Both sleep and wakefulness are modulated by an endogenous biological clock located in the suprachiasmatic nuclei (SCN) of the anterior hypothalamus. Beyond driving the body to fall asleep and to wake up, the biological clock also modulates waking behavior, as reflected in sleepiness and cognitive performance, generating circadian rhythmicity in almost all neurobehavioral variables investigated to date [1,2]. Theoretical conceptualizations of the daily temporal modulation of sleep and wakefulness (and to a lesser extent the modulation of waking cognitive functions) have been instantiated in the two-process mathematical model of sleep regulation [3,4] and its mathematical variants [5]. The two-process model of sleep regulation has been applied to the temporal profiles of sleep [6,7] and wakefulness [8]. The model consists of a sleep homeostatic process (S) and a circadian process (C), which interact to determine the timing of sleep onset and offset, as well as the stability of waking neurocognitive functions [1,2,9]. The homeostatic process represents the drive for sleep that increases during wakefulness (as can be observed when wakefulness is maintained beyond habitual bedtime into the night and subsequent day) and decreases during sleep (which represents recuperation obtained from sleep). When this homeostatic drive increases above a certain threshold, sleep is triggered; when it decreases below a different threshold, wakefulness is invoked. The circadian process represents daily oscillatory modulation of these threshold levels. It has been suggested that the circadian system actively promotes wakefulness more than sleep [10], although this hypothesis is not presently universally accepted. The circadian drive for wakefulness may be experienced as spontaneously-enhanced alertness in the early evening even after a sleepless night. Sleep deprivation, however, can elevate homeostatic pressure to the point that waking cognitive functions are degraded even at the time of the peak circadian drive for wakefulness [11]. There are robust individual differences in both the sleep homeostatic and circadian processes, pointing to genetic underpinnings.
This review begins with a discussion of the genetic basis of circadian rhythms and the timing of sleep. It next briefly discusses the genetic basis of sleep in healthy adults, including observed individual variability in sleep parameters. Next, it discusses individual differences in the context of the total absence of and the restriction of sleep as a phenotype, and recent studies using candidate gene approaches to identify genes that may relate to such responses. Finally, future areas of research are discussed.
Genetics of Individual Differences in Circadian Rhythms and the Timing of Sleep
There are genetic underpinnings of individual differences in the circadian system, which are important for the timing of sleep. Morningness-eveningness (i.e., the tendency to be an early “lark” or a late “owl”) is perhaps the most frequently used measure of interindividual variation in circadian rhythmicity. Morning- and evening-type individuals differ endogenously in the circadian phase of their biological clocks [12,13]. Self-report measures, such as the Horne-Östberg morningness-eveningness questionnaire [MEQ; 14] and its variants [e.g., 15], and more recent scales such as the Munich ChronoType Questionnaire [16,17], which differentiates timing of activities on workdays versus free days, are the most commonly utilized measures of circadian phase preference, mainly because of their convenience and cost effectiveness.
Age influences morningness-eveningness as shown in laboratory studies [18] and more naturalistic population-based settings [see reviews, 17,19]. In addition, gender also affects morningness-eveningness, whereby women show a greater skew toward morningness than men [17,20,21]. This circadian phase preference difference is an enduring trait, with a significant genetic basis [22–24]. Thus, chronotype represents a circadian rhythmicity phenotype in humans [25].
The genetic basis of morningness-eveningness in the general population has been investigated in several core circadian genes, with mixed results [26]. For example, the 3111C allele of the CLOCK gene 5'-UTR region has been associated with eveningness and delayed sleep timing in some studies [27,28] but not others [29–31]. Similarly, the variable number tandem repeat (VNTR) polymorphism in PERIOD3 (PER3), another core clock gene, has been linked to diurnal preference, but not uniformly so [32–37]. Both the 111G polymorphism in the 5'-untranslated region of PERIOD2 (PER2) and the T2434C polymorphism of PERIOD1 (PER1) also have been associated with morning preference [38,39]. Since morningness-eveningness represents a continuum, this trait is likely polygenic, influenced by several genes, each contributing to the determination of circadian phase preference. Thus, further studies investigating other clock genes, as well as replication of the PER and CLOCK findings, are needed to establish precisely the molecular genetic components of behavioral circadian phase preference.
Individual differences in morningness-eveningness (chronotype) can manifest into extreme cases classified as primary circadian rhythm sleep disorders (CRSDs), with altered phase relationships of the biological clock to the light-dark cycle, including alterations in sleep timing [40,41]. Thus, extreme eveningness is believed to result in CRSD, delayed sleep phase type (typically referred to as a disorder and abbreviated as DSPD; [41]), while extreme morningness can manifest as CRSD, advanced sleep phase type [40] (typically referred to as a disorder and abbreviated as ASPD; [41]). The extent to which these phase-displacement disorders reflect differences in endogenous circadian period, entrainment, amplitude, coupling and other aspects of clock neurobiology has received recent attention.
The genetic basis of DSPD and ASPD has been investigated in recent years, with both disorders having links to core clock genes [26]. DSPD, the most common circadian rhythm sleep disorder in the general population, is characterized by an inability to fall asleep at the desired and “normal” time of day; the average onset of sleep in DSPD occurs in the early morning (3 am–6 am), and the average wake up time occurs in the late morning to early afternoon (11 am–2 pm) [41]. DSPD also may be characterized by a longer than normal tau (25.38 hours) [42]. The VNTR polymorphism in PER3 is associated with DSPD in large sample studies [32,33,35], and the 3111C allele of the CLOCK gene 5'-UTR region also has been related to DSPD [27]. A specific haplotype of PER3, which includes the polymorphism G647, is also associated positively with DSPD [35], while the N408 allele of casein kinase I epsilon (CK1ε) may protect against the development of DSPD [43].
ASPD is a rare disorder characterized by 3 to 4-hour advanced sleep onsets and wake times relative to desired, normal times [41,44]. It may be characterized by a shorter than normal tau (23.3 hours) [45]. In one study, ASPD was associated with a mutation in PER2 [46], although this mutation is not found in all families with this disorder [47]. Another study has implicated mutations in casein kinase I delta (CK1δ) in ASPD [48]. Future studies on additional core clock genes are needed to determine other mutations which may underlie this disorder.
Morningness-eveningness and differences in circadian phase preference are reflected in the diurnal course of neurobehavioral variables [as reviewed in 49]—some people perform consistently better in the morning, whereas others are more alert and perform better in the evening. How genetic variants underlying morningness-eveningness and disorders of chronotype affect performance and alertness under normal and sleep-deprived conditions remains a new field of investigation. Recent studies [50–52] have shown that the longer, 5-repeat allele of the VNTR polymorphism in PER3, a clock gene linked to diurnal preference and DSPD, may be associated with higher sleep propensity both at baseline and after total sleep deprivation (TSD), and worse cognitive performance following TSD and higher sleep propensity during chronic partial sleep deprivation (PSD) (see below for a more detailed discussion of these studies). The role of other clock gene polymorphisms such as the 3111C allele of the CLOCK gene in response to TSD and to chronic PSD remain unknown and are worthy of investigation.
Collectively, six core clock genes (PER1, PER2, PER3, CLOCK, CK1δ, CK1ε) have thus far been associated with interindividual differences in diurnal preference or its extreme variants. This is an active area of research with promising implications for objectively detecting individual differences in circadian disorders and determining situations and lifestyles that adversely affect the timing of sleep and sleep homeostasis.
