Abstract
Uniparental parthenotes are considered an unwanted byproduct of in vitro fertilization. In utero parthenote development is severely compromised by defective organogenesis and in particular by defective cardiogenesis. Although developmentally compromised, apparently pluripotent stem cells can be derived from parthenogenetic blastocysts. Here we hypothesized that nonembryonic parthenogenetic stem cells (PSCs) can be directed toward the cardiac lineage and applied to tissue-engineered heart repair. We first confirmed similar fundamental properties in murine PSCs and embryonic stem cells (ESCs), despite notable differences in genetic (allelic variability) and epigenetic (differential imprinting) characteristics. Haploidentity of major histocompatibility complexes (MHCs) in PSCs is particularly attractive for allogeneic cell-based therapies. Accordingly, we confirmed acceptance of PSCs in MHC-matched allotransplantation. Cardiomyocyte derivation from PSCs and ESCs was equally effective. The use of cardiomyocyte-restricted GFP enabled cell sorting and documentation of advanced structural and functional maturation in vitro and in vivo. This included seamless electrical integration of PSC-derived cardiomyocytes into recipient myocardium. Finally, we enriched cardiomyocytes to facilitate engineering of force-generating myocardium and demonstrated the utility of this technique in enhancing regional myocardial function after myocardial infarction. Collectively, our data demonstrate pluripotency, with unrestricted cardiogenicity in PSCs, and introduce this unique cell type as an attractive source for tissue-engineered heart repair.
Introduction
Unisexual reproduction by parthenogenesis is commonly observed in fish, amphibians, and reptiles (1); however, mammals lack this capacity (2, 3). In humans, spontaneous activation of unfertilized oocytes is a rare event that has been identified as the cause for ovarian teratoma formation (4). In vitro, parthenogenetic activation of mammalian oocytes can be stimulated chemically, leading to the development of diploid nonembryonic blastocysts (5–8), and apparently pluripotent stem cells have been derived from the resulting blastocoel inner cell mass (9–11). Uniparental parthenogenetic stem cells (PSCs) exhibit self-renewal capacity and clonogenic proliferation in vitro, but show abnormal embryonic and extraembryonic development as a consequence of differential expression of imprinted genes in vivo (6, 12–14). Ectodermal lineage specification appears to be least affected in vitro (5, 15) and in vivo (14), while endodermal and mesodermal cell lineages have been reported to be developmentally compromised in parthenotes (6, 12–14).
Given the enormous efforts to develop cell-based strategies to repair failing hearts (16), exploring the capacity for mesoderm formation in and cardiomyocyte derivation from PSCs appears warranted. The utility of PSCs in cell-based organ repair may, moreover, be facilitated by: (a) the availability of unfertilized oocytes from uncompleted in vitro fertilization procedures typically owing to oocyte immaturity or lack of sperm (17), (b) the high efficiency of PSC derivation (18), and (c) the widely haploidentical genomes of PSCs (8, 19). Major histocompatibility complex (MHC) haploidentity is particularly interesting, as it would increase cell acceptance in allogeneic applications and provide a realistic rationale for therapeutic cell banking (20, 21).
A key concern associated with cell-based organ, and in particular heart repair is the limited cell retention observed after intracoronary or intramyocardial delivery (22). To address this concern and introduce sustained myocardial support, tissue engineering technologies are presently being explored (23). A fundamental challenge in cardiac tissue engineering is the provision of sufficiently large cell populations with appropriate cardiomyocyte content and quality. Whether recent developments in stem cell differentiation (24, 25) and selection (26–29) can overcome this limitation has yet to be investigated.
Here we demonstrate that PSCs exhibit properties similar to other pluripotent stem cells, including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs). This encompasses the ability to: (a) derive bona fide cardiomyocytes; (b) enrich PSC-derived cardiomyocytes (PCMs) using 3 different technologies (i.e., FACS, antibiotic selection in genetically modified PSCs, and directed differentiation); and (c) construct engineered heart muscle (EHM) with the structural and functional properties of native myocardium for subsequent utilization in heart muscle repair. Moreover, we provide evidence for immunological acceptance of PSC allografts in related and unrelated recipients with matching MHCs.
Results
PSCs exhibit properties similar to other pluripotent stem cells.
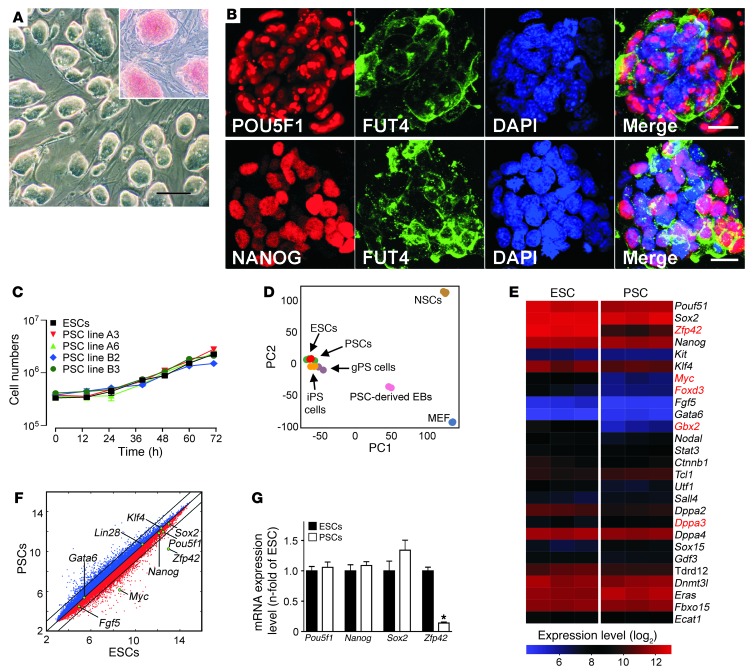
We generated 12 PSC lines from 63 nontransgenic blastocysts, and 2 PSC lines from 30 αMHC-EGFP transgenic blastocysts. The αMHC-EGFP transgene used the cardiomyocyte-restricted α-myosin heavy chain (Myh6; also known as αMHC) promoter to drive expression of an EGFP reporter (27). We first compared morphology and alkaline phosphatase activity (Figure 1A), presence of stemness factors (Figure 1B), and growth kinetics (Figure 1C), and found no differences in PSCs or the well-characterized pluripotent R1-ESC line (30). Principal component analysis (PCA) of transcriptome data from PSCs, ESCs, iPSCs, and germline-derived pluripotent stem cells (gPSCs) demonstrated a remarkable similarity between the investigated cell types (Figure 1D). Interestingly, only 5 (Zfp42, Myc, Foxd3, Gbx2, Dppa3) of the 27 annotated stemness-related transcripts were differentially expressed in PSCs (A3) and ESCs (R1) at a similar passage number (Figure 1E). Notably, of the 6 established reprogramming factors (Pou5fl, Sox2, Klf4, Myc, Nanog, Lin28; refs. 18, 31) only Myc showed lower transcript abundance in PSC line A3 versus ESC line R1 (Figure 1F). A lower abundance of Zfp42 in PSCs versus ESCs (Figure 1G) was anticipated because of reported differences in pluripotency-related gene expression in Sv129-derived versus C57BL/6-derived stem cells (32).
Figure 1. Basic characterization of PSCs.
(A) Undifferentiated PSCs cultured on MEFs formed ESC-like colonies with alkaline phosphatase activity (red – inset). Scale bar: 100 μm. (B) Immunofluorescence labeling of POU5F1, NANOG, and FUT4 (also known as SSEA1) in undifferentiated PSC colonies. Scale bars: 20 μm. (C) Growth kinetics of ESC line R1 and PSC lines A3, A6, B2, and B3 (n = 3 per group and time point; data represent means ± SEM; cell-doubling time: 16–17 hours). (D) PCA of global gene expression profiles of pluripotent cells (PSCs, ESCs, iPSCs, and gPSCs) and somatic cells (MEFs and neural stem cells [NSCs]). The respective Gene Expression Omnibus accession numbers are: GSE11274 (includes MEF and gPSCs); GSE10806 (includes NSC and iPSCs); and GSE30868 (includes ESCs [R1], PSCs [A3] and PSC-derived EBs [A3 EB culture day 15]). Each microarray experiment is represented by a dot, which is positioned in 2D space according to its similarity or degree of variance to all samples analyzed. PC1 and PC2 show variances of 47% and 20%, respectively. (E) Heat map of 80 annotated imprinted genes expressed in ESCs (R1 passage 25; n = 3) and PSCs (A3 passage 25; n = 3). Each gene is represented by a single row of colored boxes. Red represents transcript levels above median; blue represents transcript levels below median. (F) Comparison of global gene expression in ESCs and PSCs with key reprogramming factors highlighted. (G) qPCR of selected stemness factors (n = 4 per group; data represent means ± SEM).
PSCs display characteristic genomic and epigenetic properties.
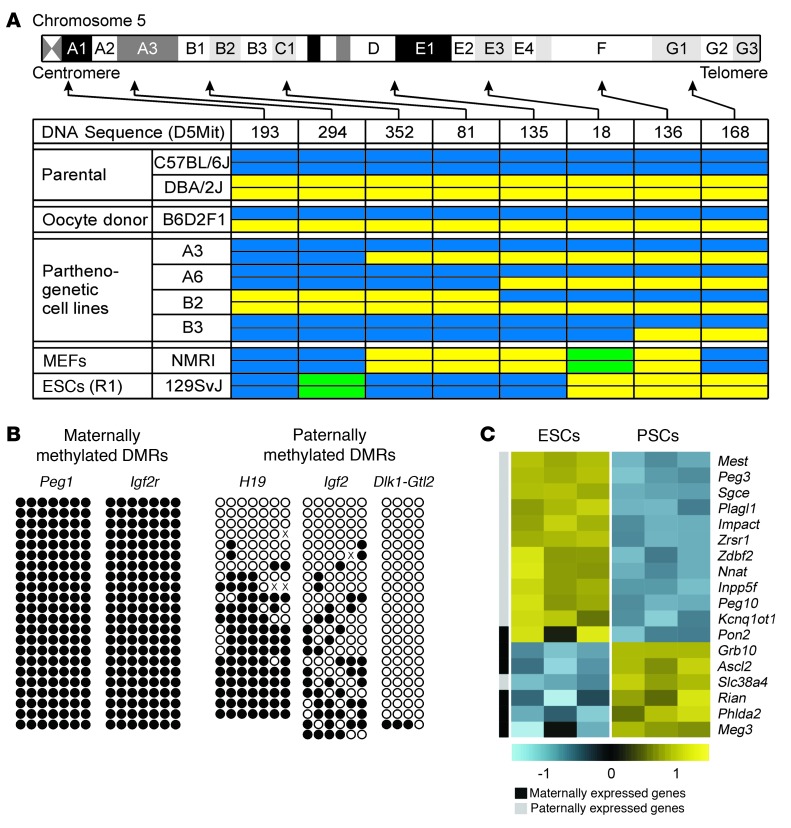
The hallmarks of PSC identity are largely haploidentical genomes and a lack of differential methylation of imprinted genomic loci (differentially methylated regions, referred to herein as DMRs) (19). PCR-based microsatellite marker analyses of chromosome 5 demonstrated the anticipated variable genomic constitution of PSCs, with high homozygosity for centromeric markers and an increasing degree of heterozygocity with increasing distance from the centromeres (Figure 2A). This characteristic feature of PSCs results from homologous recombination prior to chromosomal segregation in meiosis I (33). Giemsa-stained metaphase spreads suggested normal karyotypes in the different PSC lines (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI66854DS1). High-resolution spectral karyotyping analyses at passage 24–25 did, however, indicate the expected karyotype variability with clones containing regular (40, XX), but also abnormal [40,X, Der(X)T(XA.1;12D1) — confirmed by array CGH (passage 5 versus passage 24); 40,X,–X,+1; 39,X,–X] karyotypes (Supplemental Figure 1B). The frequently observed partial or complete loss of 1 X chromosome is a common observation in female pluripotent stem cells, but does not appear to affect cell differentiation in vitro (9). As anticipated, maternally methylated DMRs controlling Peg1 and Igf2r transcription were hypermethylated (100%), and the paternally methylated DMRs controlling Dlk1 and Gtl2 transcription (IG-DMR) were hypomethylated (3%) in PSCs at passage 25 (Figure 2B). In contrast, paternally methylated DMRs of H19 (CTCF1) and Igf2 (DMR2) displayed 59% and 33% CpG methylation, respectively, which is in agreement with recent reports on variable methylation of these loci in PSCs (34) and ESCs (35). Transcriptome profiling identified differences in 18 of 80 annotated imprinted genes, with most paternally and maternally expressed imprints displaying lower and higher transcript abundance in PSCs versus ESCs, respectively (Figure 2C).
