Abstract
The derivation of induced pluripotent stem (iPS) cells from individuals of genetic disorders offers new opportunities for basic research into these diseases and the development of therapeutic compounds. Severe congenital neutropenia (SCN) is a serious disorder characterized by severe neutropenia at birth. SCN is associated with heterozygous mutations in the neutrophil elastase [elastase, neutrophil-expressed (ELANE)] gene, but the mechanisms that disrupt neutrophil development have not yet been clarified because of the current lack of an appropriate disease model. Here, we generated iPS cells from an individual with SCN (SCN-iPS cells). Granulopoiesis from SCN-iPS cells revealed neutrophil maturation arrest and little sensitivity to granulocyte-colony stimulating factor, reflecting a disease status of SCN. Molecular analysis of the granulopoiesis from the SCN-iPS cells vs. control iPS cells showed reduced expression of genes related to the wingless-type mmtv integration site family, member 3a (Wnt3a)/β-catenin pathway [e.g., lymphoid enhancer-binding factor 1], whereas Wnt3a administration induced elevation lymphoid enhancer-binding factor 1-expression and the maturation of SCN-iPS cell-derived neutrophils. These results indicate that SCN-iPS cells provide a useful disease model for SCN, and the activation of the Wnt3a/β-catenin pathway may offer a novel therapy for SCN with ELANE mutation.
Keywords: apoptosis, unfolded protein response, SCN disease model
Severe congenital neutropenia (SCN) is a heterogeneous bone marrow (BM) failure syndrome characterized by severe neutropenia at birth, leading to recurrent infections by bacteria or fungi (1). SCN patients reveal an arrest in neutrophil differentiation in the BM at the promyelocyte or myelocyte stage (1), as well as a propensity to develop myelodysplastic syndrome and acute myeloid leukemia (2). Current treatment by high-dose granulocyte-colony stimulating factor (G-CSF) administration induces an increase in the number of mature neutrophils in the peripheral blood of most SCN patients (3). Although this treatment is curative for the severe infections, there is a concern that high-dose G-CSF may increase the risk of hematologic malignancy in these individuals (4).
Several genetic mutations have been identified in SCN patients. Approximately 50% of autosomal-dominant SCN cases were shown to have various heterozygous mutations in the gene encoding neutrophil elastase [elastase, neutrophil-expressed (ELANE)] (5, 6), a monomeric, 218-amino acid (25 kDa) chymotryptic serine protease (7) that is synthesized during the early stages of primary granule production in promyelocytes (8, 9). However, the mechanism(s) causing impaired neutrophil maturation in SCN patients remains unclear due to the current lack of an appropriate disease model.
Results and Discussion
In the present study, we generated induced pluripotent stem (iPS) cells from the BM cells obtained from an SCN patient with a heterologous ELANE gene mutation (exon 5, 707 region, C194X) (SCN-iPS cells) to provide the basis for an SCN disease model. The patient who donated BM cells recurrently suffered from severe infections without exogenous G-CSF administration, but the G-CSF administration once a week prevented his repeated infection. The SCN-iPS cells continued to show embryonic stem cell morphology after >20 passages and also expressed pluripotent markers (Fig. S1A). The silencing of exogenous genes and the capability to differentiate into three germ layers by teratoma formation were confirmed for each of the three SCN-iPS cell clones (Fig. S1 B and C). Furthermore, the same ELANE gene mutation that was present in the patient persisted in the SCN-iPS cells (Fig. S1D). The SCN-iPS cells, as well as control iPS cells that were generated from healthy donors, had the normal karyotype (Fig. S1E) (10, 11) and no mutations in the mutation-sensitive region of the G-CSF receptor gene (12).
