Abstract
Despite advances in clinical therapy, metastasis remains the leading cause of death in breast cancer patients. Mutations in mitochondrial DNA, including those affecting complex I and oxidative phosphorylation, are found in breast tumors and could facilitate metastasis. This study identifies mitochondrial complex I as critical for defining an aggressive phenotype in breast cancer cells. Specific enhancement of mitochondrial complex I activity inhibited tumor growth and metastasis through regulation of the tumor cell NAD+/NADH redox balance, mTORC1 activity, and autophagy. Conversely, nonlethal reduction of NAD+ levels by interfering with nicotinamide phosphoribosyltransferase expression rendered tumor cells more aggressive and increased metastasis. The results translate into a new therapeutic strategy: enhancement of the NAD+/NADH balance through treatment with NAD+ precursors inhibited metastasis in xenograft models, increased animal survival, and strongly interfered with oncogene-driven breast cancer progression in the MMTV-PyMT mouse model. Thus, aberration in mitochondrial complex I NADH dehydrogenase activity can profoundly enhance the aggressiveness of human breast cancer cells, while therapeutic normalization of the NAD+/NADH balance can inhibit metastasis and prevent disease progression.
Introduction
Despite advances in clinical therapy, metastasis is still the leading cause of death in breast cancer patients (1). A clearer understanding of molecular mechanisms that drive metastasis will help to develop more effective therapies (2). Our present study focused on metabolism as an essential driver of tumor growth and metastasis, potentially common to all breast cancer types. Normal cells primarily use mitochondrial oxidative phosphorylation (OXPHOS) for energy production, whereas cancer cells depend on aerobic glycolysis (the Warburg effect) to generate energy and glycolytic intermediates for enhanced growth (3, 4). Tumor cells also generate high levels of reduced forms of NAD+, NADH, and NADPH as important cofactors and redox components (4, 5). These altered metabolic activities can be linked to mitochondrial dysfunction that inhibits OXPHOS, increases ROS, promotes uncontrolled growth, and causes DNA damage that further supports a metastatic phenotype (6, 7). Mitochondrial dysfunctions can be caused by mutations in mitochondrial DNA (mtDNA) or nuclear genes encoding mitochondrial proteins (6, 8) that are essential for the respiratory chain/OXPHOS system. Due to the lack of protective histones and limited DNA repair (8), mtDNA mutations occur at high rates and were found in tumors including breast cancer (6, 9–14), which suggests that defects in OXPHOS might contribute to tumorigenesis.
By combining the nuclear genome of a recipient cell with the mitochondrial genome of a donor cell using cybrid technology, mitochondria from the triple-negative aggressive breast cancer cell lines MDA-MB-435 (15) and MDA-MB-231 facilitated tumor progression and metastasis in nonmetastatic tumor cells (7, 10). The donor cell lines harbor mtDNA mutations in tRNAs, in the noncoding D-loop region (9, 10), and in mitochondrial complex I subunit genes (10). These defects suggest a role of mtDNA mutations and complex I in tumor progression. Therefore, these cell lines are excellent models for defining a specific role of complex I activity in tumor growth and metastatic aggressiveness.
Complex I is the gatekeeper of the respiratory chain and catalyzes the first step of NADH oxidation. It elevates the NAD+/NADH ratio and translocates protons across the inner mitochondrial membrane, which ultimately leads to energy production. mtDNA mutations in genes encoding complex I subunits are found in malignancies including breast cancer (6, 11–14, 16). However, it is largely unknown how alterations in complex I and the cellular NAD+/NADH redox balance affect tumorigenesis and metastasis.
We used a unique approach to define contributions of complex I activity to breast cancer progression, based on expression of the yeast NADH dehydrogenase Ndi1 in human tumor cells. Ndi1 encodes a single protein that faces the inner mitochondrial matrix and oxidizes NADH from the Krebs cycle. Unlike mammalian complex I, Ndi1 is rotenone insensitive (17). Ndi1 contains 26 N-terminal residues for mitochondrial import (17), can be functionally expressed in mammalian cells (18, 19), and does not cause an immune response (20). Ndi1 restores complex I function (18) in diseased cells, e.g., in neurons of Parkinson’s disease (21) and optic neuropathy (22); protects cardiomyocytes from ischemic reperfusion injury (23); and increases lifespan in Drosophila (24). Recently, it was shown that Ndi1 expression in complex I–deficient tumor cells can reduce soft agar colony formation (25).
We used Ndi1 to investigate a cause-and-effect relationship between aberrant mitochondrial complex I activity and malignant progression in breast cancer. Moreover, we analyzed metabolic alterations caused by mitochondrial complex I malfunction and translated the information gained into a novel therapeutic approach against breast cancer progression.
Results
Enhancement of complex I activity in human breast cancer cells inhibits tumor growth and metastasis.
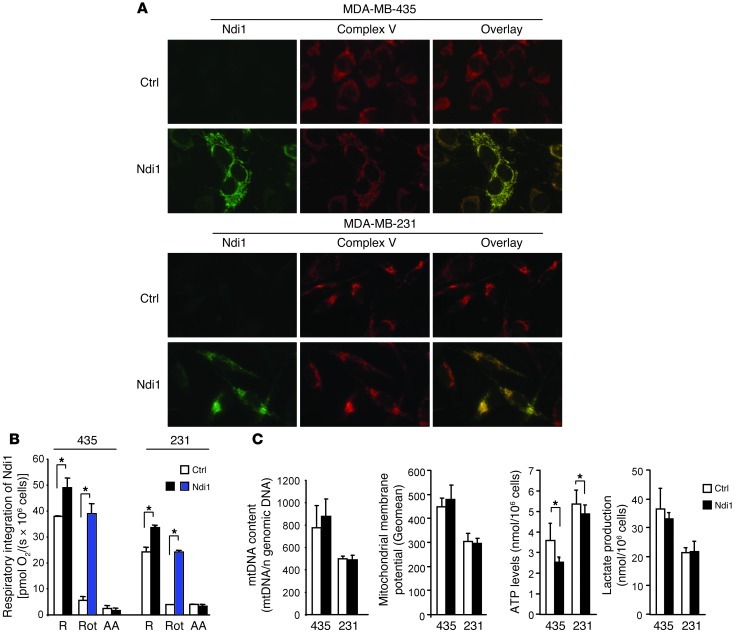
Complex I function in MDA-MB-435 and MDA-MB-231 cells was enhanced by stable transduction with Ndi1. The encoded enzyme was expressed by 90% of the cells, localized to mitochondria (Figure 1A) without altering the stoichiometry of mitochondrial complexes (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI64264DS1), and significantly enhanced mitochondrial respiration in the intact tumor cells (Figure 1B). Confirming the functional contribution of Ndi1, respiration in Ndi1-expressing MDA-MB-435 and MDA-MB-231 cells was inhibited by the mammalian complex I antagonist rotenone by only 20% and 28%, respectively, in contrast to 85% and 84% in controls. In all cases, oxygen consumption was fully blocked by the complex III inhibitor antimycin A (Figure 1B), which indicates that the measurements specifically reported mitochondrial respiration. Ndi1 also increased OXPHOS-uncoupled respiration, as shown by the oligomycin-inhibited respiratory rate (Supplemental Figure 2A), and enhanced complex I–mediated respiration in permeabilized cells upon addition of the complex I substrates malate and glutamate (Supplemental Figure 2, B and C). Ndi1 did not change the maximal capacity of the mitochondrial electron transfer system (Supplemental Figure 2A). Resistance to rotenone and susceptibility to flavone affirmed the contribution of Ndi1 (Supplemental Figure 2, B and C). In both tumor cell models, Ndi1 did not alter mtDNA content or mitochondrial membrane potential (Figure 1C). Reflecting overall energy metabolism and balance between glycolysis and mitochondrial OXPHOS, Ndi1 expression decreased ATP levels, whereas lactate production was unchanged in both breast cancer cell models (Figure 1C). These results demonstrated full functional integration of Ndi1 NADH dehydrogenase into the respiratory chain of 2 human cancer cell models, causing enhanced mitochondrial complex I activity without a major effect on overall energy balance.
Figure 1. Enhancement of mitochondrial complex I activity by integration of Ndi1: metabolic characterization.
(A) Ndi1 expressed in MDA-MB-435 and MDA-MB-231 human cancer cells upon lentiviral transduction localized to mitochondria, as shown by dual label immunocytochemistry. Control cells (Ctrl) were transduced with empty vector. Shown are Ndi1 (green), mitochondrial complex V (red), and merged (yellow) staining patterns of the same representative fields. Original magnification, ×20. (B) Ndi1 was integrated into the mitochondrial respiratory chain and enhanced tumor cell respiration. Respiration of MDA-MB-435 and MDA-MB-231 (denoted “435” and “231,” respectively) control and Ndi1-expressing cells, measured by oxygen consumption. R, routine respiration; Rot, rotenone (mammalian complex I inhibitor); AA, antimycin A (complex III inhibitor). All parameters were measured by high-resolution respirometry in intact cells. (C) Effects of Ndi1 expression on cellular metabolism in MDA-MB-435 and MDA-MB-231 cells. mtDNA content was analyzed by quantitative real-time PCR and referenced to nuclear genomic DNA. Mitochondrial membrane potential was analyzed by flow cytometry and expressed as geometric mean of the signal. ATP levels were measured by ATP-dependent luciferase activity. Lactate production was measured by fluorometry. Data are mean ± SEM (n = 3). *P < 0.05, unpaired 2-tailed Student’s t test.