Figure 2. Characteristic genetic and epigenetic differences in ESCs and PSCs.
(A) Overview of chromosome 5 genotype demonstrating the expected variable allelic disparity in PSC lines (A3, A6, B2, B3). Color codes: C57BL/6J allele, blue; DBA/2J allele, yellow; alternative allele, green. (B) Bisulfite genomic sequencing of DMRs of imprinted genes in PSC line A3. Each row represents a cloned allele and each circle a CpG site (black circles represent methylated and white circles represent unmethylated CpG sites). (C) Differentially expressed imprinted genes in ESCs (R1; n = 3) versus PSCs (A3; n = 3). Each gene is represented by a single row of colored boxes. Color codes: yellow represents transcript levels above median; blue represents transcript levels below median.
PSCs are capable of differentiating into ectodermal, endodermal, and mesodermal cells in vitro and in vivo.
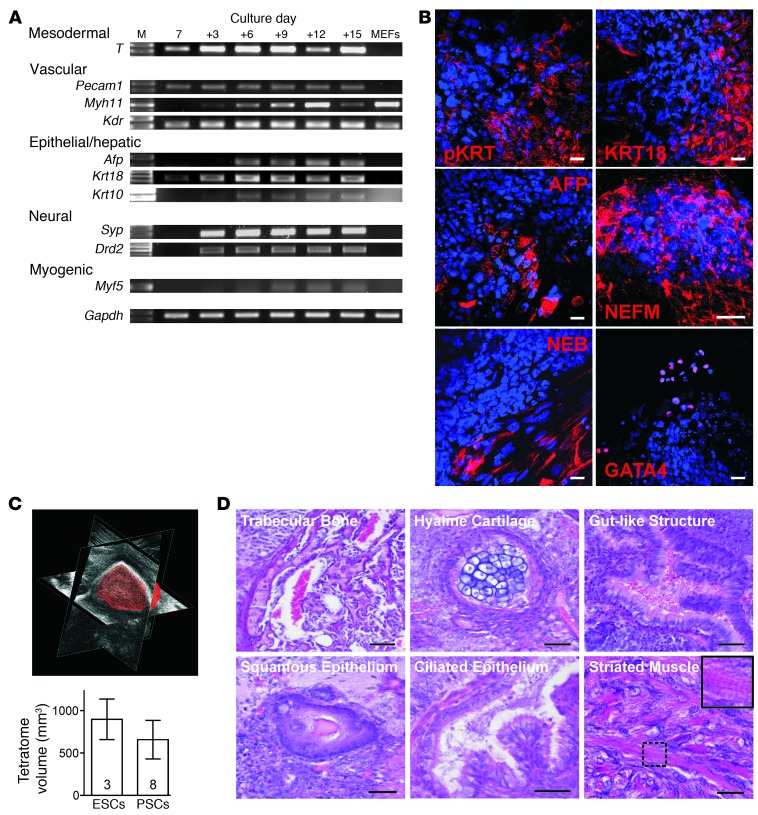
We tested 2 selected nontransgenic PSC lines (B2 and B3) and the 2 transgenic (A3 and A6) PSC lines for their multilineage differentiation capability in embryoid body (EB) cultures using (a) RT-PCR for mesodermal, vascular, epithelial/hepatic, neural, and myogenic markers (Figure 3A); (b) immunofluorescence staining for endodermal (KRT, AFP), ectodermal (NEFM), and mesodermal (NEB, GATA4) markers (Figure 3B); and (c) teratoma formation after subcutaneous injection in SCID mice (Figure 3, C and D). Collectively, these data confirmed similar differentiation and growth potential in the tested uniparental PSCs and the widely used biparental R1-ESCs.
Figure 3. Multilineage potential of PSCs in vitro and in vivo.
(A) RT-PCR analyses of lineage-specific transcripts in differentiating PSCs (PSC line B2). MEFs served as controls. Culture day 7; i.e., 2 days of hanging drop followed by 5 days in suspension culture. (+): Subsequent adhesion culture days. M, 100 bp DNA marker (full uncut gels are shown in the Supplemental Material). (B) EB culture day 22 (PSC line B3): pancytokeratin (pKRT), cytokeratin-18 (KRT18), α-fetoprotein (AFP), neurofilament protein M (NEFM), nebulin (NEB), GATA4. Respective lineage markers (red); nuclei (blue). Scale bars: 20 μm. (C and D) Comparison of teratoma size and composition 3 weeks after s.c. injection of ESCs and PSCs in SCID mice. Teratoma volume was quantified by ultrasonic biomicroscopy (representative image in C; data represent means ± SEM). H&E staining revealed multilineage potential of PSCs, including the capacity to generate cross-striated muscle (inset in D shows magnification of smaller boxed area). Scale bars: 50 μm.
PSCs are largely MHC haploidentical, making them attractive for allotransplantation.
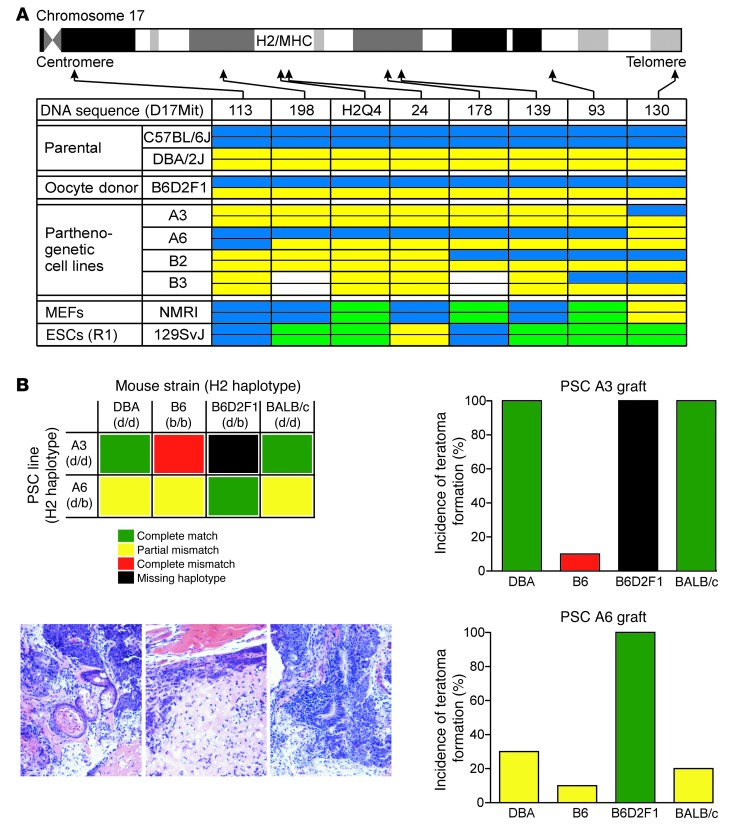
We first analyzed the H2 locus, encoding for MHC, on chromosome 17 in the 4 well-characterized PSC lines (A3, A6, B2, B3) using a PCR microsatellite screen. Similar to chromosome 5 (Figure 2A), we observed the characteristic allelic disparity in the PSC genomes. Three of the 4 lines (A3 transgenic; B2 and B3 nontransgenic) were H2 haploidentical; the A6 line was heterozygotic for the H2 locus (Figure 4A). MHC haploidentity is considered immunologically advantageous in allotransplantation (20, 21). We screened for acceptance of PSC allografts using a teratoma formation assay (33); in this model, teratoma formation after i.m. injection of pluripotent stem cells indicates immunological acceptance. We specifically investigated the hypothesis that MHC-haploidentical donor cells (A3 – H2d/d) would be tolerated if implanted in related (DBA – H2d/d) and unrelated (BALB/c – H2d/d) allogeneic recipients with a complete MHC match; moreover, we hypothesized that allografts with a missing, but not mismatching, H2 haplotype (H2d/d A3-PSCs in H2d/b B6D2F1 recipients) would not be rejected (Figure 4B). In agreement with our hypotheses, we found that the A3 line (H2d/d) was not rejected in DBA (H2d/d), B6D2 (H2d/b), or BALB/c (H2d/d) mice; in contrast, a small teratoma was observed in only 1 of 10 B6 mice (H2b/b) with A3 PSC (H2d/d) implants. MHC-heterozygotic A6 PSCs (H2d/b) were not rejected if implanted in B6D2F1 (H2d/b) mice. In contrast, only 3 of 10 DBA (H2d/d) mice, 1 of 10 B6 mice (H2b/b), and 2 of 10 BALB/c (H2d/d) mice developed teratoma after A6 PSC (H2d/b) implantation.
Figure 4. H2-haploidentical PSCs as preferred allografts.
(A) Allele contribution on chromosome 17 (MHC locus is between 18.4 and 20.3 cM) in A3, A6, B2, and B3 PSC lines. Allele color codes: C57BL/6J, blue; DBA/2J, yellow; alternative alleles, green; no PCR product detected, white. (B) Schematic of transplant conditions (upper left); representative teratoma histologies (bottom left); teratoma formation after grafting of H2-haploidentical A3 PSC grafts in indicated recipient mice; n = 10 per group (upper right); teratoma formation after grafting of H2-heterozygous A6 PSC grafts in indicated recipient mice; n = 10 per group (bottom right).
PSCs and ESCs exhibit a similar potential to differentiate into cardiomyocytes.
