Abstract
The xeroderma pigmentosum group D (XPD) helicase is a subunit of transcription/DNA repair factor, transcription factor II H (TFIIH) that catalyzes the unwinding of a damaged DNA duplex during nucleotide excision repair. Apart from two canonical helicase domains, XPD is composed of a 4Fe-S cluster domain involved in DNA damage recognition and a module of uncharacterized function termed the “ARCH domain.” By investigating the consequences of a mutation found in a patient with trichothiodystrophy, we show that the ARCH domain is critical for the recruitment of the cyclin-dependent kinase (CDK)-activating kinase (CAK) complex. Indeed, this mutation not only affects the interaction with the MAT1 CAK subunit, thereby decreasing the in vitro basal transcription activity of TFIIH itself and impeding the efficient recruitment of the transcription machinery on the promoter of an activated gene, but also impairs the DNA unwinding activity of XPD and the nucleotide excision repair activity of TFIIH. We further demonstrate the role of CAK in downregulating the XPD helicase activity within TFIIH. Taken together, our results identify the ARCH domain of XPD as a platform for the recruitment of CAK and as a potential molecular switch that might control TFIIH composition and play a key role in the conversion of TFIIH from a factor active in transcription to a factor involved in DNA repair.
Keywords: rare disease, regulation of gene expression
The xeroderma pigmentosum group D (XPD) gene encodes a 5′–3′ helicase (XPD) that harbors mutations in patients suffering from three rare autosomal recessive diseases, xeroderma pigmentosum (XP), trichothiodystrophy (TTD), and Cockayne syndrome (CS) (1, 2). XP is characterized by a deficit of the nucleotide excision repair (NER) pathway, leading to sun sensitivity and susceptibility to skin cancer. TTD is characterized by sulfur-deficient brittle hair and a variety of neuroectodermal symptoms (3). XPD is the founding member of a family of DNA helicases conserved in archaea and eukaryotes. All family members share a four-domain organization including a conserved (Fe-S) cluster-binding domain that is essential for the helicase activity and a module of uncharacterized function named the ARCH domain by its arch-shape structure (4–7). Although archeal XPD homologs are monomers and have no known stable interactors, eukaryotic XPD homologs are part of the general transcription/DNA repair factor transcription factor II H (TFIIH), a multisubunit complex made up of 10 subunits (reviewed in ref. 8). Low-resolution models for TFIIH have been obtained for the complex in yeast (9, 10) and for the human complex (11), showing an overall conservation of shape. Human TFIIH can be resolved into two functional and structural entities bridged by XPD: the core-TFIIH consists of XPB, p62, p52, p44, p34, and p8, whereas the cyclin-dependent kinase (CDK)-activating kinase (CAK) subcomplex contains CDK7, cyclin H, and ménage a trois 1 (MAT1). XPD interacts with the p44 core-TFIIH subunit and with MAT1, a subunit of CAK involved in the regulation of CDK 7 transcription activity (12–15).
TFIIH first was identified as a basal transcription factor and subsequently was shown to play a key role in DNA repair. The xeroderma pigmentosum group B (XPB) helicase is involved in promoter opening during transcription initiation, whereas XPD allows strand separation around the DNA lesion in the context of DNA repair by NER (16, 17). Biochemical and genetic studies have shown that both XPB and XPD ATPase activities are needed to open up DNA around a damaged site (18, 19). Recent data showed that only XPB ATPase activity of is required for opening and remodeling of DNA in NER and transcription, and its helicase is devoted to promoter escape in transcription (20, 21). XPD helicase activity, on the other hand, plays a minor role in transcription but is necessary for NER (19, 22). Once recruited to the DNA damage/ xeroderma pigmentosum group C (XPC)/HR23B complex, CAK dissociates from core-TFIIH and XPD upon the arrival of xeroderma pigmentosum group A (XPA) and other NER factors, thus promoting incision/excision of the damaged oligonucleotide and repair of the DNA (23). The CDK7 subunit of TFIIH phosphorylates residues Ser5 and Ser7 from the C-terminal domain (CTD) of the rpb1 RNA polymerase II (RNA Pol II) subunit and functions in promoter-proximal pausing and termination (24–26). In addition to its role in basal transcription, CDK7 also participates in the transactivation of several hormone-dependent genes by phosphorylating nuclear receptors (NRs) including retinoic acid receptors (27, 28), the estrogen receptor (29), peroxisome proliferator-activated receptor (30), and the androgen receptor (31). The importance of NR phosphorylation by TFIIH in transcription has been highlighted in studies using cells from patients bearing mutations in the XPD subunit of TFIIH or in the xeroderma pigmentosum group G (XPG) endonuclease that established the consequence of hormonal/transcriptional dysfunctions in XP-CS/TTD phenotypes. In patient cell lines, NRs are hypophosphorylated, and the ligand-dependent response is decreased (28, 32).
Most mutations in TFIIH that originate XP, TTD, and XP/CS both disturb the regulatory interactions between TFIIH components and affect the catalytic activities of the complex. For example, mutations in p8/TTD-A weaken interactions with the p52 core-TFIIH subunit, leading to a reduced intracellular TFIIH concentration and a defect in NER, a common feature of TTD cells (33, 34). The F99S mutation in XPB weakens the XPB/p52 interaction and thus the resulting decrease in the XPB ATPase activity (21). Similarly, the R683W and R722W mutations found in XPD patients weaken its interaction with p44, another TFIIH subunit, and consequently disrupt its helicase activation function (35, 36).
We have combined in vitro reconstituted assays with cell-based approaches to provide insights on the relationships between the structure and function for XPD and the architecture of TFIIH. Based on a detailed characterization of mutations identified in cell lines derived from patients suffering from TTD, we show that the ARCH domain of XPD plays a key role in DNA recognition and in the strand-displacement activity of the helicase and identify this domain as a platform for the recruitment of CAK and thus the regulation of TFIIH transcription and NER activities.
Results
Composition of Reconstituted Mutant TFIIH.
To address the role of the ARCH domain of human XPD, we take advantage of the C259Y mutation, which to our knowledge is the only mutation identified in this domain and which is associated with the R722W allele in patients TTD12PV and TTD15PV (Fig. 1A). Fibroblasts from these patients show a drastic reduction of the capacity to perform DNA-repair synthesis, with unscheduled DNA synthesis levels that are 20% of normal, and poor survival after UV irradiation (37).
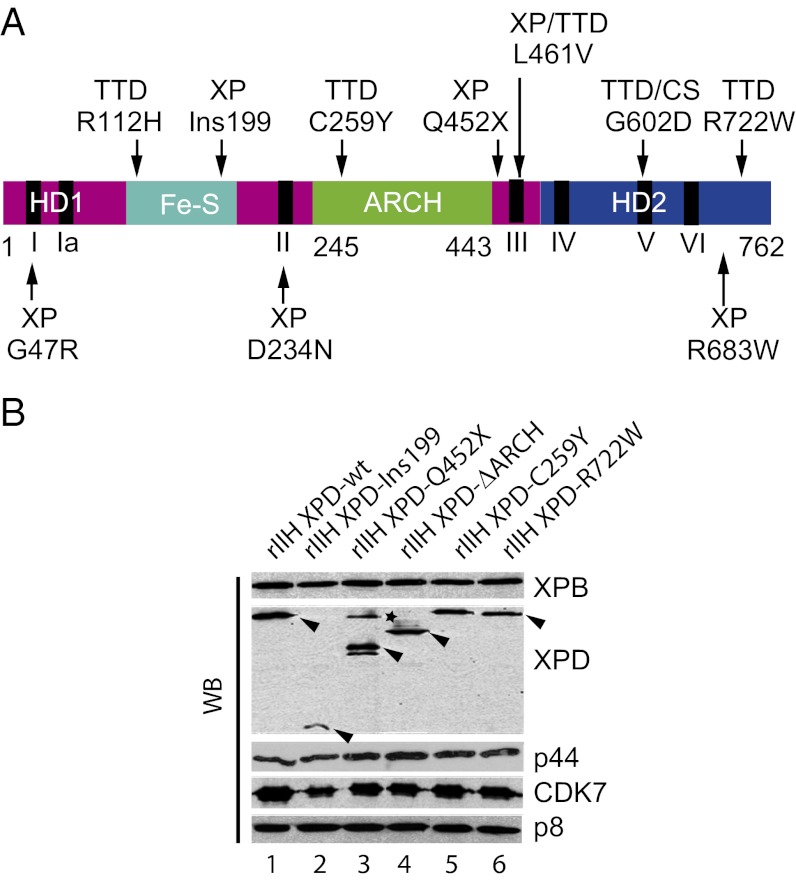
Fig. 1.
TTD and XP mutations in the XPD TFIIH subunit. (A) Schematic representation of XPD. Helicase motor domains HD1 and HD2 are shown in magenta and blue, respectively, the Fe-S iron sulfur-containing domain is in cyan, and the ARCH domain is in green. Black bars indicate the helicase motifs (I, Ia, II, II, IV, V, and VI). Positions of mutations flanking the ARCH domain and diseases associated with the mutations are shown. (B) Production of rIIH. The 10 subunits of human TFIIH, including either wild-type or mutated XPD, were coexpressed in insect cells using the baculovirus expression system, and complexes were immunoprecipitated using an antibody directed against the p44 subunit of the core-TFIIH in low-salt conditions (buffer B containing 75 mM KCl). After elution with a synthetic peptide recognized by Ab-p44, equal amounts of purified rIIHs were analyzed by SDS/PAGE with 12% (wt/vol) polyacrylamide followed by Western blot (WB) analysis with antibodies directed against XPB, the N-terminus of XPD, p44, CDK7, or p8. Arrowheads indicate the theoretical molecular weight of each XPD mutated form. The asterisk indicates a nonspecific band.