We first compared the hematopoietic differentiation from SCN-iPS cells with that from control iPS cells that were generated from healthy donors. SCN-iPS and control iPS cells were cocultured with a 15-Gy-irradiated murine stromal cell line (the AGM-S3 cell line), as reported (13). After 12 d, the cocultured cells were harvested, and the CD34+ cells separated from these cells (SCN-iPS–CD34+ and control iPS–CD34+ cells, respectively) were cultured in a hematopoietic colony assay by using a cytokine mixture (Materials and Methods). The number and size of the erythroid (E) and mixed-lineage (Mix) colonies derived from SCN-iPS–CD34+ cells (1 × 104 cells) were nearly identical to those of the corresponding colonies derived from control iPS–CD34+ cells (E colonies: SCN-iPS cells, 11.0 ± 3.0, and control iPS cells, 11.4 ± 3.9; Mix colonies: SCN-iPS cells, 25.1 ± 7.2, and control iPS cells, 17.4 ± 4.0) (Fig. 1 B and C and Fig. S2 A and B). However, the number of myeloid colonies derived from SCN-iPS–CD34+ vs. control iPS–CD34+ cells was significantly lower (SCN-iPS cells, 47.4 ± 19.5; control iPS cells, 127.8 ± 17.9; P < 0.01), and the size of the colonies was also smaller (Fig. 1 A and D). In particular, only a few SCN-iPS cell-derived granulocyte (G) colonies—myeloid colonies consisting of only granulocytes—were detected (Fig. 1A). SCN-iPS cell-derived granulocyte–macrophage (GM) colonies—myeloid colonies consisting of macrophages/monocytes with/without granulocytes—contained a few immature myeloid cells in addition to macrophages/monocytes, whereas control iPS cell-derived GM colonies included a substantial number of mature, segmented, and band neutrophils (Fig. 1D).
Fig. 1.
Impaired neutrophil development from SCN-iPS cells. (A–C) A hematopoietic colony assay was performed by using 1 × 104 CD34+ cells derived from three SCN-iPS cell clones (SPN0101, SPN0102, and SPN0103) and three control iPS cell clones (controls 1, 2, and 3) in the presence of a cytokine mixture. Colonies were sorted as myeloid (A), erythroid (B), and mixed-lineage (Mix) (C). Data are shown as mean ± SD. (D) Photographs of colonies (Left; 100×) and cells in a GM colony (Right; 400×; May–Grünwald–Giemsa staining). (E) A hematopoietic colony assay with dose escalation of G-CSF was performed by using 1 × 105 CD34+ cells derived from SCN-iPS and control iPS cells. Filled and open bars indicate small colonies consisting of <100 cells and large colonies consisting of >100 cells, respectively. Data are shown as the average of three independent experiments. (F) Photographs of a small colony derived from SCN-iPS cells (SPN0102) in the presence of 10 ng/mL G-CSF, large colonies derived from SCN-iPS cells in the presence of 1,000 ng/mL G-CSF, and large colonies derived from control iPS cells (control 1) in the presence of 10 ng/mL G-CSF. (Scale bars, 200 μm.)
We also found that Mix colonies derived from SCN-iPS cells, but not control iPS cells, contained immature myeloid cells and few mature neutrophils (Fig. S2 C and D). Next, we conducted a hematopoietic colony assay using various concentrations of G-CSF alone instead of the cytokine mixture to examine the G-CSF dose dependency of neutrophil differentiation from SCN-iPS and control iPS–CD34+ cells. For all concentrations of G-CSF used (1–1,000 ng/mL), the SCN-iPS cell-derived myeloid colonies were significantly lower in number and smaller in size than the control iPS cell-derived myeloid colonies (Fig. 1E). Myeloid colony formation from control iPS cells reached a plateau at ∼1–10 ng/mL G-CSF, whereas the number and size of those from SCN-iPS cells gradually increased with increasing concentrations of G-CSF. However, the values observed for SCN-iPS cells did not reach those for the control iPS cells, even at the highest dose of G-CSF used (1,000 ng/mL). Furthermore, large colonies consisting of >100 cells derived from SCN-iPS cells were only found with higher concentrations of G-CSF (Fig. 1F). Thus, granulopoiesis initiated from SCN-iPS cells was relatively insensitive to G-CSF, reflecting the inadequate in vivo response of neutrophils to G-CSF in SCN patients (14, 15). Therefore, these results support the applicability of the SCN-iPS cells established herein as a disease model for SCN.