Genetics of Sleep
Sleep is a highly complex trait that involves many genes and their interactions with environmental factors. In humans, research dating back to as early as the 1930s employing twins has indicated a strong genetic basis underlying the regulation of normal sleep, including sleep duration, sleep onset, sleep quality, and sleep homeostasis [reviewed in 53–55]. In addition, in 2008, two studies in normal sleepers found strong heritability of the sleep EEG power spectrum, underscoring prior studies indicating that while the sleep EEG is consistent across nights in the same individual, it differs among individuals [56,57]. The genetic nature of sleep EEG is also observed across a variety of frequencies indicating trait-like features [58–63]. Moreover, waking EEG patterns are also highly heritable [reviewed in 55]. Notably, familial linkage studies on EEG traits are currently lacking [reviewed in 53, 64].
More recently, candidate gene studies have investigated the role of specific genes in the regulation of sleep. For example, a point mutation in the DEC2 gene, believed to function in the circadian clock as a repressor of Clock/Bmal1 is associated with a short sleep duration phenotype (average 6.25h vs. 8.06h of self-reported sleep) that is characterized by an earlier non-workday habitual sleep offset time, with normal onset time, in 2 adults [65]. Moreover, the insertion of this point mutation into mice also decreased sleep time without affecting tau. Future studies should determine the role of this DEC2 mutation in individuals undergoing sleep deprivation and in studies using EEG and SWA physiological sleep assessments for sleep duration, sleep homeostasis and other related variables. By contrast, a recent study found that a polymorphism within intron 8 of the dopamine transporter 1 (DAT1) gene failed to correlate with the inter-individual variability of basal PSG sleep architecture, including slow-wave sleep latency, REM sleep latency, sleep efficiency, or sleep stage percentages (stages 1, 2, SWS or REM) in a group of unrelated healthy men [66].
Candidate Gene Studies of Sleep Deprivation
Beyond these studies, which assess habitual sleep or one night of baseline sleep, candidate gene studies have been used to study basal (fully-rested) sleep and responses to sleep loss. This approach was motivated by the results of studies that indicated that there are stable phenotypic individual differences in response to sleep deprivation.
Subjects undergoing TSD display differential vulnerability to sleep loss, demonstrating robust inter-individual differences in response to the same laboratory conditions, as measured by various physiological and subjective sleep measures and neurobehavioral tasks sensitive to sleep loss [67–72]. Approximately a third of healthy adults are highly vulnerable to the neurobehavioral effects of sleep deprivation, another third are vulnerable, and the remaining third are much less vulnerable. These stable (phenotypic) differences in neurobehavioral responses to sleep deprivation are not reliably accounted for by demographic factors (e.g., age, sex, IQ), by baseline functioning, by various aspects of habitual sleep timing, by circadian chronotype, or by any other investigated factor [73–76].
Such differential vulnerability extends to chronic PSD—a condition associated with a wide range of serious health consequences and experienced by millions of people on a consecutive and daily basis [77,78]—in which sleep is restricted to 3–7 hours time in bed per night [52,75,79,80].
At present, it remains unknown whether the same individuals vulnerable to the adverse effects of acute TSD are also vulnerable to chronic PSD. The few reports comparing responses to both acute TSD and chronic PSD have used small sample sizes (9–13 subjects) and limited assessments [75,81,82], and only one [81] has systematically studied the same subjects in both types of sleep loss.
The stable, trait-like interindividual differences observed in response to TSD [68,70,76,83]—with intraclass correlations (which express the proportion of variance in the data that is explained by systematic interindividual variability) ranging from 58–92% for neurobehavioral measures [70,76]—strongly suggest an underlying genetic component. Despite this link, however, relatively little is known about the genetic basis of differential vulnerability in healthy subjects undergoing deprivation. Furthermore, as mentioned above, because of reported differences in sleep homeostatic, physiological, and behavioral responses to chronic PSD and acute TSD [75,81,82], it is likely that specific candidate genes play different roles in the degree of vulnerability and/or resilience to the sleep homeostatic and neurobehavioral effects of acute TSD and chronic PSD. These compelling questions have produced a rapidly emerging and promising field of scientific investigation; recent studies have thus far focused on a number of select candidate genes, which are reviewed below.
PERIOD3 VNTR Polymorphism
Three related studies investigated the role of the variable number tandem repeat (VNTR) polymorphism of the circadian gene PERIOD3 (PER3)—which shows similar allelic frequencies in African Americans and Caucasians [84,85] and is characterized by a 54-nucleotide coding region motif repeating in 4 or 5 units—in response to TSD using a small group of the same subjects specifically recruited for the homozygotic versions of this polymorphism. Compared with the 4-repeat allele (PER34/4; 14 subjects), the longer, 5-repeat allele (PER35/5; 10 subjects) was associated with higher sleep propensity including SWA in the sleep EEG both before and after TSD and worse cognitive performance, as assessed by a composite score of 12 tests, following TSD [50]. A subsequent report—using the same 24 subjects—clarified that the PER35/5 overall performance deficits were selective: they only occurred on certain executive function tests, and only at 2–4 hours following the melatonin rhythm peak, from approximately 6–8 am [51]. Such performance differences were hypothesized to be mediated by sleep homeostasis [50,51]. Another publication using the same subjects showed that PER35/5 subjects had more slow-wave sleep and elevated sympathetic predominance and a reduction of parasympathetic activity during baseline sleep [86]. These studies found no significant differences in the melatonin and cortisol circadian rhythms, PER3 mRNA levels, or in a self-report morningness-eveningness measure [50,51], although another study using these same subjects found PER3 expression and sleep timing were more strongly correlated in PER35/5 subjects [87].
A recent neuroimaging study found that 27 healthy subjects categorized according to homozygosity for the PER3 VNTR genotype (15 PER34/4subjects, 12 PER35/5 subjects) showed markedly different cerebral blood flow profiles using blood oxygenation level dependent functional magnetic resonance imaging (BOLD fMRI) and corresponding differences in vulnerability of executive function performance in response to TSD [88]. More studies examining the relationship of the neural mechanisms mediating trait-like differential vulnerability to sleep deprivation with selective candidate genes (beyond the PER3 VNTR polymorphism) are warranted.
The PER3 findings in TSD may not generalize to responses to chronic PSD. My colleagues and I recently evaluated whether the PER3 VNTR polymorphism contributed to sleep homeostatic responses and cumulative neurobehavioral deficits during chronic PSD in PER34/4, PER34/5 and PER35/5 healthy adults [52]. During chronic PSD, PER35/5 subjects had slightly but reliably elevated sleep homeostatic pressure as measured by NREM SWE compared with PER34/4 subjects (Figure 1). The PER34/4, PER34/5 and PER35/5 genotypes also demonstrated large, but equivalent cumulative increases in sleepiness and cumulative decreases in cognitive performance and physiological alertness, with increasing daily inter-subject variability in all genotypes. In contrast to the aforementioned data in TSD [50,51], the PER3 VNTR variants did not differ on baseline sleep measures or in their physiological sleepiness, cognitive, executive functioning or subjective responses to chronic PSD. Thus, the PER3 VNTR polymorphism does not appear to be a genetic marker of differential vulnerability to the cumulative neurobehavioral effects of chronic PSD. It remains possible, however, that the PER35/5 genotype may contribute to differential neurobehavioral vulnerability to acute TSD because it involves wakefulness at a specific circadian time in the early morning hours (6–8 am), when subjects in the PSD study were asleep [52].