We first examined the consequences of the C259Y mutation on TFIIH composition and activities. Recombinant TFIIH complexes (rIIH), resulting from coinfection by baculoviruses expressing either wild-type (XPD-wt) or mutant (XPD-C259Y and XPD-R722W) proteins were produced in insect cells. Sf21 cells were coinfected first with a virus for expression of the six core-TFIIH subunits, with a second virus for the three CAK subunits, and a third virus for the production of XPD. Recombinant complexes were immunopurified from infected cell extracts using an antibody directed against the p44 core-TFIIH subunit under physiological salt conditions to prevent dissociation of weakly associated subunits, and bound complexes were eluted with a competitor peptide. Western blot analysis of immunopurified complexes shows that the C259Y mutation does not affect the composition and stoichiometry of rIIH XPD-C259Y, which is comparable to that of rIIH XPD-wt or rIIH XPD-R722W. Immunopurification by the p44 antibody precipitated not only the p44, XPB, and p8 subunits of core-TFIIH but also the XPD and CDK7, a component of the CAK subcomplex (Fig. 1B compare lanes 1, 5, and 6). For further insights, we also engineered an XPD variant (XPD-∆ARCH) in which residues 248–438 that correspond to the entire ARCH domain were deleted and replaced by a short linker peptide. Deletion of the ARCH domain had no visible effect on the composition and stoichiometry of the immunopurified complex (Fig. 1B, lane 4); this result was not surprising, because previous reports had shown that deletion of entire domains does not necessarily affect TFIIH composition (34, 38). We also analyzed XPD-Ins199 and XPD-Q452X, two mutant forms located on either side of the ARCH domain (36). Consistent with previous experiments, in rIIH XPD-Q452X, where the XPD ARCH domain is present but the CTD is lacking, the level of CDK7 is similar to that observed in the control (compare lanes 1 and 3). The frameshift at position 199 in XPD-Ins199 leads to a protein lacking the ARCH domain and the C-terminal part of the protein. In rIIH XPD-Ins199, XPD as well as CDK are sub-stoichiometric, but XPB and p8 are not affected (compare lanes 1 and 2), suggesting that the ARCH domain, deleted in the Ins199 but not in the Q452X mutant, might be required for a stable association of CAK with core-TFIIH. The reduced amount of XPD and CDK7 could result directly from the impaired association of CAK with XPD or from an altered association of XPD with core-TFIIH, which, as a consequence, might affect recruitment of CAK.
DNA Repair and Helicase Activities of XPD Mutants.
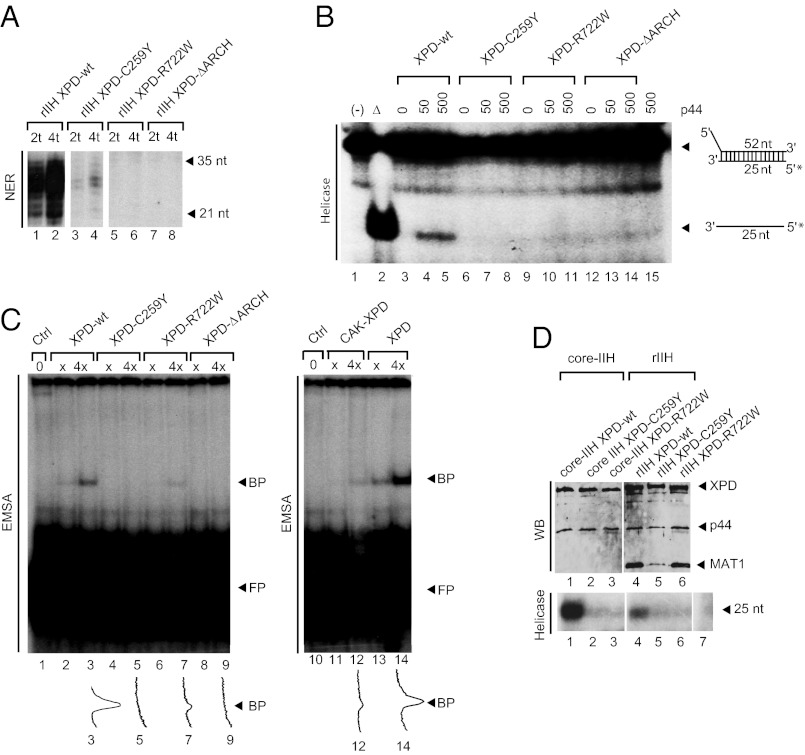
We tested the different rIIH recombinant complexes in a dual-incision assay that contains the XPC-HR23B, XPA, RPA, XPG, and ERCC1-XPF factors and a closed circular plasmid with a single 1,3-intrastrand d(GpTpG) cisplatin-DNA crosslink as template (35). Both deletion of the ARCH domain and the C259Y mutation impair incision: Although rIIH XPD-ΔARCH is totally inactive (Fig. 2A, lanes 7 and 8), low but significant residual incision activity is detectable with rIIH XPD-C259Y (lanes 3 and 4). Note that rIIH XPD-R722W, the complex harboring the mutation found in the second allele in TTD12PV and TTD15PV patients, is unable to potentiate dual incision of the damaged DNA (lanes 5 and 6), suggesting that C259Y is a causative mutation i.e., is responsible for the molecular phenotype.
Fig. 2.
DNA repair and helicase activities of rIIHs. (A) In vitro double-incision assay. Increasing amounts of immunopurified rIIH were added to an incision/excision assay using recombinant NER factors, and the reaction was analyzed by electrophoresis followed by autoradiography. “t” corresponds to ∼25 ng of rIIH with p44 as reference. (B) The 5′–3′ helicase activity of XPD variants. Equivalent amounts of each Flag-purified XPD variant (∼100 ng) were added to a 5′-strand extension probe obtained by annealing a 52-nt single-strand DNA to a 5′ 32P-labeled 25-nt single-stranded DNA in the presence of increasing amounts of p44 (0, 50, or 500 ng). Single- and double-stranded DNA are separated by electrophoresis on a 14% (wt/vol) polyacrylamide gel and analyzed by autoradiography (lanes 3–15). The symbols “−” and “Δ” indicate the native and denatured probes, respectively (lanes 1 and 2). (C) DNA-binding activity. Increasing amounts of XPD variants (Left, lanes 1–9) and CAK-XPD (Right, lanes 10–14) were incubated with the labeled 5′-strand extension probe shown in B, and the resulting nucleoprotein complexes were analyzed by electrophoresis using a 6% (wt/vol) polyacrylamide gel followed by autoradiography with XPD as reference. Densitometric analysis of lanes 3, 5, 7, 9, 12, and 14 are shown as below. “×” corresponds to 100 ng of XPD. BP, bound probe; FB, free probe. (D) 5′–3′ helicase activities of rIIH complexes. Insect cells were infected with a set of baculoviruses overexpressing the subunits of TFIIH including either wild-type or mutated Flag-tagged XPD, and complexes were immunoprecipitated using an antibody directed against the Flag epitope in buffer B. After elution with the Flag synthetic peptide, core-TFIIH/XPD (core-IIH, lanes 1–3) and Holo-TFIIH (rIIH, lanes 4–6) were analyzed by Western blot analysis (WB), and equivalent amounts of complex (∼200 ng) were tested for their 5′–3′ helicase activities (Helicase). The negative control is shown in lane 7. Densitometric analysis suggests a fivefold decrease of Holo-TFIIH (lane 4) 5′–3′ helicase activity compared with that of core-TFIIH (lane 1) in the case of XPD-wt.
We then evaluated the helicase 5′–3′ activity of purified XPD using a double-stranded oligonucleotide substrate with a 5′ single-strand extension (Fig. 2B). Experiments were performed in presence of the p44 core-TFIIH subunit, known to interact specifically with and to regulate XPD helicase activity (39). In contrast to XPD-wt, which is able to displace the 25-bp 32P-labeled oligonucleotide in the presence of p44, neither XPD-C259Y nor XPD-ΔARCH exhibit detectable helicase activity, even with the addition of a large excess of p44 (Fig. 2B, compare lanes 6–8 and lanes 12–14 with lanes 3–5). We next investigated interactions with the same substrate DNA using an EMSA and tested whether mutations in the ARCH domain impair DNA binding. Although the XPD-R722W mutant has partially retained the capacity to form a stable complex with DNA (Fig. 2C, compare lanes 6 and 7 with lanes 2 and 3), neither XPD-C259Y nor XPD-ΔARCH is able to shift the helicase DNA probe (Fig. 2C, compare lanes 4 and 5 and lanes 8 and 9). Because TFIIH subunits are involved in an intricate protein–protein interaction network that is likely to modulate its catalytic activities, we also analyzed the 5′–3′ helicase activity of XPD when associated with core-TFIIH and in the context of Holo-TFIIH (composed of core-TFIIH, XPD, and CAK). In accordance with previous observations on isolated subunits (13), we indeed found that the 5′–3′ helicase activity of Holo-TFIIH is reduced significantly compared with that of core-TFIIH (Fig. 2D, compare lanes 1 and 4). However, none of the mutant complexes exhibited detectable helicase activity (Fig. 2D, Lower, compare lane 1 with lanes 2 and 3 and lane 4 with lanes 5 and 6). Taken together, these results show that the ARCH domain is required for the unwinding reaction and is involved in recognition of the DNA substrate. Comparison of the helicase activity of core-TFIIH with that of Holo-TFIIH suggests that the presence of CAK represses XPD helicase activity. In agreement with this observation, gel-shift experiments performed with CAK/XPD instead of XPD do not lead to the formation of a stable nucleoprotein complex detectable under our experimental conditions (Fig. 2C, compare lanes 11 and12 with lanes 13 and 14).
XPD Mutations Impair in Vitro Transcription Activity of TFIIH.