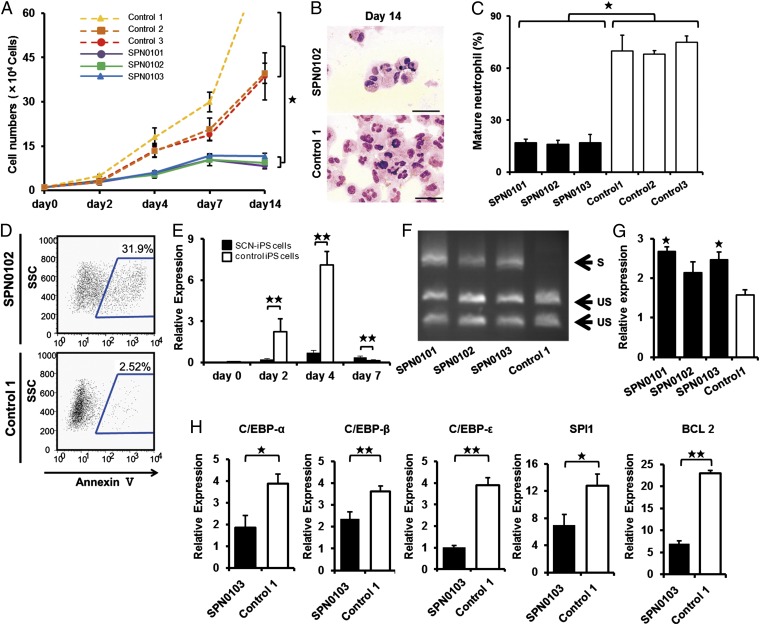
To examine neutrophil development from SCN-iPS cells in more detail, SCN-iPS and control iPS–CD34+ cells (1 × 104 cells each) were cocultured in suspension with AGM-S3 cells in the presence of neutrophil differentiation medium (SI Materials and Methods). The number of nonadherent cells derived from SCN-iPS–CD34+ cells was lower than that from control iPS–CD34+ cells on day 14 of culture (SCN-iPS cells, 9.77 × 104 ± 1.65 × 104 cells; control iPS cells, 52.48 × 104 ± 23.13 × 104 cells; P < 0.05) (Fig. 2A). The proportion of mature neutrophils among the nonadherent cells was also significantly lower for SCN-iPS cells relative to control iPS cells on day 14 (SPN-iPS cells, 15.53% ± 4.33%; control iPS cells, 71.285 ± 3.30%; P < 0.05) (Fig. 2 B and C), indicating that myeloid cells derived from SCN-iPS cells revealed the maturation arrest in the neutrophil development. We then examined a possibility that the maturation arrest in SCN-iPS cell-derived myeloid cells might be caused by their apoptosis. In flow cytometric analysis, SCN-iPS cell-derived myeloid cells contained a significantly higher proportion of annexin V-positive cells than control iPS-derived myeloid cells on day 7 of culture, suggesting that the maturation arrest in myeloid cells derived from SCN-iPS cells might be caused by their apoptosis (Fig. 2D).
Fig. 2.
Analysis of impaired neutrophil development from SCN-iPS cells. (A) Total number of nonadherent cells in the suspension culture of 1 × 104 CD34+ cells derived from SCN-iPS and control iPS cells. Data are shown as mean ± SD. ★P < 0.01. (B) Photographs of nonadherent cells derived from SCN-iPS (SPN0103) and control iPS cells (control 1) on day 14 of culture (400×; May–Grünwald–Giemsa staining; scale bars, 50 μm.) (C) Filled and open bars show the proportion of mature neutrophils among the cells derived from SCN-iPS (filled bars) and control iPS (open bars) cells on day 14 of suspension culture. Data are shown as mean ± SD. ★P < 0.05. (D) Flow cytometric analysis of annexin V expression on cultured cells from SCN-iPS cells (SPN0102) or control iPS cells (control 1) on day 7. (E) Sequential qRT-PCR analysis of the relative expression of ELANE mRNA [ELANE/hypoxanthine–guanine phosphoribosyltransferase (HPRT) expression]. Data obtained from independent experiments using three SCN-iPS cell clones (SPN0101, SPN0102, and SPN0103) and three control iPS cell clones are shown as mean ± SD. ★★P < 0.01. (F and G) CD34+ cells derived from SCN-iPS or control iPS cells were cultured in neutrophil differentiation medium (see text). On day 7, nonadherent cells were collected and analyzed. (F) Representative gel showing spliced (S) and unspliced (US) XBP-1 bands on day 7. (G) qRT-PCR analysis of the relative mRNA expression (target/HPRT expression) of BiP on day 7. Data are shown as mean ± SD. ★P < 0.05; different from control 1). (H) qRT-PCR analysis of the relative mRNA expression (target / HPRT expression) of C/EBP-α, C/EBP-β, C/EBP-ɛ, SPI1, and BCL2 genes in non-adherent cells derived from SCN-iPS cells (filled bars, SPN0103) and control iPS cells (open bars, control 1) on day 2 of suspension culture. Data are shown as the mean ± the s.d. (★★P < 0.01, ★P < 0.05).
