Abstract
In mammals, the prototypical endoplasmic reticulum (ER) stress sensor inositol-requiring enzyme 1 (IRE1) has diverged into two paralogs. IRE1α is broadly expressed and mediates the unconventional splicing of X-box binding protein 1 (XBP1) mRNA during ER stress. By contrast, IRE1β is expressed selectively in the digestive tract, and its function remains unclear. Here, we report that IRE1β plays a distinctive role in mucin-secreting goblet cells. In IRE1β−/− mice, aberrant mucin 2 (MUC2) accumulated in the ER of goblet cells, accompanied by ER distension and elevated ER stress signaling such as increased XBP1 mRNA splicing. In contrast, conditional IRE1α−/− mice showed no such ER distension but a marked decrease in spliced XBP1 mRNA. mRNA stability assay revealed that MUC2 mRNA was greatly stabilized in IRE1β−/− mice. These findings suggest that in goblet cells, IRE1β, but not IRE1α, promotes efficient protein folding and secretion in the ER by optimizing the level of mRNA encoding their major secretory product, MUC2.
Keywords: inflammatory bowel disease, unfolded protein response
The endoplasmic reticulum (ER) is the site of synthesis and maturation of proteins destined for secretion. In the ER, chaperones and folding enzymes facilitate productive folding and assembly of nascent polypeptides (1). However, not all nascent peptides can fold properly in this process, particularly in cells that display high levels of secretion. When cells synthesize amounts of secretory or membrane proteins that exceed the folding and/or processing capacity of the ER, unfolded proteins accumulate in the ER; this condition is called ER stress. Under ER stress, signaling pathways, collectively termed the unfolded protein response (UPR), are activated to restore the ER to its normal state (2–5). This recovery is achieved by translational attenuation, refolding of unfolded proteins and degradation of irreversibly unfolded proteins via the ER-associated degradation (ERAD) pathway (6). ER chaperones, such as immunoglobulin heavy chain binding protein (BiP) (2, 7), and ERAD components are induced transcriptionally by the UPR (8). In the initial step of the UPR, ER stress is sensed and a signal is transmitted across the ER membrane (5, 9). Several ER transmembrane proteins act as ER stress sensors. The oldest among these, inositol-requiring enzyme 1 (IRE1), is conserved from yeast to mammals and possesses a luminal sensor domain (10–14) and a cytosolic effector domain (15, 16). In yeast Ire1p’s effector is a highly sequence-specific RNase that participates in the regulated first step of an unconventional splicing event that activates the Homologous to Atf/Creb 1 (HAC1) mRNA to encode a potent transactivator of UPR target genes (8, 17–19).
Mammals have two IRE1 paralogs, IRE1α (20) and IRE1β (21), which seem to possess nonoverlapping physiological roles, although these paralogs show a high degree of sequence similarity to each other (22). IRE1α is a ubiquitously expressed gene whose deletion results in early embryonic lethality (23). We demonstrated that this lethality was caused by failure in placenta development by generating nonlethal conditional knockout mice that express IRE1α only in the placenta (24). In contrast, the expression of IRE1β is restricted to the gastrointestinal tract, and its knockout mice are phenotypically normal, apart from hypersensitivity to experimental colitis (25). It is unclear whether these dramatic differences merely reflect different patterns of expression or whether the two proteins have different molecular activities.
X-box binding protein 1 (XBP1) is an animal homolog of yeast HAC1, and IRE1α is required for the unconventional splicing of its mRNA in most animal tissues (26, 27). The overlapping phenotypes of IRE1α knockout and XBP1 knockout suggest that XBP1 mRNA splicing is IRE1α’s essential function. However, animal IRE1 seems to possess additional activities, such as a contribution to the relatively promiscuous degradation of membrane-associated mRNAs observed in ER stressed cells (28, 29). The gut presents an interesting context for these two functions of IRE1. The importance of XBP1 mRNA splicing is supported by the association of rare alleles of XBP1 with inflammatory bowel disease and by the dramatic defect in Paneth cells observed in mice with intestine-specific deletion of XBP1 (30). Together with the observation that IRE1β is competent to splice XBP1 mRNA both in vitro and in vivo (27), these observations suggest that IRE1β evolved to enhance the capacity to splice XBP1 mRNA in response to stress in the ER of intestinal cells. However, a role for IRE1β in other RNA processing events is suggested by the observations that, in HeLa cells, ectopic expression of IRE1β led to cleavage of 28S rRNA and apoptosis (22), and in IRE1β knockout mice the mRNA encoding microsomal transfer protein is stabilized, promoting chylomicron secretion from the intestine (31).
Here we report on a detailed exploration of IRE1β’s expression and its molecular mechanism of action. Our study has led to the discovery of a hitherto unanticipated role for IRE1β in ER homeostasis of goblet cells that is mediated by the posttranscriptional metabolism of mucin mRNA.
Results
Localization of IRE1β.