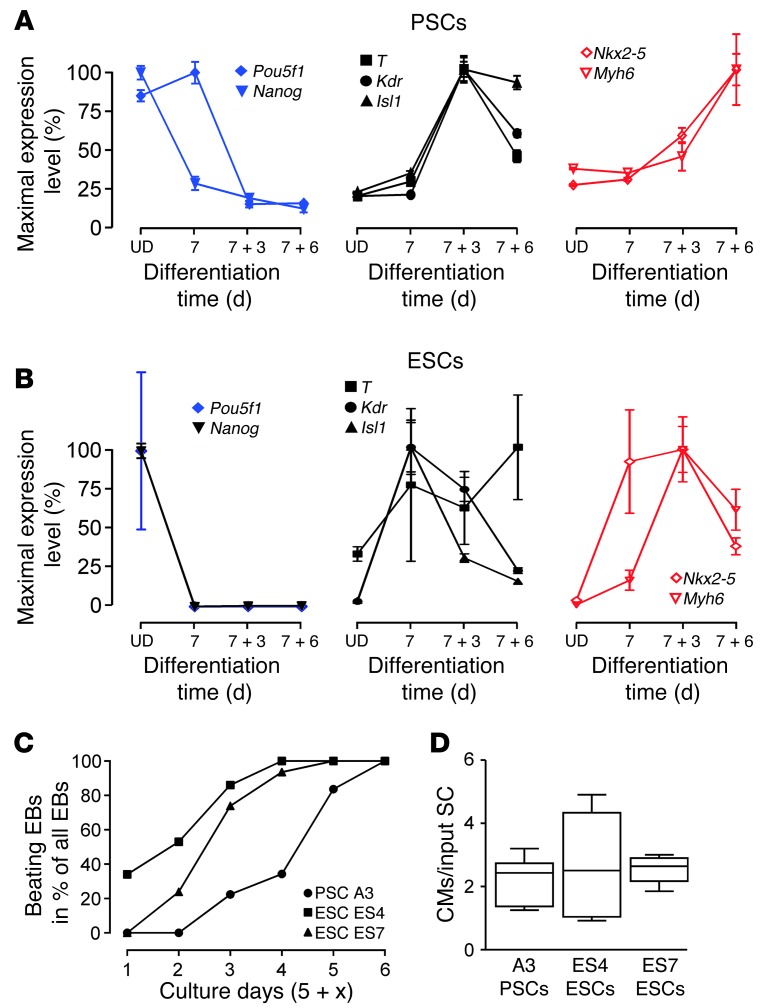
Given (a) the utility of the cardiomyocyte-restricted EGFP reporter for myocyte isolation and in vivo tracing; (b) the immunological advantage of an MHC-haploidentical cell line; and (c) a similar cardiomyogenic potential in all investigated PSC lines, we chose PSC line A3 for further in-depth analyses of cardiomyocyte generation and myocardial restoration. To scrutinize the process of cardiomyocyte differentiation in PSCs, we compared molecular markers for: (a) undifferentiated cells (Pou5f1 and Nanog); (b) mesodermal commitment (i.e., T, Isl1, Kdr); and (c) myocardial specification (i.e., Nkx2-5, Myh6) by quantitative PCR (qPCR) in A3 PSC and R1 ESC EB cultures (Figure 5, A and B). These data confirmed the anticipated downregulation of Pou5f1 and Nanog, a transient upregulation of T, Kdr, and Isl1, and a late elevation of Nkx2-5 and Myh6. In addition, we determined the percentage of EBs exhibiting spontaneous contractile activity and found all to contain beating areas by culture day 11 (analyzed EB numbers: 46, 43, and 67 in ES4, ES7, and PSC-A3 cultures; Figure 5C). Cardiomyocyte yield per input stem cell was on average 2.5 ± 0.3 (n = 16), with no differences in ESC lines (ES4 and ES7) and PSC line A3 (Figure 5D); note that we derived these cell lines from the same transgenic mouse model (αMHC-EGFP) (27) to exclude potentially confounding mouse background bias. Collectively, these data confirmed that, despite an apparent 2- to 3-day delay in PSC differentiation, there were no essential differences in absolute cardiomyocyte derivation efficiency in PSC and ESC cultures.
Figure 5. Mesodermal differentiation and cardiogenicity.
qPCR for pluripotency (Pou5f1), (cardio)mesoderm (T, Kdr, Isl1), and cardiac (Nkx2-5, Myh6) markers in undifferentiated (UD) cells and differentiating EBs in (A) PSC (n = 4–5 per group) and (B) ESC (n = 3–4 per group) cultures (data represent means ± SEM). Decrease in Nkx2-5 and Mhy6 in ESCs on culture day 7 + 6 was the consequence of nonmyocyte overgrowth. (C) Enumeration of beating EBs at the indicated culture days (n = 2–3 per time point; data represent means). (D) Quantity of EGFP-positive cardiomyocytes (CMs) per input stem cell (SC; 400 ESCs or PSCs per EB; cells from 5 single EBs were pooled for analytic FACS; n = 5–6) on culture day 5 + 10 (box plots indicate median with interquartile range; whiskers indicate minimal and maximal values). PSC line A3 and ESC lines ES4 and ES7 were generated from the same αMHC-EGFP mouse strain.
PSCs can differentiate into terminally mature cardiomyocyte subtypes.
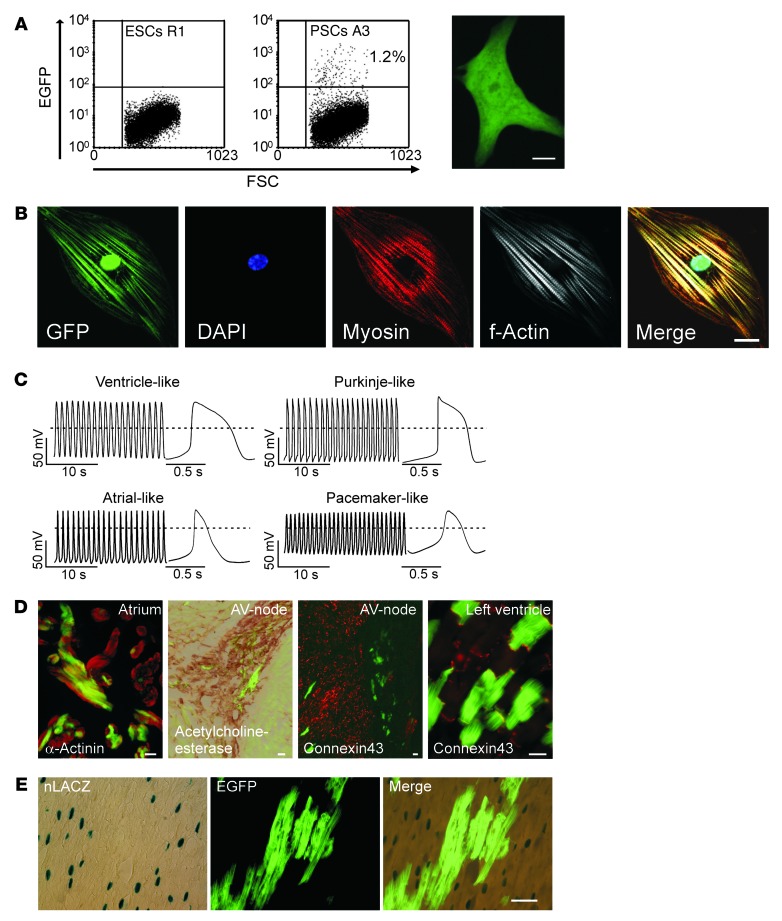
Expression of the lineage-restricted αMHC-EGFP reporter was further exploited to FACS-purify PCMs from EB cultures for subsequent morphological and functional analyses (Figure 6A and Supplemental Video 1). This revealed the presence of cardiac-specific structural proteins organized within well-developed sarcomeres (Figure 6B) and electrophysiological properties of distinct cardiomyocyte subtypes (ref. 36, Figure 6C, and Supplemental Table 1), including: (a) ventricle-like cells (n = 49) with a low maximal diastolic potential (MDP), a fast maximal upstroke-velocity (max dV/dt), a large action potential amplitude (APA), and a pronounced plateau phase; (b) Purkinje-like cells (n = 13) with a faster max dV/dt, the presence of a notch, and also a plateau phase; (c) atrial-like cells (n = 11) lacking a significant plateau phase; and (d) pacemaker-like cells (n = 15) with less negative MDP, a slower max dV/dt, a smaller APA, and no defined plateau phase. Fourteen of the 102 cells studied exhibited intermediate action potential (AP) phenotypes. Pharmacologic interventions stratified these cells as being dependent on Na+ or Ca2+ influx in phase 0 of the AP (Supplemental Figure 2). Blastocyst injection experiments confirmed the capacity of PSCs to develop into mature (Figure 6D) and functionally integrated (Supplemental Figure 3) cardiomyocyte subtypes within different anatomic regions of the developing heart. Importantly, the formation of PCMs following blastocyst injection occurred largely independently from cell fusion events (Figure 6E).
Figure 6. Cardiomyocyte differentiation from PSCs in vitro and in vivo.
(A) FACS of GFP-positive PCMs (culture day 7 + 15); nontransgenic ESCs served as the control cell population (right panel illustrates a PCM 24 hours after plating; also refer to Supplemental Video 1). (B) Morphology of FACS-purified PCMs (EGP-positive) assessed by confocal laser scanning microscopy. (C) Distinct APs in FACS-purified PCMs. (D) Epifluorescence images and histochemical staining of atrial, atrioventricular (AV) nodal (acetylcholinesterase-positive/connexin43-negative), and ventricular regions in chimeric adult mouse heart (generated by blastocyst injection of αMHC-EGFP PSCs, A3 line). (E) Heart section from a chimeric mouse after injection of αMHC-EGFP PSCs (A3 line) into αMHC-nLacZ blastocysts. Light microscopy after X-gal staining (left panel); epifluorescence image of the same section showing PSC-derived EGFP-positive cardiomyocytes (middle panel); merged image did not show cells double-positive for EGFP and nLacZ, ruling out overt fusion events (right panel). Scale bars: (A) 10 μm, (B and D) 20 μm, (E) 50 μm.
PCMs survive and integrate into recipient myocardium after intramyocardial injection.