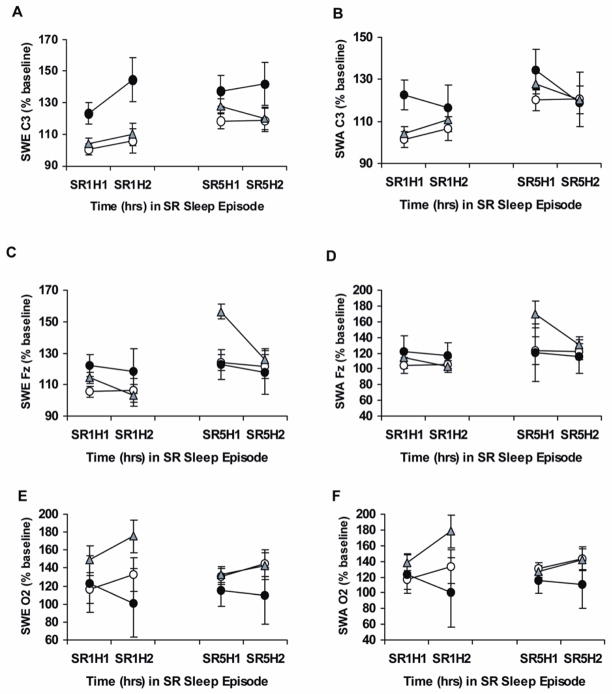
Figure 1.
This figure shows slow-wave energy and slow-wave activity during chronic partial sleep deprivation for the PER3 genotypes. Mean (± SEM) hourly slow-wave energy (SWE) and slow-wave activity (SWA) as a percentage of baseline at the same corresponding hour derived from the C3 (A, B), Fz (C, D) or O2 (E, F) channels at partial sleep deprivation/restriction night 1 (SR1) and partial sleep deprivation/restriction night 5 (SR5) for hour 1 (H1) and hour 2 (H2) in PER34/4 (open circles), PER34/5 (gray triangles) and PER35/5 (closed circles) subjects. SWE derived from C3 (but not from Fz or O2) was significantly higher during chronic partial sleep deprivation in PER35/5 compared with PER34/4 and PER34/5 subjects. From Goel N, Banks S, Mignot E, et al. PER3 polymorphism predicts cumulative sleep homeostatic but not neurobehavioral changes to chronic partial sleep deprivation. PLoS ONE 2009;4:e5874, with permission.
DQB1*0602 Allele
The human leukocyte antigen DQB1*0602 allele is closely associated with narcolepsy, a sleep disorder characterized by excessive daytime sleepiness, fragmented sleep, and shortened REM latency, although it is neither necessary nor sufficient for its development [89,90].
In one large study, DQB1*0602 positive healthy sleepers showed shorter nighttime REM sleep latency, greater sleep continuity, and more REM sleep, but no differences in daytime sleepiness [89]. Positivity for DQB1*0602 also was related to more sleep-onset REM sleep periods and greater REM sleep duration during naps [91]. Thus, DQB1*0602 positive subjects displayed subclinical presentations of some sleep features that were reminiscent of narcolepsy.
My colleagues and I recently evaluated whether DQB1*0602 was a novel biomarker of differential vulnerability to homeostatic, sleepiness and neurobehavioral deficits during chronic PSD in healthy sleepers positive and negative for DQB1*0602 [79]. DQB1*0602 positive subjects showed decreased sleep homeostatic pressure with differentially steeper declines (Figure 2), and greater sleepiness and fatigue during baseline. During chronic PSD, positive subjects displayed SWE elevation comparable to negative subjects (Figure 3), despite higher sleepiness and fatigue. DQB1*0602 positive subjects also had more fragmented sleep during baseline and PSD and showed differentially greater REM sleep latency reductions and smaller stage 2 reductions, along with differentially greater increases in fatigue. Both groups demonstrated comparable cumulative decreases in cognitive performance and increases in physiological sleepiness to chronic PSD, and did not differ on executive function tasks.
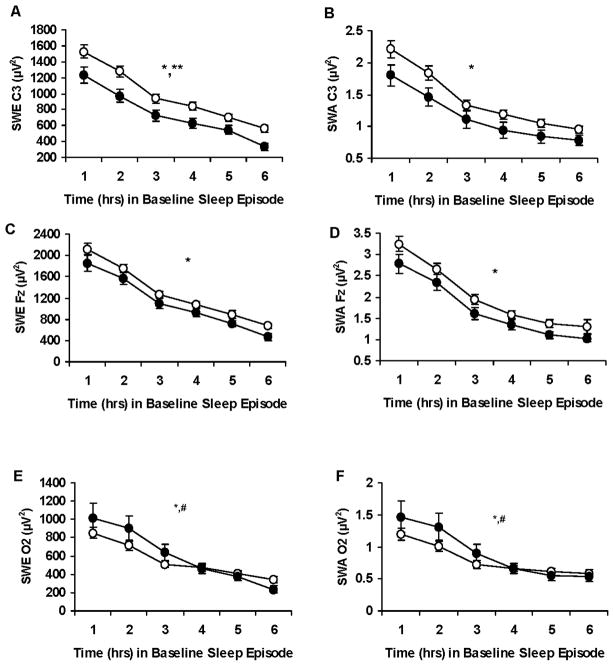
Figure 2.
Hourly slow-wave energy and slow-wave activity during baseline for the DQB1*0602 groups. Mean (± SEM) hourly slow-wave energy (SWE) and slow-wave activity (SWA) derived from the C3 (A, B), Fz (C, D) or O2 (E, F) channels during baseline for DQB1*0602 negative subjects (open circles) and DQB1*0602 positive subjects (closed circles). SWE derived from C3 was lower in DQB1*0602 positive subjects (denoted by **, p <0.05); SWA derived from C3 and SWE and SWA derived from the Fz channel showed similar trends. As expected, SWE and SWA showed a typical pattern of dissipation across the baseline night in all 3 channels for both groups (denoted by *, p <0.05); moreover, DQB1*0602 positive subjects demonstrated sharper declines in sleep pressure derived from the O2 channel during the first few hours of the night than DQB1*0602 negative subjects (denoted by #, p <0.05). In some records, EEG signal quality was insufficient or contained too much artifact for reliable power spectral analysis. Thus, the final sample sizes were: for C3, DQB1*0602 negative (n = 68) and positive (n = 24) subjects; for Fz, DQB1*0602 negative (n = 70) and positive (n = 28) subjects; for O2, DQB1*0602 negative (n = 74) and positive (n = 27) subjects. From Goel N, Banks S, Mignot E, et al. DQB1*0602 predicts interindividual differences in physiologic sleep, sleepiness and fatigue. Neurology 2010;75:1509–19, with permission.