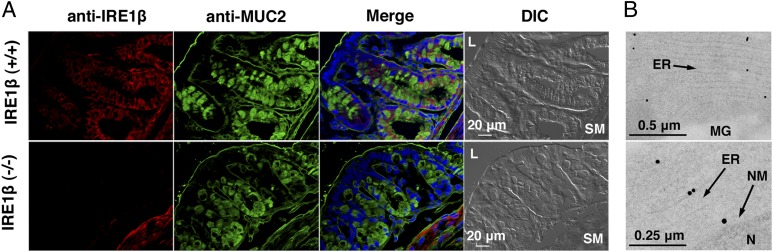
RNAs were extracted from mouse tissues and analyzed by Northern blot (Fig. S1A). Signals for IRE1β mRNA were detected in tissues comprising the digestive tract: stomach, duodenum, small intestine, cecum, and most strongly, colon. The signals were weaker in the duodenum and small intestine, and none were detected in tissues other than the digestive tract. In contrast, IRE1α mRNA was detected in all tissues tested. Analyses in human tissues gave comparable results (Fig. S1C). For detailed analyses, we raised an antibody against the putative cytosolic domain of mouse IRE1β. As expected, the results of a Western blot with this antibody were consistent with those obtained in the Northern blot (Fig. S1B) and confirmed a preceding study (25). To determine which cells express IRE1β in the digestive tract, immunofluorescence microscopy was performed on cryosections of mouse colon, the tissue that expressed IRE1β most abundantly (Fig. 1A). Specific staining for IRE1β was seen in goblet cells, whereas other cells, such as absorptive cells, were not stained. Nonspecific signals were seen in smooth muscle cells of both IRE1β+/+ and IRE1β−/− mice. Specific staining of goblet cells was also observed in small intestine (Fig. S2).
Fig. 1.
IRE1β is expressed and localized in the ER of goblet cells. (A) After deglycosylation, cryosections of mouse colons were stained with the antibody raised against the cytosolic region of IRE1β and Cy3-conjugated anti-guinea pig IgG as the secondary antibody. MUC2 was stained with anti-MUC2 (R-12) and FITC-conjugated anti-goat IgG. MUC2-positive cells (goblet cells) were also stained with anti-IRE1β antibody. DNA was stained with DAPI. Staining in smooth muscle was judged to be nonspecific, because IRE1β−/− and IRE1β+/+ smooth muscle both exhibited the same staining pattern. Differential interference contrast (DIC) images are shown, far right. L, intestinal lumen; SM, smooth muscle. (B) Ultrathin sections of mouse colon were treated with anti-IRE1β and gold particle-labeled anti-guinea pig IgG. Gold particles are seen only on the outside of the ER because this antibody was raised against the cytosolic region of IRE1β. Less (Upper) and more magnified images (Lower) are shown. MG, mucin granule; N, nucleus; NM, nuclear membrane.
Goblet cells are specialized to secrete mucins, which contribute to the protective mucus gel barrier between the epithelium and the harsh environment of the lumen (32). Mucins are a family of highly O-glycosylated proteins, and mucin 2 (MUC2) is the most prominent protein secreted from goblet cells in the colon (33). The specific expression of IRE1β in goblet cells suggested that IRE1β might have some relation with mucin production. In this context it was important to determine the intracellular location of endogenous IRE1β in goblet cells, so we performed immunoelectron microscopy (Fig. 1B). Low-magnification images showed that the signals for IRE1β colocated with ER membranes (Fig. 1B, Upper), and higher magnification localized the IRE1β signals to the cytosolic surface of the ER and of the outer nuclear membrane (Fig. 1B, Lower). This seems reasonable because IRE1β is regarded as a type I transmembrane protein (22), and the antibody we used was raised against the cytosolic (C-terminal) region of IRE1β. No signals were detectable at the surface of the inner nuclear membrane. These data indicate that IRE1β is primarily localized in the ER membrane in goblet cells.
Enhanced ER Stress in Goblet Cells of IRE1β Knockout Mice.
