Significance
Although ALS is a motor neuron disease, processes within glial cells contribute significantly to motor neuron-specific degeneration. Using a mouse model of ALS, we identified cell autonomous and nonautonomous changes in gene expression in motor neurons cocultured with glia. We also found a remarkable concordance between the cell culture data and expression profiles of whole spinal cords and acutely isolated spinal cord cells during disease progression in this model. We identified changes in the expression of specific genes and signaling pathways that may contribute to motor neuron degeneration in ALS, among which are TGF-β signaling pathways.
Keywords: motor neuron gene expression in ALS, reactive astrocyte gene expression in ALS, G93A mouse model of ALS, cell intrinsic and extrinsic effects on gene expression
Abstract
ALS results from the selective and progressive degeneration of motor neurons. Although the underlying disease mechanisms remain unknown, glial cells have been implicated in ALS disease progression. Here, we examine the effects of glial cell/motor neuron interactions on gene expression using the hSOD1G93A (the G93A allele of the human superoxide dismutase gene) mouse model of ALS. We detect striking cell autonomous and nonautonomous changes in gene expression in cocultured motor neurons and glia, revealing that the two cell types profoundly affect each other. In addition, we found a remarkable concordance between the cell culture data and expression profiles of whole spinal cords and acutely isolated spinal cord cells during disease progression in the G93A mouse model, providing validation of the cell culture approach. Bioinformatics analyses identified changes in the expression of specific genes and signaling pathways that may contribute to motor neuron degeneration in ALS, among which are TGF-β signaling pathways.
ALS is a late-onset, fatal neurodegenerative disease caused by the selective loss of upper and lower motor neurons in the brain and spinal cord and progressive paralysis of voluntary muscles; death ultimately results from respiratory failure (reviewed in ref. 1). Most ALS cases (∼90%) are sporadic, with an unknown cause, whereas the remaining cases are of familial origin (reviewed in ref. 2), among which ∼20–25% are caused by dominantly inherited mutations in the SOD1 gene; this gene encodes a cytosolic Cu/Zn superoxide dismutase (3). Overproducing pathogenic alleles of human SOD1 in mice and rats leads to late-onset progressive motor neuron degeneration, strikingly similar to the human disease (4–7). Because the pathological progression in both sporadic and familial ALS is indistinguishable (8, 9), insights derived from studies of the SOD1 mouse model are thought to be informative for both sporadic and familial ALS pathology.
The fundamental pathological basis for ALS remains to be determined along with the specific insults that target motor neurons for death. Mutant SOD1 genes are expressed ubiquitously in humans and mice and when expressed exclusively in mouse motor neurons, are not sufficient to cause disease (10–12). An important insight into this enigma was provided by the observation that the presence of mutant SOD1 within neighboring nonneuronal cells contributes to motor neuron toxicity and thereby, disease onset and progression (reviewed in ref. 13). The principal nonneuronal cell types implicated in motor neuron death in ALS are astrocytes, microglia, and oligodendrocytes; in vivo approaches focused on excising the mutant transgene from microglia and astrocytes in SOD1-based ALS mouse models have shown that disease onset and/or progression are affected (reviewed in ref. 13). There is increasing evidence that the presence of the mutant SOD1 protein in these nonneuronal cell types contributes significantly to ALS disease progression in the ALS mouse model. Evidence that astrocytes also play a negative role in human ALS was provided by a recent study showing that astrocytes generated from postmortem spinal cords from SOD1 or sporadic ALS patients adversely affect the viability of cultured ES cell-derived mouse motor neurons (14).
The question of extrinsic vs. intrinsic effects on gene expression in motor neurons in ALS in vivo has been difficult to address using laser capture microdissection (LCMD), because only the cell soma is captured, excluding the dendritic arbor as well as the axon. Moreover, LCMD is limited to neuronal cells, because it is not possible to cleanly capture glial cell bodies from among the neuropil in the spinal cord. Other approaches involve studies of entire spinal cords, which are heterogeneous and do not provide cell type-specific information. Thus, either approach alone yields an incomplete picture. Consequently, it has thus far not been possible to relate progressive gene expression changes in motor neurons to changes in gene expression in the surrounding glial cells in whole-animal studies. A potential solution to this problem is to make use of cell culture models to study how glial cells adversely affect motor neuron viability.
In previous studies, we (15) and others (6) established a cell culture system to study astrocyte/motor neuron interactions. This approach involves the generation of motor neurons by in vitro differentiation of ES cells derived from mice harboring the human SOD1G93A transgene. These ES cell-derived motor neurons are cocultured with either ES cell-derived or primary glial cells. Remarkably, motor neurons in this coculture system recapitulate aspects of the abnormal pathology characteristic of ALS in humans as well as in transgenic mice (15). In addition, SOD1G93A mutant glia can adversely affect the viability of mutant as well as WT motor neurons in vitro and in vivo (14, 16).
Although each approach to the analysis of ALS disease mechanisms (postmortem ALS patient samples, mouse models, and cell culture) has its limitations, an integrated approach that combines whole-animal and cell culture analyses could provide novel insights into the pathways leading to motor neuron-specific degeneration. Here, we describe an adaptation of this in vitro model to capture both cell autonomous and nonautonomous changes in neuronal and glial cell gene expression. We studied FACS-purified ES cell-derived motor neurons in sandwich cultures (17), in which motor neurons plated on glass coverslips are cultured over primary glial cells as a function of time. We then used separate RNA sequencing (RNA-Seq) analysis of the two cell types to identify changes in gene expression profiles intrinsic to each cell type (cell autonomous effects) or mediated by the cocultured cell (cell nonautonomous effects).
In parallel, we used RNA-Seq to determine expression profiles of a longitudinal series of whole spinal cords from the same G93A mouse model of ALS over the course of the disease. These data, combined with gene expression data from acutely isolated glia, microglia, and oligodendrocytes from control and mutant SOD1 mouse spinal cords and ALS patient postmortem human spinal cord samples, may lead to the identification of common features that will provide an ALS disease signature and thereby, identify potential ALS drug targets.
Results
Of critical importance in understanding ALS disease mechanisms is to distinguish between primary causal events and secondary consequences that accelerate disease progression and understand the relative contributions of cell autonomous and cell nonautonomous changes in gene expression. To address these issues, we used a strategy that aims to, first, relate disease progression in cell culture to disease progression in animal models; second, identify and distinguish between early and late events; and third, dissect the contributions of glial cells to motor neuron death. We studied in vitro and in vivo (whole spinal cords) mouse models in parallel and combined them such that the strengths of each model complement the weaknesses of the other model.
Overview of Methodology.