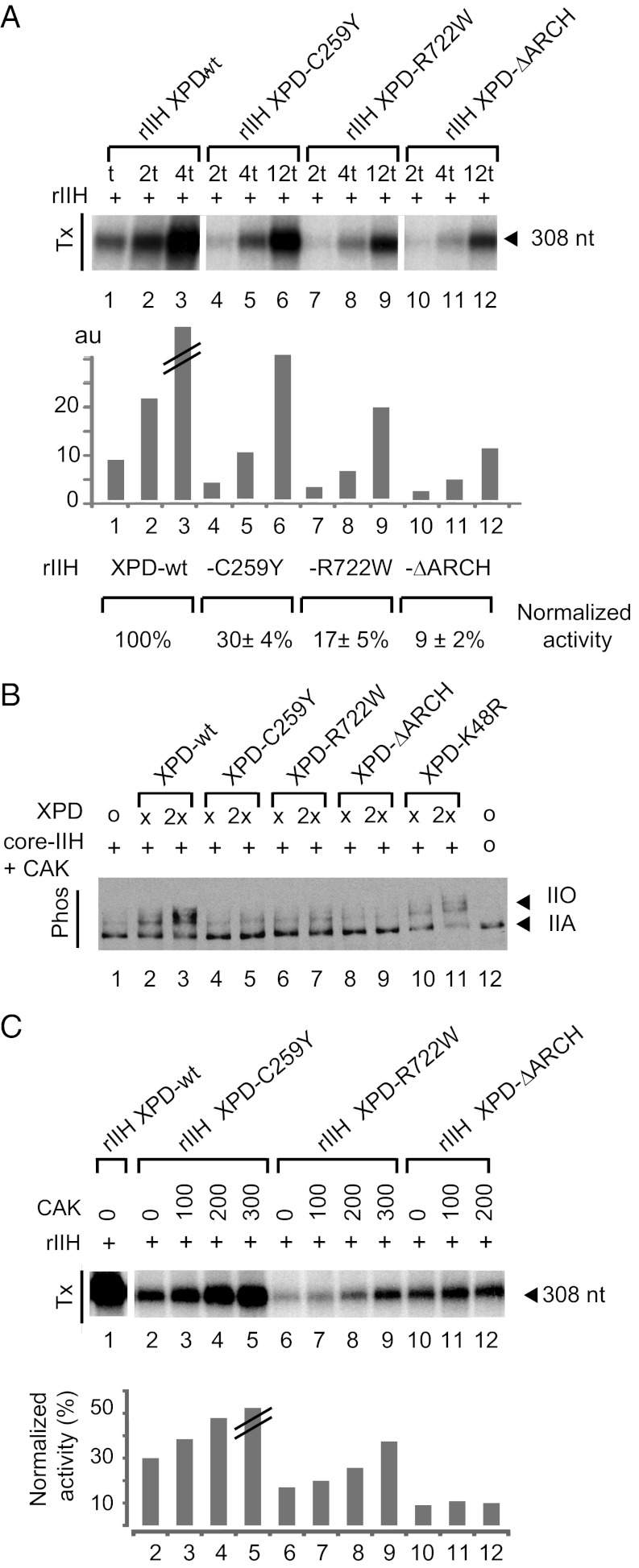
The rIIHs complexes next were tested for their transcription activity using an in vitro reconstituted assay in the presence of recombinant TBP, TFIIA, TFIIB, TFIIE, and TFIIF in addition to purified RNA Pol II and a linearized DNA template containing the adenovirus major late promoter. rIIH XPD-C259Y and rIIH XPD-R722W significantly impair RNA synthesis relative to wild type (Fig. 3A, compare lanes 1–3 with lanes 4–6 and 7–9).
Fig. 3.
Transcription and CTD kinase activity of rIIHs. (A) Basal transcription activity. Increasing amounts of purified rIIH with different XPD variants were added to an in vitro reconstituted transcription system lacking TFIIH. Transcripts were analyzed by electrophoresis followed by autoradiography and quantification. The length of the corresponding transcript is indicated on the left. Data from a representative experiment are shown in the histogram. For each mutant, the transcription activity from three independent experiments and normalized to wild-type rIIH is indicated. “//” indicates saturation; “t” corresponds to ∼25 ng of rIIH with p44 as reference; au, arbitrary units. (B) To evaluate the RNA Pol II kinase activity of the TFIIH variants, purified core-TFIIH (200 ng), CAK (100 ng), and XPD (x or 2×) mutants were mixed in an in vitro assay containing all the basal transcription factors and the AdMLP. Arrows indicate hypophosphorylated (IIA) and hyperphosphorylated (IIO) forms of RNA Pol II. “×” corresponds to ∼100 ng of XPD. (C) The transcription activity of rIIHs containing the different XPD variants was assessed in presence of increasing amounts of purified recombinant CAK. “+” corresponds to 50 ng of rIIH-wt, -C259Y, and -R722W and 150 ng of rIIH-ΔARCH. Wild-type rIIH was used as control. The signals were quantified and normalized to wild type assuming activities of 30, 17, and 9% for rIIH-C259Y, -R722W, and -ΔARCH, respectively.
Because CAK phosphorylation of the RNA Pol II CTD is required for the transition from initiation to elongation and for further promoter escape (40, 41), we investigated how mutations in XPD impact the phosphorylation status of the polymerase. Addition of XPD-wt to an in vitro transcription system that contains all the basal transcription factors in addition to core-TFIIH and CAK results in a shift from the lower-molecular-weight RNA Pol IIA (the nonphosphorylated form) to higher-molecular-weight RNA Pol IIO (the hyperphosphorylated form; Fig. 3B, lanes 1–3). Interestingly, no phosphorylation of RNA Pol II CTD is observed in the presence of XPD-C259Y or XPD-R722W (Fig. 3B, compare lanes 2 and 3 with lanes 4 and 5 and with lanes 6 and 7), whereas TFIIH containing XPD-K48R (which carries a mutation in the helicase motif I) allows CTD phosphorylation (lanes 10 and 11) (22). We then wondered whether an excess of CAK could enhance the transcription activity of recombinant TFIIH harboring the XPD mutations. Addition of CAK to purified recombinant rIIH XPD-C259Y leads to a twofold stimulation of RNA synthesis and a partial recovery of TFIIH transcription activity (Fig. 3C, lanes 2–5). Addition of CAK to rIIH-R722W also stimulates RNA synthesis, but only when a large excess of CAK is used (Fig. 3C, compare lanes 6–9 with lanes 2–5). The addition of CAK to rIIH-ΔARCH did not result in any stimulation (Fig. 3C, lanes 10–12). Taken together, these results establish that the C259Y mutation impairs the initiation of TFIIH basal transcription but in a manner that differs from the deletion of the entire ARCH domain and from the R722W mutation, which impairs the interaction with the p44 core-TFIIH subunit (39).
ARCH Domain Is a Platform for CAK Anchoring.
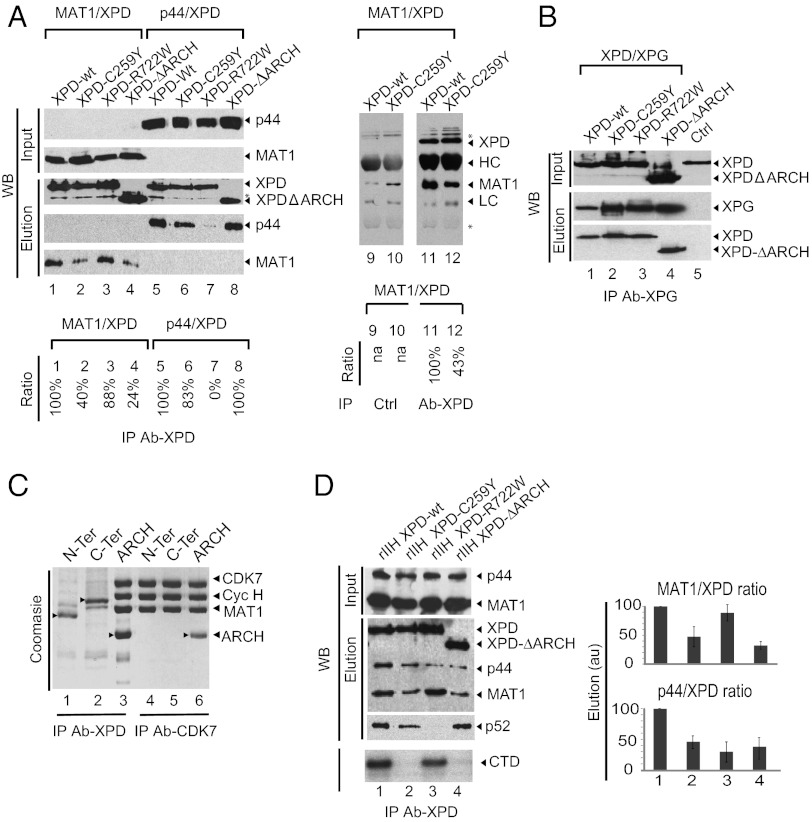
To explain why the C259Y mutation in XPD affects TFIIH transcription activity, we first tested pairwise interactions between mutant XPDs and p44 from core-TFIIH or XPDs with MAT1 from CAK, the two TFIIH subunits that interact tightly with the helicase (12, 39). Proteins were coexpressed in insect cells, immunoprecipitated in 250 mM NaCl using either the Flag-tag fused to the N-terminal extremity of XPD or an anti-XPD antibody, and analyzed by Western blot. Mutation C259Y in the ARCH domain and deletion of the entire module have no visible effect on the association of XPD with p44 (Fig. 4A, compare lanes 6 and 8 with lane 5) but significantly affect the MAT1/XPD interaction (compare lanes 2 and 4 with lane 1 and with lanes 11 and 12), suggesting that the ARCH domain is involved in anchoring CAK to XPD. Point mutation R722W, in contrast, impairs the p44/XPD interaction (compare lane 7 with lane 5) but does not modify its interaction with MAT1 (compare lane 3 with lane 1). In a second set of experiments, we analyzed interactions between XPD variants and the XPG endonuclease that associates strongly with TFIIH and stabilizes the interaction between core-TFIIH, CAK, and XPD (32). Pull down from cell extracts of Sf9 cells expressing XPG and XPD showed that the two proteins interact (Fig. 4B, lane 1) and that all XPD mutants, including XPD-ΔARCH, have retained the capacity to bind immobilized XPG (Fig. 4B, lanes 2 and 4), although to a lesser extent than wild-type XPD (Fig. 4B, lane 1). Mutations in either the ARCH domain and/or in the C-terminal end of XPD affect the ability to interact with other partners but in general do not abolish binding.