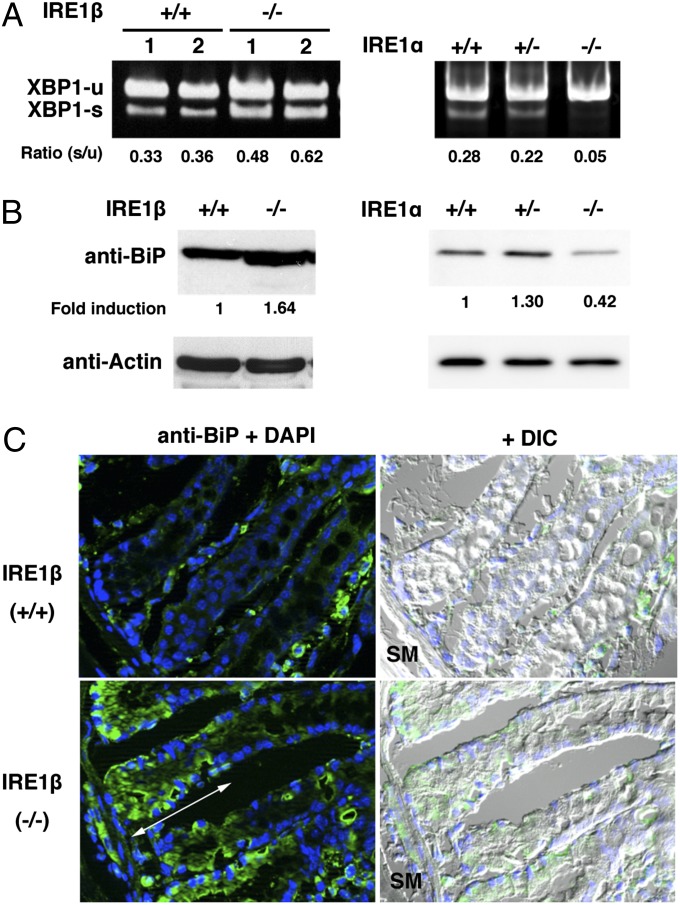
To confirm a previous report that ER stress occurs in the colons of IRE1β−/− mice (25), XBP1 mRNA was analyzed by RT-PCR (Fig. 2A). Although some amount of XBP1 mRNA is spliced in wild-type (IRE1β+/+, IRE1α+/+) colon, the ratio of spliced to unspliced XBP1 mRNA was higher in IRE1β−/− than in wild-type mice. This suggests that IRE1α was activated more strongly in IRE1β−/− colons. Another ER stress marker, BiP, was also examined by Western blot (Fig. 2B), and its level was found to be 1.6-fold higher in IRE1β−/− colon than in wild-type colon. The restricted expression of IRE1β predicts that enhanced ER stress would be most conspicuous in the goblet cells, and we used immunohistofluorescence microscopy to test this prediction. Goblet cells differentiate from stem cells in the bottom of the crypt (34), and differentiated cells mature during a 3- to 5-d migration (35) toward the lumen. In cryosections treated with anti-BiP antibody (Fig. 2C), IRE1β−/− goblet cells near the bottom of the crypt (indicated by the double-headed arrow) stained strongly, whereas staining in IRE1β+/+ goblet cells was faint. These data suggest that the lack of IRE1β promotes ER stress specifically in immature goblet cells.
Fig. 2.
ER stress in goblet cells of IRE1β−/− mice. (A) The ratio of spliced to unspliced XBP1 mRNA is elevated in IRE1β−/− colon but decreased in IRE1α−/− colon. RNAs were extracted from two individual colons of wild-type (IRE1β+/+, IRE1α+/+), IRE1β−/−, IRE1α+/−, or IRE1α−/− mice. After RT-PCR, the intensity of each band was measured, and the ratio (s/u) of spliced to unspliced XBP1 mRNA was calculated. (B) Increase of BiP in IRE1β−/− colons. Western blotting was performed with anti-BiP antibody for colon lysates from wild-type (IRE1β+/+, IRE1α+/+), IRE1β−/−, IRE1α+/−, or IRE1α−/− mice. Signal intensity was normalized using β-actin, and fold induction relative to the level for the IRE1β+/+ sample was calculated. (C) Increase of BiP in IRE1β−/− goblet cells. Cryosections of IRE1β+/+ or IRE1β−/− colons were stained with anti-BiP (green) antibody and DAPI (blue). Double-headed arrow indicates the region where BiP-induced goblet cells were prominent. The “+ DIC” image depicts superimposed immunofluorescence and DIC images.
In case of IRE1α conditional knockout mouse (IRE1α−/−, IRE1β+/+) that expresses IRE1α only in placenta (24), XBP1 mRNA was hardly spliced in its colon (Fig. 2A). This suggests that IRE1β is not responsible for the splicing of XBP1 mRNA at least in weak ER stress condition. It should be noted that the expression of BiP was also decreased in IRE1α conditional knockout colon (Fig. 2B), implying that some of the basal expression of BiP may be maintained by XBP1 in colon.
IRE1β Knockout Immature Goblet Cells Display a Distended ER and Aberrant Mucin Accumulation.
Overlapping series of electron micrographs were taken to visualize whole (Fig. 3 A and B) or a part (Fig. 3G) of crypts. Although the morphology of mature goblet cells near the lumen (Fig. 3 A and B, Right) appeared to be similar in IRE1β+/+ and IRE1β−/− mice, granule-like structures (indicated by arrows) in immature cells located near the bottom of the crypt differed visibly in the two genotypes (Fig. 3 C and E). After observing higher-magnification micrographs (Fig. 3 D and F and Fig. S3A), we concluded that these structures in IRE1β−/− goblet cells correspond to a distended ER rather than the large mucin granules seen in IRE1β+/+ goblet cells for the following reasons. First, the observed distended ER was continuous with the normal-shaped ER (arrow in Fig. S3A). Second, on their surfaces, ribosomes were attached. Finally, signals for BiP were also observed in a normal-shaped ER in wild-type goblet cells and in the distended ER in IRE1β −/− goblet cells by immunoelectron microscopy (Fig. S3 B and C).
Fig. 3.
