent role for unoccupied insulin and IGF-1 receptors in the control of apoptosis. Sci Signal. 2010;3(151):ra87. doi: 10.1126/scisignal.2001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mourali J, et al. Anaplastic lymphoma kinase is a dependence receptor whose proapoptotic functions are activated by caspase cleavage. Mol Cell Biol. 2006;26(16):6209–6222. doi: 10.1128/MCB.01515-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldschneider D, Mehlen P. Dependence receptors: A new paradigm in cell signaling and cancer therapy. Oncogene. 2010;29(13):1865–1882. doi: 10.1038/onc.2010.13. [DOI] [PubMed] [Google Scholar]
- 16.Mehlen P, Delloye-Bourgeois C, Chédotal A. Novel roles for Slits and netrins: Axon guidance cues as anticancer targets? Nat Rev Cancer. 2011;11(3):188–197. doi: 10.1038/nrc3005. [DOI] [PubMed] [Google Scholar]
- 17.Cho KR, et al. The DCC gene: Structural analysis and mutations in colorectal carcinomas. Genomics. 1994;19(3):525–531. doi: 10.1006/geno.1994.1102. [DOI] [PubMed] [Google Scholar]
- 18.Thiebault K, et al. The netrin-1 receptors UNC5H are putative tumor suppressors controlling cell death commitment. Proc Natl Acad Sci USA. 2003;100(7):4173–4178. doi: 10.1073/pnas.0738063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehlen P, Fearon ER. Role of the dependence receptor DCC in colorectal cancer pathogenesis. J Clin Oncol. 2004;22(16):3420–3428. doi: 10.1200/JCO.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 20.Bernet A, et al. Inactivation of the UNC5C Netrin-1 receptor is associated with tumor progression in colorectal malignancies. Gastroenterology. 2007;133(6):1840–1848. doi: 10.1053/j.gastro.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castets M, et al. DCC constrains tumour progression via its dependence receptor activity. Nature. 2012;482(7386):534–537. doi: 10.1038/nature10708. [DOI] [PubMed] [Google Scholar]
- 22.Coissieux MM, et al. Variants in the netrin-1 receptor UNC5C prevent apoptosis and increase risk of familial colorectal cancer. Gastroenterology. 2011;141(6):2039–2046. doi: 10.1053/j.gastro.2011.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitamant J, et al. Netrin-1 expression confers a selective advantage for tumor cell survival in metastatic breast cancer. Proc Natl Acad Sci USA. 2008;105(12):4850–4855. doi: 10.1073/pnas.0709810105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delloye-Bourgeois C, et al. Interference with netrin-1 and tumor cell death in non-small cell lung cancer. J Natl Cancer Inst. 2009;101(4):237–247. doi: 10.1093/jnci/djn491. [DOI] [PubMed] [Google Scholar]
- 25.Delloye-Bourgeois C, et al. Netrin-1 acts as a survival factor for aggressive neuroblastoma. J Exp Med. 2009;206(4):833–847. doi: 10.1084/jem.20082299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bouzas-Rodriguez J, et al. Neurotrophin-3 production promotes human neuroblastoma cell survival by inhibiting TrkC-induced apoptosis. J Clin Invest. 2010;120(3):850–858. doi: 10.1172/JCI41013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shibayama E, Koizumi H. Cellular localization of the Trk neurotrophin receptor family in human non-neuronal tissues. Am J Pathol. 1996;148(6):1807–1818. [PMC free article] [PubMed] [Google Scholar]
- 28.Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci USA. 2000;97(18):10014–10019. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 30.Rhee I, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416(6880):552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 31.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 32.Stupack DG, Puente XS, Boutsaboualoy S, Storgard CM, Cheresh DA. Apoptosis of adherent cells by recruitment of caspase-8 to unligated integrins. J Cell Biol. 2001;155(3):459–470. doi: 10.1083/jcb.200106070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bardelli A, et al. Mutational analysis of the tyrosine kinome in colorectal cancers. Science. 2003;300(5621):949. doi: 10.1126/science.1082596. [DOI] [PubMed] [Google Scholar]