Fig. 4.
The ARCH domain is a recruitment platform for CAK. (A) Pairwise interactions between XPD variants and MAT1 or p44. Sf9 insect cells were coinfected with viruses that allow the expression of Flag-tagged XPD variants and a virus that allows the expression of MAT1 or p44. Proteins were immunopurified with the anti-Flag M2 resin (Left) or were immunoprecipitated with an anti-XPD antibody (Right) in the presence of 250 mM NaCl (buffer C). Input and purified complexes were analyzed by anti-p44, anti-MAT1, or anti-Flag Western blot. The asterisk indicates a nonspecific band. (B) Pairwise interactions between XPD variants and XPG. Sf9 insect cells were coinfected with viruses that allow the expression of Flag-tagged XPD variants and a virus that allows the expression of c-myc–tagged XPG. Immunopurification was performed with anti–c-Myc resin in buffer C. Input and the purified complexes were analyzed by Western blot with anti-Flag antibody. (C) Interactions between XPD fragments and CAK. Sf9 insect cells were coinfected with a virus that allows the expression of the CAK subcomplex with CDK7 C-terminal Strep-tag fusion and with viruses that allow the expression of N-terminal XPD (residues 1–245), C-terminal XPD (residues 443–762), or ARCH (residues 245–443) with an N-terminal Flag tag. The capacity of XPD fragments to bind CAK and to copurify was tested using the Flag-tag (lanes 1–3) or Strep-tag II (lanes 4–6) for the purification of the complexes in buffer C. Purified proteins were analyzed using PAGE with 12% (wt/vol) polyacrylamide followed by Coomassie staining. Proteins are indicated by arrowheads. (D) Consequences of XPD mutations on association with CAK and core-TFIIH. Insect cells were infected with a set of baculoviruses overexpressing the subunits of TFIIH including either wild-type or mutated Flag-tagged XPD, and complexes were immunoprecipitated using an antibody directed toward the Flag epitope in buffer C. After elution with the Flag synthetic peptide, immunopurified complexes (rIIHs) were analyzed by Western blot analysis (WB, Upper) and were tested for their capacity to phosphorylate the CTD of RNA Pol II (autoradio, Lower). Experiments were performed in triplicate. Histograms represent estimated ratios (in arbitrary units, au) between MAT1 (a representative CAK subunit) and XPD or p44 (a representative core-TFIIH subunit).
Next, to characterize further the CAK/XPD interface, XPD deletion fragments corresponding to the N-terminal region (residues 1–277), the ARCH domain (residues 245–443), or the C-terminal region (residues 444–760) of XPD (Fig. 1A) were coexpressed with the three subunits of CAK in insect cells. Complexes were affinity purified using either the Flag-tag fused to the N terminus of XPD constructs or the C-terminal Strep-tag from the CDK7 component of CAK. Both Flag-XPD and CDK7-Strep pull-down experiments followed by Coomassie staining show that the XPD fragment corresponding to the ARCH domain is able to coimmunoprecipitate with the three subunits of CAK (Fig. 4C, lanes 3 and 6). This is not the case for fragments corresponding to the N- and C-terminal regions of XPD, which do not associate with CAK (Fig. 4C, lanes 1 and2 and lanes 4 and 5), showing that the ARCH module indeed is required for stable association with CAK and can be considered a recruitment platform.
To investigate the consequences of XPD mutations on association with CAK and core-TFIIH, recombinant complexes were coexpressed in insect cells and immunopurified using the epitope Flag fused to the N terminus of XPD. These experiments were performed in the presence of 250 mM NaCl and 0.1% Nonidet P-40 to eliminate subunits that might be nonspecifically coimmunopurified, because the experiments presented above at lower ionic strength and in the absence of detergent showed that neither the C259Y or R722W point mutations nor the deletion of the ARCH domain affects the composition of the purified complex (Fig. 1B). Analysis of CDK7 kinase activity using a peptide that mimics the RNA Pol II CTD shows that the immunopurified complexes rIIH XPD-C259Y and rIIH XPD-ΔARCH are unable to phosphorylate their substrate because they contain only limited amounts of active CAK (Fig. 4D, compare lanes 1 and 3 with lanes 2 and 4). Western blot analysis shows that these immunoprecipitated complexes contain detectable amounts of p52, a core-TFIIH subunit, but the complex harboring the R722W mutation does not. A quantitative interpretation of Western blot experiments shows that mutations in the ARCH domain, and particularly the deletion of the entire domain, affect association not only with CAK but also with the p44 core-TFIIH subunit (Fig. 4D, histogram). This result suggests that subunit associations are not limited to a set of strong pairwise interactions and implies additional weaker contacts. These secondary interactions may anchor XPD positively within the complex or regulate catalytic activities, perhaps explaining why CTD phosphorylation is extremely low but the XPD/MAT1 interaction is not totally abolished (Fig. 4D, lanes 2 and 4). Together, these observations establish the role of the ARCH domain as a platform for recruitment of CAK and highlight the intricacy of the protein–protein interaction network within TFIIH.
Incidence of XPD Mutations on Transactivation Mediated by NRs.
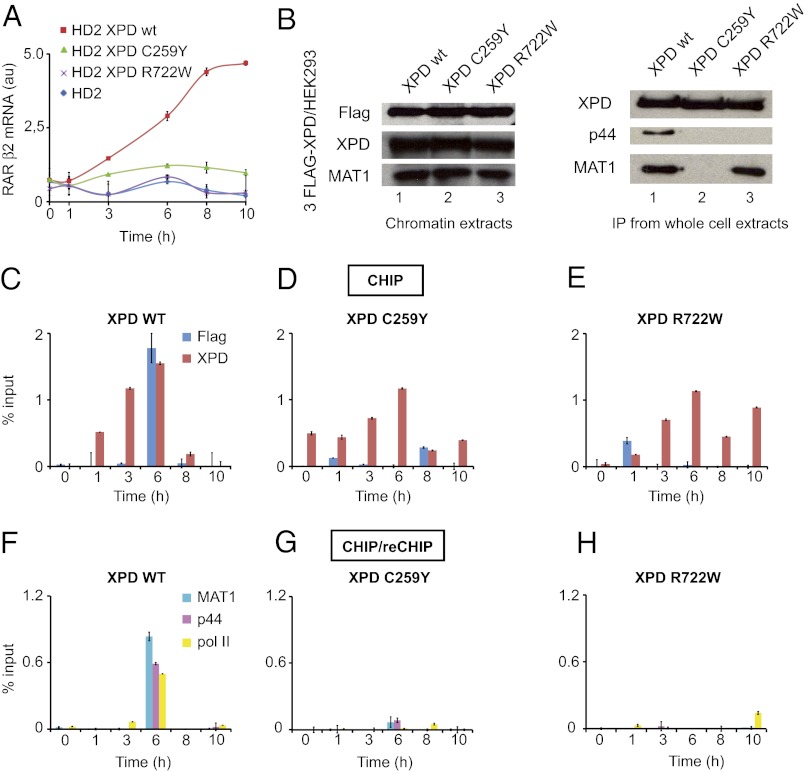
Knowing that mutations in XPD can affect ligand-dependent transactivation mediated by nuclear hormone receptors (28), we tested the effect of the C259Y mutation on the capacity of TFIIH to transactivate the RARβ2 gene in response to all-trans retinoic acid (t-RA). The established cell line HD2 harboring the causative XPD-R683 point mutation (42) was transfected with a plasmid expressing XPD-wt, XPD-C259Y, or XPD-R722W, treated with t-RA, and the concentration of RARβ2 mRNA was quantified by quantitative PCR (qPCR) (Fig. 5A). Overexpression of XPD-wt allows a significant transcriptional activation upon ligand induction compared with HD2 control. This correction is specific to XPD-wt; neither XPD-C259Y nor XPD-R722W was able to increase the level of RARβ2 mRNA significantly, demonstrating that, although not lethal, both the C259Y and R722W mutations strongly affect TFIIH function in vivo.
Fig. 5.
Implications of XPD in retinoic acid-dependent recruitment of TFIIH on the RARβ2 promoter. (A) Relative RARβ2 mRNA expression monitored by qPCR from XPD-transfected HD2 cells (XPD-wt, -C259Y, -R722W) treated with 1 µM t-RA. The effect of XPD mutations on nuclear hormone receptors mediated transactivation. The XPD-deficient cell line HD2 was transfected with an empty plasmid (blue diamond) or with a plasmid expressing XPD-wt (red square), XPD-C259Y (green triangle), or XPD-R722W (magenta cross) and was treated with t-RA. Transcription of the RARβ2 mRNA was quantified by qPCR. (B) HEK293 Flp-IN cells were stably transfected with 3Flag-XPD (XPD-wt, -C259Y, -R722W). Chromatin extracts were prepared and analyzed for Flag, XPD, and MAT1 by Western blot (Left). Whole-cell extracts were immunoprecipitated using the Flag tag and analyzed for XPD, p44, and MAT1 (Right). (C–H) ChIP/ReChIP monitoring the coimmunoprecipitation of the RARβ2 promoter using Flag or XPD antibodies (C–E) or a combination of antibodies against Flag/MAT1, Flag/p44, or Flag/pol II (F–H ) from 3Flag-XPD–transfected HEK293 stable cell lines (XPD-wt, XPD-C259Y, XPD-R722W) treated with t-RA (1 µM).