We next examined ELANE mRNA expression levels in nonadherent cells derived from SCN-iPS vs. control iPS cells (Fig. 2E). ELANE expression was significantly lower in nonadherent cells derived from SCN-iPS vs. control iPS cells on days 2 and 4 of culture (P < 0.01), as reported (16, 17). However, the former was a little higher than the latter on day 7 (P < 0.01). This result may be explained by the existence of SCN-iPS cell-derived myeloid cells arrested at an early stage along the neutrophil differentiation pathway even on day 7 of culture. We also examined the expression of proteinase 3 and azurocidin, which comprise a family of closely related genes encoding neutrophil granule proteins along with ELANE, and found these genes were more highly expressed on day 4 (Fig. S3).
It has been reported that induction of the endoplasmic reticulum stress (ER) response and the unfolded protein response (UPR) has been advanced as a potential explanation for the molecular pathogenesis of SCN (18, 19). Thus, we examined activation of the UPR by X-box binding protein 1 (XBP-1) mRNA splicing on day 7. As shown in Fig. 2F, SPN-iPS cells induced XBP-1 mRNA splicing. We also found the up-regulation of BiP (also known as GRP78 or HSPA5) (Fig. 2G). These results suggested that ER stress response and UPR might be involved in the pathogenesis in SCN.
To examine further the differences in gene expression between the two cell types, a microarray analysis was carried out by using CD34+ cells derived from SCN-iPS and control iPS cells (three clones of each) in suspension culture on day 2. At this early time point, differences in cell number and morphology were not yet readily discernible between SCN-iPS and control iPS cells, as shown in Fig. 2A. However, the microarray analysis revealed a differential expression of various genes between the two cell types. Transcription factor genes, which were related to neutrophil development [e.g., CCAAT/enhancer-binding protein (C/EBP)-α (20), C/EBP-β (21), C/EBP-ɛ (22), and SPI1 (also known as PU.1) (23)], were all down-regulated in SCN-iPS cells. B-cell chronic lymphocytic leukemia/lymphoma 2, which regulates cell death under ER stress through the core mitochondrial apoptosis pathway (24), was also down-regulated (Fig. 3A). These findings were confirmed by quantitative reverse-transcriptional PCR (qRT-PCR), as shown in Fig. 2H.
Fig. 3.
Effects of Wnt3a on neutrophil development from SCN-iPS cells. (A) Heat map showing differential gene expression among SCN-iPS and control iPS cells on day 2. Red, high gene expression; blue, low gene expression compared with gene expression in control 3. (B) qRT-PCR analysis of the relative mRNA expression (target/HPRT expression) of Wnt3a on day 2. Filled and open bars indicate experiments using SCN-iPS cells (SPN0101, SPN0102, and SPN0103) and control iPS cells (controls 1, 2, and 3), respectively. Data are shown as mean ± SD. ★P < 0.05. (C) qRT-PCR analysis of the relative expression (target/HPRT expression) of genes regulated by the Wnt3a/β-catenin pathway (LEF-1, survivin, and cyclin D1) in SCN-iPS cells (filled bars, SPN0103) vs. control iPS cells (open bars, control 1) on day 2 of suspension culture. Data are shown as mean ± SD. ★★P < 0.01; ★P < 0.05. (D) Proportion of mature neutrophils among the cells derived from SCN-iPS cells (SPN0102) on day 14 of suspension culture with dose escalation of Wnt3a. Data are shown as mean ± SD. ★★P < 0.01. (E) Photographs of nonadherent cells on day 7 of suspension culture with or without Wnt3a (500 ng/mL) (400×; May–Grünwald–Giemsa staining).