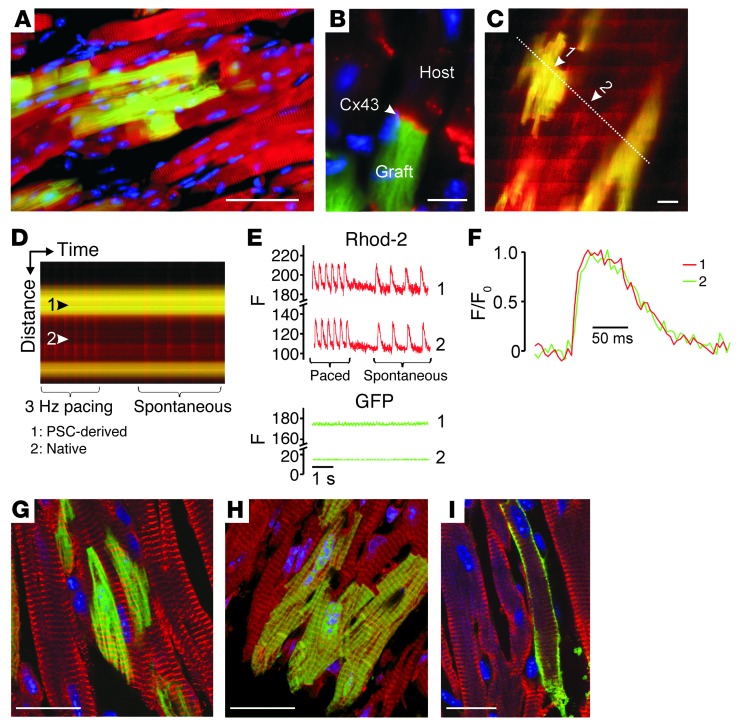
We next investigated whether PCMs would also integrate after intramyocardial injection into adult mouse heart. Three weeks after injection, PCM grafts displayed a rod-shaped morphology with regular cross-striations (Figure 7A) and connexin43-positive junctional complexes between engrafted and native cardiomyocytes (Figure 7B), making them indistinguishable from native cardiomyocytes (with the exception of the EGFP signal in PCMs). In agreement with these morphological observations, two-photon laser scanning fluorescence microscopy (27) revealed synchronous AP-evoked Ca2+ transients in adjacent EGFP-positive and EGFP-negative cardiomyocytes (Figure 7, C–E), indicating electrical coupling of PCMs to the recipient myocardium. Moreover, similar Ca2+ transient kinetics in engrafted and host cardiomyocytes indicated the capacity of the in vitro–“generated” PCMs to develop mature Ca2+-handling properties if exposed to the appropriate in vivo environment (Figure 7F). Similar observations were made in chimeric mice generated from blastocysts injected with undifferentiated A3-PSCs (Supplemental Figure 3C). Additional immunofluorescence staining confirmed that PCMs were well integrated into the recipient myocardium (Figure 7, G–I).
Figure 7. Retention and functional integration of PCMs after intramyocardial injection.
(A and B) Immunofluorescent labeling of α-actinin (red, A) and connexin43 (red, B) in adult ventricular mouse heart tissue 3 weeks after injection of PCMs (EGFP, green; nuclei, blue). (C–F) Two-photon laser scanning microscopy of intracellular Ca2+ transients in adult mouse hearts after injection of PCMs: 2D-scan (C) and line-scan (D) images of stimulated (3 Hz) and spontaneous Ca2+ transients. Arrow 1, EGFP-positive cell; arrow 2, EGFP-negative cell; the dotted line indicates the location of the line scan. Bands of increased rhod-2 fluorescence intensity reflect AP-induced Ca2+ transients. (E) Plots of rhod-2 and GFP line-scan data in the EGFP-expressing cardiomyocyte 1 and the GFP-negative (native) cardiomyocyte 2 as a function of time. (F) Superimposed tracings of AP-evoked changes in rhod-2 fluorescence as a function of time from cardiomyocytes 1 (green) and 2 (red). For each cell, the relative changes in fluorescence were normalized such that 0 represents the prestimulus fluorescence intensity (F0), and 1 represents the peak fluorescence intensity. Additional immunofluorescence analyses confirmed the maturity of engrafted PCMs: A3 line; EGFP-positive; (G) α-actinin (red); (H) actin (red); (I) troponin (red); nuclei (blue; DAPI). Scale bars: (A) 100 μm, (B, C, G–I) 20 μm.
EHM from PCMs.
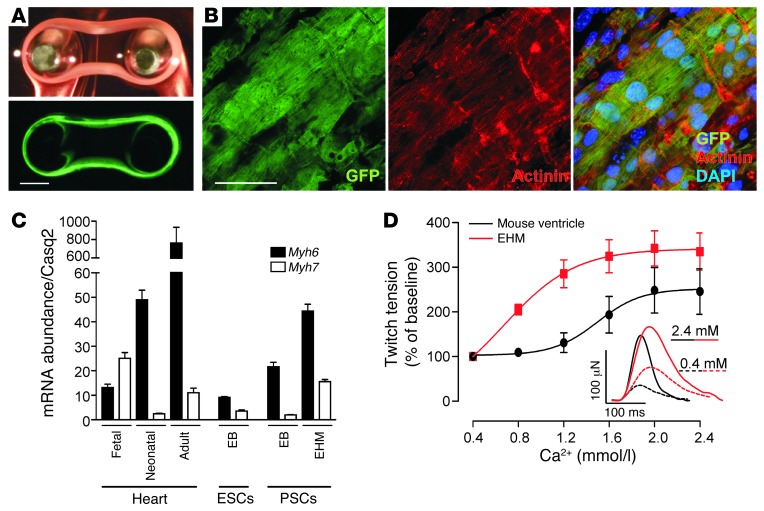
The level of spontaneous myocyte differentiation in pluripotent stem cell culture is notoriously low (Figure 6A) (26). Any large-scale in vitro or in vivo application such as tissue engineering and cardiac muscle repair requires augmentation of this process. We exploited 2 different strategies to reach the necessary scale and myocytic purity, namely: (a) directed differentiation (24), and (b) genetic selection after stable introduction into the A3 line of a second transgene encoding for neomycin resistance (neoR) under Myh6 promotor control (26). Either protocol provided the necessary cell quality [cardiomyocyte content: (a) 50–60%, ref. 24; and (b) >95%, ref. 26] and quantity (1.5 × 106 cells/EHM) to enable EHM generation (Figure 8A and Supplemental Video 2). The nonmyocytes in protocol (a) were predominantly smooth muscle cells and fibroblasts (24). Genetically selected cells from protocol (b) had to be supplemented with nonmyocytes (25% fibroblasts) to yield contractile EHM. Cells were suspended in hydrogels and cast in circular molds to facilitate the formation of EHM. Irrespective of the cell protocol used, the EHM developed spontaneous contractions within 3 days after casting. Immunofluorescence stainings for sarcomeric actinin after an additional 5 days of culture on custom-made holders to maintain the EHM under constant load demonstrated the presence of highly mature cardiomyocytes with regular sarcomeric cross-striations and anisotropic syncytial arrangement (Figure 8B). Accordingly, Myh6 transcript abundance was markedly higher in EHM cultures compared with ESC (4.9 ± 0.4-fold; P < 0.05 vs. EHM) or PSC (2.1 ± 0.2-fold; P < 0.05 vs. EHM) EB cultures, and reached similar levels as identified in neonatal mouse heart tissue (Figure 8C). Interestingly, EHM and adult myocardium exhibited similarly low levels of Myh7 transcripts (high levels of Myh7 transcripts are typically associated with immature or injured myocardium in rodents). Contractile properties of EHM were comparable to native myocardium with, however, higher calcium sensitivity (EHM [n = 8] vs. myocardium [n = 4]: 0.82 ± 0.03 vs. 1.52 ± 0.06 mmol/l; P < 0.05) and slower contraction kinetics (contraction time: 65 ± 3 ms vs. 38 ± 2 ms; P < 0.05; relaxation time: 69 ± 4 ms vs. 38 ± 2 ms; P < 0.05; Figure 8D and Supplemental Table 2).
Figure 8. Tissue-engineered myocardium from PSC derivatives.
(A) Light microscopic (top panel) and epifluorescence images (bottom panel) of an EHM (culture day 8; also refer to Supplemental Video 2). (B) Confocal laser scanning microscopy of a cardiomyocyte bundle in EHM. EGFP (green); α-actinin (red); nuclei (blue, DAPI). (C) Transcript abundance of Myh6 and Myh7 (normalized to Casq2) in native myocardium (fetal, neonatal, adult), EB cultures (ESC and PSC), and EHM (n = 3–4 per group; data represent means ± SEM). (D) Concentration response curve under increasing extracellular [Ca2+] for EHM (n = 8) and native mouse myocardium (ventricular wedge; n = 4). Data represent means ± SEM (also refer to Supplemental Table 2). Single isometric contractions of EHM and native heart muscle (inset) at low (0.4 mmol/l) and high (2.4 mmol/l) extracellular [Ca2+]. Scale bars: (A) 1 mm, (B) 50 μm.
Therapeutic utility of EHM.
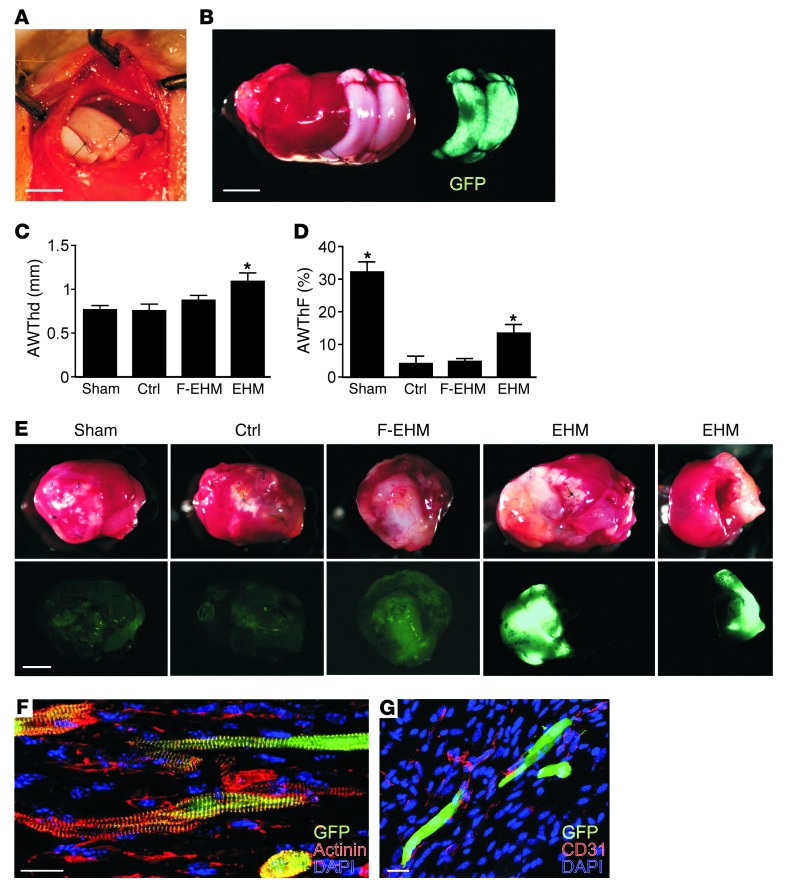
To examine whether the therapeutic application of EHM is in principle feasible, we tested EHM from the A3-PSC line (H2d/d) in immune-competent DBA (H2d/d) mice after induction of myocardial infarction (MI) by permanent left coronary artery ligation (Figure 9A). The EGFP transgene allowed for easy identification of PCMs in EHM grafts (Figure 9B). EHM-“treated” hearts (n = 7), in contrast to control groups (control, MI but no EHM graft [n = 8]; F-EHM, MI with formaldehyde-fixed nonviable EHM graft [n = 8]), exhibited thicker anterior walls (EHM vs. control: 1.3 ± 0.1 mm vs. 0.8 ± 0.1 mm; P < 0.05) (Figure 9C and Supplemental Table 3) and enhanced systolic anterior wall thickening (EHM vs. control: 13 ± 2% vs. 4 ± 2%; P < 0.05) (Figure 9D and Supplemental Table 3). Visual inspection clearly identified the grafts in situ and distinguished the viable EHM grafts from the nonviable F-EHM grafts by their strong EGFP fluorescence (EGFP fluorescence was destroyed by the fixation procedure in F-EHM; Figure 9E). Morphological analyses confirmed the presence of well-structured PCMs (Figure 9F) surrounded by CD31-positive capillaries (Figure 9G).