ChIP assays then were undertaken to investigate whether mutations affect the recruitment of TFIIH on activated RARβ2 promoter using isogenic stable cell lines engineered by site-specific integration of transgenes into HEK293 cells using an FLP-FRT system. In addition to endogenous XPD, these cells express either XPD-wt or a mutant protein (XPD-C259Y or XPD-R722W) fused to an N-terminal Flag peptide (Fig. 5B, Left). The genomic DNA fragments bound to XPD were immunoprecipitated with antibodies directed against either the Flag epitope or the XPD protein and were analyzed further by qPCR. After 6 h of treatment with t-RA and synthesis of RARβ2 mRNA (43), the recruitment of TFIIH containing the Flag-XPD at the RARβ2 promoter was evaluated. Using antibodies directed against either Flag-XPD or the XPD subunit itself demonstrated that both integrate TFIIH and are recruited together with RNA Pol II at the RARβ2 promoter in XPD-wt/HEK293 cells (Fig. 5C). We next observed that mutants XPD-C259Y and XPD-R722W are not recruited efficiently at the RARβ2 promoter (Fig. 5 D and E), explaining the inability of this mutated protein to restore the RARβ2 expression (Fig. 5A). Further ChIP/reChIP experiments confirm that the immunoprecipitated Flag-XPD complex contains the MAT1 and p44 TFIIH subunits when XPD-wt is used (Fig. 5 F–H). HEK293 Flp-In cells express endogenous XPD which competes with ectopically expressed mutants and impairs the assembly of mutant complexes at detectable levels. Interestingly, and in agreement with our previous conclusions obtained with recombinant complexes, pull-down experiments from whole-cell extracts prepared from HEK293 Flp-In expressing XPD-R722W show that the XPD-R722W mutation does not affect coimmunoprecipitation of XPD with the MAT1 CAK subunit but does affect coimmunoprecipitation of XPD with the p44 core-protein (Fig. 5B, Right). The C259Y mutation affects the interaction with both core-TFIIH and CAK. Taken together, our data show that the XPD-C259Y and XPD-R722W mutations are detrimental for RNA synthesis in a cellular context because the inability of mutants to integrate a functional TFIIH complex impairs the assembly of functional transactivation complexes.
Discussion
Helicases are modular proteins that use ATP to bind and/or remodel nucleic acid complexes. Involved in virtually all aspects of RNA and DNA metabolism, these motor proteins are built around a conserved helicase core consisting of two similar RecA-like motor domains with additional accessory domains, i.e., N- or C-flanking regions or inserts within the core. Specific regulation or complementary catalytic activities often are provided by interactions with protein partners, and several helicases are components of large macromolecular complexes.
Structural Considerations.
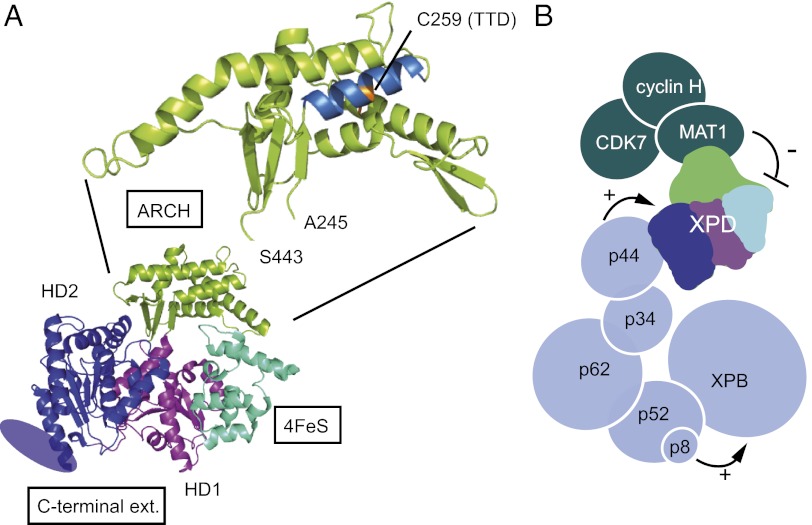
The XPD helicase illustrates the molecular organization of these motor proteins and the key role of accessory domains for regulation. Apart from its motor domains HD1 and HD2, human XPD is composed of a 4Fe-S cluster domain, the ARCH domain, and a specific C-terminal extension required for association with the p44 core-TFIIH subunit (Fig. 6A) (5–7). The 4Fe-S cluster domain contains four redox-sensitive cysteines that are thought to play a role in DNA damage recognition (44) and, together with the ARCH domain, forms a small tunnel large enough to accommodate single-stranded DNA (5–7, 45).
Fig. 6.
XPD structure and TFIIH architecture. (A) Modular organization of XPD. Thermoplasma acidophilum XPD (Protein Data Bank ID code 2VSF) is composed of four structural domains: two RecA-like helicase domains (HD1 in blue and HD2 in magenta), one domain that contains an iron/sulfur group (in cyan), and a domain described as the ARCH domain (in green). The C-terminal extension of eukaryotic XPD that is required for interaction with the p44 core-TFIIH subunit and for which no structural data are available is represented by an ellipse. Residue 259 mutated into a tyrosine in the TTD12PV patient cell line maps onto the first helix of the ARCH domain, and its side chain points into the center of the helical bundle. (B) Schematic of subunit architecture of TFIIH. Each subunit is represented by a circle with the radius of a sphere corresponding to its molecular weight. Interactions between XPB, p52, and p8/TTD-A stimulate XPB ATPase activity and consequently favor binding of TFIIH to damaged DNA. Interactions between p44 and XPD stimulate the helicase activity, allowing unwinding of DNA around the lesion and subsequent double incision by the endonucleases XPF-ERCC1 and XPG. XPD helicase activity, dispensable for transcription initiation but required for NER, is repressed when CAK is associated with TFIIH.
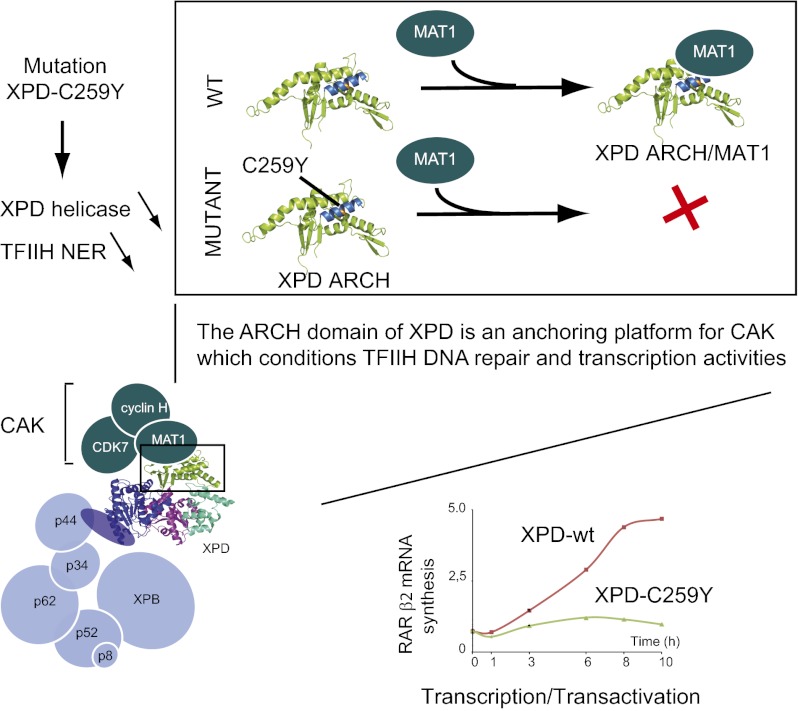
We have shown that the ARCH domain of human XPD is a platform for CAK anchoring and is required for the stability of TFIIH. The ARCH domain interacts specifically with the MAT1 subunit of CAK and has been identified as the minimal region required for a stable association of XPD with the kinase complex. In the context of TFIIH, deletion of the ARCH domain impairs association not only with CAK but also with core-TFIIH, as demonstrated by quantitative XPD pull-down experiments performed in the presence of medium salt concentration and of limited amounts of nonionic detergent to dissociate weakly associated subunits. In this work, we largely took advantage of the C259Y mutation, which impedes the MAT1/XPD interface. Residue C259 maps onto the first helix of the ARCH domain, and its side chain points into the center of the helical bundle where it packs with other residues that form the core of the domain (Fig. 6A). Mutation of C259 into a tyrosine probably destabilizes or unfolds the protein locally, as is consistent with the reduced thermal stability of a mutant XPD homolog from Sulfolobus tokodaii. We have shown that mutation C259Y in human XPD affects the XPD/MAT1 interface and thus the stability of mutant TFIIH. In TTD12PV fibroblasts, this mutation is associated with the R722W allele that leads to destabilization of the XPD/p44 interaction (39) and as a consequence also impacts the stability of TFIIH, explaining the low intracellular concentration of TFIIH, a common feature of TTD-derived cell lines (37, 46, 47).
Functional Implications.
We have shown that the C259Y mutation affects TFIIH in vitro and in vivo DNA repair and transcription activities but does so in a manner that differs from deletion of the entire ARCH domain or from the R722W point mutation. The R722W mutation, which affects the interface with p44, abolishes XPD helicase activity but partially retains the capacity to bind DNA. In contrast, the C259Y mutation totally impairs DNA binding, suggesting direct involvement of the ARCH domain in DNA recognition and thus in the unwinding reaction. In favor of this hypothesis, mutagenesis and structural data obtained on archeal XPD suggest that the translocated DNA strand protrudes through the pore formed by HD1, the iron-sulfur cluster, and the ARCH domain (45). Because the helicase loads on the DNA bubble in vivo without a DNA end available, these domains must be mobile to allow strand passage, and the C259Y mutation might affect this mobility. In Sulfolobus tokodaii, mutation of the residue corresponding to C259 in human has no measurable consequences on in vitro helicase activity but affects the DNA-dependent ATPase activity (6) which is connected to the conformational changes required for loading helicases on the DNA bubble.