Notably, the down-regulation of the genes in SCN-iPS cells related to and regulated by the wingless-type mmtv integration site family, member 3a (Wnt3a)/β-catenin pathway [e.g., Wnt3a, lymphoid enhance-binding factor (LEF)-1, BIRC5 (also known as survivin), and cyclin D1] was also uncovered by microarray analysis and qRT-PCR (Fig. 3 A–C and Fig. S4). Therefore, we examined the effect of enhancement of Wnt3a/β-catenin signaling by exogenous Wnt3a addition on the neutrophil development of CD34+ cells derived from SCN-iPS and control iPS cells. Although Wnt3a did not stimulate the survival, proliferation, and differentiation of CD34+ cells derived from both iPS cells in the absence of cytokines stimulating myelopoiesis including G-CSF, the addition of Wnt3a to the neutrophil differentiation medium induced a dose-dependent increase in the percentage of mature neutrophils among the cultured cells, as shown in Fig. 3 D and E. Furthermore, when Wnt3a was added concurrently with 1,000ng/mL G-CSF, the proportion of mature neutrophils increased more than it did with Wnt3a or 1,000 ng/mL G-CSF alone, reaching a value comparable with that observed for control iPS cells (Fig. 4 A and B).
Fig. 4.
Effects of Wnt3a in combination with high-dose G-CSF. (A) Filled and open bars show the proportion of mature neutrophils among the cells derived from SCN-iPS cells (SPN0101) on day 14 of suspension culture in the presence of neutrophil differentiation medium containing 10 ng/mL G-CSF (GCSF 10 ng/mL); 500 ng/mL Wnt3a and 10 ng/mL G-CSF (Wnt3a+GCSF 10 ng/mL); 1,000 ng/mL G-CSF (GCSF 1,000 ng/mL); or 500 ng/mL Wnt3a and 1,000 ng/mL G-CSF (Wnt3a + GCSF 1,000 ng/mL); and that from control iPS cells (controls 1 and 2) cultured in the neutrophil differentiation medium containing 10 ng/mL G-CSF, respectively. Data are shown as mean ± SD. ★★P < 0.01; ★P < 0.05. (B) The proportion of mature neutrophils among the cells derived from three SCN-iPS cell clones (SPN0101, SPN0102, and SPN0103) on day 14 of suspension culture in the presence of neutrophil differentiation medium containing 10 ng/mL G-CSF (GCSF 10 ng/mL); 500 ng/mL Wnt3a and 10 ng/mL G-CSF (Wnt3a+GCSF 10 ng/mL); or 500 ng/mL Wnt3a and 1,000 ng/mL G-CSF (Wnt3a + GCSF 1,000 ng/mL). Data are shown as mean ± SD. ★★P < 0.01; ★P < 0.05. (C) Filled and open bars show the relative expression (target/HPRT expression) of LEF-1 mRNA in SCN-iPS cells (SPN0101) on day 2 of suspension culture in the presence of differentiation medium containing the same combinations of Wnt3a and G-CSF as shown in A and that from control iPS cells (control 2), respectively. Data are shown as mean ± SD. ★★P < 0.01; ★P < 0.05. (D) Filled and open bars show the relative expression (target/HPRT expression) of C/EBP-α, BIRC5, or cyclin D1 mRNA in SCN-iPS cells (SPN0101) on day 2 of suspension culture in the presence of differentiation medium containing the same combinations of Wnt3a and G-CSF as shown in A and that from control iPS cells (control 2), respectively. Data are shown as mean ± SD. ★★P < 0.01; ★P < 0.05. (E) Total cell numbers of nonadherent cells in suspension cultures of 1 × 104 CD34+ cells derived from control iPS cells (control 2; red broken line) and SCN-iPS cells (SPN0101) in the presence of neutrophil differentiation medium (black line) and those from SCN-iPS cells in the presence of neutrophil differentiation medium containing 500 ng/mL Wnt3a (yellow line) or 1,000 ng/mL G-CSF (black line). Data are shown as mean ± SD. ★★P < 0.05.