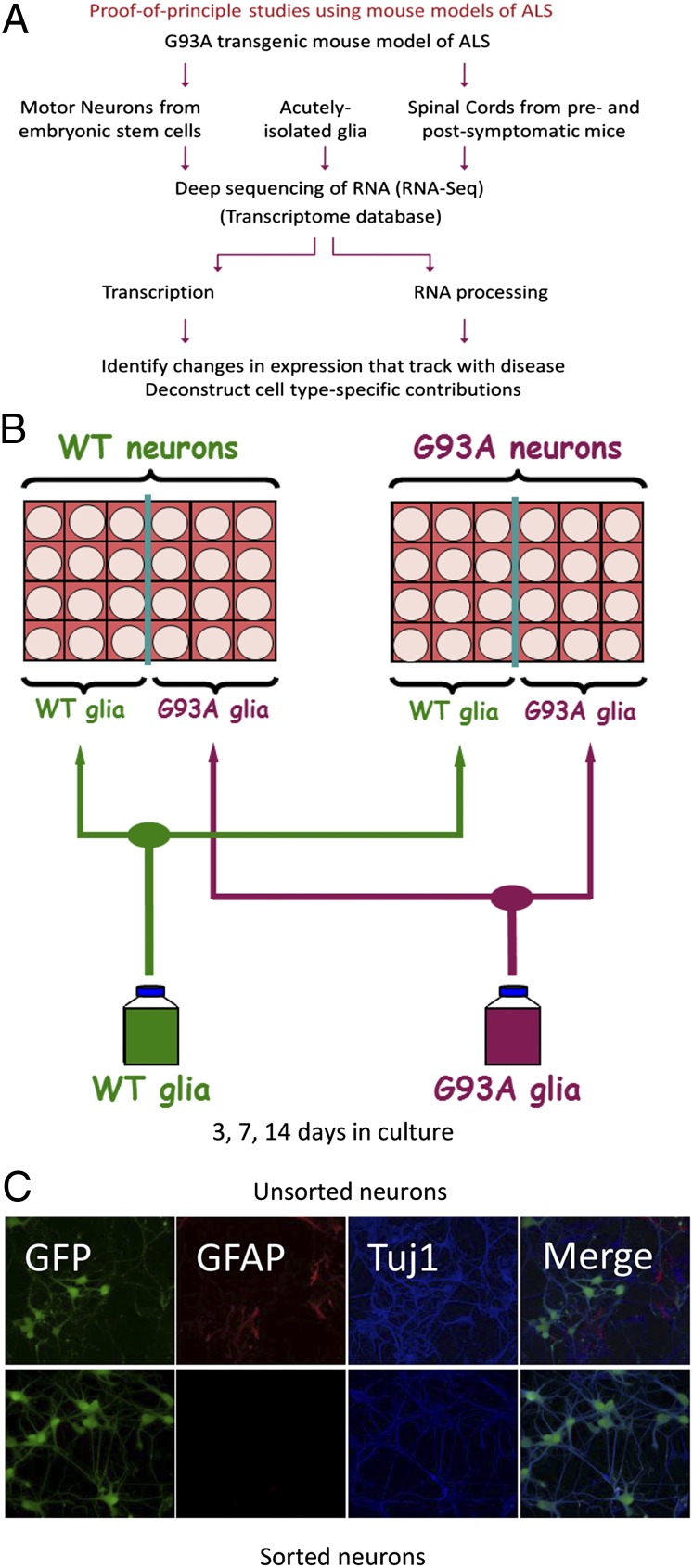
A schematic of our analytical framework is shown in Fig. 1A. We used the well-characterized G93A mouse model of ALS in our studies: a model that carries multiple copies of the human SOD1G93A transgene (5). To control for effects of overexpressing SOD1, we examined mice that are transgenic for multiple copies of the WT allele of the human SOD1 gene (these mice do not display ALS-like disease despite expressing comparable amounts of human SOD1 as the G93A mice) (5). Our in vivo samples consisted of a longitudinal series of whole spinal cords collected over the lifetime of adult pre- and postsymptomatic ALS mice. Our in vitro model used primary WT or G93A glia cultured with motor neurons derived from WT or G93A ES cells, which also expressed GFP under the control of the motor neuron-specific Mnx1 promoter that encodes the transcription factor Hb9.
Fig. 1.
(A) Schematic representation of analytical strategy to study ALS disease mechanisms. The hSOD1G93A-based mouse model of ALS was used as a source for cells and tissue. A parallel analysis was carried out with glia/motor neuron cultures, acutely isolated cells, and whole-spinal cord samples. The cell culture samples are ES cell-derived motor neurons cultured with primary glial cells. Whole spinal cords were isolated in a longitudinal series at 4, 8, 12, and 17 wk (symptom onset is at ∼12 wk). WT ES-derived motor neurons, glia, and whole spinal cords were obtained from transgenic animals overexpressing the nonmutated SOD1 protein. RNA was extracted from the samples and subjected to deep sequencing (RNA-Seq). RNA-Seq data were incorporated into a transcriptome database and queried for changes in gene expression. In addition, published transcriptional profiles (obtained using microarrays) of acutely isolated astroglia, oligodendrocytes, and oligodendrocyte progenitor cells were also incorporated into the transcriptome database and used for comparative analyses. (B) Schematic representation of the experimental design for the analysis of sorted sandwich cultures. FACS-purified ES cell-derived motor neurons (transgenic for WT or G93A alleles of human SOD1) were plated over primary cortical glial cultures derived from WT or G93A neonatal mice and grown in culture for 3, 7, or 14 d. FACS-purified neurons were plated on coverslips that were then inverted over glia in 24-well plates such that one-half of the wells contained WT glia, whereas the other one-half contained G93A glia. WT and G93A motor neurons were sorted and plated in parallel along with WT and G93A glia. Cells were plated such that both sets of neurons were exposed to glia derived from the same starting population and both sets of glia were exposed to the same batch of WT or G93A neurons. (C) Comparison of FACS-purified (Lower) with unpurified (Upper) neurons in sandwich culture. ES cell-derived neurons were plated in sandwich cultures over glia and grown in culture for 7 d, after which time samples were fixed and stained for GFP, GFAP, and Tuj1.
As depicted in Fig. 1B, we plated WT or G93A astrocytes in sandwich cultures (17) with FACS-purified WT or G93A motor neurons and cultured them over a time course. We also plated FACS-purified ES cell-derived motor neurons (transgenic for WT or G93A alleles of human SOD1) over primary cortical glial cultures derived from WT or G93A neonatal mice and cultured these neurons for 3, 7, or 14 d. We plated the WT or G93A FACS-purified neurons on glass coverslips coated with poly-D-lysine and laminin; we then inverted the coverslips over glia in 24-well plates such that one-half of the wells contained WT glia, whereas the other one-half contained G93A glia. WT and G93A motor neurons were sorted and plated in parallel along with WT and G93A glia. Cells were plated such that both sets of neurons were exposed to glia derived from the same starting population, and both sets of glia were exposed to the same batch of WT or G93A neurons. The rationale for this arrangement was to provide internal controls to anchor replicate experiments. This arrangement made it possible to assess reproducibility by comparing the changes in one experiment with the changes in another experiment. At specific time points after plating, the two cell types were separated from each other (by simply lifting off the neuron-containing coverslips) and recovered for RNA or imaging studies. A comparison between unsorted and sorted neurons in sandwich culture, stained using GFP, GFAP (for astrocytes), and neuronal β-III tubulin (Tuj1; for all neurons), showed that FACS purification resulted in a nearly homogeneous population of motor neurons, because the vast majority of the recovered cells positive for Tuj1 were also positive for GFP (Fig. 1C).
RNA-Seq Analysis Reveals a Complex Interplay Between Glia and Motor Neurons.