XPD helicase activity is not required for the transcription activity, and the limited capacity of TFIIH containing XPD-C259Y and XPD-R722W to stimulate transcription/transactivation probably results from the perturbed XPD/MAT1 and XPD/p44 interactions. We can speculate that these mutations lead to an inactive conformation and to the mispositioning of TFIIH within transcription/transactivation complexes. This mispositioning can explain the inability of the CDK7 subunit of CAK to phosphorylate the CTD of RNA Pol II (and thus the transcription defect observed in vitro) as well as the absence of TFIIH recruitment on the RARβ2 promoter in response to t-RA. The XPD-C259Y and -R722W alleles affect different interfaces within TFIIH, thus rationalizing the observation that the addition of CAK overcomes the inhibition of TFIIH/XPD-R722W only when a large excess of CAK is used. Interestingly, deletion of the entire ARCH domain leads to an enzyme that does not respond at all to addition of CAK. Taken together, these data provide insights into the XPD alleles C259Y and R722W and show that the ARCH domain of XPD not only plays a direct role in catalysis but also constitutes a platform for the recruitment of CAK.
Negative Regulation Mechanism.
XPB, XPD, and CDK7 exert their enzymatic activities in a complex network of interactions essential to ensure the correct functioning of TFIIH in transcription and repair. Fine-tuning mechanisms exerted by core-TFIIH subunits have been proposed to explain regulation of XPB ATPase and XPD helicase activities (Fig. 6B). Interactions among XPB, p52, and p8/TTD-A stimulate XPB ATPase activity and consequently favor the binding of TFIIH to damaged DNA (21). Interactions between p44 and XPD stimulate the helicase activity (35, 38), allowing unwinding of DNA around the lesion and subsequent double incision by the endonucleases XPF-ERCC1 and XPG. Our data provide evidence for a negative regulation mechanism that involves the CAK module of TFIIH and cross-talk among core-TFIIH, XPD, and CAK. Following previous observations with isolated subunits (13), we compared the 5′–3′ catalytic activities of wild-type core- and Holo-TFIIH complexes and show that the XPD helicase activity, which is dispensable for transcription initiation but is required for NER (19, 22), is repressed when CAK is associated with TFIIH. Underlining the crucial role of the ARCH platform domain in the anchoring of CAK, it was shown recently that NER is driven by dissociation of CAK from core-TFIIH, a mechanism that allows TFIIH to switch from its transcription to its repair function (23). In favor of such a model in which CAK negatively regulates NER, CDK7 also has been shown to inhibit repair by phosphorylation of one or more NER factors (48).
Non-TFIIH XPD Complexes.
The XPD helicase functions as a component of TFIIH but also can be found associated with non-TFIIH complexes that play roles in processes other than transcription and DNA repair. The MMXD complex is composed of XPD, MMS19, MIP18, Ciao1, and ANT2 and is involved in chromosome segregation and nuclear-shape formation (49). XPD also associates with CAK in the absence of the other TFIIH subunits, regulating the localization of CAK on its substrate and therefore its activity (50). How do mutations in XPD impact the functions of the different non-TFIIH complexes? Identification of the XPD ARCH domain as the minimal region required for the association of CAK with XPD suggests that mutations in ARCH that affect the association between CAK and XPD should impact the intracellular distribution of CAK and of the CAK/XPD complex. Because XPD/CAK plays a role in the coordination and progression of mitosis during late-division steps (51, 52), mutations that impair association between CAK and XPD are likely to affect cell-cycle regulation and could provide further insights to our understanding of XP, XP/CS, and TTD.
In conclusion, the present study underlines the essential roles of regulatory domains from motor proteins in the context of transcription regulation and maintenance of genome integrity. Using the human XPD helicase, a subunit of the transcription/DNA repair factor TFIIH, we have shown that its ARCH domain not only is involved in DNA binding and catalysis but also constitutes an essential component of the intricate protein–protein interaction network that enables the assembly of functional transcription/transactivation complexes. Introduction of a TTD mutation identified in a TTD patient impairs the helicase activity of XPD and the NER activity of TFIIH. We also have demonstrated that the ARCH domain is critical for the recruitment of the CAK kinase module of TFIIH and that alteration of the interface with the MAT1 CAK subunit both decreases the in vitro basal transcription activity of TFIIH and impedes an efficient recruitment of the transactivation complex on the promoter of the activated RARβ2 gene. Interestingly, comparison of the 5′–3′ catalytic activities of wild-type core- and Holo-TFIIH complexes showed that XPD is repressed when CAK is associated with TFIIH, suggesting that the ARCH domain also participates in the fine tuning of TFIIH enzymatic activities.
Methods
Construction of Baculoviruses, Protein Production, and Purification.
The cDNAs encoding XPD-wt (residues 1–762), XPD variants [K48R, C259Y, R722W, Ins199PP, Q452X, N-terminal (residues1–245), C-terminal (residues 443–762), ARCH (residues 245–443), and ∆ARCH], and XPG were cloned into pAC8F (53). Variant XPD-∆ARCH, designed on the basis of the crystal structures of archeal XPD homologs (4–7) and analysis of multiple sequence alignments, results from the deletion of residues 248–438 of human XPD that were replaced by the hexapeptide SGASAS. The mutations generating a frame shift after a 2-aa PP insertion at position 199 (Ins199PP) and the premature stop codon at position 452 (Q452X) have been described previously (36). All resulting transfer vectors were recombined with baculovirus DNA (BaculoGold DNA; Pharmingen) in Sf9 cells to generate viruses for the production of proteins in fusion with the Flag peptide (DYKDDDDK). In the case of XPG, the protein also is fused to a C-terminal c-Myc peptide (AEEQKLISEEDLLRKRREQLKHKLE). Baculoviruses overexpressing p44 and MAT1 were described previously (22). Two multigene-expressing viruses were used; the first, VCAK, allows expression of CDK7 with a C-terminal Strep-tag II together with cyclin H and MAT1 (54); the second, Vcore, allows coexpression of the core-TFIIH subunits p8, p34, p44, p52, p62, and XPB. Vcore was generated from a transfer vector resulting from Cre/LoxP fusion of three vectors (55): (i) pFL, in which XPB, p34, and p44 were cloned; (ii) pUCDM, in which p52 and p62 were cloned; and (iii) pSPL, in which p8 was cloned.
For the production of proteins and complexes, Sf21 insect cells grown in suspension (typically 250 mL) were infected/coinfected with the appropriate viruses/combinations of viruses, were collected 48 h postinfection, and were resuspended in 10 mL of buffer A [20 mM Tris⋅HCl (pH 8.0), 150 mM KCl, 1 mM DTT] supplied with Complete protease inhibitor mixture (Roche). Cells were disrupted using a Vibra-Cell (Sonics) sonicator (3-mm probe at 20% intensity for 30 s). After centrifugation at 14,000 × g for 30 min at 4 °C, the lysate was incubated for 2 h with 200 µL of Protein A Sepharose beads crosslinked to the M2 anti-Flag antibody (Sigma) for purification of Flag-XPD, to the 1H5 anti-p44 antibody (directed against residues 1–17 of human p44) for purification of core-IIH and rIIH, or with StrepTactin Sepharose (IBA) for purification of CAK. After extensive washing in buffer A and equilibration in buffer B [50 mM Tris⋅HCl (pH 8.0), 75 mM KCl, 20% (vol/vol) glycerol, 0.1% Nonidet P-40, 1 mM DTT], proteins were eluted by competition with 2 column volume (CV) of buffer B containing the appropriate synthetic peptide at 0.5 mg/mL for immunoprecipitations or 2.5 mM d-desthiobiotin (Sigma) for elution from StrepTactin Sepharose (IBA).
Monoclonal antibodies against the TFIIH subunits XPB (1B3), XPD (2F6), p52 (1D11), p44 (1H5), CDK7 (2F8), MAT1 (2D3), p8 (1D1), and XPG (1B5) were obtained from Institut de Génétique et de Biologie Moléculaire et Cellulaire facilities. The antibody directed against the Flag peptide was obtained from Sigma.
In Vitro Transcription and Phosphorylation Assays.
Run-off transcription assays were performed as previously described (22, 56) using reaction mixes containing the adenovirus major late promoter sequence (AdMLP) EcoRI–SalI DNA template (50 ng), TFIIB (15 ng), TFIIE (160 ng), TFIIF [500 ng of the phenyl fraction,from Pol II and the GTF purification scheme (55)], TBP (30 ng), endogenous RNA Pol II [10 µg of the 1 M DEAE fraction (55)], and the different rIIHs (see the legend of Fig. 3 for concentrations estimated according to quantitative Western blot analysis using the 1H5 anti-p44 antibody). RNA Pol II phosphorylation was carried out as a classical runoff transcription except that ATP was added to a final concentration of 5 mM, the amount of purified RNA Pol II polymerase was adjusted (typically reduced by a factor 3), and rIIH was replaced by a mixture of purified core-IIH, CAK, and XPD, which allows the preparation of a premix containing all components but XPD variants. Hypophosphorylated (IIA) and hyper phosphorylated (IIO) forms of RNA Pol II were resolved by SDS/PAGE with 12% (wt/vol) polyacrylamide and detected by Western blot using the monoclonal antibody (7C2) directed against the CTD.
To analyze the CDK7 kinase activity, purified complexes (typically 500 ng) were added to a reaction mixture containing 1 µg of bacterially expressed GST-fused human CTD in 20 mM Tris⋅HCl (pH 7.5), 250 mM NaCl, 10 mM MgCl2, 20% (vol/vol) glycerol, and 0.005 µM 0.5 µCi [γ-32P] in a total reaction volume of 20 µL for 30 min at 25 °C. Reactions were stopped by the addition of 5 µL concentrated Laemmli sample buffer, and the amount of phosphate incorporated was analyzed by autoradiography after separation on a 12% (wt/vol) SDS-polyacrylamide gel.