The ER is distended only in goblet cells of IRE1α+/+β−/− mice (this is the same mouse line shown as IRE1β−/−, but here we describe IRE1β−/− as IRE1α+/+β−/− to be clearly understandable). Colons from IRE1α+/+β+/+ (A, C, and D), IRE1α+/+β−/− (B, E, and F), and IRE1α−/−β+/+ (G and H) mice were fixed, stained, and observed by electron microscope as described in SI Materials and Methods. High-magnification images revealed that the ER of IRE1α+/+β−/− in early-stage goblet cells (E and F) was distended, whereas the ER of IRE1α+/+β+/+ (C and D) and IRE1α−/−β+/+ (H) goblet cells in the same stage showed normal structure. A series of low-magnification images depicts an entire (A and B) or a part (G) of crypt, and the double-headed arrow indicates the region where ER-distended goblet cells existed. The black and white arrows in A and C and B and E indicate mucin granule and ER, respectively.
The region of the crypt in which these ER-distended goblet cells occurred (double-headed arrow in Fig. 3B) was comparable to the region displaying ER-stressed goblet cells (Fig. 2C) in IRE1β−/− colon. In contrast, goblet cells in IRE1α conditional knockout mice did not display ER distension (Fig. 3H). These findings suggest that IRE1β is involved in ER homeostasis in goblet cells.
What causes the ER to distend so dramatically in immature goblet cells of IRE1β−/− mice? Because goblet cells are specialized to produce large amounts of the O-glycosylated protein mucin, it seemed plausible that the ER distension was attributable to excessive mucin accumulation. To examine this possibility, we first tried to detect mucin with soybean agglutinin (SBA), which binds to serine- or threonine-linked N-acetylgalactosamine (Fig. S4). Compared with goblet cells in IRE1β+/− colon, those in IRE1β−/− colon were more strongly stained. Moreover, the SBA-stained regions in IRE1β−/− goblet cells were costained with an anti-calreticulin antibody, suggesting that O-glycosylated proteins accumulated in the ER of IRE1β−/− goblet cells rather than the mucin granules.
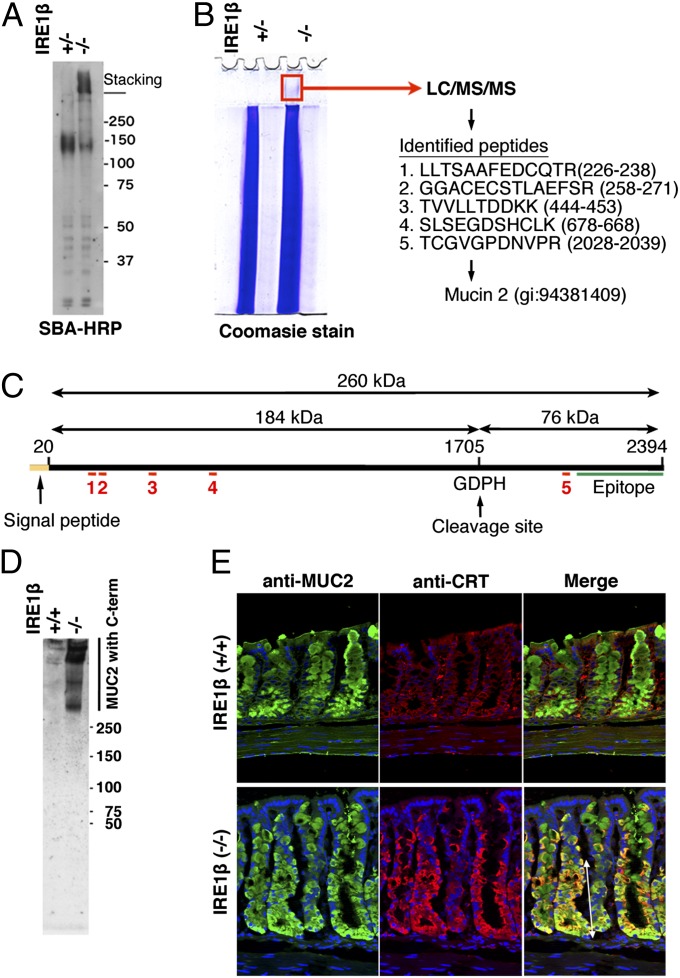
O-glycosylated proteins were also investigated by Western blot using SBA (Fig. 4A), which yielded diffuse bands at around 150 kDa and in the stacking gel. The latter was only visible in the IRE1β−/− lysate and thus correlates with the abnormal ER staining observed by fluorescence microscopy in the IRE1β−/− tissue (Fig. S4). To characterize the proteins in this region of the gel, it was excised from the stacking gel and analyzed by mass spectrometry (Fig. 4B). We identified five tryptic fragments that matched with the sequence of mouse MUC2; one of them mapped to the C-terminal region of this protein, and the other four to the N-terminal region (Fig. 4C). Although we cut out and analyzed the corresponding region of the IRE1β+/− lane in the same way, no MUC2-related peptides were identified.
Fig. 4.