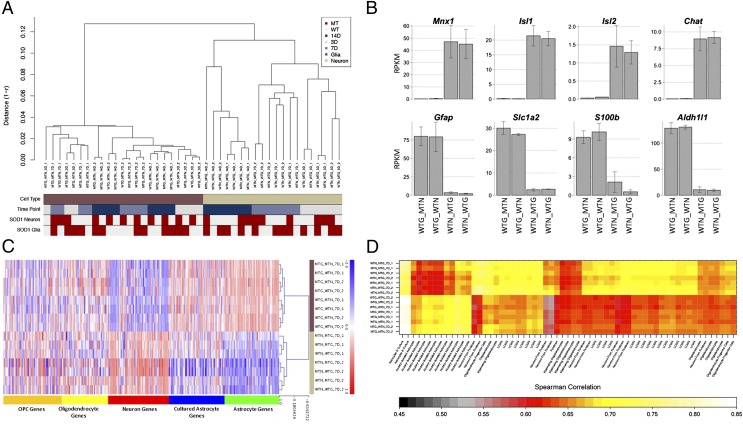
We extracted RNA from glia and neurons that were grown in sandwich culture and analyzed it by ultra-high throughput sequencing (RNA-Seq) with the Illumina platform. Hierarchical clustering based on genome-wide expression levels in all samples across all time points showed that the sorted sandwich samples clustered by cell type, time in culture, and genotype. As shown in Fig. 2A, topmost in the hierarchy of differentiators between the samples was cell type, because glial samples are more similar to each other than they are to any neuronal samples and vice versa. Within a particular cell type, time in culture was a greater differentiator between samples than genotype—for example, neurons grown in culture for 7 d, from two independent experiments, were more similar to each other than they were to neurons grown in culture for 3 or 14 d, regardless of whether they were WT or mutant; this result held true for all three time points tested (Fig. 2A). Interestingly, at each time point, cell autonomous effects dominate over nonautonomous effects (i.e., expression profiles in mutant neurons were more similar to each other than they were to WT neurons, regardless of which glial genotype they were grown over) (Fig. 2A). Thus, there seems to be a hierarchy of changes in the sorted sandwich system that governs the relatedness of samples to each other (cell type > time point > genotype of individual cell type > genotype of juxtaposed cell type), which in turn, suggests that, with this system, we can detect cell type-specific changes in mutant neurons and glia as a function of time and identify cell autonomous and nonautonomous effects on gene expression.
Fig. 2.
(A) Hierarchical clustering of sandwich culture RNA-Seq data. Expression levels of all genes [in reads per kilobase per million mapped reads (RPKM)] were used to assess similarity between RNA-Seq datasets (nearest neighbor Pearson’s correlation coefficient). The clustering was overlaid with different phenotype information: cell type (brown, glia; sand, neurons), time point (gray, 3 d in vitro; light blue, 7 d in vitro; dark blue, 14 d in vitro), and genetic background of the SOD1 transgene separately for neurons and glia (light gray, WT; red, G93A). WTG_MTN, WT glia grown under mutant neurons; WTG_WTN, WT glia grown under WT neurons; WTN_MTG, WT neurons grown over mutant glia; WTN_WTG, WT neurons grown over WT glia; 14D, 7D, and 3D, 14, 7, and 3 d in culture; 1 and 2 refer to replicate experiments. (B) Cell type specificity of sorted sandwich samples. Mean expression levels (in RPKM) ± SD of known cell type-specific genes were mapped in the neuronal and glial RNA-Seq data. Cell type-specific genes are expressed at different levels within the cell type in question as reflected in the scale on the y axis. (C) Heat map representation of hierarchical clustering for cell type-specific transcripts. Expression levels (z score-transformed log2 RPKM) of cell type-specific signature genes modified from Cahoy et al. (18) and Lei et al. (19) were used to cluster RNA-Seq data (complete linkage Spearman’s correlation). Genes are grouped according to their overexpression in specific cell types. The blue, white, and red color scheme represents low, medium, and high relative expression, respectively (right bar). We compared the transcript expression of our cell types (brown, glia; sand, motor neurons) to progenitor (dark yellow) and mature oligodendrocytes (light yellow), neurons (red), cultured astroglia (blue), and astrocytes (green) genes. (D) Matrix of correlation coefficients comparing the similarities for RNA-Seq with microarray-based transcriptional profiles of sandwich culture samples and samples from publicly available datasets. Spearman’s correlation coefficients were computed using all mappable genes across different platforms and visually represented in a color-coded heat map (colored bar). White and yellow indicate the highest correlation between cell types, whereas black and red show poor and intermediate correlation, respectively. Different cell types include cultured astroglia, isolated astrocytes, myelinating oligodendrocytes, laser-captured motor neurons, neurons (forebrain), and oligodendrocyte progenitor cells.
Sorted Sandwich System Maintains Cell Type Specificity.
We used a battery of tests to verify the cell type specificity of our data. To assess the extent of cross-contamination between neuronal and glial samples, we used reads mapping to codon 93 of human SOD1 mRNA. We reasoned that reads mapping to the G93A mutation in RNA samples from WT neurons grown over mutant glia could arise only from contamination of the neuronal sample with glial RNA (Fig. S1); similarly, reads aligning to the WT codon in RNA samples from mutant glia grown under WT neurons could arise only from contamination of the glial samples with neuronal RNA (Fig. S1). Using this measure, we estimate <5% contamination between glia and motor neurons. As shown in Fig. 2B, we also verified cell type-specific expression of known neuronal and glial markers: known neuronal genes [e.g., genes encoding transcription factors Hb9 (Mnx1), Islet-1 and -2, and choline acetyl transferase (Chat)] were enriched in neuronal samples, and known glial genes were enriched in glial samples (e.g., genes encoding glial glutamate transporter Eaat2, Gfap, the calcium binding protein S100b, and the 10-formyltetrahydrofolate dehydrogenase Aldh1L1). In addition to this group of well-known neuronal and glial genes, we examined the expression levels in our sandwich culture samples of the 317 cell type-specific genes (listed in Dataset S1) derived from published microarray gene expression profiles of various cell types in the CNS, including cultured astrocytes, acutely isolated astrocytes, oligodendrocytes, and cortical neurons (18, 19). The expression levels of each of these neuron-, astrocyte-, and oligodendrocyte-enriched genes in our glial and neuronal samples are depicted in the matrix in Fig. 2C. As shown in Fig. 2C, astrocyte-enriched genes were expressed at much higher levels in sandwich culture astrocytes compared with neurons, and neuron-enriched genes were reciprocally expressed at much higher levels in sandwich culture neurons compared with astrocytes. We also compared the transcriptome-wide RNA-Seq profiles of WT and G93A mutant (MT) glia and neurons in our cultures with these published microarray profiles of CNS cell types. The similarity (Spearman correlation) (Materials and Methods) of the expression profiles of WT and MT glia and neurons to each of these CNS cell types is depicted in Fig. 2D. Based on these gene expression profiles, the glia in the sorted sandwich system were more similar to acutely isolated astrocytes than to any other CNS cell type, whereas the ES cell-derived motor neurons most resembled LCMD motor neurons. Thus, the sorted sandwich system recapitulates known cell type-specific expression patterns in vitro and can reasonably form the basis for more detailed genotype-specific intrinsic and extrinsic effects on motor neuron viability.
Differences Between WT and Mutant Glia in Culture.
Although the glia in our experiments were more similar to acutely isolated astrocytes than to any other CNS cell type, there was a clear difference in the degree of their relatedness to cultured vs. acutely isolated astrocytes. This difference in the transcriptional profiles of cultured vs. acutely isolated astrocytes was previously noted (18); that is, astrocytes in culture are thought to be reactive. To determine whether astrocytes in our sandwich cultures are reactive in a way that is relevant to disease, we first defined cultured astrocyte-specific genes and then asked whether these genes were differentially expressed in WT (no reactive astrocytes) vs. MT (enriched reactive astrocytes) spinal cords. Cultured astrocyte-specific genes were defined as those genes that were expressed at a fivefold or greater level in cultured astroglia compared with all acutely isolated astroglia samples (P < 0.0001) (data from ref. 18) (Fig. 3A, red dots); 159 genes met these criteria (genes are listed in Dataset S1). Fig. 3B depicts empirical cumulative distribution functions (ECDFs) of these cultured astrocyte genes in spinal cord samples over time, where each curve represents the fraction of these 159 genes that has a given expression ratio in MT compared with WT spinal cords. ECDFs, thus, allow the visualization of shifts in the behavior of a defined set of genes. The blue curve in Fig. 3B, which represents the MT to WT spinal cord comparison at 120 d, is shifted to the right compared with the red curve in Fig. 3B, which represents the MT to WT spinal cord comparison at 30 d, indicating that, at 120 d, a greater number of these 159 cultured astrocyte-specific genes is expressed at higher levels in MT spinal cords than at 30 d. The curves are shifted progressively to the right during disease progression, indicating that a large fraction of cultured astrocyte-specific genes are up-regulated with age in MT spinal cords. These changes were reproducible at all time points (Fig. S2). We conclude that astrocytes in culture are reactive in a manner that is relevant to disease.