The reduced expression of LEF-1 (as regulated by the Wnt3a/β-catenin pathway) reportedly plays a critical role in the defective maturation of neutrophils in SCN patients (25). Therefore, we next examined LEF-1 mRNA expression in SCN-iPS–CD34+ cells cultured in the presence of Wnt3a, G-CSF (1,000 ng/mL), or both. Wnt3a and G-CSF both enhanced LEF-1 mRNA expression, but the most significant increase was observed in the presence of Wnt3a plus G-CSF. LEF-1 expression in SCN-iPS–CD34+ cells in response to Wnt3a plus G-CSF was almost the same as that in control iPS–CD34+ cells (Fig. 4C). These results substantiate the importance of LEF-1 in neutrophil development and the pathogenesis of SCN, as shown (25). Moreover the administration of Wnt3a led to up-regulate C/EBP-α, cyclin D1, and BIRC5/survivin in addition to LEF-1 in the presence of G-CSF (Fig. 4D). These results suggested that the up-regulation of LEF-1 expression might promote granulopoiesis by increasing the expressions of cyclin D1, BIRC5/survivin, and C/EBP-α and its binding to LEF-1 in accordance with the previous report (25). Interestingly, Wnt3a did not stimulate the proliferation of myeloid cells, whereas 1,000 ng/mL G-CSF did to a certain extent (Fig. 4E). Hence, Wnt3a was capable of stimulating the maturation of impaired neutrophils in the presence of G-CSF, but not the proliferation of myeloid cells from SCN-iPS cells.
Importantly, aside from providing new insights into the mechanisms behind impaired neutrophil development in SCN patients, the present study demonstrates that agents activating the Wnt3a/β-catenin pathway are potential candidates for new drugs for SCN with mutations in the ELANE gene. Because endogenous G-CSF is readily increased in SCN patients (26), these activating agents may be viable alternatives to exogenous G-CSF treatment.
Materials and Methods
Additional information is available in SI Materials and Methods.
Generation of Human iPS Cells.
BM fibroblasts from a patient with SCN and skin dermal fibroblasts from a healthy donor were acquired after obtaining informed consent after getting the approval by the Ethics Committee of the Institute of Medical Science, University of Tokyo, in accordance with the Declaration of Helsinki. The SCN patient presented with a heterozygous mutation in the ELANE gene in the 707 region of exon 5. SCN-iPS cells were established from the SCN-BM fibroblasts by transfection with the pMX retroviral vector, as described (10). This vector expressed the human transcription factors OCT3/4, SOX2, KLF4, and c-MYC. Control iPS cell clones, control 1 (TkDN4-M) and control 3 (201B7), were gifts from K. Eto and S. Yamanaka (Kyoto University, Kyoto), respectively (10, 11). Control 2 (SPH0101) was newly generated from another healthy donor’s skin dermal fibroblasts by using the same methods.
Hematopoietic Colony Assay.
A hematopoietic colony assay was performed in an aliquot of culture mixture, which contained 1.2% methylcellulose (Shin-Etsu Chemical), 30% (vol/vol) FBS, 1% (vol/vol) deionized fraction V BSA, 0.1 mM 2-mercaptoethanol (2-ME), α-minimum essential medium, and a cytokine mixture consisting of 100 ng/mL human stem cell factor (hSCF) (Wako), 100 ng/mL fusion protein 6 [FP6; a fusion protein of interleukin (IL)-6 and IL-6 receptor] (a gift from Tosoh), 10 ng/mL human IL-3 (hIL-3) (a gift from Kirin Brewery), 10 ng/mL human thrombopoietin (hTPO) (a gift from Kirin Brewery), 10 ng/mL human G-CSF (a gift from Chugai Pharmaceutical), and 5 U/mL human erythropoietin (a gift from Kirin Brewery). For dose escalation experiments, various concentrations (0, 1, 10, 100, and 1,000 ng/mL) of G-CSF were used instead of the cytokine mixture described above. Colony types were determined according to established criteria on day 14 of culture by in situ observations under an inverted microscope (IX70; Olympus) (27).