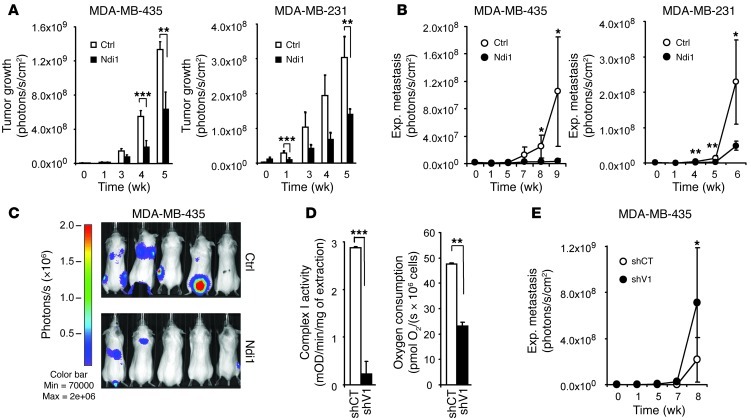
To investigate whether enhancement of tumor cell complex I activity affects tumorigenicity and metastasis, Ndi1-expressing F-luc–tagged MDA-MB-435 and MDA-MB-231 cells were followed in female SCID mice by noninvasive bioluminescence imaging. Ndi1 expression strikingly affected the malignant phenotype of the tumor cells, as tumor growth rates in the mammary fat pad and metastasis from the bloodstream were significantly reduced (Figure 2, A–C).
Figure 2. Mitochondrial complex I activity modulates tumor growth and metastasis.
(A) Ndi1 expression inhibited mammary fat pad tumor growth of MDA-MB-435 and MDA-MB-231 cells. Control cells were transduced with empty vector (n = 6). (B) Ndi1 expression inhibited lung colonization (experimental metastasis) by MDA-MB-435 or MDA-MB-231 cells after i.v. injection. Control cells were transduced with empty vector (n = 6). (C) Ndi1 expression inhibited multiorgan experimental metastasis, as indicated by noninvasive bioluminescence imaging 7 weeks after i.v. injection of 2.5 × 105 MDA-MB-435 control or Ndi1-expressing cells (n = 5). (D) Knockdown of complex I subunit NFUFV1 expression inhibited complex I activity and respiratory capacity in MDA-MB-435 cells. NDUFV1-knockdown (shV1) and control (shCT) cells were compared. Complex I was immunocaptured from cell lysates, analyzed based on oxidation of NADH to NAD+, expressed as mean OD/min/mg protein (n = 3). Routine mitochondrial respiration, corrected for residual oxygen consumption due to oxidative side reactions, was measured in intact MDA-MB-435 control and NDUFV1-knockdown cells by high-resolution respirometry (n = 3). (E) NDUFV1 knockdown increased lung colonization activity in MDA-MB-435 cells. NDUFV1-knockdown and control cells were compared (n = 8). Data are mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, nonparametric Mann-Whitney test (A, B, and E) or unpaired 2-tailed Student’s t test (D).
To corroborate a role of aberrant complex I activity in metastatic progression, we challenged our findings with an independent reverse approach, asking whether interference with tumor cell complex I function enhances metastatic aggressiveness. To this end, we knocked down NDUFV1 in MDA-MB-435 cells. NDUFV1 encodes the 51-kDa complex I subunit, which belongs to the minimal assembly required for catalysis and carries the NADH binding site (26). Mutations of this gene cause complex I deficiency in severe neurological diseases such as Leigh syndrome (26). NDUFV1 knockdown (Supplemental Figure 3) reduced complex I activity by 92% and cellular respiration by 51% and significantly enhanced the metastatic activity of the already aggressive MDA-MB-435 cell line (Figure 2, D and E).
These results provided definitive evidence that specific modulation of tumor cell complex I function can significantly alter tumor growth and metastatic activity. Therefore, complex I mutations found in primary tumors of breast cancer patients (6, 11–14, 16) may play a key role in disease progression.
Inhibition of metastatic activity by enhancement of complex I function depends on autophagy.
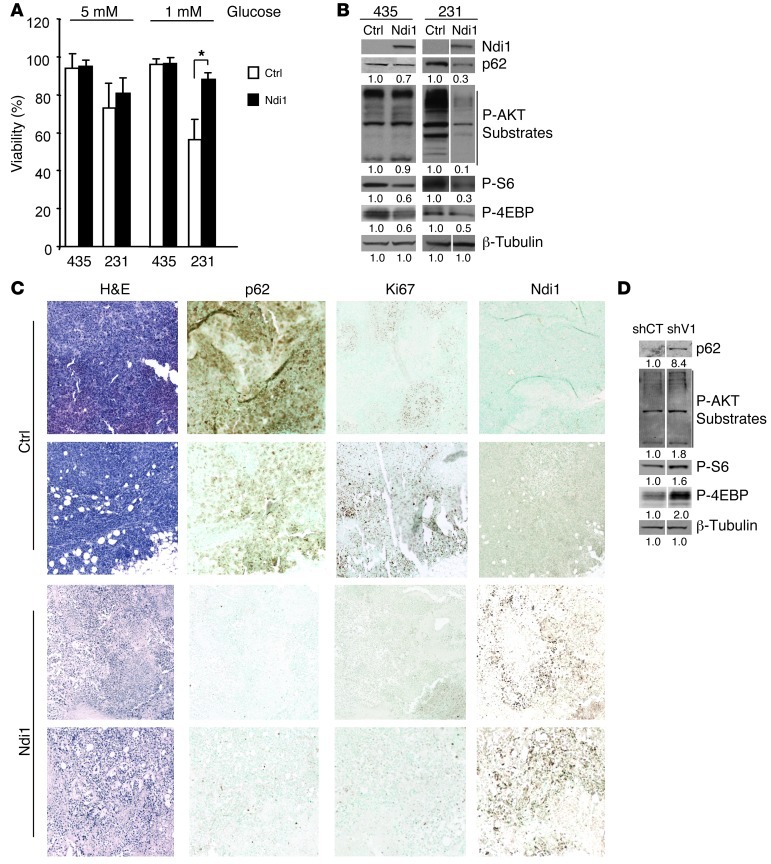
To investigate how complex I activity affects tumor growth and metastasis, we first analyzed effects of Ndi1 expression on tumor cell viability and growth. While Ndi1 did not affect proliferation in vitro (Supplemental Figure 4, A and B), Ndi1 increased resistance to glucose deprivation in MDA-MB-231 cells (Figure 3A) and suppressed tumorigenicity in both cell lines (Figure 2A). A similar paradoxical phenomenon was previously described and linked to the ability of tumor cells to undergo autophagy (27–29), without indication at the time that mitochondrial complex I might be involved. Following the hypothesis that breast cancer cell complex I activity can affect autophagy, we analyzed the effects of Ndi1 expression on p62, a ubiquitin-binding scaffolding protein that is eliminated during autophagy (27, 30). Ndi1 expression increased p62 degradation (Figure 3B), which indicated that enhancement of mitochondrial complex I activity induced autophagy. Following signaling pathways that regulate autophagy, we analyzed effects of Ndi1 on mTORC1 activity and AKT, one of mTOR’s main regulators. mTORC1 activity was measured by phosphorylation of mTORC1-related substrates S6 and 4EBP1; AKT activity was based on levels of phospho-AKT substrates. Ndi1 clearly reduced AKT activity in MDA-MB-231 cells and inhibited mTORC1 in both cell lines (Figure 3B), which suggested that complex I activity can induce autophagy by regulating mTORC1 signaling. This process likely involves AKT in MDA-MB-231 cells and alternative mechanisms in MDA-MB-435 cells.
Figure 3. Mitochondrial complex I activity regulates mTORC1 and autophagy.
(A) Ndi1 expression enhanced resistance to glucose deprivation in MDA-MB-231 cells, shown after 72 hours of incubation in medium with 5 versus 1 mM glucose. Viability was measured by flow cytometry (non–sub-G0/G1 population). n = 3 independent analyses. *P < 0.05, unpaired 2-tailed Student’s t test. (B) Ndi1 expression influenced mTORC1 activity and autophagy. Western blot analysis for p62, phospho-AKT substrates, and the mTORC1 kinase–related substrates phospho-S6Ser240/244 and phospho-4EBPThr37/46 in MDA-MB-435 or MDA-MB-231 control and Ndi1-expressing cells. β-Tubulin was used as protein loading control. Signal quantification, measured by infrared imaging (total of detectable bands) and expressed relative to control, is shown below. Results are representative of 5 independent experiments. Lanes were run on the same gel but were noncontiguous (white lines). (C) H&E staining and p62, Ki67, or Ndi1 expression in mammary fat pad tumors 5 weeks after implanting 2.5 × 105 MDA-MB-435 control versus Ndi1-expressing cells into SCID mice. 2 representative tumors of 6 are shown per group. Original magnification, ×10. (D) Inhibition of complex I activity through NDUFV1 knockdown affected mTORC1 activity and p62 elimination. Western blot analysis for p62, phospho-AKT substrates, phospho-S6Ser240/244, and phospho-4EBPThr37/46, comparing NDUFV1-knockdown versus control MDA-MB-435 cells. Signal quantification, measured by infrared imaging (total of detectable bands) and expressed relative to control, is shown below. Results are representative of 3 independent experiments. Lanes were run on the same gel but were noncontiguous (white lines).
Importantly, levels of p62 and Ki67 were strongly decreased in primary tumors expressing Ndi1 (Figure 3C), which indicates that enhancement of complex I increases tumor cell autophagy and inhibits proliferation in vivo. This conclusion was strongly supported by enhancement of AKT and mTORC1 activities and clear inhibition of p62 elimination by NDUFV1 knockdown, which reduced complex I functionality in the tumor cells (Figure 3D) without affecting their proliferation (Supplemental Figure 4C). These results demonstrated that tumor cell complex I activity can regulate tumor cell mTORC1 signaling and autophagy in vitro and in vivo.