Fig. 3.
(A) Two-way plot of genes differentially expressed in astrocytes in culture (data from ref. 18). Microarray signal intensities from Affymetrix microarrays for astrocytes in culture is plotted vs. those signal intensities for acutely isolated astrocytes. Red dots represent genes up-regulated greater than fivefold in astrocytes in culture relative to acutely isolated astrocytes (P < 0.001); blue dots represent genes down-regulated in astrocytes in culture relative to acutely isolated astrocytes. Gray dots represent genes that were expressed at comparable levels in both samples. Marginal diagonal lines indicate the cutoff thresholds. (B) Empirical cumulative distribution functions of cultured/reactive astrocyte genes in spinal cord samples. Variation in fold changes in expression of cultured/reactive astrocyte genes across the different spinal cord comparisons is shown. Colors indicate the following comparisons: red, MT vs. WT spinal cords at 30 d; yellow, MT vs. WT spinal cords at 60 d; green, MT vs. WT spinal cords at 90 d; blue, MT vs. WT spinal cords at 120 d. Note that most of the 159 genes are up-regulated as disease progresses.
To identify the functional categories enriched among the genes up-regulated in cultured (reactive) astrocytes, we used the web-based bioinformatics tool DAVID (20, 21). Among the set of cultured astrocyte-specific genes, those genes encoding secreted glycoproteins and components of the ECM were highly enriched along with genes involved in cell growth and migration and genes encoding cytoskeletal proteins (listed in Dataset S1).
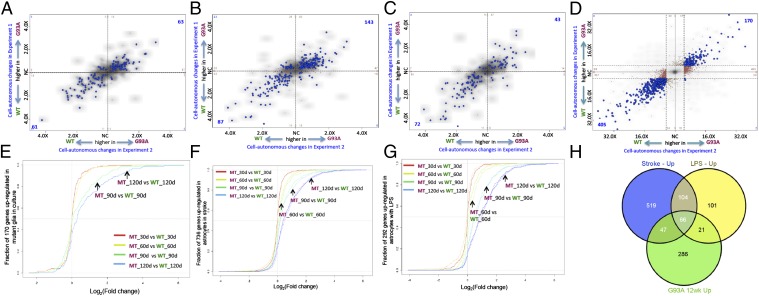
We next asked whether genes specific to cultured astrocytes were differentially expressed in the MT and WT glia in sorted sandwich cultures. As shown in Fig. 4A, we overlaid cultured astrocyte-specific genes onto four-way comparison plots of cell autonomous changes in glia in sandwich culture. Relative expression in MT compared with WT glia in one experiment is plotted against relative expression in a replicate experiment, and therefore, the changes in one experiment are compared with changes in another experiment. Genes that were up-regulated in MT glia in both experiments are in the upper right quadrant of Fig. 4A, and genes that were down-regulated in MT glia in both experiments are in the lower left quadrant of Fig. 4A. Thus, the sorted sandwich reveals the difference between reactive WT and MT glia. One hundred twenty-four cultured astrocyte-specific genes were differentially expressed in WT and MT glia in our experiments; of these genes, 63 genes were expressed at higher levels in MT glia, whereas 61 genes were expressed at higher levels in WT glia (listed in Dataset S1). Because reactive astrocytosis is context dependent (i.e., the nature and extent of the astrocytic response is influenced by the nature and duration of the stimulus (22, 23), we also assessed how reactive astrocytosis generated by different inducers compared with the response in ALS. We overlaid genes up-regulated in acutely isolated reactive astrocytes as a result of stroke or in response to LPS (23) onto four-way comparison plots of cell autonomous changes in ALS astrocytes in culture as discussed above. As shown in Fig. 4 B, stroke and C, LPS, genes that are up-regulated in acutely isolated reactive astrocytes in response to either stroke or LPS were differentially expressed in reactive ALS astrocytes in culture. Taken together, the results shown in Fig. 4 A–C indicate that reactive astrocyte genes are differentially expressed between WT and MT ALS astrocytes in culture.
Fig. 4.
Differential expression of reactive astrocyte genes in G93A astrocytes in culture. Various types of reactive astrocyte-specific genes are overlaid onto four-way comparison plots of cell autonomous changes in glia in sandwich culture. (A) Cultured astrocyte-specific genes. (B) Genes up-regulated in astrocytes as a result of stroke. (C) Genes up-regulated in astrocytes in response to LPS. Fold changes from expression in one experiment are plotted against changes from a replicate experiment. Blue dots represent changed genes that meet the cutoff for statistical significance (P < 0.001) in both experiments. Blue dots in the upper right quadrant correspond to genes expressed at higher levels in MT glia in both experiments; blue dots in the lower left quadrant correspond to genes expressed at higher levels in WT glia (also lower levels in MT glia) in both experiments. (D) Four-way comparison plot showing all cell autonomous changes in G93A astrocytes in culture. Fold changes in expression from one experiment are plotted against changes from a replicate experiment. Colored dots represent changed genes that meet the cutoff for statistical significance (fold change > 1.5, P < 0.0001); blue dots represent genes that meet the cutoffs in both and thus, represent transcripts that are reproducibly changed in both experiments. Brown or tan dots meet the cutoffs in one experiment but not the other. Thus, blue dots in the upper right quadrant correspond to genes up-regulated in G93A MT glia in both experiments, and blue dots in the lower left quadrant correspond to genes down-regulated in MT glia in both experiments. Although the genes represented by the brown and tan dots met the statistical criteria in one of two experiments, they nevertheless exhibited the same trend in the second experiment—most of the brown and tan dots are close to the diagonal. NC, no change. (E) Empirical cumulative distribution functions in spinal cord samples of genes up-regulated in reactive mutant astrocytes in sorted sandwich cultures. Variation in fold changes in expression of reactive mutant astrocyte genes across the different spinal cord comparisons is shown. (F) Empirical cumulative distribution functions in spinal cord samples of genes up-regulated in reactive astrocytes in stroke. Variation in fold changes in expression of reactive astrocyte genes across the different spinal cord comparisons is shown. Solid curves represent spinal cords from males, whereas dotted lines represent spinal cords from females. (G) Empirical cumulative distribution functions in spinal cord samples of genes up-regulated in reactive astrocytes in response to LPS. Variation in fold changes in expression of reactive astrocyte genes across the different spinal cord comparisons is shown. Solid curves represent spinal cords from males, whereas dotted lines represent spinal cords from females. (H) Overlap between genes up-regulated in astrocytes in response to either stroke or LPS and genes up-regulated in G93A spinal cords (P < < 0.0001).
Genes Up-Regulated in Mutant Astrocytes in Culture Are Progressively Up-Regulated in Mutant Spinal Cords.
The expression of 575 genes was altered >1.5-fold between WT and MT glia (P < 0.0001) as shown in the four-way comparison plot in Fig. 4D; we selected a subset of genes representing a range of expression levels and validated the changes by quantitative PCR (Fig. S3 and Table S1).
We then asked whether the genes up-regulated in reactive mutant astrocytes in sorted sandwich cultures were affected in mutant spinal cords over time. Fig. 4E depicts ECDFs of these 170 genes up-regulated in reactive mutant astrocytes in spinal cord samples over time; the curves are shifted progressively to the right, indicating that the levels of these genes continued to increase with age in MT spinal cords. That is, the presence of the G93A mutant SOD1 protein in the cultured astrocytes leads to changes in gene expression in culture that are similar to the changes that occur in the spinal cords of mice undergoing disease progression.