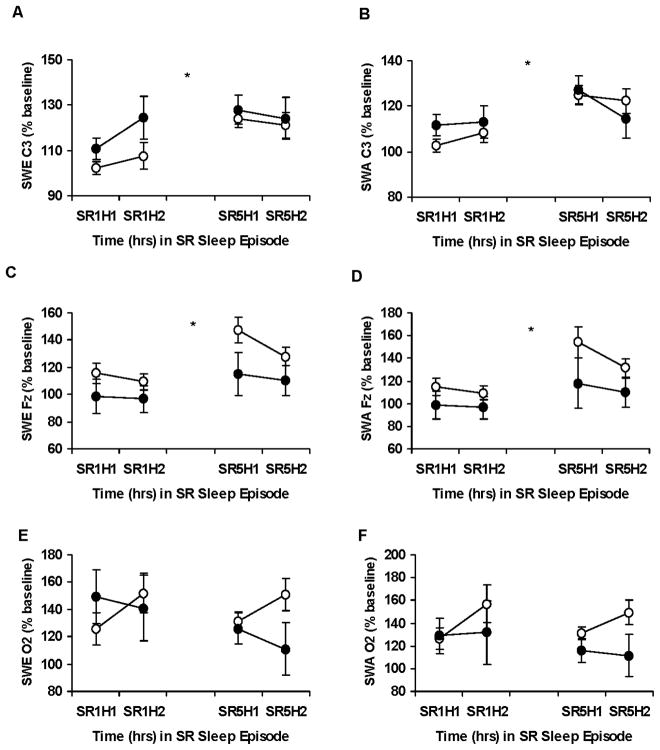
Figure 3.
Slow-wave energy and slow-wave activity during chronic partial sleep deprivation for the DQB1*0602 groups. Mean (± SEM) hourly slow-wave energy (SWE) and slow-wave activity (SWA) as a percentage of baseline at the same corresponding hour derived from the C3 (A, B), Fz (C, D) or O2 (E, F) channels at partial sleep deprivation/restriction night 1 (SR1) and partial sleep deprivation/restriction night 5 (SR5) for hour 1 (H1) and hour 2 (H2) for DQB1*0602 negative subjects (open circles) and DQB1*0602 positive subjects (closed circles). SWE and SWA increased from SR1 to SR5 for the C3 and Fz channels (denoted by *, p <0.05). There were no group differences or differential changes across nights. In some records, EEG signal quality was insufficient or contained too much artifact for reliable power spectral analysis. Thus, the final sample sizes were: for SR1 and SR5 C3, DQB1*0602 negative (n = 72) and positive (n = 28) subjects; for SR1 and SR5 Fz, DQB1*0602 negative (n = 72) and positive (n = 27) subjects; for SR1 and SR5 O2, DQB1*0602 negative (n = 72) and positive (n = 26) subjects. From Goel N, Banks S, Mignot E, et al. DQB1*0602 predicts interindividual differences in physiologic sleep, sleepiness and fatigue. Neurology 2010;75:1509–19, with permission.
Thus, DQB1*0602 is associated with inter-individual differences in sleep homeostasis, physiological sleep, sleepiness and fatigue, but not cognitive responses, during baseline and PSD. DQB1*0602 may be a genetic marker for predicting such individual differences in both basal (fully-rested) and sleep loss conditions; moreover, its positivity in healthy subjects may represent a continuum of some sleep-wake features of narcolepsy. The influence of the DQB1*0602 allele on sleep homeostatic and neurobehavioral responses has not yet been examined in healthy subjects undergoing acute TSD or replicated in an independent sample of individuals undergoing chronic PSD.
Catechol-O-Methyltransferase (COMT) Val158Met Polymorphism
The valine158methionine (Val158Met) polymorphism of the catechol-O-methyltransferase (COMT gene), replaces valine (Val) with methionine (Met) at codon 158 of the COMT protein. As a result of this common substitution, activity of the COMT enzyme, which modulates dopaminergic catabolism in the prefrontal cortex (PFC), is reduced 3-to-4-fold in COMT Met carriers compared with Val carriers, translating into more dopamine availability at the receptors and higher cortical dopamine concentrations [92]. This COMT polymorphism functionally predicts less efficient prefrontal cortex functioning and poor working memory performance in healthy subjects [93–96] who have the high-activity Val allele.
In sleep and neurodegenerative disorders, the COMT Val158Met polymorphism has been tied to daytime sleepiness. Val/Val female narcoleptic patients fell asleep two times faster than the Val/Met or Met/Met genotypes during the multiple sleep latency test (MSLT) while the opposite was true for males [97]. Met/Met also narcoleptic patients showed more sleep onset
REM periods during the MSLT while Val/Val subjects showed less sleep paralysis [97] and were more responsive to modafinil’s stimulating effects [98]. Met/Met and Val/Met Parkinson’s disease subjects demonstrated higher subjective daytime sleepiness than Val/Val subjects [99].
In healthy men, the COMT Val158Met polymorphism is associated with sleep physiology. In acute TSD, the polymorphism predicted interindividual differences in brain alpha oscillations in wakefulness and 11–13 Hz EEG activity in wakefulness, rapid-eye movement (REM) and non-REM sleep [100]. It also modulated the effects of the wake-promoting drug modafinil on subjective well-being, sustained vigilant attention and executive functioning, and on 3.0–6.75 Hz and >16.75 Hz activity in non-REM sleep, but was not associated with subjective sleepiness, slow-wave activity or slow-wave sleep changes in recovery sleep following TSD or at baseline [101,102]. Current studies are underway to investigate whether the COMT Val158Met polymorphism contributes to sleep homeostatic and cumulative neurobehavioral responses during basal (fully-rested) conditions and during chronic PSD in Met/Met, Val/Met and Val/Val healthy adult sleepers.
Adenosine-Related Polymorphisms
Other studies have investigated the role of select adenosine-related candidate genes in individual differences and in response to acute TSD. Rétey et al. [103] found that the 22G A polymorphism of the adenosine deaminase gene (ADA) was associated with enhanced slow-wave sleep and NREM SWA, contributing to interindividual variability in baseline sleep. Specifically, individuals with the G/A genotype (7 subjects) showed 30 minutes more slow-wave sleep than subjects with the G/G genotype (7 subjects) and consistent with this finding, SWA was higher in G/A than G/G subjects. Notably, this study did not test responses of these individuals to sleep deprivation and thus it remains unknown whether these genotypes show differential sleep homeostatic responses under evoked phenotypic deprivation conditions. This group also found that the c.1083T>C polymorphism of the adenosine A2A receptor gene (ADORA2A) related to objective and subjective differences in the effects of caffeine on NREM sleep after TSD [104] and associated with individual differences in various measures of baseline EEG during sleep and wakefulness [103]. While promising, replication of these data in independent samples is needed; in addition, the role of these two genetic variants in response to chronic PSD has not yet been established.
Thus, a number of common genetic polymorphisms involved in circadian, sleep-wake, and cognitive regulation appear to underlie inter-individual differences in basal (fully-rested) sleep parameters and homeostatic regulation of sleep in response to sleep deprivation (both chronic restriction and acute total sleep deprivation) in healthy adults.
Genome-wide Association Studies (GWAS) of Human Sleep
To date, only one study has employed a genome-wide association approach to examine phenotypic-genotypic interactions in healthy human sleepers [105]. Moderate heritability estimates for self-rated sleepiness (29%; assessed by the Epworth Sleepiness Scale) and for habitual sleep duration (17%) and habitual bedtime (22%), assessed by a standard questionnaire used in the Sleep Heart Health Study, were found in 749 subjects. The genome-wide analysis revealed that habitual bedtime and sleep duration were modulated by genetic loci containing circadian clock-related genes including casein kinase 2A2 (CSNK2A2), prokineticin 2 (PROK2) and CLOCK. Furthermore, genes encoding NPSRl and PDE4D were identified as possible mediators of habitual bedtime and subjective sleepiness, respectively. While intriguing, these data need to be replicated and extended to studies that include physiological measures of sleep.