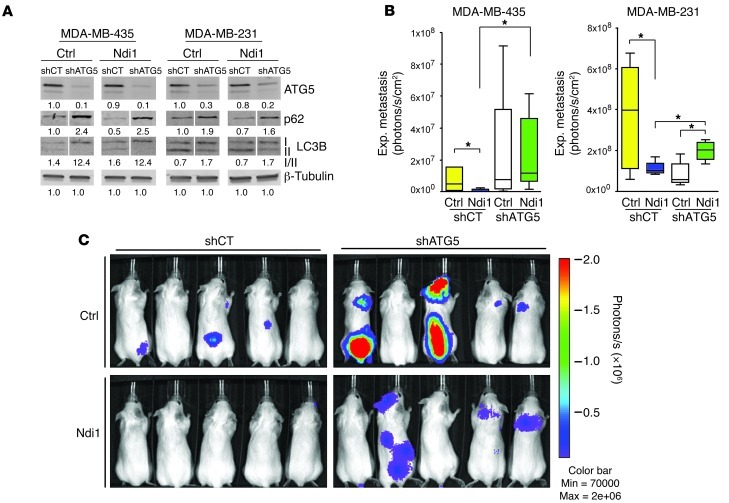
To analyze directly whether complex I–mediated regulation of breast cancer metastasis involves control of tumor cell autophagy, we knocked down ATG5, a protein required for autophagy induction (31). Targeting ATG5 in control and Ndi1-expressing MDA-MB-435 and MDA-MB-231 cells by stable transduction with shRNA (Figure 4A and Supplemental Figure 5A) inhibited autophagy, as shown by p62 and LC3BI accumulation (Figure 4A), without affecting proliferation in vitro (Supplemental Figure 5B). Importantly, although basal levels of autophagy facilitated metastasis in MDA-MB-231 cells, ATG5 knockdown abolished the inhibitory effect of Ndi1-mediated complex I enhancement on metastatic organ colonization (Figure 4, B and C). This was seen primarily in the lungs for MDA-MB-231 and in multiple organs for MDA-MB-435, including lung, liver, bone, brain, and adrenal glands. These findings affirm the conclusion that inhibition of metastasis through enhancement of mitochondrial complex I activity depends on autophagy induction in 2 aggressive tumor cell models.
Figure 4. Metastasis inhibition by enhanced complex I activity depends on autophagy.
(A) ATG5 knockdown (shATG5) inhibited autophagy in MDA-MB-435 and MDA-MB-231 control and Ndi1-expressing cells, as shown by p62 and LC3BI accumulation. Signal quantification of ATG5, p62 signal, and LC3BI/II ratios, measured by infrared imaging (total of detectable bands) and expressed relative to control, is shown below. β-Tubulin served as protein loading control. Lanes were run on the same gel but were noncontiguous (white lines). (B) ATG5 knockdown blocked the antimetastatic effect of Ndi1 in MDA-MB-435 and MDA-MB-231 cells. Lung colonization was measured by ex vivo lung imaging 7 weeks after i.v. injection of 2.5 × 105 tumor cells (n = 8 per group). Boxes denote interquartile range; lines within boxes denote median; whiskers denote minima and maxima. *P < 0.05, nonparametric Mann-Whitney test. (C) ATG5 knockdown enhanced multiorgan metastasis and reversed metastasis inhibition by Ndi1 in MDA-MB-435 cells. Shown is noninvasive bioluminescence imaging of 5 representative mice per group at 7 weeks after tail vein injection of 2.5 × 105 MDA-MB-435 control or Ndi1-expressing cells, with or without ATG5 knockdown.
NAD+ level modulation by complex I and alteration in NAD+ synthesis and recycling pathways regulate AKT/mTORC1 activity, autophagy, and metastatic activity.
To further investigate the mechanism by which enhancement of complex I activity inhibits tumor progression, we next ruled out alterations in ROS or NADPH production as major underlying causes. Ndi1 expression in MDA-MB-435 and MDA-MB-231 cells did not significantly alter ROS levels or NADPH-reducing equivalents (Supplemental Figure 6, A and B). Likewise, NDUFV1 knockdown did not significantly affect ROS (Supplemental Figure 6C), which suggests that enhancement of complex I activity by Ndi1 inhibits tumorigenicity and metastasis in a ROS-independent manner.
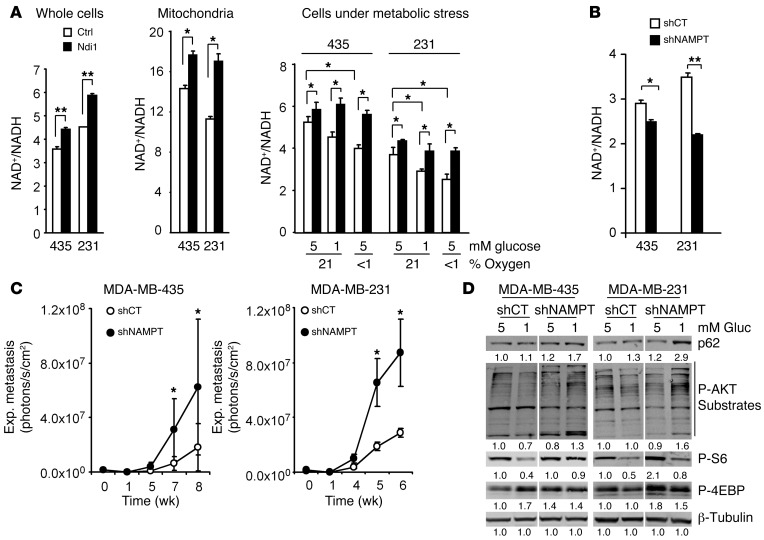
A major function of mammalian complex I and Ndi1 is NADH dehydrogenase activity. Ndi1 expression in MDA-MB-435 and MDA-MB-231 cells increased NAD+/NADH ratios in whole-cell extracts and purified mitochondria, particularly under metabolic stress induced by glucose or oxygen deprivation (Figure 5A). In culture, an Ndi1-mediated increase in NAD+/NADH ratios was measurable during exponential cell growth (Supplemental Figure 7, A and B). Conversely, disturbance of endogenous complex I activity via NDUFV1 knockdown reduced NAD+/NADH ratios (Supplemental Figure 8). We reasoned that modulation of the cellular redox potential (NAD+/NADH ratio) by mitochondrial complex I activity can regulate the metastatic activity of tumor cells. To test this hypothesis using an independent approach, we experimentally decreased NAD+/NADH ratios in MDA-MB-435 and MDA-MB-231 cells and analyzed the effect on metastasis in vivo.
Figure 5. NAD+ level modulation by complex I and NAD+ synthesis and recycling pathways regulate AKT/mTORC1 activity, autophagy, and metastasis.
(A) Ndi1 expression enhanced NAD+/NADH balance. NAD+/NADH ratios in whole-cell or mitochondrial extracts of MDA-MB-435 or MDA-MB-231 control versus Ndi1-expressing cells. Ndi1 stabilized NAD+/NADH ratios, especially under metabolic stress induced by glucose deprivation and hypoxia. NAD+/NADH ratios under stress were measured in whole-cell extracts after 48 hours of culture. (B) Interference with NAD+ synthesis and recycling pathways reduced NAD+/NADH ratios. Knockdown of NAMPT (shNAMPT) in MDA-MB-435 and MDA-MB-231 cells decreased NAD+/NADH ratios (whole-cell extracts after 48 hours growth in 5 mM glucose and normoxia). (C) NAMPT knockdown increased lung colonization activity in MDA-MB-435 and MDA-MB-231 cells (n = 6 per group). (D) NAMPT knockdown affected mTORC1 activity and p62 elimination. Western blot analysis for p62, phospho-AKT substrates, phospho-S6Ser240/244, and phospho-4EBPThr37/46 in MDA-MB-435 and MDA-MB-231 NAMPT-knockdown versus control cells. β-Tubulin served as protein loading control. Signal quantification, measured by infrared imaging (total of detectable bands) and expressed relative to control, is shown below. Lanes were run on the same gel but were noncontiguous (white lines). Results are representative of 3 independent experiments. (A–C) Data are mean ± SEM. *P < 0.05, **P < 0.01, unpaired 2-tailed Student’s t test (A and B) or nonparametric Mann-Whitney test (C).
To decrease the NAD+/NADH ratio in the tumor cells, we disturbed the NAD+ synthesis and recycling pathway by targeting nicotinamide phosphoribosyltransferase (NAMPT). NAMPT is essential for utilization and recycling of nicotinamide (NAM) and for biosynthesis of NAD+ (32). Interference with NAMPT expression through stable transduction with shRNA (Supplemental Figure 9A) reduced cellular NAD+/NADH ratios (Figure 5B), independently of the growth state of the tumor cells (Supplemental Figure 10, A and B) and without affecting cell proliferation in vitro (Supplemental Figure 9B). In vivo, however, NAMPT knockdown significantly enhanced metastasis in MDA-MB-435 and MDA-MB-231 cells (Figure 5C), indicative of a cause-and-effect relationship between reduced NAD+/NADH ratios and metastatic activity. The increased metastatic activity due to interference with cellular NAD+/NADH ratios involved altered mTORC1 activity and autophagy. NAMPT-knockdown cells were analyzed under normal conditions and glucose deprivation. Under metabolic stress due to glucose deprivation (1 mM glucose), NAMPT knockdown induced p62 accumulation and enhanced AKT and mTORC1 activities (Figure 5D), which indicated that NAD+ levels can regulate AKT/mTORC1 signaling and autophagy pathways in MDA-MB-435 and MDA-MB-231 cells.
Although our results showed that downregulation of NAMPT expression enhanced metastatic activity, chemical NAMPT inhibitors have previously been suggested for anticancer therapy (33). To investigate this potential discrepancy, we analyzed effects of FK866, a noncompetitive inhibitor of NAMPT (34), in vitro and in vivo. In vitro, FK866 decreased cellular NAD+/NADH ratios in MDA-MB-435 and MDA-MB-231 cells to levels similar to those seen after NAMPT knockdown (Supplemental Figure 11A), indicative of an effect of this drug on NAD metabolism. Treatment of NAMPT-knockdown cells with FK866 reduced NAD+/NADH levels further, likely due to a dose effect of NAMPT inhibition (Supplemental Figure 11A). In vivo, FK866 did reduce metastatic activity over a 42-day observation period (Supplemental Figure 11B). This in vivo effect might be related to cytotoxic properties of the drug. While genetic interference with tumor cell NAMPT expression did not affect cell growth or cause cell death in vitro (Supplemental Figure 9B), FK866 treatment induced S-G2/M cell cycle arrest and necrosis (Supplemental Figure 11, C and D). The cytotoxic effects of FK866 were enhanced in NAMPT-knockdown cells. These results indicate that this drug, originally identified as a cell proliferation inhibitor in a high-throughput screen, exerts effects on cell viability. Concentrations of FK866 that reduced NAD+/NADH ratios in the tumor cells as much as NAMPT knockdown did caused significant cell death in vitro, whereas genetic NAMPT interference did not. Thus, the cytotoxic effects of the drug may not be related to NAMPT inhibition alone.