Interestingly, a fraction of the genes up-regulated in acutely isolated reactive astrocytes responding to stroke or LPS was also progressively up-regulated in MT SOD1 spinal cords (Fig. 4 F and G). However, despite this overlap, each paradigm (stroke, LPS, and ALS) involves a distinct series of transcriptional changes (23) (Fig. 4H). Fig. 5A lists the regulatory pathways that are affected in ALS compared with stroke and LPS. Consistent with the plots shown in Fig. 4 A–C, pathways perturbed in acutely isolated reactive astrocytes responding to stroke or LPS are differentially regulated in reactive ALS astrocytes.
Fig. 5.
(A) Pathway enrichment analysis in ALS vs. stroke and LPS. Significant pathway activation is represented in red, whereas inhibition is in blue as computed with pathway guide. (B) Functional analysis of cell autonomous and nonautonomous changes in neurons.
Genes Involved in the Response to Tissue Insult or Injury Are Dysregulated in Mutant Glia.
We used DAVID (20, 21) to analyze the functional categories [gene ontology (GO) terms for biological process, molecular function, and cellular component; pathways from KEGG (24) and BIOCARTA (25); and PIR (26) keywords, sequence features, and protein domains from INTERPRO (27) and SMART (28)] of genes differentially expressed between WT and MT glia (all blue dots in Fig. 4D). As listed in Dataset S2, we found that the most significantly affected cluster of biological processes affected in MT glia in culture consisted of the GO terms representing response to wounding (GO:0009611), inflammatory response (GO:0006954), and defense response (GO:0006952). Only ∼27% of 47 total genes that comprised this cluster were up-regulated in MT glia, whereas the remaining ∼73% of genes were down-regulated, suggesting that MT glia are compromised in their ability to respond to tissue insult or injury (Dataset S2). Consistent with these changes, also enriched among the set of differentially expressed glial genes were those genes involved in cell adhesion (GO:0045785, GO:0030155, GO:0010811, and GO:0010810), ECM binding (GO:0050840), and cell migration (GO:0006935, GO:0042330, and GO:0007626) (Dataset S2). In addition, a vast majority (44) of 52 genes grouped into the cluster associated with chemotaxis and locomotory behavior was down-regulated in MT glia (Dataset S2).
A large fraction of differentially expressed genes encoded signaling glycoproteins and components of the ECM. Among ECM genes down-regulated in MT glia were Osteopontin and Sparcl1, matricellular proteins known to be expressed by astrocytes in response to injury (29). These genes were also among the most highly expressed genes in these glial cultures (Dataset S3). Other highly expressed injury/stress response-secreted genes that are down-regulated in MT glia include Sepp1, which has been implicated as an extracellular antioxidant, Apolipoprotein D, which is known to be transcriptionally up-regulated in response to and involved in mechanisms regulating protection from oxidative stress (30–32), and Pleiotrophin, which has been shown to be a trophic factor for spinal motor neurons, protecting against chronic excitotoxic injury and causing increased outgrowth of motor axons out of spinal cord explants (33).
Functional Analysis of Cell Autonomous and Nonautonomous Changes in Neurons.
Pathway enrichment analyses of changes in neuronal gene expression showed that many of the pathways altered in astrocytes were also affected in neurons (Fig. 5B) (e.g., focal adhesion, complement and coagulation cascade, regulation of actin cytoskeleton, cytokine–cytokine receptor interaction, and ECM–receptor interaction were all affected in both glia and neurons). Thus, the presence of mutant G93A SOD1 elicits similar changes in both cell types, suggesting that similar pathways are perturbed in both cell types.
As shown in Fig. 6A (adapted from KEGG) (Fig. S4 shows constituent pathways), these pathways form a higher-order network of interactions (e.g., ECM–receptor interactions and TGF-β signaling reciprocally affect each other) (reviewed in ref. 34), ECM–receptor interactions and cytokine–receptor interactions feed into the focal adhesion pathway, which in turn, regulates the actin cytoskeleton by integrin-linked kinase signaling (KEGG), and there is cross-talk between the TGF-β and MAPK signaling pathways (reviewed in refs. 35 and 36). As highlighted in Fig. 6 B and C, these pathways showed concordant time-dependent changes in neurons; moreover, they were up-regulated in WT neurons grown over mutant glia, whereas they were down-regulated in mutant neurons compared with WT neurons over the same glia. These observations reveal a profound and dynamic intercellular communication between motor neurons and glia as a consequence of the presence of the mutant SOD1 protein in one cell type or the other.
Fig. 6.
(A) Network of interactions among pathways perturbed in neurons (Fig. S4). (B) Time-dependent concordant cell autonomous changes in this pathway subset. Total accumulation (a measure of the extent of perturbation in each pathway) is shown for cell autonomous changes in neurons at 3, 7, and 14 d. (C) Comparison between cell nonautonomous and cell autonomous changes in concordantly changed pathways. Total accumulation (a measure of the extent of perturbation in each pathway) is shown for cell autonomous changes in neurons on the left and nonautonomous changes on the right. (D) Functional categories of genes dysregulated in neurons.
The distribution into categories of genes dysregulated in neurons is shown in Fig. 6D. Genes encoding cell surface proteins and those genes involved in signal transduction, transport/trafficking or cytoskeleton, protein turnover, and stress or injury response constituted the majority of changes. These categories were consistent with the results of the pathway analyses described above. Changes in cell surface and signal transduction components, taken together with the changes in glial gene expression, likely indicate that neuron–glia communication is disrupted in the sorted sandwich system. Thus, cell autonomous changes in glia as well as cell autonomous and nonautonomous changes in neurons apparently converge to dysregulate an interconnected network of pathways that culminates in the dysregulation observed in the sandwich culture system.
Some of these changes are encapsulated in Fig. 7, which depicts representative changes in neuronal and glial cell surface molecules as well as their linked intracellular signaling components (a complete list of all changed genes in neurons can be found in Dataset S3). Many of the genes shown were affected both autonomously and nonautonomously. For example, ECM components as well their cell surface receptors derived from glia were down-regulated, and their respective neuron-derived receptors and ECM molecules were up-regulated in WT neurons grown over these mutant glia compared with neurons grown over WT glia. In contrast to WT neurons over mutant glia, mutant neurons over the same glia had comparatively lower levels of these same ECM ligands and receptors. Consistent with these cell surface changes in ECM–receptor interactions, downstream consequences attendant on disrupting this signaling were also evident in the sorted sandwich system (shown by the network of arrows in Fig. 7) [e.g., disruption of focal adhesion components and effects on the actin cytoskeleton through integrin-linked signaling (for instance, levels of actinin, filamin, integrin-linked kinase, vinculin, phosphorylable myosin light chain, and myosin light chain kinase are all disrupted)].
Fig. 7.
Diagrammatic representation of some of the cell–cell communication elements and interaction networks perturbed in sorted sandwich cultures. N, neurons (upper one-half of the figure); G, glia (lower one-half of the figure). Representative changes in neuronal and glial cell surface molecules as well as their linked intracellular signaling components are shown. Down-regulated genes are indicated by blue boxes, whereas up-regulated genes are shown as red boxes. Both cell autonomous (lower one-half of the box representing a gene) and nonautonomous changes (upper one-half of the box representing a gene) in neurons are shown.