Future Directions
With the exception of two recent studies [52,79], all candidate gene studies involving sleep physiological and neurobehavioral responses to sleep loss have used small sample sizes (14–24 subjects) and have only examined homozygotic individuals [50,51, 100,101,103,104]. Larger sample sizes and assessment of phenotype-genotype relationships in both homozygous and heterozygous individuals are needed to definitively determine whether such candidate genes involved in regulation of sleep-wake, circadian and cognitive functions are associated with inter-individual neurobehavioral responses to sleep loss across an entire population. This is particularly critical since individuals are necessarily categorized into different genotypes, reducing sample sizes in each subgroup. Future candidate gene studies therefore must consistently use sample sizes in the hundreds, rather than tens, to detect statistically reliable differences across genotypes.
In addition, replication of findings in independent samples is needed to determine whether findings are genuine and are not due to chance; ideally, studies should also be replicated in different ethnic groups to increase generalizability of the findings. GWAS studies using physiological sleep measures as outcomes are also needed to assess basal (fully rested) individual differences as well as responses to sleep deprivation; however, these will likely require obtaining data in several laboratories, given the expense, time and effort needed to conduct such rigorous studies.
In conclusion, there is an active area of inquiry searching for other potential genetic markers of basal sleep measures and of sleep homeostatic and neurobehavioral differential vulnerability to sleep deprivation. Among other advantages, identification of such markers will provide a viable means to determine those individuals in the general population who may need more habitual sleep or who may need to prevent or mitigate sleep deprivation through lifestyle choices and effective interventions and countermeasures (e.g., caffeine, naps, etc).
Synopsis
The circadian biological clock interacts with sleep homeostatic drive. This paper reviews the genetic underpinnings of sleep timing, sleep duration and sleep homeostasis in healthy adult sleepers. Stable, trait-like (phenotypic) individual differences in circadian and sleep measures as well as in neurobehavioral performance have motivated recent studies utilizing candidate gene approaches to predict baseline (fully-rested conditions) responses as well as responses to sleep loss (acute total and chronic partial sleep deprivation). Results from this growing database point to several important circadian and noncircadian genetic biomarkers involved in differential vulnerability to sleep loss. Active searching for other potential genetic biomarkers, using candidate gene and GWAS approaches, will allow for effective prediction of response and utilization of countermeasures in response to sleep loss.
Acknowledgments
The writing of this paper was supported by National Space Biomedical Research Institute through NASA NCC 9-58, NIH NR004281 and CTRC UL1RR024134, and by a grant from the Institute for Translational Medicine and Therapeutics’ (ITMAT) Transdisciplinary Program in Translational Medicine and Therapeutics. The project described was supported in part by Grant Number UL1RR024134 from the National Center for Research Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
Footnotes
The author has nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Van Dongen HPA, Dinges DF. Investigating the interaction between the homeostatic and circadian processes of sleep-wake regulation for the prediction of waking neurobehavioral performance. J Sleep Res. 2003;12:181–87. doi: 10.1046/j.1365-2869.2003.00357.x. [DOI] [PubMed] [Google Scholar]
- 2.Goel N, Van Dongen HPA, Dinges DF. Circadian rhythms in sleepiness, alertness, and performance. In: Kryger MH, Dement WC, Roth T, editors. Principles and Practice of Sleep Medicine. 5. Philadelphia: Elsevier; 2011. pp. 445–55. [Google Scholar]
- 3.Achermann P, Dijk DJ, Brunner DP, et al. A model of human sleep homeostasis based on EEG slow-wave activity; quantitative comparison of data and simulations. Brain Res Bull. 1993;31:97–113. doi: 10.1016/0361-9230(93)90016-5. [DOI] [PubMed] [Google Scholar]
- 4.Borbély AA. A two process model of sleep regulation. Hum Neurobiol. 1982;1:195–204. [PubMed] [Google Scholar]
- 5.Mallis MM, Mejdal S, Nguyen TT, et al. Summary of the key features of seven biomathematical models of human fatigue and performance. Aviat Space Environ Med. 2004;75:A4–14. [PubMed] [Google Scholar]
- 6.Borbély AA. A two-process model of sleep regulation. Hum Neurobiol. 1982;1:195–204. [PubMed] [Google Scholar]
- 7.Daan S, Beersma DGM, Borbély AA. Timing of human sleep: Recovery process gated by a circadian pacemaker. Am J Physiol. 1984;246:R161–78. doi: 10.1152/ajpregu.1984.246.2.R161. [DOI] [PubMed] [Google Scholar]
- 8.Achermann P, Borbély AA. Simulation of daytime vigilance by the additive interaction of a homeostatic and a circadian process. Biol Cybern. 1994;71:115–21. doi: 10.1007/BF00197314. [DOI] [PubMed] [Google Scholar]
- 9.Khalsa SBS, Jewett ME, Duffy JF, et al. The timing of the human circadian clock is accurately represented by the core body temperature rhythm following phase shifts to a three-cycle light stimulus near the critical zone. J Biol Rhythms. 2000;15:524–30. doi: 10.1177/074873040001500609. [DOI] [PubMed] [Google Scholar]
- 10.Edgar DM, Dement WC, Fuller CA. Effect of SCN lesions on sleep in squirrel monkeys: Evidence for opponent processes in sleep-wake regulation. J Neurosci. 1993;13:1065–79. doi: 10.1523/JNEUROSCI.13-03-01065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doran SM, Van Dongen HPA, Dinges DF. Sustained attention performance during sleep deprivation: evidence of state instability. Arch Ital Biol. 2001;139:253–67. [PubMed] [Google Scholar]
- 12.Kerkhof GA, Van Dongen HPA. Morning-type and evening-type individuals differ in the phase position of their endogenous circadian oscillator. Neurosci Lett. 1996;218:153–6. doi: 10.1016/s0304-3940(96)13140-2. [DOI] [PubMed] [Google Scholar]
- 13.Baehr EK, Revelle W, Eastman CI. Individual differences in the phase and amplitude of the human circadian temperature rhythm: with an emphasis on morningness-eveningness. J Sleep Res. 2000;9:117–27. doi: 10.1046/j.1365-2869.2000.00196.x. [DOI] [PubMed] [Google Scholar]
- 14.Horne JA, Östberg O. A self-assessment questionnaire to determine morningness–eveningness in human circadian rhythms. Int J Chronobiol. 1976;4:97–110. [PubMed] [Google Scholar]