Dual-Incision, Helicase, and Gel-Shift Experiments.
NER dual-incision assays were performed using a plasmid with a single 1,3-intrastrand d(GpTpG) (50 ng) in a buffer containing 50 mM Hepes-KOH (pH 7.8), 5 mM MgCl2, 1 mM DTT, 0.3 mM EDTA, 10% (vol/vol) glycerol, 2.5 μg BSA, 50 mM KCl, and 2 mM ATP. Reaction mixtures (10 µL) containing XPG (5 ng), XPF/ERCC1 (15 ng), XPC/ hHR23B (10 ng), RPA (50 ng), XPA (25 ng), and increasing amounts of rIIH complexes (see the legend of Fig. 2A for concentrations estimated according to quantitative Western blot analysis using the 1H5 anti-p44 antibody) were incubated at 30 °C for 90 min and analyzed as previously described (35).
DNA unwinding reaction substrate was prepared by annealing two oligonucleotides: AL (TTTTTTTTTTTTTTTTTTTTTTTTTTTCGAGCACCGCTGCGGCTGCACCGGC) and BC (GCCGGTGCAGCCGCAGCGGTGCTCG). Before annealing, BC was labeled using [γ-32P] ATP and T4 polynucleotide kinase (New England BioLabs) and was purified using Micro Bio-Spin clean-up columns (Bio-Rad). Reaction was performed for 40 min at 25 °C by adding the immunopurified helicase (see the legend of Fig. 2 B and D for concentrations estimated and adjusted according to the Western blot analysis using the M2 anti-Flag antibody) to the DNA probe at 10 nM in a buffer containing 20 mM Tris⋅HCl (pH 8.0), 75 mM KCl, 4 mM MgCl2, 1 mM DTT, 4 mM ATP, and 0.1 mg/mL BSA with a total reaction volume of 20 µL. Reaction was stopped by adding 20 mM EDTA, 14% glycerol, 0.2% SDS, and 0.028% bromophenol to the reaction mixture. Analyses were performed by migration in 14% (wt/vol) polyacrylamide gel (acrylamide/bis-acrylamide ratio: 33/1) and autoradiography.
The helicase assay probe described above was used for gel-shift experiments. Increasing amounts (see the legend of Fig. 2C for concentrations estimated according to Western blot analysis using the M2 anti-Flag antibody) of immunopurified XPD variants or TFIIH complexes were incubated with the probe in the absence of ATP for 15 min at 25 °C in 20 mM Tris⋅HCl (pH 8.0), 75 mM KCl, 4 mM MgCl2, 1 mM DTT, and 0.1 mg/mL BSA. Analyses were performed by migration in 6% (wt/vol) polyacrylamide gel (acrylamide/bis-acrylamide ratio: 29/1) and autoradiography.
Protein-Interaction Assays.
Wild-type or mutated XPD complexes were coexpressed by coinfecting Sf9 insect cell monolayers (typically 20 × 106 cells) with the appropriate baculoviruses and were resuspended in 1 mL of buffer A. Cell extracts were prepared as described above. Pull downs were performed in buffer C [50 mM Tris⋅HCl (pH 8.0), 250 mM NaCl, 0.1% Nonidet P-40, 1 mM DTT] using 30 µL of Protein A Sepharose cross-linked with M2 anti-Flag (Sigma) or 9E10 anti–c-Myc antibodies or StrepTactin Sepharose (IBA). Pulled-down complexes were eluted by competition in nondenaturing conditions and were analyzed on SDS PAGE followed by Coomassie staining or Western blot experiments.
Cell Culture and Reagents.
Human fibroblast HD2 (42) cells derived from patients and HEK293 Flp-In cells (Invitrogen) were cultured in DMEM with 4.5 g/L glucose and 862 mg/L Glutamax-I (Life Technologies) supplemented with penicillin (100 UI/mL), streptomycin (100 µg/mL), and 10% (vol/vol) FCS at 37 °C in the presence of 5% (vol/vol) CO2. HD2 cells were transfected with a pSG5 plasmid containing XPD-wt, -C259Y or -R722W with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Stable cells lines were obtained after transfection of HEK293 Flp-In cells with a pcDNA5FRT plasmid containing a 3Flag-tagged version of XPD (wild type or mutants), pOG44, and JetPEI according to the manufacturer’s instructions. After selection with hygromycin (150 µg/mL), cells were diluted to establish subclonal populations. For the transactivation experiment, cells were incubated with red phenol-free medium containing 10% (vol/vol) charcoal-treated FCS and 40 mg/mL gentamycin 1 d before treatment with 10 µM t-RA (Biomol).
Quantitative RT-PCR.
Total RNA was isolated using a GenElute Mammalian Total RNA Miniprep kit (Sigma) and reverse transcribed with SuperScript II reverse transcriptase (Invitrogen). q PCR was done using the Lightcycler 480 SYBR Green I Master and the Lightcycler 480 (Roche). The primer sequences for RARβ2 and GADPH genes used in real-time PCR are available upon request. RARβ2 mRNA levels represent the ratio between values obtained from treated and untreated cells normalized against the housekeeping GADPH mRNA.
ChIP.
Cells were crosslinked at room temperature for 10 min with 1% (vol/vol) formaldehyde. Chromatin was prepared (57) and sonicated on ice 30 min using a Bioruptor (Diagenode). Samples were immunoprecipitated with antibodies at 4 °C for 6 h, and Protein G Sepharose beads (Upstate) were added, incubated overnight at 4 °C, and sequentially washed. For ChIP/ReChIP experiments, after the immunoprecipitation with Flag M2 Sepharose beads (Sigma) and washes, protein–DNA complexes were eluted with a competitive peptide (PD157). Then samples were immunoprecipitated with antibodies at 4 °C overnight, and Protein G Sepharose beads were added, incubated for 4 h at 4 °C, and sequentially washed. The complexes were eluted, and DNA fragments were purified using the QiAquick PCR purification kit (QIAGEN) and were analyzed by real-time PCR using a set of primers targeting the transcription initiation start (from −80 to −30) of the RARβ2 promoter region.
Acknowledgments
We thank Nathalie Troffer-Charlier and Isabelle Kolb-Cheynel (Institut de Génétique et de Biologie Moléculaire et Cellulaire Baculovirus Facility) for insect cell products; Jean-Marie Garnier for help with cloning; and Emmanuel Compe, Anne Catherine Dock-Bregeon, and Patrick Schultz for fruitful advice and constructive discussions. This work was funded by the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, the University of Strasbourg, the Alsace Region, and by Grants ANR-08-GENO-042-02 and ANR-12-BSV8-0015-01 from the Agence Nationale de la Recherche; Grant INCA-2008-041 from the Institut National du Cancer, the Association pour la Recherche sur le Cancer (ARC, subvention fixe and programme ARC); SPINE2 (Structural Proteomics In Europe) complexes Grant LSHG-CT-2006-031220 from the European Commission; and by a personal advanced grant from the European Commission (to J.-M.E.). W.A. was supported by the Ministere de l’Education Nationale de la Recherche et de Technologie and ARC, L.R. by La Ligue contre le Cancer, and A.M.-R. by the Fondation pour la Recheche Médicale.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.R.L. is a guest editor invited by the Editorial Board.
See Author Summary on page 2705 (volume 110, number 8).