Interestingly, several of these dysregulated genes are known to affect TGF-β signaling—these genes include ECM components like the fibrillins and Decorin as well as fibulins and latent TGF-β binding protein (reviewed in refs. 34, 35, and 37–39); the TGF-β type II receptor (TGFβ-RII) was also dysregulated. As shown in the schematic in Fig. 6A, TGF-β signaling is, in turn, known to influence several of the other pathways that were also perturbed in the sorted sandwich system; several of these pathways were also affected in G93A spinal cords (Fig. 5). Of note, the principal impacts on the TGF-β pathway seemed to be at the level of the cell surface (Fig. S4) (KEGG pathway diagram of TGF-β signaling changes in neurons) in the sandwich culture system. TGF-β is known to be a modulator of reactive astrogliosis and reduce neuronal damage in the context of acute local injury that involves a transient inflammatory response (reviewed in refs. 22 and 40; 41). In the case of chronic neurodegenerative disease, exogenous TGF-β2 has been shown to improve motor performance in G93A mice (42). In addition, dysfunction of TGF-β signaling has been implicated in Alzheimer’s disease (AD): levels of TGFβ-RII are reduced in brains of AD patients and correlate with pathological hallmarks of disease, and reduced neuronal TGF-β signaling in mice has been shown to result in age-dependent degeneration (43). Accordingly, we asked whether the receptor TGFβ-RII was also dysregulated in G93A spinal cords.
TGFβ-RII Levels Are Dysregulated in G93A Spinal Cords and Correlate with Reactive Astrogliosis.
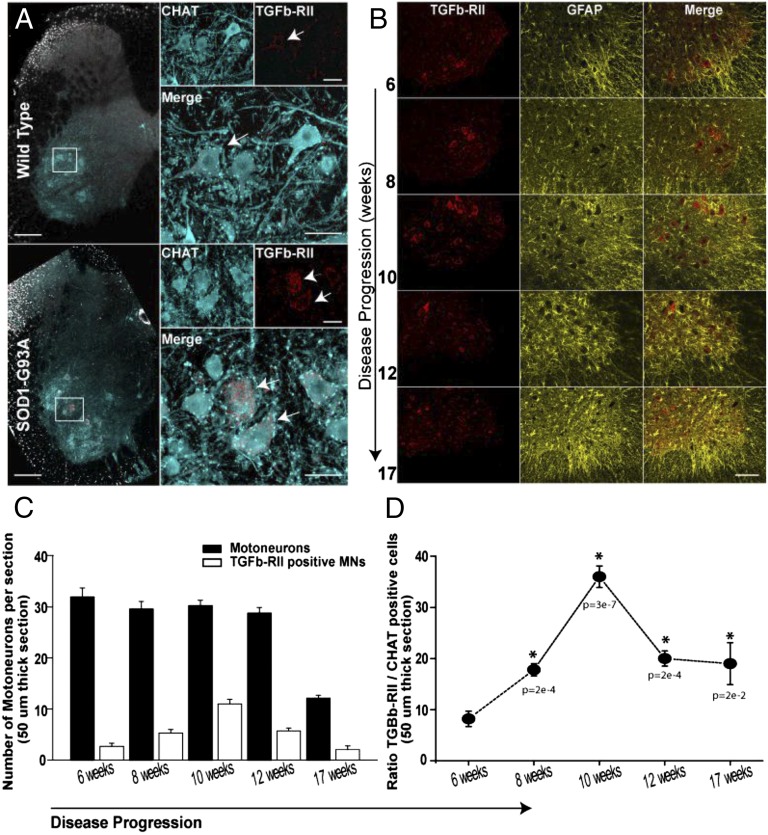
Compared with WT mice, TGFβ-RII immunoreactivity was more intense in the ventral horns of G93A lumbar spinal cords, and the signal seemed to be predominantly in motor neurons (Fig. 8A). In addition, although the immunoreactivity in WT samples seemed to be diffusely distributed throughout the soma, motor neurons in G93A lumbar spinal cords displayed a more punctate or clumped pattern (Fig. 8A). An analysis of the time course of TGFβ-RII expression, shown in Fig. 8B (Fig. S5), revealed that elevated TGFβ-RII levels in G93A motor neurons were observed as early as at 6 wk, well before symptom onset in this mouse model, which typically occurs around 12 wk (5). Interestingly, both the intensity of TGFβ-RII immunoreactivity and the number of motor neurons that are immunoreactive for the receptor showed a progressive increase, with a peak at 10 wk; relatively fewer TGFβ-RII immunoreactive motor neurons were observed in 12- and 17-wk lumbar spinal cords (Fig. 8B; quantified in Fig. 8 C and D). Moreover, as also shown in Fig. 8B, the increase in TGFβ-RII immunoreactivity seemed to track with the progression of reactive astrogliosis (as monitored by immunostaining for GFAP). The intensity of GFAP immunoreactivity increased over the same time course as signal for TGFβ-RII, GFAP-positive astrocytes surrounded the motor neurons with intense TGFβ-RII signal, and these astrocytes seemed thicker and more hypertrophic over time.
Fig. 8.
Increased expression of TGFβ-RII in ALS-afflicted motor neurons. Images in A show lumbar segments (L2–L5) of spinal cords from WT (Upper) and SOD1G93A (Lower) mice immunostained against TGFβ-RII (red), which mainly colocalized with ventral horn- ChAT–positive neurons (cyan, motor neurons). Upper Right (high magnification view of the boxed region) illustrates the expression level (weak) of TGFβ-RII (arrow) in WT motor neurons (cyan in merge). Lower Right illustrates the expression level (high) of TGFβ-RII (red and arrows in merge) in aged-matched SOD1G93A motor neurons (cyan). Note that, in the mutant neurons, TGFβ-RII (arrows) was typically observed as large filamentous aggregates throughout the cell (red and arrows). Images in B show the prevalence of TGFβ-RII–positive motor neurons during the disease progression. Abundance of reactive astrocytes (Center); immunoreactivity against GFAP was used as an indicator of the disease condition. (C) Quantification of TGFβ-RII–positive motor neurons (white bars) at different stages of the disease. Black bars indicate the total number of motor neurons. (D) Graph summarizes the prevalence of TGFβ-RII–positive motor neurons during the disease. Data displayed as average ± SEM; *P < 0.05, ANOVA test.
Discussion
Although the pathophysiology of ALS involves the selective dysfunction and ultimate death of motor neurons, processes within neighboring nonneuronal cells significantly contribute to the development of motor neuron-specific degeneration. To study in more detail how nonneuronal cells adversely affect motor neuron viability, we collected gene expression data from isolated mutant and WT glial and neuronal cells over a time course in cell coculture as well as from spinal cords of both WT and SOD1G93A mutant mice. We computationally mined these gene expression profiles in an effort to understand the underlying complexity of cell intrinsic and extrinsic effects on motor neuron survival in ALS.
We found that a large fraction of changes in gene expression involve genes that encode cell surface proteins and those genes involved in cellular responses to stress or injury. The cumulative results of our computational analyses of gene expression changes in neurons and glia revealed that neuron–glia communication is profoundly disrupted by mutant glia in the sorted sandwich system. Moreover, these perturbations in neuronal and glial gene expression apparently converge to perturb an interconnected network of pathways that culminate in the altered regulation and ultimate loss of motor neuron viability observed in the sandwich culture system.
TGF-β signaling is one such pathway that is perturbed in the sorted sandwich system. This pathway also seemed to be affected in G93A spinal cords as early as 6 wk of age, well before the onset of symptoms, and we found that these changes correlate with the progression of reactive astrocytosis. We note that decreased activity of a positive regulator of TGF-β signaling was recently implicated in an increased susceptibility to ALS (44). Thus, our combined in vivo and in vitro studies have identified a signaling pathway that may play an important role in the pathophysiology of astrocyte/motor neuron interactions in ALS.