- 15.Smith CS, Reilly D, Midkiff K. Evaluation of three circadian rhythm questionnaires with suggestions for an improved measure of morningness. J Appl Psychol. 1989;74:728–38. doi: 10.1037/0021-9010.74.5.728. [DOI] [PubMed] [Google Scholar]
- 16.Roenneberg T, Wirz-Justice A, Merrow M. Life between clocks: daily temporal patterns of human chronotypes. J Biol Rhythms. 2003;18:80–90. doi: 10.1177/0748730402239679. [DOI] [PubMed] [Google Scholar]
- 17.Roenneberg T, Kuehnle T, Juda M, et al. Epidemiology of the human circadian clock. Sleep Med Rev. 2007;11:429–38. doi: 10.1016/j.smrv.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Duffy JF, Dijk D-J, Klerman EB, et al. Later endogenous circadian temperature nadir relative to an earlier wake time in older people. Am J Physiol. 1998;275:R1478–87. doi: 10.1152/ajpregu.1998.275.5.r1478. [DOI] [PubMed] [Google Scholar]
- 19.Foster RG, Roenneberg T. Human responses to the geophysical daily, annual and lunar cycles. Curr Biol. 2008;18:R784–94. doi: 10.1016/j.cub.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Adan A, Natale V. Gender differences in morningness–eveningness preference. Chronobiol Int. 2002;19:709–20. doi: 10.1081/cbi-120005390. [DOI] [PubMed] [Google Scholar]
- 21.Randler C. Gender differences in morningness–eveningness assessed by self-report questionnaires: A meta-analysis. Pers Indiv Differ. 2007;43:1667–75. [Google Scholar]
- 22.Hur Y-M. Stability of genetic influence on morningness-eveningness: a cross-sectional examination of South Korean twins from preadolescence to young adulthood. J Sleep Res. 2007;16:17–23. doi: 10.1111/j.1365-2869.2007.00562.x. [DOI] [PubMed] [Google Scholar]
- 23.Hur Y-M, Bouchard TJ, Jr, Lykken DT. Genetic and environmental influence on morningness–eveningness. Pers Indiv Differ. 1998;25:917–25. [Google Scholar]
- 24.Vink JM, Groot AS, Kerkhof GA, et al. Genetic analysis of morningness and eveningness. Chronobiol Int. 2001;18:809–22. doi: 10.1081/cbi-100107516. [DOI] [PubMed] [Google Scholar]
- 25.Van Dongen HPA, Kerkhof GA, Dinges DF. Human circadian rhythms. In: Sehgal A, editor. Molecular Biology of Circadian Rhythms. New York: John Wiley and Sons; 2004. pp. 255–69. [Google Scholar]
- 26.Takahashi JS, Hong HK, Ko CH, et al. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–75. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katzenberg D, Young T, Finn L, et al. A CLOCK polymorphism associated with human diurnal preference. Sleep. 1998;21:569–76. doi: 10.1093/sleep/21.6.569. [DOI] [PubMed] [Google Scholar]
- 28.Mishima K, Tozawa T, Satoh K, et al. The 3111T/C polymorphism of hClock is associated with evening preference and delayed sleep timing in a Japanese population sample. Am J Med Genet B Neuropsychiatr Genet. 2005;133:101–4. doi: 10.1002/ajmg.b.30110. [DOI] [PubMed] [Google Scholar]
- 29.Robilliard DL, Archer SN, Arendt J, et al. The 3111 Clock gene polymorphism is not associated with sleep and circadian rhythmicity in phenotypically characterized human subjects. J Sleep Res. 2002;11:305–12. doi: 10.1046/j.1365-2869.2002.00320.x. [DOI] [PubMed] [Google Scholar]
- 30.Iwase T, Kajimura N, Uchiyama M, et al. Mutation screening of the human Clock gene in circadian rhythm sleep disorders. Psychiatry Res. 2002;109:121–28. doi: 10.1016/s0165-1781(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 31.Pedrazzoli M, Louzada FM, Pereira DS, et al. Clock polymorphisms and circadian rhythms phenotypes in a sample of the Brazilian population. Chronobiol Int. 2007;24:1–8. doi: 10.1080/07420520601139789. [DOI] [PubMed] [Google Scholar]
- 32.Archer SN, Robilliard DL, Skene DJ, et al. A length polymorphism in the circadian clock gene PER3 is linked to delayed sleep phase syndrome and extreme diurnal preference. Sleep. 2003;26:413–15. doi: 10.1093/sleep/26.4.413. [DOI] [PubMed] [Google Scholar]
- 33.Pereira DS, Tufik S, Louzada FM, et al. Association of the length polymorphism in the human PER3 gene with the delayed sleep-phase syndrome: Does latitude have an influence upon it? Sleep. 2005;28:29–32. [PubMed] [Google Scholar]
- 34.Jones KH, Ellis J, von Schantz M, et al. Age-related change in the association between a polymorphism in the PER3 gene and preferred timing of sleep and waking activities. J Sleep Res. 2007;16:12–6. doi: 10.1111/j.1365-2869.2007.00561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ebisawa T, Uchiyama M, Kajimura N, et al. Association of structural polymorphisms in the human period3 gene with delayed sleep phase syndrome. EMBO Rep. 2001;2:342–46. doi: 10.1093/embo-reports/kve070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viola AU, Archer SN, James LM, et al. PER3 polymorphism predicts sleep structure and waking performance. Curr Biol. 2007;17:613–18. doi: 10.1016/j.cub.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 37.Groeger JA, Viola AU, Lo JC, et al. Early morning executive functioning during sleep deprivation is compromised by a PERIOD3 polymorphism. Sleep. 2008;31:1159–67. [PMC free article] [PubMed] [Google Scholar]
- 38.Carpen JD, Archer SN, Skene DJ, et al. A single-nucleotide polymorphism in the 5'-untranslated region of the hPER2 gene is associated with diurnal preference. J Sleep Res. 2005;14:293–97. doi: 10.1111/j.1365-2869.2005.00471.x. [DOI] [PubMed] [Google Scholar]
- 39.Carpen JD, von Schantz M, Smits M, et al. A silent polymorphism in the PER1 gene associates with extreme diurnal preference in humans. J Hum Genet. 2006;51:1122–25. doi: 10.1007/s10038-006-0060-y. [DOI] [PubMed] [Google Scholar]
- 40.The International Classification of Sleep Disorders: Diagnostic and Coding Manual. 2. Westchester: American Academy of Sleep Medicine; 2005. [Google Scholar]
- 41.Sack RL, Auckley D, Auger RR, et al. Circadian rhythm sleep disorders: part II, advanced sleep phase disorder, delayed sleep phase disorder, free-running disorder, and irregular sleep-wake rhythm. An American Academy of Sleep Medicine review. Sleep. 2007;30:1484–501. doi: 10.1093/sleep/30.11.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campbell SS, Murphy PJ. Delayed sleep phase disorder in temporal isolation. Sleep. 2007;30:1225–28. doi: 10.1093/sleep/30.9.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takano A, Uchiyama M, Kajimura N, et al. A missense variation in human casein kinase I epsilon gene that induces functional alteration and shows an inverse association with circadian rhythm sleep disorders. Neuropsychopharmacology. 2004;29:1901–09. doi: 10.1038/sj.npp.1300503. [DOI] [PubMed] [Google Scholar]
- 44.Reid KJ, Chang AM, Dubocovich ML, et al. Familial advanced sleep phase syndrome. Arch Neurol. 2001;58:1089–94. doi: 10.1001/archneur.58.7.1089. [DOI] [PubMed] [Google Scholar]