Suspension Culture and Neutrophil Differentiation Assay.
CD34+ cells (1 × 104 cells) were cocultured with irradiate confluent AGM-S3 cells in neutrophil differentiation medium containing Iscove’s modified Dulbecco’s medium, 10% FBS, 3 mM L-glutamine, 1 × 10−4 M 2-ME, 1 × 10−4 M nonessential amino acids solution, 100 ng/mL hSCF, 100 ng/mL FP6, 10 ng/mL hIL-3, 10 ng/mL hTPO, and 10 or 1,000 ng/mL human G-CSF. Wnt3a (10, 100, or 500 ng/mL) (R&D) was then added. The medium was replaced with an equivalent volume of fresh medium every 4 d. Living, nonadherent cells were counted following 0.4% trypan blue staining.
PCR primer.
All primer sets used in this study are shown in Table S1.
Statistical Analysis.
All data are presented as mean ± SD. P < 0.05 was considered significant. Statistical analyses were performed by using Prism software (GraphPad).
Supplementary Material
Acknowledgments
We thank the individual with SCN who participated in this study; K. Eto for providing control iPS cells (control 1; TkDN4-M); S. Yamanaka for providing control iPS cells (control 3; 206B7); and E. Matsuzaka and S. Hanada for technical assistance. This work was supported by in part by Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) Grants-in-Aid (to Y.E.) and Project for Realization of Regenerative Medicine (MEXT) Grants-in-Aid (to K.Tsuji).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.Q.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217039110/-/DCSupplemental.
References
- 1.Zeidler C, Germeshausen M, Klein C, Welte K. Clinical implications of ELA2-, HAX1-, and G-CSF-receptor (CSF3R) mutations in severe congenital neutropenia. Br J Haematol. 2009;144(4):459–467. doi: 10.1111/j.1365-2141.2008.07425.x. [DOI] [PubMed] [Google Scholar]
- 2.Freedman MH, et al. Myelodysplasia syndrome and acute myeloid leukemia in patients with congenital neutropenia receiving G-CSF therapy. Blood. 2000;96(2):429–436. [PubMed] [Google Scholar]
- 3.Dale DC, et al. A randomized controlled phase III trial of recombinant human granulocyte colony-stimulating factor (filgrastim) for treatment of severe chronic neutropenia. Blood. 1993;81(10):2496–2502. [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg PS, et al. Severe Chronic Neutropenia International Registry The incidence of leukemia and mortality from sepsis in patients with severe congenital neutropenia receiving long-term G-CSF therapy. Blood. 2006;107(12):4628–4635. doi: 10.1182/blood-2005-11-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xia J, et al. Prevalence of mutations in ELANE, GFI1, HAX1, SBDS, WAS and G6PC3 in patients with severe congenital neutropenia. Br J Haematol. 2009;147(4):535–542. doi: 10.1111/j.1365-2141.2009.07888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horwitz MS, et al. Neutrophil elastase in cyclic and severe congenital neutropenia. Blood. 2007;109(5):1817–1824. doi: 10.1182/blood-2006-08-019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hajjar E, Broemstrup T, Kantari C, Witko-Sarsat V, Reuter N. Structures of human proteinase 3 and neutrophil elastase—so similar yet so different. FEBS J. 2010;277(10):2238–2254. doi: 10.1111/j.1742-4658.2010.07659.x. [DOI] [PubMed] [Google Scholar]
- 8.Fouret P, et al. Expression of the neutrophil elastase gene during human bone marrow cell differentiation. J Exp Med. 1989;169(3):833–845. doi: 10.1084/jem.169.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pham CT. Neutrophil serine proteases: Specific regulators of inflammation. Nat Rev Immunol. 2006;6(7):541–550. doi: 10.1038/nri1841. [DOI] [PubMed] [Google Scholar]
