Abstract
Ataxia-telangiectasia (A-T) is an autosomal recessive neurodegenerative disorder characterized by radiosensitivity, genomic instability, and predisposition to cancer. A-T is caused by biallelic mutations in the ataxia-telangiectasia mutated (ATM) gene, but heterozygous carriers, though apparently healthy, are believed to be at increased risk for cancer and more sensitive to ionizing radiation than the general population. Despite progress in functional and sequencing-based assays, no straightforward, rapid, and inexpensive test is available for the identification of A-T homozygotes and heterozygotes, which is essential for diagnosis, genetic counseling, and carrier prediction. The oncosuppressor p53 prevents genomic instability and centrosomal amplification. During mitosis, p53 localizes at the centrosome in an ATM-dependent manner. We capitalized on the latter finding and established a simple, fast, minimally invasive, reliable, and inexpensive test to determine mutant ATM zygosity. The percentage of mitotic lymphoblasts or PBMCs bearing p53 centrosomal localization clearly discriminated among healthy donors (>75%), A-T heterozygotes (40%–56%), and A-T homozygotes (<30%). The test is specific for A-T, independent of the type of ATM mutations, and recognized tumor-associated ATM polymorphisms. In a preliminary study, our test confirmed that ATM is a breast cancer susceptibility gene. These data open the possibility of cost-effective, early diagnosis of A-T homozygotes and large-scale screenings for heterozygotes.
Introduction
Ataxia-telangiectasia (A-T) is a rare autosomal recessive multisystemic syndrome characterized by progressive cerebellar degeneration, telangiectasias, immunodeficiency, recurrent infections, insulin-resistant diabetes, premature aging, radiosensitivity, and high risk for malignancy, particularly leukemia and lymphoma in children and epithelial cancers in surviving adults (1, 2). No effective disease-modifying therapy is presently available, though early supportive symptomatic medications and aggressive physical stimulation can minimize infections and delay neurological symptoms (3).
In A-T, mutations in the ATM (ataxia-telangiectasia mutated) gene result in a lack or inactivation of the ATM protein, a ubiquitous serine/threonine kinase of 3056 amino acids mainly involved in the maintenance of genomic stability (4–7). The ATM gene contains 66 exons spanning approximately 160 Kb of genomic DNA (8), and a large variety (>600) of mutations occurs across the full-length transcript without hotspots. In addition, like other large genes, ATM possesses many polymorphisms that must be distinguished from mutations (9, 10).
A-T is present throughout the world, with an incidence of 1 in 10,000 to 100,000 newborns (11). However, because of the difficulties in diagnosis, particularly in those children who die at a young age, A-T might actually be more frequent than currently estimated. The theoretical frequency of A-T mutant allele heterozygosity (A-T carriers) has been calculated as 1.4%–2% of the general population, though slight variations can be encountered in different countries (2, 11, 12). A-T heterozygotes are usually asymptomatic and largely considered healthy carriers, but compared with the general population, they have been reported to be more vulnerable to ionizing radiation (IR) and to have a higher risk of ischemic heart disease (2) and breast, stomach, and lung cancers (4). Though definitive information on these susceptibilities is not yet available, regular surveillance of A-T carriers is considered part of A-T management in carrier families.
A-T diagnosis is based on the combination of clinical features with laboratory tests showing high levels of serum alpha-fetoprotein, cell sensitivity to IR, and reduced or absent levels of ATM protein. None of these tests, alone or in combination, is sufficiently sensitive and specific for early differential diagnosis, genetic counseling, and carrier prediction (3). Because of the complex genomic organization of the ATM gene, its direct sequencing is not yet cost effective, particularly for large-scale surveys of A-T carriers. A further level of complexity is established by the fact that a majority of the ATM mutations in A-T patients are protein truncations or splice junction variants, while missense mutations in evolutionarily conserved residues are more common in breast cancer (BC) patients than in controls (13). Functional assays that are able to distinguish between deleterious and neutral alterations in A-T carriers are thus necessary. In the past 3 decades, an intense effort has therefore been dedicated to developing rapid and reliable methods for identifying A-T homozygotes and heterozygotes. However, with the exception of gene sequencing, none of the assays presently available is unambiguously diagnostic without the support of clinical symptoms or can identify A-T carriers in the absence of a direct intratest comparison (14–21).