As reviewed in ref. 40, TGF-β seemed to be effective in reducing neuronal damage in the context of acute local injury with a transient inflammatory response (such as found in animal models of stroke). TGF-β signaling may also be protective in the context of chronic neurodegenerative disease, where inflammation is persistent. For example, exogenous TGF-β2 has been shown to be temporarily beneficial in G93A mice (42). In addition, altered TGF-β signaling has been implicated in AD: levels of TGFβ-RII are reduced in brains of AD patients, and this reduction correlates with pathological hallmarks of disease. In addition, reduced neuronal TGF-β signaling in mice has been shown to result in age-dependent neurodegeneration (43). In vivo studies directed to understanding how changes in the TGF-β pathway in specific cell types affect inflammation and reactive gliosis will be necessary to understand the role of TGF-β signaling in chronic neurodegenerative diseases.
The expression profile of astrocytes grown in culture is distinct from the profile of acutely isolated astrocytes, and growing astrocytes in culture is thought to make them reactive (18). To ascertain whether astrocytes in culture are reactive in a way that is relevant to disease, we carried out a series of bioinformatics analyses. Using a set of published microarray gene expression profiles of various WT CNS cell types (18), we first defined a set of cultured astrocyte-specific genes and then asked whether this set of genes is altered in ALS spinal cords. We found that genes up-regulated in cultured astrocytes (relative to acutely isolated astrocytes) are also progressively up-regulated in ALS spinal cords. This observation suggests that astrocytes in culture are reactive in a manner relevant to disease and that reactive astrocytosis in ALS may be a graded rather than an all-or-none response. In addition, reactive G93A astrocytes share some similarities but are also distinct from reactive WT astrocytes responding to stroke or LPS treatment.
Our analyses of astrocyte gene expression also show that, although whole spinal cords are a heterogeneous sample comprised of many different cell types, we can computationally dissect the contributions of these various cell types. Using microarray profiles of acutely isolated CNS cell types, we can define cell type-specific gene expression fingerprints and then use these fingerprints to probe the whole-spinal cord expression profiles. Extending this methodology to other cell types, we have found that the oligodendrocyte progenitor cell signature progressively increases over the time course of the disease. This observation is consistent with recent data showing that oligodendrocyte progenitor cells increase in the spinal cords of G93A ALS mice over the course of the disease (45). In addition, analyses with microglial gene expression profiles from G93A mouse spinal cords show a similarly progressive increase in genes up-regulated in G93A microglia; thus, it is also possible to examine the relative timing of activation of the various glial cell types (e.g., whether the activation of one cell type precedes the activation of another cell type). As new models of ALS emerge, based on newly identified disease genes, it will be possible to undertake similar analyses with a time series from these mice to assess the degree of universality in the glial response in ALS.
Materials and Methods
Glia Dissection.
Glia monolayers were obtained essentially as described previously (14). After monolayers were confluent (generally in 7 d), cells were replated on 24-well multiwell dishes (details in SI Materials and Methods).
Differentiation of ES Cells into Motor Neurons and Sandwich Culture.
Mouse ES cells were differentiated into motor neurons according to methods previously described (14, 46). Dissociated embryoid bodies were FACS-purified and plated on poly-D-lysine/laminin coverslips with paraffin wax feet (described in ref. 17). The coverslips were inverted over a layer of primary glia cells in 24-well dishes. Cells were harvested at different time points after plating (7, 14, and 21 d). RNA was extracted immediately, or the tubes were stored at −80 °C (additional details in SI Materials and Methods).
RNA Extraction and cDNA Library Preparation.
RNA was extracted using a combination of TRIzol (Invitrogen) and RNeasy columns (Qiagen). Libraries were prepared using either standard protocols from Illumina or the Nextera transposase-mediated method (47) and sequenced using the Illumina platform (additional details in SI Materials and Methods).
RNA-Seq Data Analysis.
We used ExpressionPlot (48) to analyze sequence data and generate the two- and four-way plots and ECDFs (additional details in SI Materials and Methods).
Clustering Analyses.
We used the R\Bioconductor statistical framework (49, 50) to preprocess expression values and cluster the samples as detailed in SI Materials and Methods.
Functional Analyses.
To perform functional enrichment analysis, we used Ingenuity Pathway Analysis (Ingenuity Systems; www.ingenuity.com) and Pathway Guide (Advaita Corporation; www.advaitacorporation.com) (51, 52) as detailed in SI Materials and Methods.
Immunofluorescence Studies: Immunocytochemistry Analyses.
We performed immunocytochemistry analyses essentially as described previously (14). We used the following primary antibodies: mouse monoclonal anti-Tuj1 (Covance), rabbit anti-GFP (Alexa488-conjugated; Molecular Probes), or rabbit anti-GFAP (Chemicon). As secondary antibodies, we used donkey antibodies specific for rabbit conjugated to Cy3 (1:100, 2 h; Jackson ImmunoResearch) and/or donkey antibodies specific for mouse conjugated to Cy5 antibodies (1:100, 2 h; Jackson ImmunoResearch). Samples were imaged using Olympus FV 1000 (additional details in SI Materials and Methods).
Immunofluorescence Studies: Immunohistochemistry Analyses.
Immunolabeling.
Cryoprotected sections from lumbar spinal cord were incubated overnight with specific primary antibodies: anti-ChAT (goat: 1/100; Invitrogen), anti–TGFβ-RII (rabbit: 1/250; Novus Biologicals), or anti-GFAP (rabbit: 1/500; DAKO or mouse: 1/200; Millipore). Sections were incubated with species-specific secondary antibodies (Alexa488–anti-goat: 1/1,000, Alexa594–anti-rabbit: 1/1,000, and Alexa594–anti-mouse: 1/500; Invitrogen). Samples were imaged using an Olympus FluoView FV1000 Confocal Microscope (additional details in SI Materials and Methods).
Quantification of motor neurons.
Cell counts were obtained by tracking (ImageJ; National Institutes of Health) ChAT-positive motor neurons from optical stacks covering the entire ventral horn. The ChAT motor neurons expressing TGFβ-RII were also annotated in these stacks. Prevalence was determined by dividing TGFβ-RII motor neurons by all ChAT neurons. A minimum of three sections per age was analyzed. Average cell counts were obtained from five SOD1G93A animals. One-way ANOVA with P < 0.05 was used for statistical comparisons. Results were expressed as mean ± SE.
Supplementary Material
Acknowledgments
We thank Jason Gertz and Tim Reddy for help with the bioinformatics and members of T.M. laboratory for critical discussions and comments on the manuscript. H.P.P. was supported by a grant from the ALS Association and funds from the Robert Packard Center for ALS Research at Johns Hopkins and Project ALS (P2ALS) consortium. RNA sequencing costs and support for P.G., B.A.F., and S.O. were provided by the ALS Therapy Alliance (ATA) and HudsonAlpha funds. This work was also supported by National Institutes of Health Director’s Pioneer Award 8DP1NS082099-06 (to T.M.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE43879 (accession no. GSE43879).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222361110/-/DCSupplemental.