NAD+ precursor treatment inhibits metastasis and spontaneous breast cancer progression.
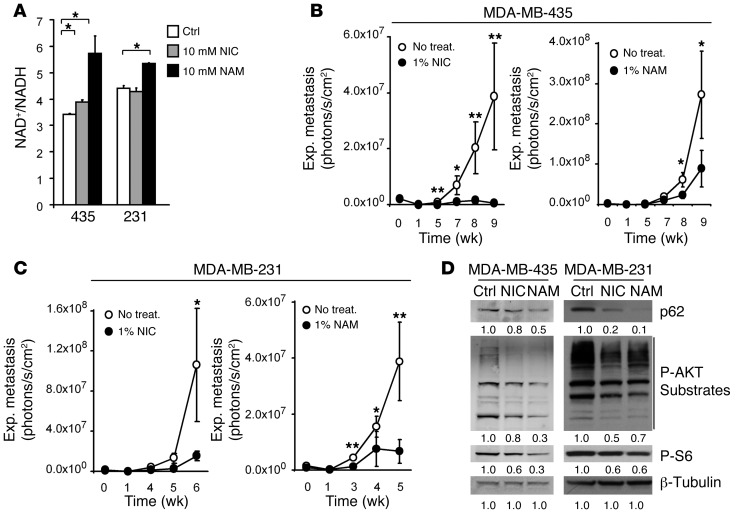
Having demonstrated that reduced but nonlethal NAD+/NADH ratios enhanced metastatic activity, we reasoned that increasing the NAD+/NADH balance might interfere with metastatic progression. To test this hypothesis, MDA-MB-435 and MDA-MB-231 cells were treated with the NAD+ precursors nicotinic acid (NIC) or NAM. NAM can induce stronger increases in NAD+ levels than NIC, because at high concentrations, NAM not only increases NAD+ synthesis, but can also inhibit NAD+ consumption by NAD+-dependent enzymes (33). In MDA-MB-231 and MDA-MB-435 cells, NAD+/NADH ratios were significantly enhanced by NIC or NAM (Figure 6A), independent of the proliferation state of the tumor cells at the time of treatment (Supplemental Figure 12, A and B). Most importantly, while not affecting cell growth in vitro (Supplemental Figure 13A), treatment of experimental animals with NAD+ precursors given in the drinking water strongly reduced lung metastasis of both cell lines after i.v. injection (Figure 6, B and C). Moreover, this treatment also clearly interfered with multiorgan metastasis of MDA-MB-435 cells, in which even brain lesions were notably reduced (Supplemental Figure 13B). This striking antimetastatic effect was confirmed in an independent cell model that preferably seeds brain metastases from the bloodstream. MDA-MB-453 cells, a HER2+ cell line derived from a pericardial effusion of a breast cancer patient with brain lesions (35), were injected into the left cardiac ventricle. Brain metastasis was significantly reduced in animals receiving NAM treatment (Supplemental Figure 14, A and B).
Figure 6. NAD+ precursor treatment inhibits metastatic activity.
(A) NAD+ precursor treatment enhanced the NAD+/NADH ratio in cultured MDA-MB-435 and MDA-MB-231 parental cells. NAD+/NADH levels were measured after 3 days of cell treatment with 10 mM NIC or NAM in complete medium. n = 3 independent experiments. (B and C) NAD+ precursor treatment of experimental mice inhibited lung metastasis. Lung colonization by MDA-MB-435 (B) or MDA-MB-231 (C) parental cells (2.5 × 105 i.v. each) in mice treated with NIC or NAM (1% in the drinking water ad libitum throughout the experiment). Controls received no treatment (plain drinking water at same pH). Metastatic growth was measured by repeated noninvasive bioluminescence imaging. n = 6 per group. (D) NIC or NAM treatment influenced mTORC1 activity and autophagy. Western blot analysis for p62, phospho-AKT substrates, and phospho-S6Ser240/244 in MDA-MB-435 or MDA-MB-231 parental cells with or without 48 hours of treatment with 10 mM NIC or NAM. β-Tubulin served as protein loading control. Signal quantification, measured by infrared imaging (total of detectable bands) and expressed relative to control, is shown below. Results are representative of 3 independent experiments. *P < 0.05, **P < 0.01, unpaired 2-tailed Student’s t test (A) or nonparametric Mann-Whitney test (B and C).
Next, we sought to analyze mechanisms involved in the antimetastatic activity of NAD+ precursor treatment. We found that NIC and NAM increased p62 degradation, indicative of autophagy induction, while decreasing AKT and mTORC1 activities in MDA-MB-435 and MDA-MB-231 cells (Figure 6D). Moreover, NAD+ modulation by NIC or NAM treatment significantly reduced mtDNA content, indicative of mitophagy, after a 3-day exposure without changing mitochondrial distribution within the tumor cells (Supplemental Figure 15). These results are consistent with the observed regulation of AKT/mTORC1 signaling and autophagy through enhancement of NAD+/NADH ratios in MDA-MB-435 and MDA-MB-231 cells after expression of Ndi1 to enhance mitochondrial complex I activity (Figure 3).
It has been reported that SIRT1, a NAD+-dependent deacetylase (36), can sense increased NAD+/NADH levels induced by NAM to inhibit mTORC1 and activate autophagy (37). We therefore investigated the possible involvement of SIRT1 in NAM treatment–induced autophagy in our models. We targeted SIRT1 expression in MDA-MB-435, MDA-MB-231, and MDA-MB-453 cells by stable transduction with shRNA (Supplemental Figure 16A). In the Her2+ MDA-MB-453 cell line, SIRT1 knockdown blocked NAM-mediated inhibition of mTORC1 activity and induction of autophagy. However, this was not seen in the triple-negative MDA-MB-435 and MDA-MB-231 cell lines (Supplemental Figure 16B). Thus, while NAM treatment modulated mTORC1 and autophagy in all tested cell lines, SIRT1 involvement was clearly evident in some cases, but not in others, for which additional or alternative mechanisms may be required.
Together with our results for Ndi1 expression and NAMPT knockdown, our findings demonstrated that specific modulation of NAD+/NADH ratios, either by altering mitochondrial complex I activity, by affecting NAD+ synthesis and recovery pathways, or by treating cells and experimental animals with NAD+ precursors, can markedly change metastatic activity and outcomes in vivo.
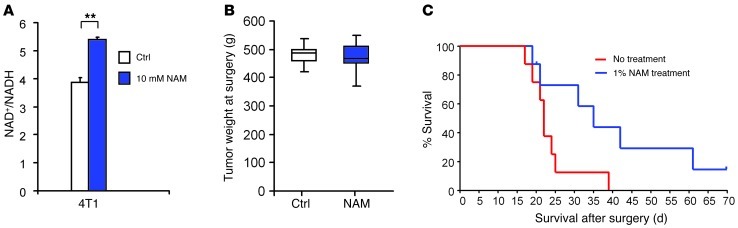
To challenge our concept that NAD+ precursor treatment can interfere with tumor growth and metastatic activity, we asked whether this therapeutic approach might inhibit spontaneous metastasis and prolong animal survival after surgical removal of a primary breast tumor. To test this hypothesis, we used the aggressive metastatic, triple-negative breast cancer cell model 4T1. In vitro, addition of NAM significantly enhanced NAD+/NADH ratios in 4T1 cells (Figure 7A), indicative of a response to NAD+ precursor treatment. To analyze therapeutic effects of NAM treatment in the animal model, F-luc–tagged 4T1 cells were injected in the fourth mammary fat pad of immunocompetent BALB/c mice, and tumors were removed surgically when they reached a volume of 300 mm3. At the time of surgical removal, mice were randomized into control (untreated water) and NAD+ precursor treatment (1% NAM in drinking water for the remainder of the experiment) groups (n = 8 each), and tumor weights were measured (Figure 7B). At the time of surgery and before NAM treatment, 1 animal in each group had spontaneous metastasis in the lungs, as measured by noninvasive bioluminescence imaging. At 1 week after tumor removal, 7 of 8 control mice and 5 of 8 NAM-treated mice had detectable metastases. After 2 weeks, 8 of 8 controls and 6 of 8 NAM-treated mice showed metastasis. At the end of the experiment (day 70 after surgery), 1 animal in the treatment group still had no detectable recurrence. Importantly, NAM treatment starting after primary tumor surgery significantly increased animal survival in this model of highly aggressive metastatic breast cancer (Figure 7C).
Figure 7. NAD+ precursor treatment after primary breast tumor removal increases animal survival.
(A) NAM treatment enhanced the NAD+/NADH ratio in cultured 4T1 murine breast carcinoma cells. NAD+/NADH levels were measured after 2 days of cell treatment with 10 mM NAM in complete medium. **P < 0.01, unpaired 2-tailed Student’s t test. n = 2 independent experiments. (B) Weight of 4T1 mammary fat pad tumors from untreated BALB/c mice. Tumors were surgically removed when their volume reached 300 mm3. Data show tumor weight distribution at time of surgery and randomization into groups (n = 8), before treatment was initiated. Boxes denote interquartile range; lines within boxes denote median; whiskers denote minima and maxima. P = 0.7480, unpaired 2-tailed Student’s t test. (C) Kaplan-Meier curves comparing survival of NAM-treated and untreated BALB/c mice after surgical removal of 4T1 mammary fat pad tumors. Mice were untreated or treated with 1% NAM in the drinking water after tumor removal (assigned as day 0). n = 8 per group. P = 0.0386, log-rank test.