Aberrant mucin accumulates in the ER in goblet cells of IRE1β−/− mice. (A) SBA-binding high-molecular-weight aggregates accumulate in IRE1β−/− colon. Lysates of mucosal epithelia were electroblotted after 8% SDS/PAGE and treated with HRP-conjugated SBA. (B) The high-molecular-weight aggregates contain MUC2. Lysates as in A were electrophoresed, and the indicated region of the stacking gel containing the IRE1β−/− lysate was cut out for mass spectrometry (LC/MS/MS) analysis. Five peptides identical to sequences in mouse MUC2 were detected. (C) Locations of the five identified peptides in the primary structure of mouse MUC2. Peptides identified by mass spectrometry are indicated as red lines, with numbers corresponding to those in B. Note that the fifth peptide maps to within the 76-kDa C-terminal region that is cleaved in the Golgi apparatus. The epitope of anti-MUC2 antibody H-300 (green line) is also shown. (D) MUC2, with its uncleaved C-terminal region, accumulates in IRE1β−/− colon. Western blotting with anti-MUC2 antibody H-300 was performed after nonreducing 2–15% gradient SDS/PAGE. High-molecular-weight protein stained intensely in IRE1β−/− colon lysate. (E) Immunofluorescence staining of MUC2 in mouse colons. Cryosections were stained with chicken anti-MUC2, goat anti-CRT antibodies, and with DAPI. IRE1β−/− goblet cells show colocalization of MUC2 and calreticulin (CRT) in the region indicated by the double-headed arrow.
Because the C-terminal 76-kDa peptide (Fig. 4C) is known to be cleaved off from MUC2 in the trans-Golgi region (36), we anticipated that the band in the stacking gel might represent the full-length MUC2 precursor that had aggregated and accumulated in the ER, and that the second major band at around 150 kDa (Fig. 4A) might represent the cleaved N-terminal portion of mature MUC2 (Fig. 4C). To confirm this, immunological analyses were performed with an anti-MUC2 antibody (H-300) that was raised against the C-terminal precursor fragment of MUC2 (Fig. 4C). After 2–15% polyacrylamide gradient gel electrophoresis and Western blotting, high-molecular-weight bands were detected only in the IRE1β−/− colon lysate (Fig. 4D). This indicates that the C-terminal peptide is rapidly removed from MUC2 after its translation in IRE1β+/+ but not IRE1β−/− goblet cells. This notion is further supported by the presence of less material with the mobility of mature mucin in the IRE1β−/− sample (Fig. 4A).
MUC2 was also localized within IRE1β−/− goblet cells by immunofluorescence microscopy (Fig. 4E). Colon tissue sections stained with anti-MUC2 and anti-calreticulin antibodies displayed coincident staining for the two antibodies in goblet cells undergoing maturation, in the lower portion of the crypt (indicated as a double-headed arrow) in IRE1β−/− sections. The region where goblet cells with MUC2-containing ER were detected corresponds to the region where the ER-distended and ER-stressed goblet cells were found (double-headed arrows in Figs. 2C and 3B). In contrast, MUC2 did not always colocalize with calreticulin in goblet cells within IRE1β+/+ crypts. These data support the idea that aberrant MUC2 accumulates in the ER of IRE1β knockout immature goblet cells.
IRE1β Promotes the Turnover of MUC2 mRNA.
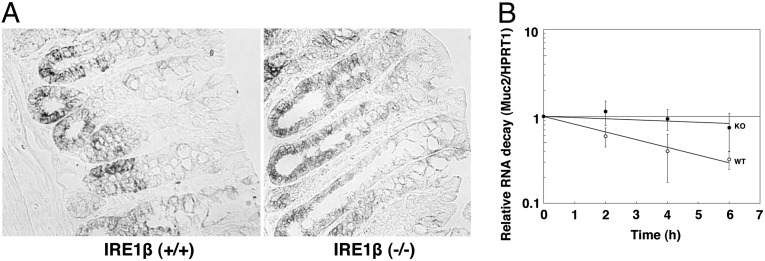
Why does MUC2 accumulate in the ER of IRE1β−/− goblet cells? One explanation might be that MUC2 protein is overproduced. Because IRE1β is known to have RNase activity, we examined whether IRE1β affects MUC2 mRNA levels. First, we analyzed MUC2 mRNA by in situ hybridization (Fig. 5A). In wild-type colon, MUC2 mRNA was observed only in goblet cells at early stages of maturation (i.e., near the bottom of the crypt). In IRE1β−/− colon, however, it was also detected in middle- to late-stage goblet cells. These results suggest that MUC2 mRNA expression is strictly regulated in wild-type cells and that this regulation fails in IRE1β−/− goblet cells.
Fig. 5.
Distribution and stability of MUC2 mRNA in colon. (A) MUC2 mRNA distribution in IRE1β+/+ and IRE1β−/− colons. Colon sections (10 μm in thickness) were fixed and then hybridized with a digoxigenin-labeled MUC2 cRNA probe (SI Materials and Methods). MUC2 mRNA appears as dark staining. (B) Stability of MUC2 mRNA in IRE1β+/+ and IRE1β−/− colons. Cells collected from IRE1β+/+ (wild-type) or IRE1β−/− (knockout) mouse colons were incubated in the medium containing α-amanitin for the indicated times. RNAs were then extracted from the cells and analyzed by quantitative RT-PCR. Relative RNA decay is expressed as MUC2 mRNA/HPRT1 mRNA ratio, and the values at time 0 were set to 1. Data presented are the averages of six independent experiments, with SD indicated by error bars.