- 10.Takayama N, et al. Transient activation of c-MYC expression is critical for efficient platelet generation from human induced pluripotent stem cells. J Exp Med. 2010;207(13):2817–2830. doi: 10.1084/jem.20100844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Germeshausen M, Ballmaier M, Welte K. Incidence of CSF3R mutations in severe congenital neutropenia and relevance for leukemogenesis: Results of a long-term survey. Blood. 2007;109(1):93–99. doi: 10.1182/blood-2006-02-004275. [DOI] [PubMed] [Google Scholar]
- 13.Ma F, et al. Novel method for efficient production of multipotential hematopoietic progenitors from human embryonic stem cells. Int J Hematol. 2007;85(5):371–379. doi: 10.1532/IJH97.06203. [DOI] [PubMed] [Google Scholar]
- 14.Konishi N, et al. Defective proliferation of primitive myeloid progenitor cells in patients with severe congenital neutropenia. Blood. 1999;94(12):4077–4083. [PubMed] [Google Scholar]
- 15.Nakamura K, et al. Abnormalities of primitive myeloid progenitor cells expressing granulocyte colony-stimulating factor receptor in patients with severe congenital neutropenia. Blood. 2000;96(13):4366–4369. [PubMed] [Google Scholar]
- 16.Skokowa J, Fobiwe JP, Dan L, Thakur BK, Welte K. Neutrophil elastase is severely down-regulated in severe congenital neutropenia independent of ELA2 or HAX1 mutations but dependent on LEF-1. Blood. 2009;114(14):3044–3051. doi: 10.1182/blood-2008-11-188755. [DOI] [PubMed] [Google Scholar]
- 17.Kawaguchi H, et al. Dysregulation of transcriptions in primary granule constituents during myeloid proliferation and differentiation in patients with severe congenital neutropenia. J Leukoc Biol. 2003;73(2):225–234. doi: 10.1189/jlb.0902427. [DOI] [PubMed] [Google Scholar]
- 18.Köllner I, et al. Mutations in neutrophil elastase causing congenital neutropenia lead to cytoplasmic protein accumulation and induction of the unfolded protein response. Blood. 2006;108(2):493–500. doi: 10.1182/blood-2005-11-4689. [DOI] [PubMed] [Google Scholar]
- 19.Grenda DS, et al. Mutations of the ELA2 gene found in patients with severe congenital neutropenia induce the unfolded protein response and cellular apoptosis. Blood. 2007;110(13):4179–4187. doi: 10.1182/blood-2006-11-057299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pabst T, et al. AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8;21) myeloid leukemia. Nat Med. 2001;7(4):444–451. doi: 10.1038/86515. [DOI] [PubMed] [Google Scholar]
- 21.Hirai H, et al. C/EBPbeta is required for ‘emergency’ granulopoiesis. Nat Immunol. 2006;7(7):732–739. doi: 10.1038/ni1354. [DOI] [PubMed] [Google Scholar]
- 22.Bedi R, Du J, Sharma AK, Gomes I, Ackerman SJ. Human C/EBP-ɛ activator and repressor isoforms differentially reprogram myeloid lineage commitment and differentiation. Blood. 2009;113(2):317–327. doi: 10.1182/blood-2008-02-139741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26(47):6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- 24.Hetz C. The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13(2):89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 25.Skokowa J, et al. LEF-1 is crucial for neutrophil granulocytopoiesis and its expression is severely reduced in congenital neutropenia. Nat Med. 2006;12(10):1191–1197. doi: 10.1038/nm1474. [DOI] [PubMed] [Google Scholar]
- 26.Mempel K, Pietsch T, Menzel T, Zeidler C, Welte K. Increased serum levels of granulocyte colony-stimulating factor in patients with severe congenital neutropenia. Blood. 1991;77(9):1919–1922. [PubMed] [Google Scholar]
- 27.Nakahata T, Ogawa M. Hemopoietic colony-forming cells in umbilical cord blood with extensive capability to generate mono- and multipotential hemopoietic progenitors. J Clin Invest. 1982;70(6):1324–1328. doi: 10.1172/JCI110734. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.