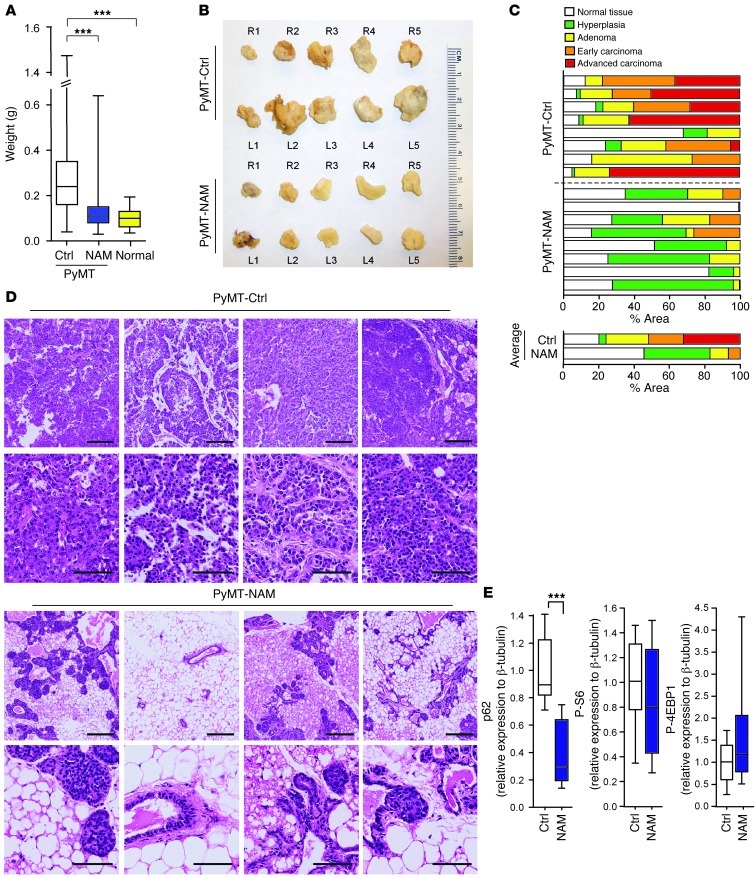
We then asked whether this therapeutic approach might generally interfere with breast cancer progression. To test this hypothesis, we used the MMTV-PyMT mouse model, in which polyoma middle T (PyMT) oncogene expression in the mammary epithelium leads to multifocal mammary tumor formation, detectable at 30 days of age, and progression to lung metastasis, detectable at 80 days, in the MT634 strain (38). The MMTV-PyMT model emulates important aspects of human breast cancer progression and replicates distinct advancing stages of breast cancer pathology, making it highly useful for preclinical therapeutic trials (39). MT634 strain MMTV-PyMT mice received 1% NAM in the drinking water beginning at weaning (day 22) and continuing throughout the experiment. Controls received water without NAM. Tumor mass, histological grade, and lung metastasis were examined on day 80. Importantly, NAD+ precursor treatment strongly interfered with oncogene-driven breast cancer progression in the MMTV-PyMT mouse model (Figure 8). Mammary fat pad tumor mass was significantly reduced in NAM-treated mice. In many cases, mammary fat pads of treated mice were normal or had minimal tumor involvement (Figure 8, A and B, and Supplemental Figure 17). Weight analysis of each individual fat pad in every mouse showed that 6.8 ± 0.6 of 10 fat pads per mouse in the control group (n = 11) were larger than the largest fat pad in PyMT-negative mice of the same strain and age. This was true for only 0.6 ± 0.4 of 10 fat pads per mouse in the NAM treatment group (n = 10; Supplemental Figure 17). The reported weights include all fat pads of each mouse, regardless of tumor presence. Importantly, histopathological analysis revealed that NAM treatment drastically inhibited breast cancer progression (Figure 8, C and D, and Supplemental Figure 18). All untreated MMTV-PyMT mice had tumors primarily containing adenoma, early and advanced carcinoma, and little normal and hyperplastic tissue. In contrast, NAM-treated mice had normal mammary fat pads; tumors, if found, were small and consisted predominantly of hyperplastic and adenoma tissue. Furthermore, whereas lung metastases were detected in 4 of 7 untreated animals, no metastases were found in NAM-treated MMTV-PyMT mice (0 of 6), as assessed by comprehensive histology in serial lung sections (data not shown). Importantly, NAM treatment induced autophagy in PyMT mammary tumors in vivo, as shown by p62 degradation in Western blot analyses of individual tumors (Figure 8E and Supplemental Figure 19). We did not detect effects on mTORC1 activity regulation, which suggests that NAM treatment in the drinking water might transiently modulate mTORC1 activity within PyMT mammary tumors in vivo.
Figure 8. NAD+ precursor treatment inhibits spontaneous breast cancer progression in MMTV-PYMT mice.
(A) NAM treatment (1% in drinking water throughout experiment, beginning at weaning) reduced mammary tumor growth. Weights of all 10 mammary fat pads from each treated mouse (n = 10), untreated control mice (n = 11), and untreated age- and strain-matched PyMT-negative mice (Normal; n = 3). Boxes denote interquartile range; lines within boxes denote median; whiskers denote minima and maxima. ***P < 0.001, nonparametric Mann-Whitney test. (B) Fat pads of untreated versus NAM-treated PyMT mice, representative of the 10 fat pad locations. (C) NAM treatment inhibited PyMT-induced breast cancer progression. Percent area of morphological stages of 8 representative fat pads from untreated (PyMT-Ctrl) or NAM-treated (PyMT-NAM) mice. Averages from each group are shown below. Scoring of hyperplasia, adenoma, and early and advanced carcinoma was performed on whole-slide scans of H&E-stained tumor sections by morphometric measurements. (D) Representative microscopic fields of 2 H&E-stained sections from 4 tumors of untreated and NAM-treated PyMT mice. Scale bars: 200 μm (top row for each treatment group); 50 μm (bottom row). (E) Quantification of Western blot analyses of mammary tumors from control and NAM-treated PyMT mice (n = 8 tumors per group). Shown is relative protein abundance of p62, phospho-S6Ser240/244, and phospho-4EBPThr37/46. Boxes denote interquartile range; lines within boxes denote median; whiskers denote minima and maxima. ***P < 0.001, unpaired 2-tailed Student’s t test.
To challenge the concept that NAD+ precursor treatment can interfere with breast cancer progression, we analyzed effects of NAM treatment in MMTV-PyMT mice that already had established spontaneous mammary tumors. Mice received 1% NAM in the drinking water, starting on day 60 when multiple palpable tumors where present in each animal. Controls received water without NAM. Tumor masses were examined on day 80. Importantly, NAD+ precursor treatment still significantly inhibited growth of already established mammary tumors, even after delayed onset of therapy (Supplemental Figure 20).
Together, these findings demonstrated that NAD+ precursor treatment can induce autophagy in vivo and effectively interfere with breast cancer progression at all stages of oncogene-driven breast cancer development.
Discussion
Our present findings establish that the tumor cell NAD+/NADH balance controlled by mitochondrial complex I can regulate breast cancer progression. We found that enhancement of complex I activity through expression of NADH dehydrogenase Ndi1 from Saccharomyces cerevisiae strongly interfered with tumor growth and metastasis, while inhibition of complex I by knockdown of the subunit NDUFV1 enhanced the metastatic potential of already aggressive breast cancer cells. Mutations in complex I were reported in human breast tumors (6, 9–12). mtDNA mutations per se are not known to cause transformation (40), but may contribute to cancer progression, as seen in lung carcinoma with EGFR mutations (41) and in oncocytic tumors with loss of complex I function (14, 42). Furthermore, oncogene activity (e.g., K-Ras transformation) can decrease mitochondrial complex I activity (43), which may support a malignant phenotype (44). Our results revealed a cause-and-effect relationship between complex I function and breast cancer progression.
Enhancement of complex I activity through Ndi1 did not alter tumor cell proliferation in vitro, but clearly reduced tumor growth and metastasis in vivo. Changing access to nutrients and oxygen within the tumor microenvironment influences cell survival and proliferation. Under those conditions, growth regulation through complex I may become critical. We found that this mechanism depended on autophagy. Enhancement of complex I through Ndi1 reduced metastasis and induced p62 elimination, whereas complex I inhibition by NDUFV1 knockdown enhanced metastasis and reduced p62 processing. The critical contribution of autophagy became clear upon ATG5 knockdown, which abrogated the antimetastatic effect of Ndi1 in both breast cancer cell models. Consistent with previous reports (45), we found that basal levels of autophagy facilitated metastasis in MDA-MB-231 cells; this was not seen in MDA-MB-435 cells. Autophagy can inhibit or promote tumorigenesis by supporting tumor cell survival under metabolic stress (27–29). Our results support a dual role of autophagy, while clearly showing that autophagy was required for mitochondrial complex I–mediated reduction of metastatic growth. One of the main regulators of autophagy is mTORC1, which links tumor growth and metabolism (46). We found that mitochondrial complex I modulated mTORC1 and its upstream regulator, AKT, in breast cancer cells, suggesting mTORC1 as a likely pathway through which complex I regulates autophagy.
Our results demonstrated that mitochondrial complex I can control breast cancer progression by regulating the cellular NAD+/NADH balance. Ndi1 expression enhanced this redox balance, particularly under metabolic stress, which indicates its involvement in strong inhibition of tumorigenicity and metastasis through Ndi1. To consolidate a role of complex I controlled NAD+/NADH ratio modulation in breast cancer progression, we experimentally altered tumor cell NAD+/NADH ratios using 2 additional independent approaches. First, we interfered with NAD+ biosynthesis and recycling pathways by knockdown of NAMPT to reduce the NAD+/NADH balance. Second, we enhanced the NAD+/NADH balance by treatment with NAD+ precursors. Importantly, decreasing NAD+/NADH ratios by knockdown of NAMPT, a rate-limiting enzyme in the NAD+ synthesis pathway that converts NAM to NAD+, significantly enhanced metastatic activity in already aggressive human breast cancer cells. NAMPT knockdown enhanced AKT/mTORC1 activities and inhibited autophagy. In apparent opposition, a recent study reported that NAMPT inhibition reduced prostate cancer cell viability (47). NAMPT was therefore suggested as a therapeutic target for inhibition progression of breast cancer (48) and other malignancies (33). We found that the chemical NAMPT inhibitor FK866 induced cell death in vitro and had tumoricidal effects in vivo. In contrast, genetic interference with NAMPT expression in breast cancer cells did not inhibit tumor cell viability and, rather than killing the tumor cells, rendered them more aggressive and metastatic in the animal models. It is possible that FK866 induces cellular stress through alternative mechanisms in addition to depleting NAD+ levels by NAMPT inhibition.