Figure 9. Proof-of-concept for the therapeutic application of MHC-matched EHM grafts.
(A) F-EHM graft sutured onto a beating heart. (B) EHM graft on infarcted myocardium (PCMs, EGFP-positive; heart was explanted directly after EHM engraftment for demonstration). (C) Anterior wall thickness in diastole (AWThd) and (D) anterior wall thickening fraction (AWThF) measured by echocardiography 2 weeks after MI and the respective surgical intervention. Sham, no MI, and no graft; control (Ctrl), MI with no graft; F-EHM, MI with formaldehyde-fixed (nonviable) EHM; EHM, MI with viable EHM graft (n = 7–8). Data represent means ± SEM; *P < 0.05 versus control (also refer to Supplemental Table 3). (E) Heart explants with F-EHM and EHM (EGFP-positive) grafts; EGFP signal (lower panels) indicates retention of viable cardiomyocytes. (F) Anisotropically arranged PCMs (α-actinin and GFP) in EHM graft. (G) PCMs surrounded by CD31-positive capillaries. Scale bars: (A, B, and E) 2 mm; (F and G) 20 μm.
Discussion
Several pluripotent stem cell species have been identified, and data on mesodermal differentiation and cardiomyocyte derivation have been reported previously in detail for ESCs (36, 37), iPSCs (38, 39), and spermatogonial stem cells (40). Most recently, the feasibility of direct reprogramming of fibroblasts into cardiomyocytes in vitro (41) and in vivo (42, 43) has been demonstrated. Thus, one may conclude that there is no need for yet another cardiogenic stem cell species. Despite the excitement about the already available stem cells and the seemingly unlimited reprogramming technologies, it must be recognized that pivotal limitations will remain unaddressed with the aforementioned cell types, especially if therapeutic applications are anticipated in disease conditions such as heart failure with a malignant short-term prognosis. Here, the key concern is timely allocation (within weeks or several months) and retention of large myocyte numbers for myocardial reconstruction. Autologous cell repair, which would certainly be ideal, appears unlikely to be the answer to this challenge because of the time-consuming quality control required for personalized therapeutics. Thus, it is understandable that well-defined allogeneic ESC derivatives continue to enter clinical trials, preferably as an off-the-shelf product in immune-privileged niches (44). The establishment of cell banks containing MHC-haploidentical stem cells has been suggested to enhance the general acceptability of stem cell grafts (20, 21). However, finding donors with these rare genotypes that also have the necessary diversity to be broadly applicable appears very ambitious. In light of this, we hypothesized that PSCs, which are by nature predominantly MHC-haploidentical, may represent attractive alternatives to biparental pluripotent stem cells, and we tested their applicability in tissue-engineered heart repair.
We first investigated in detail the fundamental biological properties of PSCs and determined that despite their unique genomic and epigenetic properties, there was no obvious difference in cardiac differentiation potential between PSCs and ESCs. This was somewhat unexpected, given the heart defects observed during the development of parthenotes in utero (14). Finally, our data suggest that potential deficits could be “overridden” in vitro, despite the persistence of unique epigenetic traits. We find this particularly interesting because it suggests that the impact of the differentially expressed genes in ESCs and PSCs was neither negative nor positive on in vitro or in vivo (in chimeric mice) cardiogenicity and subsequent cardiomyocyte maturation.
We next tested the hypothesis that MHC-haploidentical PSCs would be tolerated in related and unrelated allogeneic recipients with matching MHCs. This advances earlier investigations demonstrating acceptance in related allogeneic recipients (33). The applicability of MHC-haploidentical PSCs in unrelated, MHC-matched recipients is particularly attractive because it suggests that a small number of oocyte donors would suffice to provide the immunologically relevant MHC constellations for allogeneic applications. In fact, in silico predictions suggest that less than 100 MHC-haploidentical cell lines would be sufficient to provide cells with no MHC mismatch to more than 90% of the potential recipients in a population of approximately 100 million individuals (20, 21); in this scenario, little or no immune suppression may be necessary.
For therapeutic applications, but also for such potential applications as cellular or tissue-engineered models in studies of fundamental stem cell biology or disease processes and drug development, it is essential to enrich for cardiomyocytes and augment total cardiomyocyte numbers (45). Here, we demonstrate that PCMs can be enriched by: (a) directed differentiation (24); (b) FACS via the use of a cardiomyocyte-restricted EGFP transgene (27); or (c) administration of cytotoxic antibiotics after cardiomyocyte-restricted expression of a neomycin-resistance gene (26). Importantly, directed differentiation and neomycin selection protocols provided optimal cardiomyocyte numbers for myocardial tissue engineering; i.e., 1.5 × 106 cardiomyocyte-enriched cell populations per EHM. FACS can, in principle, also prepare the required cell numbers, but is limited by the need for high-input cell quantities to compensate for procedure-related cell damage and death.
To enable myocardial support after in vivo application, it is essential that cardiomyocyte grafts exhibit the capacity to couple electrically to recipient myocardium. Here, it is important to note that electrical integration of stem cell–derived cardiomyocytes cannot be readily anticipated (46). In light of this, we find it important to report that PCMs integrated electrically into recipient myocardium and adapted to the recipient milieu, rendering them morphologically and functionally indistinguishable from the recipient heart cardiomyocytes. Use of the cardiomyocyte-restricted EGFP transgene in PCMs was particularly helpful in these experiments to unambiguously distinguish engrafted and recipient heart cardiomyocytes. Our findings add to earlier and recent observations on electromechanical integration of ESC-derived cardiomyocytes in pig and guinea pig models (47, 48) by providing evidence at the cellular level for calcium-handling synchronization in graft and host cardiomyocytes.
We next tested the applicability of the cardiomyocyte-enriched cell pools in myocardial tissue engineering. EHM demonstrated an advanced degree of maturation compared with EB cultures and exhibited force parameters similar to those of left ventricular myocardium from adult mice. However, we observed higher calcium sensitivity and slower contraction kinetics in the EHM. This is not a peculiar observation in EHM generated from PSCs, but a general finding also in EHM from neonatal rodent cardiomyocytes (49), as well as mouse and human ESC- and iPSC-derived myocytes (50, 51). Whether limited diffusibility or fundamental differences in calcium handling account for this finding remains to be clarified. Importantly, our in vivo data demonstrate that PCMs exhibit the principle capacity to develop mature calcium handling properties if subjected to the proper in vivo environment.
Finally, we tested the concept of tissue engineering–based myocardial repair. In agreement with an earlier study on the therapeutic utility of EHM allografts from primary rat heart (23), we observed that EHM grafts were not only retained, but also enhanced regional performance of infarcted hearts. In contrast to the previous study and in accordance with our hypothesis, there was no need for specific immune suppression, likely due to the MHC matching between EHM grafts (from A3 PSC line – H2d/d) and the recipient mice (DBA – H2d/d). Further studies will, however, be necessary to establish the full potential of PSCs in MHC-matched heart repair, and, in particular, to optimize the reported tissue-engineering modality to achieve large-scale remuscularization — preferably in large animal models with human-like physiology, i.e., pig or nonhuman primate.
Taken together, PSCs have thus far attracted little attention for cell-based therapies, likely owing to the earlier observation of developmental defects in parthenotes (14). Accordingly, one of our key observations involved the capacity of PSCs to exhibit essentially normal cardiogenesis in vitro and in vivo. This, together with the ease of PSC derivation and the technical (no genetic manipulation required), ethical (no destruction of viable embryos), and immunological (MHC-haploidentity) advantages, warrants, in our view, the consideration of PSC use in heart muscle repair. Assessment of the therapeutic utility of EHM grafts will require further scrutiny, preferably in large animal models, to better address potentially limiting biological and technical issues such as hypoxia/vascularization and surgical positioning of the grafts in or on myocardial defects with clinically relevant dimensions. Potential clinical translation may benefit from the recent development of human PSCs (7). Moreover, the vast availability of unfertilized oocytes, which are otherwise discarded (17), provides a realistic option for therapeutic stem cell banking of preferably MHC-haploidentical stem cells (7, 20, 21). We conclude that our study provides a first proof-of-concept for PSC-based tissue-engineered heart repair. Once human PSCs become widely available, we assume that they will be amenable to myocardial tissue engineering just as human ESC and iPSCs are (50, 51).
Methods
Generation and culture of PSCs.
We induced superovulation in 6- to 8-week-old (C57BL/6J × DBA/2J)F1 (B6D2F1) mice by i.p. injection of 7.5 U pregnant mare serum gonadotropin (PMSG) and, 48 hours later, 7.5 U human choriogonadotropin (hCG). We harvested oviducts and collected cumulus oocyte complexes (COCs) 16 hours after hCG injection. Cumulus cells were removed with hyaluronidase (1 mg/ml for 10 minutes at room temperature). We subsequently activated oocytes with strontium chloride (10 mmol/l in Ca2+-free CZB medium in mmol/l: NaCl 82.7, KCl 4.68, KH2PO4 1.17, MgSO4 1.18, D-glucose 5.6, Na-lactate 30.1, EDTA2Na 0.1, NaHCO3 25, Na-pyruvate 0.62, glutamine 1; supplemented with 5 mg/ml BSA, and 100 U/ml penicillin and 100 μg/ml streptomycin [100 P/S]) for 6 hours and prevented second polar body extrusion with cytochalasin B (5 μg/ml). Blastocysts formed during culture in mineral oil–covered Ca2+-containing (1.71 mmol/l) CZB medium for 6 days. We transferred blastocysts onto mitomycin C (10 μg/ml) inactivated murine embryonic fibroblasts (MEFs) in KO medium (Gibco, Life Technologies) supplemented with 20% serum replacement (Gibco, Life Technologies), 1,000 U/ml leukemia inhibitory factor (LIF; Chemicon), 2 mmol/l glutamine, 1× nonessential amino acids, 50 P/S, and 100 μmol/l 2-mercaptoethanol, and manually selected inner cell mass (ICM) outgrowths after 5 days. We plated cells after trypsination on MEFs in stem cell medium: DMEM (high glucose, no pyruvate, 25 mmol/l HEPES) supplemented with 20% FCS, 1,000 U/ml LIF, 2 mmol/l glutamine, 1× nonessential amino acids, 50 P/S, 1 mmol/l Na-pyruvate, 30 μmol/l adenosine, 30 μmol/l guanosine, 30 μmol/l cytidine, 30 μmol/l uridine, 10 μmol/l thymidine, and 100 μmol/l 2-mercaptoethanol. PSC colonies were dispersed every 48–72 hours in 0.25% trypsin EDTA and split over a range of ratios from 1:2 to 1:8.