To substantiate this idea, we examined mRNA stability. Cells collected from IRE1β+/+ or IRE1β−/− colons were cultured with α-amanitin, an inhibitor of RNA polymerase II. RNA was then extracted and analyzed by quantitative RT-PCR (Fig. 5B). We used hypoxanthine phosphoribosyltransferase 1 (HPRT1) mRNA as a control, because IRE1β has little effect on mRNAs encoding cytosolic proteins (37). MUC2 mRNA from IRE1β+/+ (wild-type) cells decayed to the extent that only approximately 30% of the initial signal remained after 6 h of incubation with α-amanitin, whereas more than 80% of MUC2 mRNA from IRE1β−/− (knockout) cells remained after 6 h (P < 0.05). Thus, MUC2 mRNA was greatly stabilized in the IRE1β−/− colon.
Discussion
IRE1β knockout mice are reportedly sensitive to experimental colitis induced by dextran sodium sulfate (25). In this report, we showed that IRE1β was specifically expressed as a transmembrane protein in the ER of goblet cells. Because goblet cells are specialized for the secretion of mucins that protect the digestive tract, our study provides a plausible mechanism for this hypersensitivity. The results obtained with electron microscopy, immunohistofluorescence, Western blotting, mass spectrometry, and in situ hybridization all point to aberrant mucin accumulated in a distended ER of immature goblet cells in IRE1β knockout mice. Although the morphology of the ER seems to recover in the goblet cells that survive and mature, it is unclear whether their ability to produce correctly folded and fully glycosylated mucin also recovers. The sensitivity of IRE1β knockout mice to experimental colitis might thus reflect a qualitative defect in mucins secreted by IRE1β knockout goblet cells.
Several studies have reported ER distension in genetically modified animals. Hepatocytes in transgenic mice overexpressing mutant α1-antitrypsin showed a distended ER that accumulated the mutant protein (38). Distended ER was also observed in pancreatic cells in the double-stranded RNA-dependent protein kinase (PKR)-like ER kinase (PERK) knockout mice (39). Because PERK phosphorylates eukaryotic translation initiation factor 2α (eIF2α) to attenuate translational initiation in response to ER stress, cells in PERK knockout mice are believed to overexpress proteins. Similarly, mice expressing a mutant eIF2α that is resistant to phosphorylation by PERK displayed ER distension in pancreatic cells (40). In addition, chondrocytes in BBF2H7 (an ER-resident basic leucine zipper transcription factor)-deficient mice showed a distended ER and an accumulation of aggregated proteins caused by impaired protein transport from ER to Golgi (41). These reports indicate that overexpression of secretory proteins can induce ER distension, and it is therefore likely that mucin overexpression leads to the accumulation of aberrant mucin and ER distension in IRE1β knockout goblet cells.
We found that the expression of BiP was enhanced in the immature goblet cells of IRE1β knockout mice, suggesting that the lack of IRE1β provoked ER stress in immature goblet cells (Fig. 2C). How can it happen? Because the signal for the MUC2 mRNA was stronger in immature goblet cells than in mature goblet cells in both genotypes (Fig. 5A), the level of secretory protein production is presumably higher in immature goblet cells than in mature goblet cells. This will lead to a situation in which the ER of immature goblet cells tends to be more stressed than that of mature goblet cells. However, in the wild-type immature goblet cells, IRE1β will promote the degradation of an excess amount of MUC2 mRNA, preventing the onset of ER stress. As a result, the level of secretory protein production will be high enough to provoke ER stress in immature goblet cells of IRE1β knockout mice but not in other cells.
It is reported that IRE1α cleaves a wide variety of mRNAs besides XBP1 pre-mRNA by the mechanism called regulated Ire1-dependent decay (RIDD) under a certain stress (28, 29, 42). Authors of those reports described that the ER stress agent tunicamycin induced RIDD, but mutant IRE1α that became active upon binding to an ATP analog, 1NM-PP1, cleaved only XBP1 pre-mRNA without activating RIDD. Thus, they suggested that pathogenic conditions such as virus infection induce RIDD to protect cells (29), or severe stress activates RIDD to induce apoptosis (42). We found that splicing of XBP1 mRNA was enhanced in IRE1β knockout colon, whereas the stability of mucin mRNA was increased. This finding indicates that IRE1α cleaves XBP1 pre-mRNA but very little mucin mRNA. Hence, it is unlikely that IRE1α expresses RIDD activity in the colon. Previously we examined the difference in substrate specificity between IRE1α and IRE1β and found that the RNase activity of IRE1α against XBP1 pre-mRNA is markedly higher than that of IRE1β (43). In the present study we studied with IRE1α conditional knockout mice (24) and found the marked decrease in spliced XBP1 mRNA in their colons. Because the population of goblet cells was reported to be more than 50% in rat colon (44) and is presumably similar in mouse colon, the decrease of spliced XBP1 mRNA in the IRE1α knockout mouse conceivably occurred in both absorptive cells and IRE1β-containing goblet cells. In addition, we could observe no such ER distension in IRE1α knockout goblet cells as in IRE1β knockout cells (Fig. 3), suggesting that IRE1α is not able to compensate IRE1β deficiency. These results suggest that ER stress in colon is not sufficient for IRE1α to activate RIDD. On the other hand, rRNA in mouse colon appeared to be intact (Fig. S1A), although we reported the degradation of 28S rRNA in HeLa cells after overexpression of IRE1β (22). It may be ascribed to the difference in the origin of cells or the expression levels between cells.