Controversial effects have also been reported for ROS-inducing therapies. While some cancer treatments induce high ROS levels to kill tumor cells, nonlethal increase of ROS may facilitate tumor progression and metastasis (7, 49). A similar concept applies to our present findings, in which NDUFV1 knockdown caused complex I deficiency but did not drastically inhibit OXPHOS. While not affecting cell viability, NDUFV1 knockdown significantly enhanced metastatic activity. Thus, it should be noted that strategies aimed at killing tumor cells by interfering with mitochondrial functions or NAD+ synthesis could, if not effectively lethal, inadvertently produce even more aggressive tumor cell phenotypes. Thus, in the long run, approaches aimed at normalizing mitochondrial functions, particularly complex I activity and NAD+/NADH redox levels, could be therapeutically more effective and safer and would not interfere with normal cell function.
Having established that enhancement of NAD+/NADH levels by augmenting breast cancer cell complex I activity inhibited tumorigenicity and metastasis, we used this new concept therapeutically and hypothesized that supplementing tumor cell nutrients with NAD+ precursors, such as NIC or NAM, could interfere with breast cancer progression. NIC and NAM are used to modulate NAD+ levels and showed therapeutic effects in disorders, such as cerebral ischemia, diabetes, and cardiovascular dysfunctions (32, 33). NIC and NAM protect neurons against oxidative damage, whereas subclinical vitamin B3 deficiency is associated with genomic instability and increased cancer risk (50).
We demonstrated that enhancing NAD+ levels through NAD+ precursor treatment effectively inhibited experimental metastasis of human breast cancer cells in xenograft models. Importantly, this treatment also inhibited spontaneous metastasis and increased animal survival when the therapy was begun after surgical removal of primary tumors. Furthermore, NAD+ precursor treatment strongly interfered with oncogene-driven breast cancer development and progression in transgenic MMTV-PyMT mice.
Our results provide evidence that the mechanism involves induction of autophagy. NAD+ precursor treatment activated tumor cell autophagy in vitro as well as in vivo and included regulation of mTORC1 activity, as seen in culture. It has been reported that modulation of NAD+/NADH ratios can affect the PI3K/AKT survival pathway through inactivation of PTEN (51). Furthermore, NAD+-dependent sirtuins can modulate mTORC1 through regulation of TSC2 (52) or LKB1 activity (53, 54). Sirtuins may also affect autophagy by controlling FOXO3 degradation (55) or by directly regulating the autophagy proteins ATG5, ATG7, and ATG8 (56). We found that modulation of mTORC1 activity and autophagy by NAM involved SIRT1 activation in MDA-MB-453 cells, and likely depends on additional or alternative mechanisms in MDA-MB-435 and MDA-MB-231 cells. NAD+-dependent sirtuins and PARPs contribute to p53 function and regulate chromatin structure and genomic stability (32, 33, 50), thereby playing emerging roles in tumor progression (57). Moreover, the transcriptional corepressor CtBP responds to increased NADH under hypoxia to permit gene expression that promotes tumor cell migration (58). Importantly, BRCA1 tumor suppressor expression is inhibited by low NAD+/NADH ratios (59, 60).
The present study demonstrated that mitochondrial complex I regulation of tumor cell NAD+/NADH levels affects breast cancer growth and metastasis and translated into a new therapeutic approach for preventing breast cancer progression. This is highly relevant, as the current standard of care for cancer patients relies primarily on chemo- and radiation therapies aimed at killing the tumor cells. Evolutionary models predict that selective pressure imposed by these approaches causes survival of resistant clones that eventually reactivate the disease (61). Based on the central involvement of metabolic tumor cell alterations in cancer, therapeutic normalization of tumor cell metabolism might interfere with the expansion of residual and breakthrough clones. Thus, a combination of standard therapy with NAD+ precursor treatment may halt breast cancer progression and prevent relapse.
Methods
Cell culture.
MDA-MB-231, MDA-MB-435, MDA-MB-453, and 4T1 cells as well as their variants were stably transduced with Firefly luciferase (F-luc) using lentiviral expression vector cFUW (ubiquitin promoter; provided by B.E. Torbett, The Scripps Research Institute) to analyze tumor growth and metastasis by noninvasive bioluminescence imaging (62). Cells were grown in EMEM supplemented with nonessential amino acids, vitamins, 2 mM l-glutamine, 1 mM pyruvate, and 10% FBS (complete medium). For metabolic stress, cells were cultured in Neurobasal-A Medium (Gibco) supplemented with nonessential amino acids, vitamins, 2 mM l-glutamine, 5 mM or 1 mM glucose, and 2% dialyzed FBS in normoxia (21% oxygen) or hypoxia (<1% oxygen; Modular Incubator Chamber; Billups-Rothenberg).
Ndi1 expression.
Tumor cells were stably transduced with the Ndi1 gene from S. cerevisiae (19) subcloned into lentiviral expression vector cFUW. Transduced cells were used as pools in all experiments. Ndi1 expression was analyzed by Western analysis, and subcellular localization was analyzed by immunofluorescence, as described previously (18).
NDUFV1, NAMPT, and ATG5 knockdown.
Lentiviral vectors containing shRNA against NDUFV1 (clone TRCN0000025872), ATG5 (clone TRCN0000151963), NAMPT (clone TRCN0000116180), or nontargeting control shRNA (SHC0002) were from Sigma-Aldrich. Knockdown efficiency was quantified by real-time PCR using FastStart Universal SYBR Green Master (Rox) (Roche) and the following primers: ATG5 forward, TGGGATTGCAAAATGACAGA; ATG5 reverse, TTCCCCATCTTCAGGATCAA; NAMPT forward, GCCAGCAGGGAATTTTGTTA; NAMPT reverse, TGATGTGCTGCTTCCAGTTC; NDUFV1 forward, AAGGTTCCCTGAGTCGAGGT; NDUFV1 reverse, TTGTGAGGATCATGGCGTAA; GAPDH forward, GGGAAGGTGAAGGTCGGAGT; GAPDH reverse, TCCACTTTACCAGAGTTAAAAGCAG. Data were recorded and analyzed using an ABI-PRISM 7700 Sequence Detection System (Applied Biosystems) and Sequence Detector Software (version 2.0; SDS).
Animal experiments.
For experimental metastasis, female 6- to 8-week-old C.B-17/SCID mice were injected with F-luc–tagged cancer cells: 2.5 × 105 MDA-MB-231 or MDA-MB-435 cells i.v., or 5 × 105 MDA-MB-453 cells into the left cardiac ventricle under stereotactic guidance. For primary tumor growth, 1 × 106 MDA-MB-231 cells or 2.5 × 105 MDA-MB-435 cells were injected into the axillary mammary fat pad. For spontaneous metastasis, female 8-week-old BALB/c mice were injected with 1 × 105 F-luc–tagged 4T1 cells into the fourth mammary fat pad. Tumors were removed surgically when they reached 300 mm3 in size, and animals were euthanized when moribund to determine survival. Mice were imaged weekly (IVIS 200; Xenogen) 10 minutes after i.p. injection of d-luciferin (100 mg/kg). To assess brain metastasis from MDA-MB-453 cells, mice were imaged directly after cardiac injection to validate correct routing by verifying tumor cell signal in the brain region and lack of signal in the lungs. For ex vivo organ imaging at the end of experimental and spontaneous metastasis studies, mice were injected with luciferin 5 minutes before necropsy. For survival analyses, imaging also validated metastasis as the cause of moribundity. Bioluminescence was quantified as photons/s/cm2 in defined regions of interest using Living Image software. MMTV-PyMT mice (strain MT634; gift of W.J. Muller, McGill University, Montreal, Quebec, Canada; ref. 38) were heterozygous for the PyMT transgene and genotyped by PCR using forward primer 5′-CGGCGGAGCGAGGAACTGAGGAGAG-3′ and reverse primer 5′-TCAGAAGACTCGGCAGTCTTAGGCG-3′. For NAD+ precursor treatment, mice received 1% NIC or NAM in the drinking water, which was changed weekly. For histology, mammary fat pads and lungs were zinc fixed, sectioned serially (5 μm), and H&E stained. In the PyMT model, scoring of 4 morphological stages (hyperplasia, adenoma, early carcinoma, and advanced carcinoma; ref. 39) was performed on whole-slide scans (Leica SCN400 Digital Slide Scanner) by image analysis and morphometric measurements (Metamorph imaging software; version 7.6). Select regions were imaged using a Zeiss Axio Imager M1m microscope and AxioVision software. Lung metastases in MMTV-PyMT mice were quantified on 10 H&E-stained step sections per lung (5 μm thick, 80 μm apart).
Ndi1 integration into cellular respiration.
2 × 106 cells were collected into complete medium, and respiration was measured at 37°C in a 2-ml chamber by high-resolution respirometry using an Oroboros Oxygraph series D and DatLab software (Oroboros Instruments). Routine mitochondrial respiration, corrected for residual oxygen consumption due to oxidative side reactions, was measured in intact cells in complete medium. Functional integration of Ndi1 into tumor cell mitochondrial respiration was determined by oxygen consumption after addition of rotenone (2.5 μM) to inhibit endogenous complex I and final addition of complex III inhibitor antimycin A (2.5 μM) to terminate mitochondrial respiration.
Enzymatic analysis of mitochondrial complex I activity.
NADH oxidation to NAD+ by complex I was analyzed using an immunocapture complex I enzyme activity assay (MS-141; MitoSciences).
NAD+/NADH analysis in whole-cell and mitochondrial extracts.