In vitro differentiation of PSCs.
We generated EBs using the hanging drop method (52). Hanging drops contained 500 cells (if not indicated otherwise) in 20 μl Iscove’s medium supplemented with 20% FCS, 2 mmol/l glutamine, 1× nonessential amino acids, 50 P/S, and 100 μmol/l 2-mercaptoethanol. Under these conditions, EBs formed within 2 days. We subsequently transferred EBs into Petri dishes for a 5-day suspension culture period before final transfer to cell culture dishes to facilitate in vitro differentiation. EBs from 10 of the 12 wild-type and both transgenic PSC lines showed spontaneously contracting areas without obvious differences in cardiogenic potential. Two of the 10 wild-type PSC lines (B2 and B3) showing contracting areas in EB culture, and 1 of the 2 wild-type PSC lines (C4) showing no contractions in EB culture were randomly chosen for basic characterization. Cardiac differentiation capacity was evaluated in more detail in the 2 transgenic PSC lines (A3 and A6) and was found to be similar (A3: 45 ± 3% and A6: 38 ± 3% beating EBs on culture day 19; n = 3 per line).
Assessment of growth rate.
We plated 50,000 cells from PSC lines B2 (wild-type), B3 (wild-type), A3 (αMHC-EGFP), A6 (αMHC-EGFP), and R1 ESC on MEF feeders in 6-cm gelatin-coated dishes containing stem cell medium. We dissociated cells every 12 hours (3 dishes per cell line and time point) into single cells in 0.25% trypsin EDTA and counted the total number of cells. Cell-doubling time was calculated from the linear interval of the resulting growth curve.
Genotype analyses.
We dispersed PSC lines A3, A6, B2, B3, and R1 ESC in 0.25% trypsin EDTA and removed MEFs using a preplating step. Suspended cells were collected and DNA was extracted using standard protocols. Control DNA from mice (C57BL/6J, DBA/2J, B6D2F1) and MEFs (from NMRI mice) was isolated from liver and cultured MEFs, respectively. PCR amplification (touchdown PCR protocol: 20 cycles of 95°C for 15 seconds, 60°C–56°C for 15 seconds; 72°C for 20 seconds, and 15 cycles of 95°C for 15 seconds, 56°C for 15 seconds, and 72°C for 20 seconds) of defined microsatellite markers and electrophoretic separation in 2.5%–3% MoSieve agarose gels (Peqlab) were used to identify genotypes. Primer sequences can be found in Supplemental Table 4, as obtained from www.informatics.jax.org.
Gene array analysis.
Total RNA was extracted with an RNeasy Midi Kit (QIAGEN) with DNAse digestion using the manufacturer’s buffers and protocols. RNA quality was confirmed using an Agilent 2100 Bioanalyzer System. Probe was prepared with 1 μg RNA using the Affymetrix One-Cycle Target Labeling Kit according to the manufacturer’s instructions. Labeled RNA was fragmented by heating to 95°C for 35 minutes in 200 mmol/l Tris (pH 8.1), 500 mmol/l KOAc, 150 mmol/l MgOAc buffer. Affymetrix Mouse Genome 430 2.0 arrays were prehybridized with 200 μl MES hybridization buffer for 10 minutes at 45°C with rotation (60 rpm) in an Affymetrix Hybridization Oven 640. After removing the prehybridization solution, 10 μg of labeled cRNA in 200 μl MES hybridization buffer was applied to the array leaving a small air bubble. Arrays were incubated overnight at 45°C with rotation (60 rpm) in the hybridization chamber. Arrays were scanned using the Affymetrix GeneChip Scanner 3000. CEL files were processed using the BioConductor software package. Normalization was done using robust multichip averaging (RMA) (53). PCA was performed by prcomp in R package statistics. Differential expression was analyzed using Student’s t tests. Transcripts with a fold change greater than 2 and a P value less than 0.05 between 2 conditions were considered to be differentially expressed. Raw P values were adjusted by the Benjamini-Hochberg method (54). Data sets were submitted to the Gene Expression Omnibus database (accession number GSE30868; http://www.ncbi.nlm.nih.gov/geo).
DNA methylation analysis.
Genomic DNA from the A3-PSC line was treated with sodium bisulfite using an EpiTect Bisulfite Kit (QIAGEN) according to the manufacturer’s protocol. All genomic regions selected were then amplified using a nested primer approach. PCR amplifications were performed using SuperTaq polymerase (Ambion, Life Technologies) in a total volume of 25 μl. All PCR amplifications consisted of a total of 40 cycles of denaturation at 94°C for 30 seconds, annealing at the appropriate temperature for each target region for 30 seconds, extension at 72°C for 30 seconds with a first denaturation at 94°C for 5 minutes, and a final extension at 72°C for 10 minutes. For each primer set, 3 μl of product from the first round of PCR was used in the second round of PCR. The amplified products were verified by electrophoresis on 1% agarose gels. The PCR products were subcloned using the PCR 2.1-TOPO vector (Invitrogen) according to the manufacturer’s protocol. Reconstructed plasmids were purified using the QIAprep Spin Miniprep Kit (QIAGEN) and individual clones were sequenced (GATC Biotech). Detailed primer and annealing temperature information is provided in Supplemental Table 5.
Standard karyotyping.
PSC cultures were arrested with 0.2 μg/ml colcemid for 2 hours and dissociated with 0.25% trypsin EDTA. For hypotonic treatment, cells were subjected to 75 mmol/l KCl for 20 minutes at 37°C. After fixation with methanol/acetic acid (3:1) at room temperature, chromosome spreads were prepared by dropping the cell suspension onto prechilled slides. After drying at 90°C for 15 minutes, chromosomes were either stained directly with 10% Giemsa solution (in Weise’s phosphate buffered solution) for 5 minutes or, for GTG-banding, they were treated with 0.25% trypsin solution for 7 seconds and then stained with 10% Giemsa solution for 9 minutes.
Spectral karyotyping (SKY).
Cells were prepared as described above and dropped onto glass slides in a climate chamber (Polymer) at 22°C and 48% humidity. Hybridization with the Mouse SKY Kit (Applied Spectral Imaging) was done for 3 days according to the manufacturer’s instructions. Spectral images were acquired using an epifluorescence microscope equipped with an interferometer (SpectraCube; Applied Spectral Imaging), a custom-designed optical filter, and the SkyView software (Applied Spectral Imaging).
Array comparative genomic hybridization.
Labeling and hybridization of genomic DNA were performed according to the protocol provided by Agilent. Briefly, 3 μg of test DNA (cell line A3, passage 5) and reference DNA (cell line A3 passage 24) were double-digested with AluI and RsaI (Promega) for 2 hours at 37°C. The digested test and reference DNAs were labeled by random priming using the Agilent Genomic DNA Labeling Kit Plus, the test DNA using Cy3-dUTP, and the reference DNA using Cy5-dUTP. Labeled products were purified by Microcon YM-30 filters (Millipore), combined, and then mixed with human Cot-1 DNA (50 μg), Agilent 10X blocking agent, and Agilent 2× hybridization buffer. This solution was hybridized to Agilent’s 244K Mouse Genome comparative genomic hybridization (CGH) microarray (G4415A) at 65°C with 20 rpm rotation (hybridization oven rotator; Agilent G2530-60029) for 40 hours. Washing steps were performed according to the Agilent protocol. Microarray slides were scanned using an Agilent microarray scanner. For image analysis, default CGH settings of Feature Extraction (FE) Software 10.7.31 (Agilent) were applied. FE output files were subsequently imported into Agilent’s CGH data analysis software, Genomic workbench 5.0.14, for DNA copy number analysis using an ADM-2 (aberration detection module) algorithm set to a 6.0 threshold, and the aberration filter was set to 10 probes with a mean log2 ratio of 0.4.
RT-PCR.
Total RNA from PSCs, ESCs, EBs, and fetal (ED15.5), neonatal (postnatal days 1–3), and adult mouse hearts was isolated using Trizol reagent (Invitrogen) following the manufacturer’s instructions. One microgram (qPCR) or 2 μg (RT-PCR) RNA and oligo(dT)12–18 primers were used for cDNA synthesis with SuperScript II RNase H reverse transcriptase (Invitrogen) following the manufacturer’s instructions. One microliter of cDNA (~10–20 ng cDNA) was amplified using primer sequences and PCR conditions as previously described by Guan et al. (55). qPCR (TaqMan and SYBR Green) was performed using a TaqMan ABI Prism 7900 System (Applied Biosystems). Transcript expression levels were normalized to Gapdh or Casq2. Detailed primer and probe information is provided in Supplemental Table 6.
Alkaline phosphatase activity.
Alkaline phosphatase activity was detected with a vector red substrate kit (Vector Laboratories) according to the manufacturer’s instructions.
Confocal laser scanning microscopy.
EB or FACS-purified cells were cultured in 96-well flat-bottom plates or on glass coverslips coated with gelatin (EBs) or laminin (FACS-purified cells). Cells and EHM were fixed in PBS containing 4% formaldehyde and 1% methanol for 5 minutes or overnight, respectively, then blocked and permeabilized for 30 minutes or overnight at room temperature or 4°C, respectively, in TBS solution (pH 7.4) containing 10% FCS, 1% BSA, and 0.5% Triton-X 100, and incubated with indicated primary (overnight for 2 days at 4°C) and suitable secondary (1 hour overnight at 4°C) antibodies (Supplemental Table 7). Filamentous actin was labeled with phalloidin-Alexa 633 (3.3 U/ml). DNA was labeled with 4′,6-diamidino-2-phenylindole (DAPI; 1 μg/ml). Cells were washed extensively with TBS after each incubation step and embedded in Mowiol 4-88. Imaging was performed with Zeiss 510 META or Zeiss 710/NLO confocal laser scanning microscopes.
Teratoma assays.
PSCs were dispersed by exposure to 0.25% trypsin EDTA, resuspended in PBS, and 1 × 106 cells (in 50 μl Iscove’s medium without supplements) were either s.c. or i.m. (quadriceps) injected into immunodeficient SCID mice or immune-competent mice (DBA2, B6, B6D2F1, BALB/C), respectively. After 3 and 4 weeks, respectively, teratoma size was assessed by volumetric measurements of 3D reconstructed echographic images (Vevo770 System equipped with a 20 MHz center frequency scanhead; VisualSonics). Animals were subsequently sacrificed and the teratoma were excised, embedded in paraffin, sectioned at 4 μm, and H&E stained.
FACS.