- 45.Jones CR, Campbell SS, Zone SE, et al. Familial advanced sleep-phase syndrome: A short-period circadian rhythm variant in humans. Nat Med. 1999;5:1062–65. doi: 10.1038/12502. [DOI] [PubMed] [Google Scholar]
- 46.Toh KL, Jones CR, He Y, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–43. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 47.Satoh K, Mishima K, Inoue Y, et al. Two pedigrees of familial advanced sleep phase syndrome in Japan. Sleep. 2003;26:416–17. doi: 10.1093/sleep/26.4.416. [DOI] [PubMed] [Google Scholar]
- 48.Xu Y, Padiath QS, Shapiro RE, et al. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–44. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 49.Kerkhof GA. Inter-individual differences in the human circadian system: A review. Biol Psychol. 1985;20:83–112. doi: 10.1016/0301-0511(85)90019-5. [DOI] [PubMed] [Google Scholar]
- 50.Viola AU, Archer SN, James LM, et al. PER3 polymorphism predicts sleep structure and waking performance. Curr Biol. 2007;17:613–18. doi: 10.1016/j.cub.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 51.Groeger JA, Viola AU, Lo JC, et al. Early morning executive functioning during sleep deprivation is compromised by a PERIOD3 polymorphism. Sleep. 2008;31:1159–67. [PMC free article] [PubMed] [Google Scholar]
- 52.Goel N, Banks S, Mignot E, et al. PER3 polymorphism predicts cumulative sleep homeostatic but not neurobehavioral changes to chronic partial sleep deprivation. PLoS ONE. 2009;4:e5874. doi: 10.1371/journal.pone.0005874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dauvilliers Y, Maret S, Tafti M. Genetics of normal and pathological sleep in humans. Sleep Med Rev. 2005;9:91–100. doi: 10.1016/j.smrv.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 54.Tafti M, Maret S, Dauvilliers Y. Genes for normal sleep and sleep disorders. Ann Med. 2005;37:580–9. doi: 10.1080/07853890500372047. [DOI] [PubMed] [Google Scholar]
- 55.Tafti M. Genetic aspects of normal and disturbed sleep. Sleep Med. 2009 Sep;10(Suppl 1):S17–21. doi: 10.1016/j.sleep.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 56.Ambrosius U, Lietzenmaier S, Wehrle R, et al. Heritability of sleep electroencephalogram. Biol Psychiatry. 2008;64:344–8. doi: 10.1016/j.biopsych.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 57.De Gennaro L, Marzano C, Fratello F, et al. The electroencephalographic fingerprint of sleep is genetically determined: A twin study. Ann Neurol. 2008;64:455–60. doi: 10.1002/ana.21434. [DOI] [PubMed] [Google Scholar]
- 58.Werth E, Achermann P, Dijk DJ, et al. Spindle frequency activity in the sleep EEG: individual differences and topographic distribution. Electroencephalogr Clin Neurophysiol. 1997;103:535–42. doi: 10.1016/s0013-4694(97)00070-9. [DOI] [PubMed] [Google Scholar]
- 59.Tan X, Campbell IG, Palagini L, et al. High internight reliability of computer-measured NREM delta, sigma, and beta: biological implications. Biol Psychiatry. 2000;48:1010–9. doi: 10.1016/s0006-3223(00)00873-8. [DOI] [PubMed] [Google Scholar]
- 60.Finelli LA, Achermann P, Borbely AA. Individual ‘fingerprints’ in human sleep EEG topography. Neuropsychopharmacol. 2001;25:S57–62. doi: 10.1016/S0893-133X(01)00320-7. [DOI] [PubMed] [Google Scholar]
- 61.De Gennaro L, Ferrara M, Vecchio F, et al. An electroencephalographic fingerprint of human sleep. Neuroimage. 2005;26:114–22. doi: 10.1016/j.neuroimage.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 62.Tucker AM, Dinges DF, Van Dongen HP. Trait interindividual differences in the sleep physiology of healthy young adults. J Sleep Res. 2007;16:170–80. doi: 10.1111/j.1365-2869.2007.00594.x. [DOI] [PubMed] [Google Scholar]
- 63.Buckelmuller J, Landolt HP, Stassen HH, et al. Trait-like individual differences in the human sleep electroencephalogram. Neuroscience. 2006;138:351–6. doi: 10.1016/j.neuroscience.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 64.Andretic R, Franken P, Tafti M. Genetics of sleep. Annu Rev Genet. 2008;42:361–88. doi: 10.1146/annurev.genet.42.110807.091541. [DOI] [PubMed] [Google Scholar]
- 65.He Y, Jones CR, Fujiki N, et al. The transcriptional repressor DEC2 regulates sleep length in mammals. Science. 2009;325:866–70. doi: 10.1126/science.1174443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guindalini C, Martins RC, Andersen ML, et al. Influence of genotype on dopamine transporter availability in human striatum and sleep architecture. Psychiatry Res. 2010;179:238–40. doi: 10.1016/j.psychres.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 67.Van Dongen HP, Baynard MD, Maislin G, et al. Systematic interindividual differences in neurobehavioral impairment from sleep loss: evidence of trait-like differential vulnerability. Sleep. 2004;27:423–33. [PubMed] [Google Scholar]
- 68.Leproult R, Colecchia EF, Berardi AM, et al. Individual differences in subjective and objective alertness during sleep deprivation are stable and unrelated. Am J Physiol Regulat Integr Comp Physiol. 2003;284:R280–90. doi: 10.1152/ajpregu.00197.2002. [DOI] [PubMed] [Google Scholar]
- 69.Van Dongen HPA, Dinges DF. Sleep, circadian rhythms, and psychomotor vigilance performance. Clin Sports Med. 2005;24:237–49. doi: 10.1016/j.csm.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 70.Van Dongen HP, Maislin G, Dinges DF. Dealing with interindividual differences in the temporal dynamics of fatigue and performance: importance and techniques. Aviat Space Environ Med. 2004;75(3 Suppl):A147–54. [PubMed] [Google Scholar]
- 71.Frey DJ, Badia P, Wright KP., Jr Inter- and intra-individual variability in performance near the circadian nadir during sleep deprivation. J Sleep Res. 2004;13:305–15. doi: 10.1111/j.1365-2869.2004.00429.x. [DOI] [PubMed] [Google Scholar]
- 72.Tucker AM, Dinges DF, Van Dongen HP. Trait interindividual differences in the sleep physiology of healthy young adults. J Sleep Res. 2007;16:170–80. doi: 10.1111/j.1365-2869.2007.00594.x. [DOI] [PubMed] [Google Scholar]
- 73.Doran SM, Van Dongen HP, Dinges DF. Sustained attention performance during sleep deprivation: evidence of state instability. Arch Ital Biol. 2001;139:253–67. [PubMed] [Google Scholar]
- 74.Van Dongen HPA, Dijkman MV, Maislin G, et al. Phenotypic aspect of vigilance decrement during sleep deprivation. Physiologist. 1999;42:A-5. [Google Scholar]
- 75.Van Dongen HP, Maislin G, Mullington JM, et al. The cumulative cost of additional wakefulness: dose-response effects on neurobehavioral functions and sleep physiology from chronic sleep restriction and total sleep deprivation. Sleep. 2003;26:117–26. doi: 10.1093/sleep/26.2.117. [DOI] [PubMed] [Google Scholar]
- 76.Van Dongen HP, Baynard MD, Maislin G, et al. Systematic interindividual differences in neurobehavioral impairment from sleep loss: evidence of trait-like differential vulnerability. Sleep. 2004;27:423–33. [PubMed] [Google Scholar]