NAD+ and NADH were analyzed independently in extracts of whole cells (1 × 106) or isolated mitochondria (from 1 × 107 cells) prepared as described previously (18, 63). Concentrations were determined using a NAD/NADH fluorescence detection kit (Cell Technology Inc.).
mtDNA. mtDNA content was determined by quantitative real-time PCR of total DNA extracted from 2 × 106 cells.
PCR reactions were performed in quadruplicates using TaqMan probes specific for MT-RNR1 (mitochondrial S12 RNA) (Hs02596859_g1, FAM label) referenced to RPPH1 (RNase P, VIC label), a nuclear gene. PCR data were recorded and analyzed using the ABI-PRISM 7700 Sequence Detection System (Applied Biosystems) and Sequence Detector Software (version 2.0; SDS).
ATP measurements.
ATP was extracted from cell pellets (2 × 106 cells) in triplicates with 100 μl of ice-cold 2M perchloric acid. Extracts were left on ice for 15 minutes, mixed, and then centrifuged at 12,000 g for 5 minutes at 4°C. The supernatant was neutralized with 35 μl of 1M Bicine and 4M potassium carbonate to pH 7.5 and kept on ice for 15 minutes. After centrifugation, ATP concentrations were measured in the supernatant based on ATP-dependent luciferase activity using an ATP determination kit (Invitrogen).
Lactate production.
Lactate concentrations were measured in triplicates in conditioned media after culturing 105 cells in 6-well plates (1.5 ml medium/well) for 48 hours. 500 μl medium was deproteinized with 166 μl ice-cold 2M perchloric acid, left on ice for 15 minutes, mixed, and centrifuged at 12,000 g for 10 minutes at 4°C. Supernatants were neutralized with 58 μl of 1M Bicine and 4M potassium carbonate to pH 7 and kept on ice for 20 minutes. After centrifugation, lactate concentrations in the supernatants were determined fluorimetrically at 340 nm excitation and 466 nm emission (460 nm) before and 60 minutes after addition of lactate dehydrogenase, as described previously (5).
Analysis of mitochondrial membrane potential and cell viability.
The membrane potential–sensitive fluorochrome tetramethylrhodamine methyl ester (TMRM; Invitrogen) was used to measure mitochondrial membrane potential. Cells were incubated with 150 nM TMRM for 30 minutes at 37°C in the dark and then analyzed by flow cytometry (FACSCalibur). Data from 10,000 events per sample were collected and analyzed by measuring FL-2 fluorescence using CellQuest software. Cell viability was analyzed by measuring the percentage of the nonhypodiploid population (non–sub-G0/G1) by flow cytometry. Cells were fixed in ice-cold 70% ethanol, washed with PBS, and incubated with 50 μg/ml propidium iodide and 100 μg/ml RNAse A for 30 minutes at 37°C. Data from 20,000 events per sample were collected by measuring FL-2 fluorescence.
Immunohistochemistry.
Tumor tissues were paraformaldehyde or zinc fixed, paraffin embedded, sectioned (5 μm), transferred onto glass slides, deparaffinized in safe clear II, and rehydrated in a series of ethanol solutions. p62, Ki67, or Ndi1 staining was performed after antigen retrieval with 1 mM EDTA (pH 8), 10 mM citrate buffer, or 1 mM EDTA plus 10 mM Tris-Cl (pH 8), respectively. Sections were washed 3 times in PBS, treated with 3% H2O2 in PBS (or in methanol for Ndi1) for 15 minutes, blocked in 10% goat serum and 0.3% Triton X-100 in PBS for 1 hour, and incubated with primary antibody against p62 (Santa Cruz Biotechnology Inc.), Ki67 (BD Biosciences — Pharmingen), or Ndi1 (18) overnight (or for 1 hour for p62), followed by incubation with secondary horseradish peroxidase–, alkaline phosphatase–, or biotin-conjugated Abs (Jackson ImmunoResearch Laboratories and Vector Laboratories) for 2 hours. Signal from biotinylated antibodies was amplified using an avidin-biotin complex kit (Vector Laboratories). Horseradish peroxidase was developed with DAB substrate (BD Biosciences — Pharmingen), and nuclei were stained with Contrast Green (Kirkegaard & Perry Laboratories). The slides were washed with isopropanol and briefly incubated in SafeClear II (Fisher Scientific) before mounting in Permount (Fisher Scientific). Images were acquired with a Zeiss Axio Imager M1m microscope equipped with a digital camera, using ×20 air objectives. Digital images were analyzed with AxioVision 4.6 software (Zeiss).
Western blot analysis.
Cells were lysed with Laemmli or RIPA extraction buffer. Western blots were incubated with antibodies against Ndi1 (20), p62 (Santa Cruz Biotechnology), phospho-AKT substrates (Ser/Thr), phopho-S6Ser240/244, phospho-4E-BP1Thr37/46, LC3B (Cell Signaling Technology), ATG5 (Cell Signaling Technology), β-tubulin (Sigma-Aldrich), NDUFA9 (complex I), SDHA (complex II), UQCR2 (complex III), COXI (complex IV), ATP5A1 (complex V), or mitochondrial porin (VDAC1) (Mitosciencies). Antibody binding was detected after incubation with secondary antibodies conjugated to IRDye 800, using an Odyssey infrared imaging system (LI-COR Biosciences). Data were analyzed and quantified using Odyssey infrared imaging system application software (version 3.0).
Statistics.
Statistical comparisons between 2 groups were performed using unpaired 2-tailed Student’s t tests with unequal variance for in vitro results; nonparametric Mann-Whitney tests for in vivo results, since normal distribution could not be assumed; and Kaplan-Meier curves and log-rank test for animal survival. Statistical calculations were performed with GraphPad Prism software. A P value less than 0.05 was considered significant. Results are shown as mean ± SEM of values obtained in independent experiments.
Study approval.
Animal studies were approved by the Institutional Animal Care and Use Committee of The Scripps Research Institute. Animal work complied with NIH and institutional guidelines (The Scripps Research Institute is AAALAC accredited).
Supplementary Material
Acknowledgments
This study was supported by NIH grants R01CA112287, R01CA170737, and R01CA170140 (to B. Felding-Habermann), UL1RR025774 (to Eric J. Topol, Pilot award to B. Felding-Habermann), and R01DK053244 (to T. Yagi); CDMRP DoD grant W81XWH-08-0468 (to B. Felding-Habermann); and California Breast Cancer Research Program grants 17NB-0058, 16IB-0052, 12NB-0176, and 13NB-0180 (to B. Felding-Habermann). A.F. Santidrian received a postdoctoral fellowship from the Susan G. Komen Foundation. S.E. LeBoeuf and L.J. Gay are supported by trainee scholarships from NIH grant 5TL1RR025772-03. We thank Bruce E. Torbet for the cFUW lentiviral construct; William J. Muller for MMTV-PyMT mice; Rolf Habermann for morphometric analyses; and Mihaela Lorger, Deirdre O’Sullivan, Karin Staflin, and Jane Forsyth for helpful discussions. We gratefully acknowledge a donation from Las Patronas (La Jolla, California, USA). This is manuscript 21583 of The Scripps Research Institute.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(3):1068–1081. doi:10.1172/JCI64264.
Byoung B. Seo’s present address is: Department of Animal Resources, College of Life and Environmental Science, Daegu University, Jillyang, Gyeongsan, Gyeongbuk, Republic of Korea.