Nineteen- to 22-day-old EBs from αMHC-EGFP PSCs and R1 ESCs were carefully dispersed with 0.25% trypsin EDTA and triturated to yield single-cell suspensions. Trypsin was inactivated by the addition of medium containing 20% FCS. Cells were subsequently strained through a 30-μm mesh to remove cell aggregates and resuspended in 1 ml Ca2+- and bicarbonate-free Hanks’ solution with HEPES. EGFP-positive cells were separated using an FACSAria Cell-Sorting System (BD). Cell gating was adjusted using EGFP-negative R1 ESCs.
Patch-clamp experiments.
Laminin-coated coverslips containing 200 FACS-purified EGFP-positive PCMs were introduced into a recording chamber mounted on the stage of an inverted microscope (Axiovert 25; Zeiss) and continuously superfused with bath solution at room temperature (21°C–23°C). Spontaneous APs were evaluated using the standard patch-clamp technique in the ruptured patch whole-cell configuration. Patch pipettes were pulled from borosilicate glass (GC150-15; Clark Electromedical Instruments) with an average resistance of 3.3 ± 0.1 M⁄ (n = 102). The pipette solution contained (in mmol/l): K-glutamate 120, KCl 10, MgCl2 2, EGTA 10, HEPES 10, and Na2-ATP 2 titrated to pH 7.20 with KOH. Cells were superfused with a modified Tyrode’s solution (control solution) containing (in mmol/l): NaCl 138, KCl 4, MgCl2 1, NaH2PO4 0.33, CaCl2 2, glucose 10, and HEPES 10 titrated to pH 7.30 with NaOH. In some experiments, tetrodotoxin (1 μmol/l) or nifedipine (1 μmol/l) was added to the bath solution. Membrane potentials were recorded at a sampling rate of 5 kHz using an EPC-9 amplifier controlled by PULSE software (HEKA Elektronik). Membrane capacitance and series resistance were calculated using automated procedures. The series resistance averaged 8.9 ± 0.5 M⁄ (n = 102). APs were recorded immediately after patch rupture for at least 2 minutes and analyzed offline using Igor (WaveMetrics) to determine the MDP, APA, the maximum rise rate of the AP upstroke (max dV/dt), and the AP duration at 20%, 50%, and 90% repolarization (APD20, APD50, APD90).
Blastocyst injection.
Chimeras were generated according to standard protocols by injecting PSCs (PSC line A3) into wild-type or αMHC-nLacZ blastocysts, respectively, and then transferred into pseudopregnant SW/J surrogate mothers.
LacZ staining.
Cryosections were stained with X-gal staining solution as follows (in mmol/l): MgCl 2, K4Fe(CN)6 5, K3Fe(CN)6 5, HEPES 20, 1 mg/ml 5-bromo-4-chloro-3-indolyl-β-D-galactoside (X-gal), in PBS, adjusted to pH 7.4, at 37°C for 6 hours.
Acetylcholinesterase staining.
Air-dried (3 minutes) cryosections were fixed in ice-cold 4% neutral formalin for 15–25 minutes, washed with H2O, and incubated for 2 hours at 37°C in an aqueous staining solution (50 ml) containing 25 mg acetylthiocholine iodide, 0.82% sodium acetate, 0.6% acetic acid, 2.94% sodium citrate, 0.75% cupric sulfate, 0.137% iso-OMPA, and 0.165% potassium ferricyanide. Sections were subsequently washed in PBS (10 minutes), dried, and mounted.
Two-photon laser scanning microscopy of intracellular calcium transients.
Three weeks following intracardiac delivery of PCMs (100,000 cells per heart), recipient SCID mice received i.p. injections of heparin (200 IU) and were sacrificed via cervical dislocation. The hearts were rapidly removed and retrogradely perfused for 15 minutes at constant pressure with oxygenated Tyrode’s solution containing rhod-2/AM (10 μmol/l). Following an approximate 20-minute washout period with dye-free solution, two-photon laser scanning microscopy was performed as described previously (27). For imaging Ca2+ transients in day 15 neonatal (n = 2) and 3-month-old (n = 1) chimeric hearts, mice were given i.p. 100 IU heparin and loaded with rhod-2/AM as above. In all experiments, the excitation wavelength was 810 nm, and the rhod-2 and EGFP fluorescence signals were simultaneously collected by 2 photomultipliers equipped with 560–650 nm and 500–550 nm band pass filters, respectively. A 1.2 NA water immersion ×60 objective was used. All images were obtained during spontaneous or remote point stimulation-evoked depolarizations in the presence of the excitation-contraction uncoupler cytochalasin D (50 μmol/l).
Directed differentiation of PSC-EB cultures.
PCMs were generated using a serum-free induction protocol described previously (24). Briefly, PSCs were maintained on irradiated MEFs in serum-free ESC media (SFES) consisting of 1:1 neurobasal medium (Invitrogen) and DMEM/F12 containing 0.5× of N2/B27 supplement, 50-P/S, glutamine (2 mmol/l), BSA (0.05%), leukemia inhibitory factor (0.5% conditioned medium), BMP4 (10 ng/ml; R&D Systems), and 1-thioglycerol (150 μmol/l; Sigma-Aldrich). Cells were passaged following dissociation with TrypLE (Invitrogen). Prior to differentiation induction, cultures were depleted of MEFs by 2 passages on gelatin-coated dishes in SFES media. Feeder-depleted PSCs were dissociated and cultured (1 × 105 cells/ml) in SF differentiation (SFD) media consisting of 3:1 Iscove’s modified Dulbecco’s medium (IMDM)/F12 media containing 0.5× N2/B27 supplement, 50-P/S, 2 mmol/l glutamine (Gibco), BSA (0.05%), ascorbic acid (0.5 mmol/l), and 1-thioglycerol (4.5 μmol/l) for 2 days without additional growth factors to generate EBs. On day 2, the EBs were dissociated and allowed to reaggregate (1 × 105 cells/ml) in SFD media in the presence of human VEGF (5 ng/ml), human activin A (8 ng/ml), and human BMP4 (0.5 ng/ml; all R&D Systems) for an additional 28 hours. At this stage, EBs were harvested and cultured for an additional 3–4 days in StemPro-34 media (Invitrogen) supplemented with ascorbic acid (1 mmol/l), glutamine (2 mmol/l), human VEGF (5 ng/ml), human bFGF (10 ng/ml), and human DKK1 (150 ng/ml; all R&D Systems). Following induction, the EBs were transferred to IMDM supplemented with 10% serum (Tissue Culture Biologicals), ascorbic acid (1 mmol/l), and glutamine (2 mmol/l) for express shipment to Göttingen at ambient temperature.
EHM construction.
PSC-EBs were dispersed with 0.25% trypsin EDTA. Cells (1.5 × 106) were mixed with pH-neutralized (0.1 N NaOH) collagen type I from rat tails (0.4 mg/EHM), Engelbreth-Holm-Swarm tumor exudate (10% vol/vol; Tebu), and concentrated serum–containing culture medium (2× DMEM, 20% horse serum, 4% chick embryo extract, 200 P/S). Subsequently, the mixture was poured into casting molds (volume: 450 μl) and the EHM was cultured in EHM medium (DMEM, 10% horse serum, 2% chick embryo extract, and 100 P/S). After 3 days, the EHM was transferred to static stretch devices and cultivated for another 5 days. Twenty-five micromoles per liter cytosine-arabinoside (Ara C) was added for 2 days after transfer of the EHM onto stretch devices to prevent nonmyocyte overgrowth. Alternatively, EHM was generated from G418-purified EB cultures (double transgenic PSC line A3/αMHC-neoR) and supplemented with cardiac fibroblasts from DBA mice (25% of the total cell number).
Isometric force measurement.
After 8 days in culture, the EHMs were transferred to thermostatted organ baths (Föhr Medical Instruments), isometrically suspended, and electrically stimulated at 4 Hz. Extracellular calcium concentration was increased stepwise from 0.2 to 2.4 mmol/l. Single contractions and amplitude of contractile force (twitch tension) were evaluated with BMON2 software (Ingenieurbüro Jäckel). Alternatively, mouse ventricular wedges were cut from left ventricles of adult DBA mice and subjected to isometric force measurement under similar conditions as described above for the EHM.
MI and EHM implantation.
Mice (DBA2J) were anesthetized with isoflurane and intubated. Anesthesia was maintained by continuous inhalation of 1.5% isoflurane in room air. After left lateral thoracotomy and pericardiotomy, the left anterior descending artery (LAD) was visualized and ligated (9-0 Prolene; Ethicon). Successful ligation was confirmed by bleaching and hypokinesis of the myocardium. Fifteen to 20 minutes after LAD ligation, EHM grafts (viable or nonviable) were cut in half and sutured (9-0 Prolene; Ethicon) onto the evolving MI. Following expansion of the lungs, the thorax and skin were closed (6-0 Prolene; Ethicon). Isoflurane inhalation was stopped and the mice were ventilated until the onset of spontaneous breathing. Metamizole (1.33 mg/ml) was provided with drinking water for postoperative analgesia for 7 days. Sham surgeries were performed without LAD ligation. Control surgeries included LAD ligation and mock sutures as if EHM grafts were implanted. F-EHM was implanted on infarcted myocardium as nonviable controls. Two mice died prematurely in the sham group, and 1 mouse died prematurely in the EHM group. Two mice in the control group, 2 mice in the F-EHM group, and 1 mouse in the EHM group were excluded from analysis because they showed no clear demarcation of the anterior myocardial wall. All mice received methylprednisolone (5 mg/kg body weight per day; s.c. injections).
Echocardiography.
Heart function was assessed 14 days after the surgical intervention with a Vevo2100 System (30 MHz scan head; VisualSonics).
Statistics.
Data are presented as mean ± SEM. Statistical analyses were performed using an unpaired, 2-tailed Student’s t test or ANOVA with Dunnett’s multiple comparison test as indicated. Significance was defined as P < 0.05.
Study approval.
Animal studies were approved by the Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit (LAVES; 33.11.42502-04-020/09, -04-022/09, -05-A-994/09).
Supplementary Material
Acknowledgments
This work was supported by the DZHK (German Center for Cardiovascular Research), BMBF (German Federal Ministry of Education and Research; FKZ 01GN0827, FKZ 01GN0957), the DFG (German Research Foundation; ZI 708/7-1, 8-1 [FOR604], 10-1 [KFO155], SFB 1002, SPP1356 ZE 432/5-2, and Cluster of Excellence REBIRTH), a Klaus-Georg-und-Sigrid-Hengstberger-Forschungsstipendium, and the Forschungsförderungsfonds of the University Medical Center Hamburg-Eppendorf. We would like to thank T. Wakayama of the RIKEN Center for Developmental Biology in Kobe, Japan, for training M. Didié and W.H. Zimmermann in mouse blastocyst handling and manipulation (BMBF JPN 05/A14). We also acknowledge the excellent technical assistance of S. Feldhaus and R. Blume. Indicated antibodies were developed by D. Solter and B.B. Knowles (antibody against FUT4), and D.A. Fischman (antibody against myosin), and obtained from the Developmental Studies Hybridoma Bank.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(3):1285–1298. doi:10.1172/JCI66854.
See the related Commentary beginning on page 1010.