- 77.Banks S, Dinges DF. Behavioral and physiological consequences of sleep restriction in humans. J Clin Sleep Med. 2007;3:519–28. [PMC free article] [PubMed] [Google Scholar]
- 78.Goel N, Rao H, Durmer JS, et al. Neurocognitive consequences of sleep deprivation. Semin Neurol. 2009;29:320–39. doi: 10.1055/s-0029-1237117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goel N, Banks S, Mignot E, et al. DQB1*0602 predicts interindividual differences in physiologic sleep, sleepiness and fatigue. Neurology. 2010;75:1509–19. doi: 10.1212/WNL.0b013e3181f9615d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bliese PD, Wesensten NJ, Balkin TJ. Age and individual variability in performance during sleep restriction. J Sleep Res. 2006;15:376–85. doi: 10.1111/j.1365-2869.2006.00557.x. [DOI] [PubMed] [Google Scholar]
- 81.Drake CL, Roehrs TA, Burduvali E, et al. Effects of rapid versus slow accumulation of eight hours of sleep loss. Psychophysiology. 2001;38:979–87. doi: 10.1111/1469-8986.3860979. [DOI] [PubMed] [Google Scholar]
- 82.Rowland LM, Thomas ML, Thorne DR, et al. Oculomotor responses during partial and total sleep deprivation. Aviat Space Environ Med. 2005;76:C104–13. [PubMed] [Google Scholar]
- 83.Van Dongen HPA, Dinges DF. Sleep, circadian rhythms, and psychomotor vigilance performance. Clin Sports Med. 2005;24:237–249. doi: 10.1016/j.csm.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 84.Nadkarni NA, Weale ME, von Schantz M, et al. Evolution of a length polymorphism in the human PER3 gene, a component of the circadian system. J Biol Rhythms. 2005;20:490–99. doi: 10.1177/0748730405281332. [DOI] [PubMed] [Google Scholar]
- 85.Ciarleglio CM, Ryckman KK, Servick SV, et al. Genetic differences in human circadian clock genes among worldwide populations. J Biol Rhythms. 2008;23:330–40. doi: 10.1177/0748730408320284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Viola AU, James LM, Archer SN, et al. PER3 polymorphism and cardiac autonomic control: effects of sleep debt and circadian phase. Am J Physiol Heart Circ Physiol. 2008;295:H2156–63. doi: 10.1152/ajpheart.00662.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Archer SN, Viola AU, Kyriakopoulou V, et al. Inter-individual differences in habitual sleep timing and entrained phase of endogenous circadian rhythms of BMAL1, PER2 and PER3 mRNA in human leukocytes. Sleep. 2008;31:608–17. doi: 10.1093/sleep/31.5.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vandewalle G, Archer SN, Wuillaume C, et al. Functional magnetic resonance imaging-assessed brain responses during an executive task depend on interaction of sleep homeostasis, circadian phase, and PER3 genotype. J Neurosci. 2009;29:7948–56. doi: 10.1523/JNEUROSCI.0229-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mignot E, Young T, Lin L, et al. Nocturnal sleep and daytime sleepiness in normal subjects with HLA-DQB1*0602. Sleep. 1999;22:347–52. [PubMed] [Google Scholar]
- 90.Dauvilliers Y, Tafti M. Molecular genetics and treatment of narcolepsy. Ann Med. 2006;38:252–62. doi: 10.1080/07853890500489700. [DOI] [PubMed] [Google Scholar]
- 91.Mignot E, Lin L, Finn L, et al. Correlates of sleep-onset REM periods during the Multiple Sleep Latency Test in community adults. Brain. 2006;129:1609–23. doi: 10.1093/brain/awl079. [DOI] [PubMed] [Google Scholar]
- 92.Tunbridge EM, Harrison PJ, Weinberger DR. Catechol-o-methyltransferase, cognition, and psychosis: Val158Met and beyond. Biol Psychiatry. 2006;60:141–51. doi: 10.1016/j.biopsych.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 93.Savitz J, Solms M, Ramesar R. The molecular genetics of cognition: dopamine, COMT and BDNF. Genes Brain Behav. 2006;5:311–28. doi: 10.1111/j.1601-183X.2005.00163.x. [DOI] [PubMed] [Google Scholar]
- 94.Egan MF, Goldberg TE, Kolachana BS, et al. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci USA. 2001;98:6917–22. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dickinson D, Elvevåg B. Genes, cognition and brain through a COMT lens. Neuroscience. 2009;164:72–87. doi: 10.1016/j.neuroscience.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Barnett JH, Jones PB, Robbins TW, et al. Effects of the catechol-O-methyltransferase Val158Met polymorphism on executive function: a meta-analysis of the Wisconsin Card Sort Test in schizophrenia and healthy controls. Mol Psychiatry. 2007;12:502–09. doi: 10.1038/sj.mp.4001973. [DOI] [PubMed] [Google Scholar]
- 97.Dauvilliers Y, Neidhart E, Lecendreux M, et al. MAO-A and COMT polymorphisms and gene effects in narcolepsy. Mol Psychiatry. 2001;6:367–72. doi: 10.1038/sj.mp.4000911. [DOI] [PubMed] [Google Scholar]
- 98.Dauvilliers Y, Neidhart E, Billiard M, et al. Sexual dimorphism of the catechol-O-methyltransferase gene in narcolepsy is associated with response to modafinil. Pharmacogenomics J. 2002;2:65–8. doi: 10.1038/sj.tpj.6500088. [DOI] [PubMed] [Google Scholar]
- 99.Frauscher B, Högl B, Maret S, et al. Association of daytime sleepiness with COMT polymorphism in patients with parkinson disease: a pilot study. Sleep. 2004;27:733–6. doi: 10.1093/sleep/27.4.733. [DOI] [PubMed] [Google Scholar]
- 100.Bodenmann S, Rusterholz T, Dürr R, et al. The functional Val158Met polymorphism of COMT predicts interindividual differences in brain α oscillations in young men. J Neurosci. 2009;29:10855–62. doi: 10.1523/JNEUROSCI.1427-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bodenmann S, Xu S, Luhmann U, et al. Pharmacogenetics of modafinil after sleep loss: Catechol-O-Methyltransferase genotype modulates waking functions but not recovery sleep. Clin Pharmacol Ther. 2009;85:296–304. doi: 10.1038/clpt.2008.222. [DOI] [PubMed] [Google Scholar]
- 102.Bodenmann S, Landolt HP. Effects of modafinil on the sleep EEG depend on Val158Met genotype of COMT. Sleep. 2010;33:1027–35. doi: 10.1093/sleep/33.8.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rétey JV, Adam M, Honegger E, et al. A functional genetic variation of adenosine deaminase affects the duration and intensity of deep sleep in humans. Proc Natl Acad Sci USA. 2005;102:15676–681. doi: 10.1073/pnas.0505414102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rétey JV, Adam M, Khatami R, et al. A genetic variation in the adenosine A2A receptor gene (ADORA2A) contributes to individual sensitivity to caffeine effects on sleep. Clin Pharmacol Ther. 2007;81:692–8. doi: 10.1038/sj.clpt.6100102. [DOI] [PubMed] [Google Scholar]
- 105.Gottlieb DJ, O’Connor GT, Wilk JB. Genome-wide association of sleep and circadian phenotypes. BMC Med Genet. 2007;8(Suppl 1):S9. doi: 10.1186/1471-2350-8-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]