References
- 1.Dawood S, Broglio K, Buzdar AU, Hortobagyi GN, Giordano SH. Prognosis of women with metastatic breast cancer by HER2 status and trastuzumab treatment: an institutional-based review. J Clin Oncol. 2010;28(1):92–98. doi: 10.1200/JCO.2008.19.9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polyak K. Heterogeneity in breast cancer. J Clin Invest. 2011;121(10):3786–3788. doi: 10.1172/JCI60534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124(3215):269–270. [PubMed] [Google Scholar]
- 4.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz JP, Passonneau JV, Johnson GS, Pastan I. The effect of growth conditions on NAD+ and NADH concentrations and the NAD+:NADH ratio in normal and transformed fibroblasts. . J Biol Chem. 1974;249(13):4138–4143. [PubMed] [Google Scholar]
- 6.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25(34):4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 7.Ishikawa K, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320(5876):661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 8.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulawiec M, Owens KM, Singh KK. Cancer cell mitochondria confer apoptosis resistance and promote metastasis. Cancer Biol Ther. 2009;8(14):1378–1385. doi: 10.1158/1535-7163.MCT-08-1170. [DOI] [PubMed] [Google Scholar]
- 10.Ma Y, Bai RK, Trieu R, Wong LJ. Mitochondrial dysfunction in human breast cancer cells and their transmitochondrial cybrids. Biochim Biophys Acta. 2010;1797(1):29–37. doi: 10.1016/j.bbabio.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen L, et al. Evaluating mitochondrial DNA in patients with breast cancer and benign breast disease. J Cancer Res Clin. Oncol. 2011;137(4):669–675. doi: 10.1007/s00432-010-0912-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carew JS, Huang P. Mitochondrial defects in cancer. Mol Cancer. 2002;1:9. doi: 10.1186/1476-4598-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parrella P, et al. Detection of mitochondrial DNA mutations in primary breast cancer and fine-needle aspirates. Cancer Res. 2001;61(20):7623–7626. [PubMed] [Google Scholar]
- 14.Mayr JA, et al. Loss of complex I due to mitochondrial DNA mutations in renal oncocytoma. Clin Cancer Res. 2008;14(8):2270–2275. doi: 10.1158/1078-0432.CCR-07-4131. [DOI] [PubMed] [Google Scholar]
- 15.Chambers AF. MDA-MB-435 and M14 cell lines: identical but not M14 melanoma? Cancer Res. 2009;69(13):5292–5293. doi: 10.1158/0008-5472.CAN-09-1528. [DOI] [PubMed] [Google Scholar]
- 16.Fendt L, et al. Accumulation of mutations over the entire mitochondrial genome of breast cancer cells obtained by tissue microdissection. Breast Cancer Res Treat. 2011;128(2):327–336. doi: 10.1007/s10549-010-1092-8. [DOI] [PubMed] [Google Scholar]
- 17.De VS, Van WR, Grivell LA, Marres CA. Primary structure and import pathway of the rotenone-insensitive NADH-ubiquinone oxidoreductase of mitochondria from Saccharomyces cerevisiae. Eur J Biochem. 1992;203(3):587–592. doi: 10.1111/j.1432-1033.1992.tb16587.x. [DOI] [PubMed] [Google Scholar]
- 18.Seo BB, Kitajima-Ihara T, Chan EK, Scheffler IE, Matsuno-Yagi A, Yagi T. Molecular remedy of complex I defects: rotenone-insensitive internal NADH-quinone oxidoreductase of Saccharomyces cerevisiae mitochondria restores the NADH oxidase activity of complex I-deficient mammalian cells. Proc Natl Acad Sci U S A. 1998;95(16):9167–9171. doi: 10.1073/pnas.95.16.9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seo BB, Wang J, Flotte TR, Yagi T, Matsuno-Yagi A. Use of the NADH-quinone oxidoreductase (NDI1) gene of Saccharomyces cerevisiae as a possible cure for complex I defects in human cells. J Biol Chem. 2000;275(48):37774–37778. doi: 10.1074/jbc.M007033200. [DOI] [PubMed] [Google Scholar]
- 20.Marella M, Seo BB, Flotte TR, Matsuno-Yagi A, Yagi T. No immune responses by the expression of the yeast Ndi1 protein in rats. PLoS One. 2011;6(10):e25910. doi: 10.1371/journal.pone.0025910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barber-Singh J, Seo BB, Nakamaru-Ogiso E, Lau YS, Matsuno-Yagi A, Yagi T. Neuroprotective effect of long-term NDI1 gene expression in a chronic mouse model of Parkinson disorder. Rejuvenation Res. 2009;12(4):259–267. doi: 10.1089/rej.2009.0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marella M, Seo BB, Thomas BB, Matsuno-Yagi A, Yagi T. Successful amelioration of mitochondrial optic neuropathy using the yeast NDI1 gene in a rat animal model. PLoS One. 2010;5(7):e11472. doi: 10.1371/journal.pone.0011472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perry CN, Huang C, Liu W, Magee N, Carreira RS, Gottlieb RA. Xenotransplantation of mitochondrial electron transfer enzyme, Ndi1, in myocardial reperfusion injury. PLoS One. 2011;6(2):e16288. doi: 10.1371/journal.pone.0016288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanz A, et al. Expression of the yeast NADH dehydrogenase Ndi1 in Drosophila confers increased lifespan independently of dietary restriction. Proc Natl Acad Sci U S A. 2010;107(20):9105–9110. doi: 10.1073/pnas.0911539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma LK, Fang H, Liu J, Vartak R, Deng J, Bai Y. Mitochondrial respiratory complex I dysfunction promotes tumorigenesis through ROS alteration and AKT activation. Hum Mol Genet. 2011;20(23):4605–4616. doi: 10.1093/hmg/ddr395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pagniez-Mammeri H, Loublier S, Legrand A, Bénit P, Rustin P, Slama A. Mitochondrial complex I deficiency of nuclear origin I. Structural genes. Mol Genet Metab. 2012;105(2):163–172. doi: 10.1016/j.ymgme.2011.11.188. [DOI] [PubMed] [Google Scholar]
- 27.Mathew R, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137(6):1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathew R, White E. Autophagy in tumorigenesis and energy metabolism: friend by day, foe by night. Curr Opin Genet Dev. 2011;21(1):113–119. doi: 10.1016/j.gde.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenific CM, Thorburn A, Debnath J. Autophagy and metastasis: another double-edged sword. Curr Opin Cell Biol. 2010;22(2):241–245. doi: 10.1016/j.ceb.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4(2):151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290(5497):1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koch-Nolte F, Fischer S, Haag F, Ziegler M. Compartmentation of NAD+-dependent signalling. . FEBS Lett. 2011;585(11):1651–1656. doi: 10.1016/j.febslet.2011.03.045. [DOI] [PubMed] [Google Scholar]
- 33.Xu P, Sauve AA. Vitamin B3, the nicotinamide adenine dinucleotides and aging. Mech Ageing Dev. 2010;131(4):287–298. doi: 10.1016/j.mad.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 34.Hasmann M, Schemainda I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 2003;63(21):7436–7442. [PubMed] [Google Scholar]
- 35.Brinkley BR, Beall PT, Wible LJ, Mace ML, Turner DS, Cailleau RM. Variations in cell form and cytoskeleton in human breast carcinoma cells in vitro. Cancer Res. 1980;40(9):3118–3129. [PubMed] [Google Scholar]
- 36.Chalkiadaki A, Guarente L. Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat Rev Endocrinol. 2012;8(5):287–296. doi: 10.1038/nrendo.2011.225. [DOI] [PubMed] [Google Scholar]
- 37.Jang SY, Kang HT, Hwang ES. Nicotinamide-induced mitophagy: event mediated by high NAD+/NADH ratio and SIRT1 protein activation. . J Biol Chem. 2012;287(23):19304–19314. doi: 10.1074/jbc.M112.363747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12(3):954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin EY, Nguyen AV, Russell RG, Pollard JW. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J Exp Med. 2001;193(6):727–740. doi: 10.1084/jem.193.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yokota M, et al. Generation of trans-mitochondrial mito-mice by the introduction of a pathogenic G13997A mtDNA from highly metastatic lung carcinoma cells. FEBS Lett. 2010;584(18):3943–3948. doi: 10.1016/j.febslet.2010.07.048. [DOI] [PubMed] [Google Scholar]
- 41.Dasgupta S, et al. Mitochondrial DNA mutations in respiratory complex-I in never-smoker lung cancer patients contribute to lung cancer progression and associated with EGFR gene mutation. J Cell Physiol. 2012;227(6):2451–2460. doi: 10.1002/jcp.22980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimmermann FA, et al. Respiratory chain complex I is a mitochondrial tumor suppressor of oncocytic tumors. Front Biosci (Elite Ed). 2011;3:315–325. doi: 10.2741/e247. [DOI] [PubMed] [Google Scholar]
- 43.Baracca A, Chiaradonna F, Sgarbi G, Solaini G, Alberghina L, Lenaz G. Mitochondrial Complex I decrease is responsible for bioenergetic dysfunction in K-ras transformed cells. Biochim Biophys Acta. 2010;1797(2):314–323. doi: 10.1016/j.bbabio.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 44.Hu Y, et al. K-ras(G12V) transformation leads to mitochondrial dysfunction and a metabolic switch from oxidative phosphorylation to glycolysis. Cell Res. 2012;22(2):399–412. doi: 10.1038/cr.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tu YF, Kaipparettu BA, Ma Y, Wong LJ. Mitochondria of highly metastatic breast cancer cell line MDA-MB-231 exhibits increased autophagic properties. Biochim Biophys Acta. 2011;1807(9):1125–1132. doi: 10.1016/j.bbabio.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 46.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang B, Hasan MK, Alvarado E, Yuan H, Wu H, Chen WY. NAMPT overexpression in prostate cancer and its contribution to tumor cell survival and stress response. Oncogene. 2011;30(8):907–921. doi: 10.1038/onc.2010.468. [DOI] [PubMed] [Google Scholar]
- 48.Bajrami I, et al. Synthetic lethality of PARP and NAMPT inhibition in triple-negative breast cancer cells. EMBO Mol Med. 2012;4(10):1087–1096. doi: 10.1002/emmm.201201250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schumacker PT. Reactive oxygen species in cancer cells: live by the sword, die by the sword. Cancer Cell. 2006;10(3):175–176. doi: 10.1016/j.ccr.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 50.Kirkland JB. Niacin requirements for genomic stability. Mutat Res. 2012;733(1–2):14–20. doi: 10.1016/j.mrfmmm.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 51.Pelicano H, et al. Mitochondrial respiration defects in cancer cells cause activation of Akt survival pathway through a redox-mediated mechanism. J Cell Biol. 2006;175(6):913–923. doi: 10.1083/jcb.200512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghosh HS, McBurney M, Robbins PD. SIRT1 negatively regulates the mammalian target of rapamycin. PLoS One. 2010;5(2):e9199. doi: 10.1371/journal.pone.0009199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pillai VB, et al. Exogenous NAD blocks cardiac hypertrophic response via activation of the SIRT3-LKB1-AMP-activated kinase pathway. J Biol Chem. 2010;285(5):3133–3144. doi: 10.1074/jbc.M109.077271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lan F, Cacicedo JM, Ruderman N, Ido Y. SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1. Possible role in AMP-activated protein kinase activation. J Biol Chem. 2008;283(41):27628–27635. doi: 10.1074/jbc.M805711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kume S, et al. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest. 2010;120(4):1043–1055. doi: 10.1172/JCI41376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee IH, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105(9):3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Donmez G, Guarente L. Aging and disease: connections to sirtuins. Aging Cell. 2010;9(2):285–290. doi: 10.1111/j.1474-9726.2010.00548.x. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Q, Wang SY, Nottke AC, Rocheleau JV, Piston DW, Goodman RH. Redox sensor CtBP mediates hypoxia-induced tumor cell migration. Proc Natl Acad Sci U S A. 2006;103(24):9029–9033. doi: 10.1073/pnas.0603269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deng Y, et al. Redox-dependent Brca1 transcriptional regulation by an NADH-sensor CtBP1. Oncogene. 2010;29(50):6603–6608. doi: 10.1038/onc.2010.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Di LJ, Fernandez AG, De SA, Longo DL, Gardner K. Transcriptional regulation of BRCA1 expression by a metabolic switch. Nat Struct Mol Biol. 2010;17(12):1406–1413. doi: 10.1038/nsmb.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6(12):924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 62.Chen EI, et al. Adaptation of energy metabolism in breast cancer brain metastases. Cancer Res. 2007;67(4):1472–1486. doi: 10.1158/0008-5472.CAN-06-3137. [DOI] [PubMed] [Google Scholar]
- 63.Trounce IA, Kim YL, Jun AS, Wallace DC. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 1996;264:484–509. doi: 10.1016/s0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.