- 34.Miknyoczki SJ, et al. The neurotrophin-trk receptor axes are critical for the growth and progression of human prostatic carcinoma and pancreatic ductal adenocarcinoma xenografts in nude mice. Clin Cancer Res. 2002;8(6):1924–1931. [PubMed] [Google Scholar]
- 35.Ohta T, et al. Neurotrophin-3 expression in human pancreatic cancers. J Pathol. 1997;181(4):405–412. doi: 10.1002/(SICI)1096-9896(199704)181:4<405::AID-PATH786>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 36.Svensson T, et al. Coexpression of mRNA for the full-length neurotrophin receptor trk-C and trk-A in favourable neuroblastoma. Eur J Cancer. 1997;33(12):2058–2063. doi: 10.1016/s0959-8049(97)00290-6. [DOI] [PubMed] [Google Scholar]
- 37.Brodeur GM, Maris JM, Yamashiro DJ, Hogarty MD, White PS. Biology and genetics of human neuroblastomas. J Pediatr Hematol Oncol. 1997;19(2):93–101. doi: 10.1097/00043426-199703000-00001. [DOI] [PubMed] [Google Scholar]
- 38.Grotzer MA, et al. TrkC expression predicts good clinical outcome in primitive neuroectodermal brain tumors. J Clin Oncol. 2000;18(5):1027–1035. doi: 10.1200/JCO.2000.18.5.1027. [DOI] [PubMed] [Google Scholar]
- 39.Segal RA, Goumnerova LC, Kwon YK, Stiles CD, Pomeroy SL. Expression of the neurotrophin receptor TrkC is linked to a favorable outcome in medulloblastoma. Proc Natl Acad Sci USA. 1994;91(26):12867–12871. doi: 10.1073/pnas.91.26.12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eguchi M, et al. Fusion of ETV6 to neurotrophin-3 receptor TRKC in acute myeloid leukemia with t(12;15)(p13;q25) Blood. 1999;93(4):1355–1363. [PubMed] [Google Scholar]
- 41.Tognon C, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2(5):367–376. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 42.Yagasaki F, et al. Fusion of ETV6 to fibroblast growth factor receptor 3 in peripheral T-cell lymphoma with a t(4;12)(p16;p13) chromosomal translocation. Cancer Res. 2001;61(23):8371–8374. [PubMed] [Google Scholar]
- 43.Dubus P, et al. The detection of Tel-TrkC chimeric transcripts is more specific than TrkC immunoreactivity for the diagnosis of congenital fibrosarcoma. J Pathol. 2001;193(1):88–94. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH724>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 44.Knezevich SR, McFadden DE, Tao W, Lim JF, Sorensen PH. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat Genet. 1998;18(2):184–187. doi: 10.1038/ng0298-184. [DOI] [PubMed] [Google Scholar]
- 45.Muragaki Y, Chou TT, Kaplan DR, Trojanowski JQ, Lee VM. Nerve growth factor induces apoptosis in human medulloblastoma cell lines that express TrkA receptors. J Neurosci. 1997;17(2):530–542. doi: 10.1523/JNEUROSCI.17-02-00530.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan C, Liang Y, Nylander KD, Schor NF. TrkA as a life and death receptor: Receptor dose as a mediator of function. Cancer Res. 2002;62(17):4867–4875. [PubMed] [Google Scholar]
- 47.Lachyankar MB, et al. TrkA expression decreases the in vivo aggressiveness of C6 glioma cells. Cancer Res. 1997;57(3):532–536. [PubMed] [Google Scholar]
- 48.Dadakhujaev S, et al. Interplay between autophagy and apoptosis in TrkA-induced cell death. Autophagy. 2009;5(1):103–105. doi: 10.4161/auto.5.1.7276. [DOI] [PubMed] [Google Scholar]
- 49.Shin SK, et al. Epigenetic and genetic alterations in Netrin-1 receptors UNC5C and DCC in human colon cancer. Gastroenterology. 2007;133(6):1849–1857. doi: 10.1053/j.gastro.2007.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren J, et al. DNA hypermethylation as a chemotherapy target. Cell Signal. 2011;23(7):1082–1093. doi: 10.1016/j.cellsig.2011.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.