In addition to the mouse colon, we stained the small intestine with anti-IRE1β antibody and found the expression of IRE1β in its goblet cells (Fig. S2). Although it is reported that IRE1β plays a role in regulating microsomal triglyceride transfer protein (MTP) and in chylomicron production in enterocytes (31), we could not detect the signal for IRE1β in the enterocytes. This observation suggests that MTP mRNA is unlikely to be the primary target of IRE1β. IRE1β may mainly affect the production of mucins in the small intestine as well and affect the chyromicron production in other ways.
Our results from in situ hybridization and RNA stability assays showed a greatly increased stability of MUC2 mRNA in IRE1β knockout mice. This indicates that IRE1β normally degrades MUC2 mRNA to control levels of translatable, cytosolic mRNA despite the fact that the function of goblet cells is to produce mucin. The seemingly paradoxical role of IRE1β in maintaining mucin production may be explained as follows (Fig. S5). The folding capacity of the ER is high enough for most types of cells but may be exceeded in high-secretion cells, including goblet cells. However, goblet cells should not overexpress mucin, because doing so would lead to the accumulation of aberrant mucin in the ER, which might retard protein transport from the ER to the Golgi apparatus. The observation that IRE1β knockout goblet cells seem to contain less C-terminal–cleaved mucin than wild-type cells (Fig. 4A) supports this explanation. Maximum mucin production may therefore be achieved by regulating the rate of mucin synthesis so that it remains within the capacity of the ER. The fine-tuning of protein synthesis may thus depend on a balance between up- and down-regulation of transcripts. In this context, the present study suggests that IRE1β plays an important role in the down-regulation of MUC2 mRNA to optimize mucin protein production. This also could be a good example showing the importance of negative feedback on fine-tuning of biological activities.
We presented a possible role of IRE1β here. However, it is still unknown whether IRE1β degrades MUC2 mRNA directly. Because IRE1β has RNase activity, it may cleave mRNA. However, we cannot exclude a possibility that IRE1β decreases MUC2 mRNA by unconventional splicing of unidentified specific target mRNA like XBP1 pre-mRNA for IRE1α. Studying other tissues expressing IRE1β in addition to colon might give some hints to answer this question.
Materials and Methods
Animal experiments were carried out in accordance with the policies of the Committee on Animal Research at Nara Institute of Science and Technology. Brief methods are described in the figure legends. Detailed methods can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. L. Hendershot for anti-BiP antibody, R. Nagai for electron microscopy, M. Kuwano for mass spectrometry, Dr. T. Komatsu for help in immunohistofluorescence, H. Masuda and J. Hashimoto for technical assistance, and Prof. I. Smith (Nara Institute of Science and Technology) for critical reading of the manuscript. This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grants 24228002, 24248019 (to K.K.), and 14580699 (to A.T.), Ministry of Education, Culture, Sports, Science and Technology in Japan (MEXT) KAKENHI Grant 19058010 (to K.K.), The Uehara Memorial Foundation (K.K.), Takeda Science Foundation (K.K.), Mitsubishi Foundation (K.K.), and National Institutes of Health Grant DK047119 (to D.R., who is a Wellcome Trust Principal Research Fellow). D.N. was a JSPS fellow and a research assistant supported by the Global Center of Excellence (COE) program from MEXT.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212484110/-/DCSupplemental.