References
- 1.de Boer J, Hoeijmakers JH. Nucleotide excision repair and human syndromes. Carcinogenesis. 2000;21(3):453–460. doi: 10.1093/carcin/21.3.453. [DOI] [PubMed] [Google Scholar]
- 2.Lehmann AR. The xeroderma pigmentosum group D (XPD) gene: One gene, two functions, three diseases. Genes Dev. 2001;15(1):15–23. doi: 10.1101/gad.859501. [DOI] [PubMed] [Google Scholar]
- 3.Price VH, Odom RB, Ward WH, Jones FT. Trichothiodystrophy: Sulfur-deficient brittle hair as a marker for a neuroectodermal symptom complex. Arch Dermatol. 1980;116(12):1375–1384. doi: 10.1001/archderm.116.12.1375. [DOI] [PubMed] [Google Scholar]
- 4.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: Family matters. Curr Opin Struct Biol. 2010;20(3):313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan L, et al. XPD helicase structures and activities: Insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133(5):789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, et al. Structure of the DNA repair helicase XPD. Cell. 2008;133(5):801–812. doi: 10.1016/j.cell.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolski SC, et al. Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol. 2008;6(6):e149. doi: 10.1371/journal.pbio.0060149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Compe E, Egly JM. TFIIH: When transcription met DNA repair. Nat Rev Mol Cell Biol. 2012;13(6):343–354. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- 9.Chang WH, Kornberg RD. Electron crystal structure of the transcription factor and DNA repair complex, core TFIIH. Cell. 2000;102(5):609–613. doi: 10.1016/s0092-8674(00)00083-0. [DOI] [PubMed] [Google Scholar]
- 10.Gibbons BJ, et al. Subunit architecture of general transcription factor TFIIH. Proc Natl Acad Sci USA. 2012;109(6):1949–1954. doi: 10.1073/pnas.1105266109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schultz P, et al. Molecular structure of human TFIIH. Cell. 2000;102(5):599–607. doi: 10.1016/s0092-8674(00)00082-9. [DOI] [PubMed] [Google Scholar]
- 12.Busso D, et al. Distinct regions of MAT1 regulate cdk7 kinase and TFIIH transcription activities. J Biol Chem. 2000;275(30):22815–22823. doi: 10.1074/jbc.M002578200. [DOI] [PubMed] [Google Scholar]
- 13.Sandrock B, Egly JM. A yeast four-hybrid system identifies Cdk-activating kinase as a regulator of the XPD helicase, a subunit of transcription factor IIH. J Biol Chem. 2001;276(38):35328–35333. doi: 10.1074/jbc.M105570200. [DOI] [PubMed] [Google Scholar]
- 14.Helenius K, et al. Requirement of TFIIH kinase subunit Mat1 for RNA Pol II C-terminal domain Ser5 phosphorylation, transcription and mRNA turnover. Nucleic Acids Res. 2011;39(12):5025–5035. doi: 10.1093/nar/gkr107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helenius K, Yang Y, Alasaari J, Mäkelä TP. Mat1 inhibits peroxisome proliferator-activated receptor gamma-mediated adipocyte differentiation. Mol Cell Biol. 2009;29(2):315–323. doi: 10.1128/MCB.00347-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egly JM, Coin F. A history of TFIIH: Two decades of molecular biology on a pivotal transcription/repair factor. DNA Repair (Amst) 2011;10(7):714–721. doi: 10.1016/j.dnarep.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 17.Fuss JO, Tainer JA. XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase. DNA Repair (Amst) 2011;10(7):697–713. doi: 10.1016/j.dnarep.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sung P, Higgins D, Prakash L, Prakash S. Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes its ATPase and DNA helicase activities but not the ability to bind ATP. EMBO J. 1988;7(10):3263–3269. doi: 10.1002/j.1460-2075.1988.tb03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guzder SN, et al. DNA repair gene RAD3 of S. cerevisiae is essential for transcription by RNA polymerase II. Nature. 1994;367(6458):91–94. doi: 10.1038/367091a0. [DOI] [PubMed] [Google Scholar]
- 20.Lin YC, Choi WS, Gralla JD. TFIIH XPB mutants suggest a unified bacterial-like mechanism for promoter opening but not escape. Nat Struct Mol Biol. 2005;12(7):603–607. doi: 10.1038/nsmb949. [DOI] [PubMed] [Google Scholar]
- 21.Coin F, Oksenych V, Egly JM. Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol Cell. 2007;26(2):245–256. doi: 10.1016/j.molcel.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Tirode F, Busso D, Coin F, Egly JM. Reconstitution of the transcription factor TFIIH: Assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol Cell. 1999;3(1):87–95. doi: 10.1016/s1097-2765(00)80177-x. [DOI] [PubMed] [Google Scholar]
- 23.Coin F, et al. Nucleotide excision repair driven by the dissociation of CAK from TFIIH. Mol Cell. 2008;31(1):9–20. doi: 10.1016/j.molcel.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 24.Lu H, Zawel L, Fisher L, Egly JM, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358(6388):641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 25.Akhtar MS, et al. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell. 2009;34(3):387–393. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glover-Cutter K, et al. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol. 2009;29(20):5455–5464. doi: 10.1128/MCB.00637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rochette-Egly C, Adam S, Rossignol M, Egly JM, Chambon P. Stimulation of RAR alpha activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by CDK7. Cell. 1997;90(1):97–107. doi: 10.1016/s0092-8674(00)80317-7. [DOI] [PubMed] [Google Scholar]
- 28.Keriel A, Stary A, Sarasin A, Rochette-Egly C, Egly JM. XPD mutations prevent TFIIH-dependent transactivation by nuclear receptors and phosphorylation of RARalpha. Cell. 2002;109(1):125–135. doi: 10.1016/s0092-8674(02)00692-x. [DOI] [PubMed] [Google Scholar]
- 29.Chen D, et al. Activation of estrogen receptor alpha by S118 phosphorylation involves a ligand-dependent interaction with TFIIH and participation of CDK7. Mol Cell. 2000;6(1):127–137. [PubMed] [Google Scholar]
- 30.Compe E, et al. Dysregulation of the peroxisome proliferator-activated receptor target genes by XPD mutations. Mol Cell Biol. 2005;25(14):6065–6076. doi: 10.1128/MCB.25.14.6065-6076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chymkowitch P, Le May N, Charneau P, Compe E, Egly JM. The phosphorylation of the androgen receptor by TFIIH directs the ubiquitin/proteasome process. EMBO J. 2011;30(3):468–479. doi: 10.1038/emboj.2010.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito S, et al. XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: Implications for Cockayne syndrome in XP-G/CS patients. Mol Cell. 2007;26(2):231–243. doi: 10.1016/j.molcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 33.Giglia-Mari G, et al. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat Genet. 2004;36(7):714–719. doi: 10.1038/ng1387. [DOI] [PubMed] [Google Scholar]
- 34.Kainov DE, Vitorino M, Cavarelli J, Poterszman A, Egly JM. Structural basis for group A trichothiodystrophy. Nat Struct Mol Biol. 2008;15(9):980–984. doi: 10.1038/nsmb.1478. [DOI] [PubMed] [Google Scholar]
- 35.Dubaele S, et al. Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol Cell. 2003;11(6):1635–1646. doi: 10.1016/s1097-2765(03)00182-5. [DOI] [PubMed] [Google Scholar]
- 36.Ueda T, Compe E, Catez P, Kraemer KH, Egly JM. Both XPD alleles contribute to the phenotype of compound heterozygote xeroderma pigmentosum patients. J Exp Med. 2009;206(13):3031–3046. doi: 10.1084/jem.20091892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Botta E, et al. Analysis of mutations in the XPD gene in Italian patients with trichothiodystrophy: Site of mutation correlates with repair deficiency, but gene dosage appears to determine clinical severity. Am J Hum Genet. 1998;63(4):1036–1048. doi: 10.1086/302063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gervais V, et al. TFIIH contains a PH domain involved in DNA nucleotide excision repair. Nat Struct Mol Biol. 2004;11(7):616–622. doi: 10.1038/nsmb782. [DOI] [PubMed] [Google Scholar]
- 39.Coin F, et al. Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat Genet. 1998;20(2):184–188. doi: 10.1038/2491. [DOI] [PubMed] [Google Scholar]
- 40.Dvir A, Tan S, Conaway JW, Conaway RC. Promoter escape by RNA polymerase II. Formation of an escape-competent transcriptional intermediate is a prerequisite for exit of polymerase from the promoter. J Biol Chem. 1997;272(45):28175–28178. doi: 10.1074/jbc.272.45.28175. [DOI] [PubMed] [Google Scholar]
- 41.Akoulitchev S, Mäkelä TP, Weinberg RA, Reinberg D. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature. 1995;377(6549):557–560. doi: 10.1038/377557a0. [DOI] [PubMed] [Google Scholar]
- 42.Takayama K, et al. Defects in the DNA repair and transcription gene ERCC2 in the cancer-prone disorder xeroderma pigmentosum group D. Cancer Res. 1995;55(23):5656–5663. [PubMed] [Google Scholar]
- 43.Le May N, et al. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol Cell. 2010;38(1):54–66. doi: 10.1016/j.molcel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 44.Mui TP, Fuss JO, Ishida JP, Tainer JA, Barton JK. ATP-stimulated, DNA-mediated redox signaling by XPD, a DNA repair and transcription helicase. J Am Chem Soc. 2011;133(41):16378–16381. doi: 10.1021/ja207222t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuper J, Wolski SC, Michels G, Kisker C. Functional and structural studies of the nucleotide excision repair helicase XPD suggest a polarity for DNA translocation. EMBO J. 2012;31(2):494–502. doi: 10.1038/emboj.2011.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Botta E, et al. Reduced level of the repair/transcription factor TFIIH in trichothiodystrophy. Hum Mol Genet. 2002;11(23):2919–2928. doi: 10.1093/hmg/11.23.2919. [DOI] [PubMed] [Google Scholar]
- 47.Stefanini M, Botta E, Lanzafame M, Orioli D. Trichothiodystrophy: From basic mechanisms to clinical implications. DNA Repair (Amst) 2010;9(1):2–10. doi: 10.1016/j.dnarep.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 48.Araújo SJ, et al. Nucleotide excision repair of DNA with recombinant human proteins: Definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev. 2000;14(3):349–359. [PMC free article] [PubMed] [Google Scholar]
- 49.Ito S, et al. MMXD, a TFIIH-independent XPD-MMS19 protein complex involved in chromosome segregation. Mol Cell. 2010;39(4):632–640. doi: 10.1016/j.molcel.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 50.Aguilar-Fuentes J, Valadez-Graham V, Reynaud E, Zurita M. TFIIH trafficking and its nuclear assembly during early Drosophila embryo development. J Cell Sci. 2006;119(Pt 18):3866–3875. doi: 10.1242/jcs.03150. [DOI] [PubMed] [Google Scholar]
- 51.Chen J, Larochelle S, Li X, Suter B. Xpd/Ercc2 regulates CAK activity and mitotic progression. Nature. 2003;424(6945):228–232. doi: 10.1038/nature01746. [DOI] [PubMed] [Google Scholar]
- 52.Li X, Urwyler O, Suter B. Drosophila Xpd regulates Cdk7 localization, mitotic kinase activity, spindle dynamics, and chromosome segregation. PLoS Genet. 2010;6(3):e1000876. doi: 10.1371/journal.pgen.1000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abdulrahman W, et al. A set of baculovirus transfer vectors for screening of affinity tags and parallel expression strategies. Anal Biochem. 2009;385(2):383–385. doi: 10.1016/j.ab.2008.10.044. [DOI] [PubMed] [Google Scholar]
- 54.Fouillen L, et al. Analysis of recombinant phosphoprotein complexes with complementary mass spectrometry approaches. Anal Biochem. 2010;407(1):34–43. doi: 10.1016/j.ab.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 55.Fitzgerald DJ, et al. Protein complex expression by using multigene baculoviral vectors. Nat Methods. 2006;3(12):1021–1032. doi: 10.1038/nmeth983. [DOI] [PubMed] [Google Scholar]
- 56.Gerard M, et al. Purification and interaction properties of the human RNA polymerase B(II) general transcription factor BTF2. J Biol Chem. 1991;266(31):20940–20945. [PubMed] [Google Scholar]
- 57.Drané P, et al. Selective regulation of vitamin D receptor-responsive genes by TFIIH. Mol Cell. 2004;16:187–197. doi: 10.1016/j.molcel.2004.10.007. [DOI] [PubMed] [Google Scholar]