References
- 1.Neaves WB, Baumann P. Unisexual reproduction among vertebrates. Trends Genet. 2011;27(3):81–88. doi: 10.1016/j.tig.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Surani MA, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308(5959):548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- 3.Brevini TA, Gandolfi F. Parthenotes as a source of embryonic stem cells. Cell Prolif. 2008;41(suppl 1):20–30. doi: 10.1111/j.1365-2184.2008.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linder D, McCaw BK, Hecht F. Parthenogenic origin of benign ovarian teratomas. N Engl J Med. 1975;292(2):63–66. doi: 10.1056/NEJM197501092920202. [DOI] [PubMed] [Google Scholar]
- 5.Cibelli JB, et al. Parthenogenetic stem cells in nonhuman primates. Science. 2002;295(5556):819. doi: 10.1126/science.1065637. [DOI] [PubMed] [Google Scholar]
- 6.Allen ND, Barton SC, Hilton K, Norris ML, Surani MA. A functional analysis of imprinting in parthenogenetic embryonic stem cells. Development. 1994;120(6):1473–1482. doi: 10.1242/dev.120.6.1473. [DOI] [PubMed] [Google Scholar]
- 7.Revazova ES, et al. Patient-specific stem cell lines derived from human parthenogenetic blastocysts. Cloning Stem Cells. 2007;9(3):432–449. doi: 10.1089/clo.2007.0033. [DOI] [PubMed] [Google Scholar]
- 8.Lin G, et al. A highly homozygous and parthenogenetic human embryonic stem cell line derived from a one-pronuclear oocyte following in vitro fertilization procedure. Cell Res. 2007;17(12):999–1007. doi: 10.1038/cr.2007.97. [DOI] [PubMed] [Google Scholar]
- 9.Robertson EJ, Evans MJ, Kaufman MH. X-chromosome instability in pluripotential stem cell lines derived from parthenogenetic embryos. J Embryol Exp Morphol. 1983;74:297–309. [PubMed] [Google Scholar]
- 10.Chen Z, et al. Birth of parthenote mice directly from parthenogenetic embryonic stem cells. Stem Cells. 2009;27(9):2136–2145. doi: 10.1002/stem.158. [DOI] [PubMed] [Google Scholar]
- 11.Fundele RH, et al. Temporal and spatial selection against parthenogenetic cells during development of fetal chimeras. Development. 1990;108(1):203–211. doi: 10.1242/dev.108.1.203. [DOI] [PubMed] [Google Scholar]
- 12.Nagy A, Sass M, Markkula M. Systematic non-uniform distribution of parthenogenetic cells in adult mouse chimaeras. Development. 1989;106(2):321–324. doi: 10.1242/dev.106.2.321. [DOI] [PubMed] [Google Scholar]
- 13.Spindle A, Sturm KS, Flannery M, Meneses JJ, Wu K, Pedersen RA. Defective chorioallantoic fusion in mid-gestation lethality of parthenogenone<-->tetraploid chimeras. Dev Biol. 1996;173(2):447–458. doi: 10.1006/dbio.1996.0039. [DOI] [PubMed] [Google Scholar]
- 14.Sturm KS, Flannery ML, Pedersen RA. Abnormal development of embryonic and extraembryonic cell lineages in parthenogenetic mouse embryos. Dev Dyn. 1994;201(1):11–28. doi: 10.1002/aja.1002010103. [DOI] [PubMed] [Google Scholar]
- 15.Vrana KE, et al. Nonhuman primate parthenogenetic stem cells. Proc Natl Acad Sci U S A. 2003;100(suppl 1):11911–11916. doi: 10.1073/pnas.2034195100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laflamme MA, Murry CE. Heart regeneration. Nature. 2012;473(7347):326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bühler K, et al. DIR Annual 2010. J Reproduktionsmed Endokrinol. 2011;8(4):253–280. [Google Scholar]
- 18.Mai Q, et al. Derivation of human embryonic stem cell lines from parthenogenetic blastocysts. Cell Res. 2007;17(12):1008–1019. doi: 10.1038/cr.2007.102. [DOI] [PubMed] [Google Scholar]
- 19.Kim K, et al. Recombination signatures distinguish embryonic stem cells derived by parthenogenesis and somatic cell nuclear transfer. Cell Stem Cell. 2007;1(3):346–352. doi: 10.1016/j.stem.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Taylor CJ, Bolton EM, Pocock S, Sharples LD, Pedersen RA, Bradley JA. Banking on human embryonic stem cells: estimating the number of donor cell lines needed for HLA matching. Lancet. 2005;366(9502):2019–2025. doi: 10.1016/S0140-6736(05)67813-0. [DOI] [PubMed] [Google Scholar]
- 21.Nakajima F, Tokunaga K, Nakatsuji N. Human leukocyte antigen matching estimations in a hypothetical bank of human embryonic stem cell lines in the Japanese population for use in cell transplantation therapy. Stem Cells. 2007;25(4):983–985. doi: 10.1634/stemcells.2006-0566. [DOI] [PubMed] [Google Scholar]
- 22.Fernandes S, Kuklok S, McGonigle J, Reinecke H, Murry CE. Synthetic matrices to serve as niches for muscle cell transplantation. Cells Tissues Organs. 2012;195(1–2):48–59. doi: 10.1159/000331414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmermann WH, et al. Engineered heart tissue grafts improve systolic and diastolic function in infarcted rat hearts. Nat Med. 2006;12(4):452–458. doi: 10.1038/nm1394. [DOI] [PubMed] [Google Scholar]
- 24.Kattman SJ, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8(2):228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 25.Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10(1):16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klug MG, Soonpaa MH, Koh GY, Field LJ. Genetically selected cardiomyocytes from differentiating embryonic stem cells form stable intracardiac grafts. J Clin Invest. 1996;98(1):216–224. doi: 10.1172/JCI118769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubart M, Pasumarthi KB, Nakajima H, Soonpaa MH, Nakajima HO, Field LJ. Physiological coupling of donor and host cardiomyocytes after cellular transplantation. Circ Res. 2003;92(11):1217–1224. doi: 10.1161/01.RES.0000075089.39335.8C. [DOI] [PubMed] [Google Scholar]
- 28.Dubois NC, et al. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat Biotechnol. 2011;29(11):1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elliott DA, et al. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods. 2011;8(12):1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- 30.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci U S A. 1993;90(18):8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 32.Sharova LV, et al. Global gene expression profiling reveals similarities and differences among mouse pluripotent stem cells of different origins and strains. Dev Biol. 2007;307(2):446–459. doi: 10.1016/j.ydbio.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim K, et al. Histocompatible embryonic stem cells by parthenogenesis. Science. 2007;315(5811):482–486. doi: 10.1126/science.1133542. [DOI] [PubMed] [Google Scholar]
- 34.Do JT, et al. Generation of parthenogenetic induced pluripotent stem cells from parthenogenetic neural stem cells. Stem Cells. 2009;27(12):2962–2968. doi: 10.1002/stem.233. [DOI] [PubMed] [Google Scholar]
- 35.Mitalipov S, Clepper L, Sritanaudomchai H, Fujimoto A, Wolf D. Methylation status of imprinting centers for H19/IGF2 and SNURF/SNRPN in primate embryonic stem cells. Stem Cells. 2007;25(3):581–588. doi: 10.1634/stemcells.2006-0120. [DOI] [PubMed] [Google Scholar]
- 36.Boheler KR, Czyz J, Tweedie D, Yang HT, Anisimov SV, Wobus AM. Differentiation of pluripotent embryonic stem cells into cardiomyocytes. Circ Res. 2002;91(3):189–201. doi: 10.1161/01.RES.0000027865.61704.32. [DOI] [PubMed] [Google Scholar]
- 37.Kehat I, et al. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108(3):407–414. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zwi L, et al. Cardiomyocyte differentiation of human induced pluripotent stem cells. Circulation. 2009;120(15):1513–1523. doi: 10.1161/CIRCULATIONAHA.109.868885. [DOI] [PubMed] [Google Scholar]
- 39.Mauritz C, et al. Generation of functional murine cardiac myocytes from induced pluripotent stem cells. Circulation. 2008;118(5):507–517. doi: 10.1161/CIRCULATIONAHA.108.778795. [DOI] [PubMed] [Google Scholar]
- 40.Guan K, et al. Generation of functional cardiomyocytes from adult mouse spermatogonial stem cells. Circ Res. 2007;100(11):1615–1625. doi: 10.1161/01.RES.0000269182.22798.d9. [DOI] [PubMed] [Google Scholar]
- 41.Ieda M, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142(3):375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qian L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485(7400):593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song K, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485(7400):599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwartz SD, et al. Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet. 2012;379(9817):713–720. doi: 10.1016/S0140-6736(12)60028-2. [DOI] [PubMed] [Google Scholar]
- 45.Zimmermann WH. Embryonic and embryonic-like stem cells in heart muscle engineering. J Mol Cell Cardiol. 2011;50(2):320–326. doi: 10.1016/j.yjmcc.2010.10.027. [DOI] [PubMed] [Google Scholar]
- 46.van Laake LW, et al. Extracellular matrix formation after transplantation of human embryonic stem cell-derived cardiomyocytes. Cell Mol Life Sci. 2010;67(2):277–290. doi: 10.1007/s00018-009-0179-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kehat I, et al. Electromechanical integration of cardiomyocytes derived from human embryonic stem cells. Nat Biotechnol. 2004;22(10):1282–1289. doi: 10.1038/nbt1014. [DOI] [PubMed] [Google Scholar]
- 48.Shiba Y, et al. Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature. 2012;489(7415):322–325. doi: 10.1038/nature11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zimmermann WH, et al. Tissue engineering of a differentiated cardiac muscle construct. Circ Res. 2002;90(2):223–230. doi: 10.1161/hh0202.103644. [DOI] [PubMed] [Google Scholar]
- 50.Streckfuss-Bomeke K, et al. Eur Heart J. Comparative study of human-induced pluripotent stem cells derived from bone marrow cells, hair keratinocytes, and skin fibroblasts [published online ahead of print July 12, 2012]. doi: 10.1093/eurheartj/ehs203 . [DOI] [PubMed] [Google Scholar]
- 51.Soong PL, Tiburcy M, Zimmermann WH. Cardiac differentiation of human embryonic stem cells and their assembly into engineered heart muscle. Curr Protoc Cell Biol. 2012;Chapter 23:Unit23.8. doi: 10.1002/0471143030.cb2308s55. [DOI] [PubMed] [Google Scholar]
- 52.Wobus AM, Wallukat G, Hescheler J. Pluripotent mouse embryonic stem cells are able to differentiate into cardiomyocytes expressing chronotropic responses to adrenergic and cholinergic agents and Ca2+ channel blockers. . Differentiation. 1991;48(3):173–182. doi: 10.1111/j.1432-0436.1991.tb00255.x. [DOI] [PubMed] [Google Scholar]
- 53.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 54.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B. 1995;57:289–300. [Google Scholar]
- 55.Guan K, et al. Pluripotency of spermatogonial stem cells from adult mouse testis. Nature. 2006;440(7088):1199–1203. doi: 10.1038/nature04697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.