References
- 1.Gething MJ, Sambrook J. Protein folding in the cell. Nature. 1992;355(6355):33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 2.Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332(6163):462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- 3.Schröder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida H. ER stress and diseases. FEBS J. 2007;274(3):630–658. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- 5.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 6.Mori K. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell. 2000;101(5):451–454. doi: 10.1016/s0092-8674(00)80855-7. [DOI] [PubMed] [Google Scholar]
- 7.Normington K, Kohno K, Kozutsumi Y, Gething MJ, Sambrook J. S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell. 1989;57(7):1223–1236. doi: 10.1016/0092-8674(89)90059-7. [DOI] [PubMed] [Google Scholar]
- 8.Travers KJ, et al. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101(3):249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 9.Kimata Y, et al. Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J Cell Biol. 2007;179(1):75–86. doi: 10.1083/jcb.200704166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimata Y, Oikawa D, Shimizu Y, Ishiwata-Kimata Y, Kohno K. A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire1. J Cell Biol. 2004;167(3):445–456. doi: 10.1083/jcb.200405153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2005;102(52):18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou J, et al. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc Natl Acad Sci USA. 2006;103(39):14343–14348. doi: 10.1073/pnas.0606480103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohno K. Stress-sensing mechanisms in the unfolded protein response: Similarities and differences between yeast and mammals. J Biochem. 2010;147(1):27–33. doi: 10.1093/jb/mvp196. [DOI] [PubMed] [Google Scholar]
- 14.Oikawa D, Kimata Y, Kohno K, Iwawaki T. Activation of mammalian IRE1α upon ER stress depends on dissociation of BiP rather than on direct interaction with unfolded proteins. Exp Cell Res. 2009;315(15):2496–2504. doi: 10.1016/j.yexcr.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Lee KP, et al. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell. 2008;132(1):89–100. doi: 10.1016/j.cell.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korennykh AV, et al. The unfolded protein response signals through high-order assembly of Ire1. Nature. 2009;457(7230):687–693. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87(3):391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 18.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90(6):1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 19.Mori K, Kawahara T, Yoshida H, Yanagi H, Yura T. Signalling from endoplasmic reticulum to nucleus: Transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1996;1(9):803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- 20.Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12(12):1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang XZ, et al. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17(19):5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwawaki T, et al. Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nat Cell Biol. 2001;3(2):158–164. doi: 10.1038/35055065. [DOI] [PubMed] [Google Scholar]
- 23.Zhang K, et al. The unfolded protein response sensor IRE1α is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest. 2005;115(2):268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwawaki T, Akai R, Yamanaka S, Kohno K. Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. Proc Natl Acad Sci USA. 2009;106(39):16657–16662. doi: 10.1073/pnas.0903775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertolotti A, et al. Increased sensitivity to dextran sodium sulfate colitis in IRE1β-deficient mice. J Clin Invest. 2001;107(5):585–593. doi: 10.1172/JCI11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 27.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415(6867):92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 28.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313(5783):104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 29.Hollien J, et al. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186(3):323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaser A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134(5):743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iqbal J, et al. IRE1beta inhibits chylomicron production by selectively degrading MTP mRNA. Cell Metab. 2008;7(5):445–455. doi: 10.1016/j.cmet.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen A, Hutton DA, Pearson JP. The MUC2 gene product: A human intestinal mucin. Int J Biochem Cell Biol. 1998;30(7):797–801. doi: 10.1016/s1357-2725(98)00028-4. [DOI] [PubMed] [Google Scholar]
- 33.Van Klinken BJ, Tytgat KM, Büller HA, Einerhand AW, Dekker J. Biosynthesis of intestinal mucins: MUC1, MUC2, MUC3 and more. Biochem Soc Trans. 1995;23(4):814–818. doi: 10.1042/bst0230814. [DOI] [PubMed] [Google Scholar]
- 34.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 35.Chang WW, Leblond CP. Renewal of the epithelium in the descending colon of the mouse. I. Presence of three cell populations: vacuolated-columnar, mucous and argentaffin. Am J Anat. 1971;131(1):73–99. doi: 10.1002/aja.1001310105. [DOI] [PubMed] [Google Scholar]
- 36.Lidell ME, Johansson ME, Hansson GC. An autocatalytic cleavage in the C terminus of the human MUC2 mucin occurs at the low pH of the late secretory pathway. J Biol Chem. 2003;278(16):13944–13951. doi: 10.1074/jbc.M210069200. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura D, et al. Mammalian ER stress sensor IRE1β specifically down-regulates the synthesis of secretory pathway proteins. FEBS Lett. 2011;585(1):133–138. doi: 10.1016/j.febslet.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Carlson JA, et al. Accumulation of PiZ alpha 1-antitrypsin causes liver damage in transgenic mice. J Clin Invest. 1989;83(4):1183–1190. doi: 10.1172/JCI113999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harding HP, et al. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7(6):1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 40.Scheuner D, et al. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat Med. 2005;11(7):757–764. doi: 10.1038/nm1259. [DOI] [PubMed] [Google Scholar]
- 41.Saito A, et al. Regulation of endoplasmic reticulum stress response by a BBF2H7-mediated Sec23a pathway is essential for chondrogenesis. Nat Cell Biol. 2009;11(10):1197–1204. doi: 10.1038/ncb1962. [DOI] [PubMed] [Google Scholar]
- 42.Han D, et al. IRE1α kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138(3):562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imagawa Y, Hosoda A, Sasaka S, Tsuru A, Kohno K. RNase domains determine the functional difference between IRE1α and IRE1β. FEBS Lett. 2008;582(5):656–660. doi: 10.1016/j.febslet.2008.01.038. [DOI] [PubMed] [Google Scholar]
- 44.Morikawa Y, Inoue S, Okada T. Perinatal changes in the population of colonic goblet cells in the rat. Nippon Juigaku Zasshi. 1988;50(2):585–588. doi: 10.1292/jvms1939.50.585. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.