References
- 1.Kiernan MC, et al. Amyotrophic lateral sclerosis. Lancet. 2011;377(9769):942–955. doi: 10.1016/S0140-6736(10)61156-7. [DOI] [PubMed] [Google Scholar]
- 2.Andersen PM, Al-Chalabi A. Clinical genetics of amyotrophic lateral sclerosis: What do we really know? Nat Rev Neurol. 2011;7(11):603–615. doi: 10.1038/nrneurol.2011.150. [DOI] [PubMed] [Google Scholar]
- 3.Rosen DR, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362(6415):59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 4.Bruijn LI, et al. ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron. 1997;18(2):327–338. doi: 10.1016/s0896-6273(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 5.Gurney ME, et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264(5166):1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 6.Nagai M, et al. Rats expressing human cytosolic copper-zinc superoxide dismutase transgenes with amyotrophic lateral sclerosis: Associated mutations develop motor neuron disease. J Neurosci. 2001;21(23):9246–9254. doi: 10.1523/JNEUROSCI.21-23-09246.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong PC, et al. An adverse property of a familial ALS-linked SOD1 mutation causes motor neuron disease characterized by vacuolar degeneration of mitochondria. Neuron. 1995;14(6):1105–1116. doi: 10.1016/0896-6273(95)90259-7. [DOI] [PubMed] [Google Scholar]
- 8.Andersen PM, et al. Phenotypic heterogeneity in motor neuron disease patients with CuZn-superoxide dismutase mutations in Scandinavia. Brain. 1997;120(Pt 10):1723–1737. doi: 10.1093/brain/120.10.1723. [DOI] [PubMed] [Google Scholar]
- 9.Li TM, Alberman E, Swash M. Comparison of sporadic and familial disease amongst 580 cases of motor neuron disease. J Neurol Neurosurg Psychiatry. 1988;51(6):778–784. doi: 10.1136/jnnp.51.6.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boillée S, Vande Velde C, Cleveland DW. ALS: A disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52(1):39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Lino MM, Schneider C, Caroni P. Accumulation of SOD1 mutants in postnatal motoneurons does not cause motoneuron pathology or motoneuron disease. J Neurosci. 2002;22(12):4825–4832. doi: 10.1523/JNEUROSCI.22-12-04825.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pramatarova A, Laganière J, Roussel J, Brisebois K, Rouleau GA. Neuron-specific expression of mutant superoxide dismutase 1 in transgenic mice does not lead to motor impairment. J Neurosci. 2001;21(10):3369–3374. doi: 10.1523/JNEUROSCI.21-10-03369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ilieva H, Polymenidou M, Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol. 2009;187(6):761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haidet-Phillips AM, et al. Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat Biotechnol. 2011;29(9):824–828. doi: 10.1038/nbt.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Giorgio FP, Carrasco MA, Siao MC, Maniatis T, Eggan K. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat Neurosci. 2007;10(5):608–614. doi: 10.1038/nn1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papadeas ST, Kraig SE, O’Banion C, Lepore AC, Maragakis NJ. Astrocytes carrying the superoxide dismutase 1 (SOD1G93A) mutation induce wild-type motor neuron degeneration in vivo. Proc Natl Acad Sci USA. 2011;108(43):17803–17808. doi: 10.1073/pnas.1103141108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;1(5):2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- 18.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lei L, et al. Glioblastoma models reveal the connection between adult glial progenitors and the proneural phenotype. PLoS One. 2011;6(5):e20041. doi: 10.1371/journal.pone.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sofroniew MV. Molecular dissection of reactive astrogliosis and glial scar formation. Trends Neurosci. 2009;32(12):638–647. doi: 10.1016/j.tins.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zamanian JL, et al. Genomic analysis of reactive astrogliosis. J Neurosci. 2012;32(18):6391–6410. doi: 10.1523/JNEUROSCI.6221-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247(2002):91–101. [PubMed] [Google Scholar]
- 25. BioCarta BioCarta—Charting Pathways of Life. Available at http://www.biocarta.com/genes/index.asp.
- 26.Barker WC, et al. Protein Information Resource: A community resource for expert annotation of protein data. Nucleic Acids Res. 2001;29(1):29–32. doi: 10.1093/nar/29.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulder NJ, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35(Database issue):D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc Natl Acad Sci USA. 1998;95(11):5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eroglu C. The role of astrocyte-secreted matricellular proteins in central nervous system development and function. J Cell Commun Signal. 2009;3(3-4):167–176. doi: 10.1007/s12079-009-0078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bajo-Grañeras R, Ganfornina MD, Martín-Tejedor E, Sanchez D. Apolipoprotein D mediates autocrine protection of astrocytes and controls their reactivity level, contributing to the functional maintenance of paraquat-challenged dopaminergic systems. Glia. 2011;59(10):1551–1566. doi: 10.1002/glia.21200. [DOI] [PubMed] [Google Scholar]
- 31.Ganfornina MD, et al. ApoD, a glia-derived apolipoprotein, is required for peripheral nerve functional integrity and a timely response to injury. Glia. 2010;58(11):1320–1334. doi: 10.1002/glia.21010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ganfornina MD, et al. Apolipoprotein D is involved in the mechanisms regulating protection from oxidative stress. Aging Cell. 2008;7(4):506–515. doi: 10.1111/j.1474-9726.2008.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mi R, Chen W, Höke A. Pleiotrophin is a neurotrophic factor for spinal motor neurons. Proc Natl Acad Sci USA. 2007;104(11):4664–4669. doi: 10.1073/pnas.0603243104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munger JS, Sheppard D. Cross talk among TGF-β signaling pathways, integrins, and the extracellular matrix. Cold Spring Harb Perspect Biol. 2011;3(11):a005017. doi: 10.1101/cshperspect.a005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Massagué J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1(3):169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 36.Akhurst RJ, Hata A. Targeting the TGFβ signalling pathway in disease. Nat Rev Drug Discov. 2012;11(10):790–811. doi: 10.1038/nrd3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neill T, Schaefer L, Iozzo RV. Decorin: A guardian from the matrix. Am J Pathol. 2012;181(2):380–387. doi: 10.1016/j.ajpath.2012.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramirez F, Rifkin DB. Extracellular microfibrils: Contextual platforms for TGFbeta and BMP signaling. Curr Opin Cell Biol. 2009;21(5):616–622. doi: 10.1016/j.ceb.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doyle JJ, Gerber EE, Dietz HC. Matrix-dependent perturbation of TGFβ signaling and disease. FEBS Lett. 2012;586(14):2003–2015. doi: 10.1016/j.febslet.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flanders KC, Ren RF, Lippa CF. Transforming growth factor-betas in neurodegenerative disease. Prog Neurobiol. 1998;54(1):71–85. doi: 10.1016/s0301-0082(97)00066-x. [DOI] [PubMed] [Google Scholar]
- 41.Hamby ME, et al. Inflammatory mediators alter the astrocyte transcriptome and calcium signaling elicited by multiple G-protein-coupled receptors. J Neurosci. 2012;32(42):14489–14510. doi: 10.1523/JNEUROSCI.1256-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Day WA, Koishi K, Nukuda H, McLennan IS. Transforming growth factor-beta 2 causes an acute improvement in the motor performance of transgenic ALS mice. Neurobiol Dis. 2005;19(1-2):323–330. doi: 10.1016/j.nbd.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Tesseur I, et al. Deficiency in neuronal TGF-beta signaling promotes neurodegeneration and Alzheimer’s pathology. J Clin Invest. 2006;116(11):3060–3069. doi: 10.1172/JCI27341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iida A, et al. A functional variant in ZNF512B is associated with susceptibility to amyotrophic lateral sclerosis in Japanese. Hum Mol Genet. 2011;20(18):3684–3692. doi: 10.1093/hmg/ddr268. [DOI] [PubMed] [Google Scholar]
- 45.Kang SH, Fukaya M, Yang JK, Rothstein JD, Bergles DE. NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron. 2010;68(4):668–681. doi: 10.1016/j.neuron.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110(3):385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 47.Gertz J, et al. Transposase mediated construction of RNA-seq libraries. Genome Res. 2012;22(1):134–141. doi: 10.1101/gr.127373.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friedman BA, Maniatis T. ExpressionPlot: A web-based framework for analysis of RNA-Seq and microarray gene expression data. Genome Biol. 2011;12(7):R69. doi: 10.1186/gb-2011-12-7-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2012. [Google Scholar]
- 51.Khatri P, Draghici S, Ostermeier GC, Krawetz SA. Profiling gene expression using onto-express. Genomics. 2002;79(2):266–270. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- 52.Draghici S, Khatri P, Martins RP, Ostermeier GC, Krawetz SA. Global functional profiling of gene expression. Genomics. 2003;81(2):98–104. doi: 10.1016/s0888-7543(02)00021-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.