In the process of studying the role of the tumor suppressor p53 in response to mitotic inhibitory drugs, we discovered that, in mitosis, p53 localizes at the centrosomes in an ATM-dependent manner and monitors mitotic spindle integrity (Figure 1A) (22–24). These findings led us to propose that both the ATM and p53 proteins might contribute to the “centrosomal checkpoint,” a network that integrates cell cycle arrest and repair signals (25, 26). Although a clear mechanistic proof for this hypothesis remains to be established, we show here that p53 does not localize at the centrosomes in almost 100% of mitotic lymphoblastoid cell lines (LCLs) or PBMCs derived from A-T patients. Surprisingly, we consistently observed that in A-T heterozygous carriers, p53 localizes at the centrosomes in approximately 50% of the mitotic cells. Based on these findings, we have developed a straightforward, rapid, and inexpensive test to unambiguously diagnose A-T homozygotes and heterozygotes.
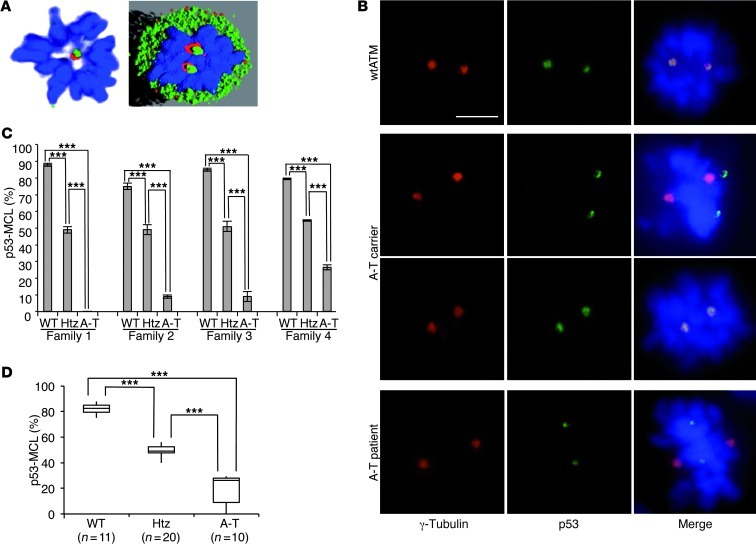
Figure 1. p53 centrosomal localization in mitotic LCLs.
(A) p53 centrosomal localization detected by confocal IF microscopy with anti-p53 (green) and anti–γ-tubulin (red) Abs. DNA was stained with DAPI (blue); representative image and its software analysis are shown in A. (B) Representative IF images of the indicated proteins in mitotic LCLs derived from the indicated donors. p53, centrosomes, and DNA are marked as in A. Scale bar = 5 μm. (C) Summary data of percentages of p53-MCL measured by 2 different operators who counted 100 metaphases per sample in quadruplicate. Data are expressed as mean ± SD. Four different families, each including 1 A-T patient (A-T), 1 parent (Htz), and 1 wtATM-carrying relative (WT) were evaluated. (D) Comparison of p53-MCL percentages, evaluated as in C, between LCLs derived from wtATM donors (n = 11), A-T carriers (n = 20), and A-T patients (n = 10). The 3 groups are significantly different. Box-and-whisker plot: horizontal lines within the box represent median values, and the bottom and top of the box show the lower and upper quartiles. ***P < 0.0001; 2-tailed Student’s t test.
Results
p53 mitotic centrosomal localization determines mutant ATM zygosity.
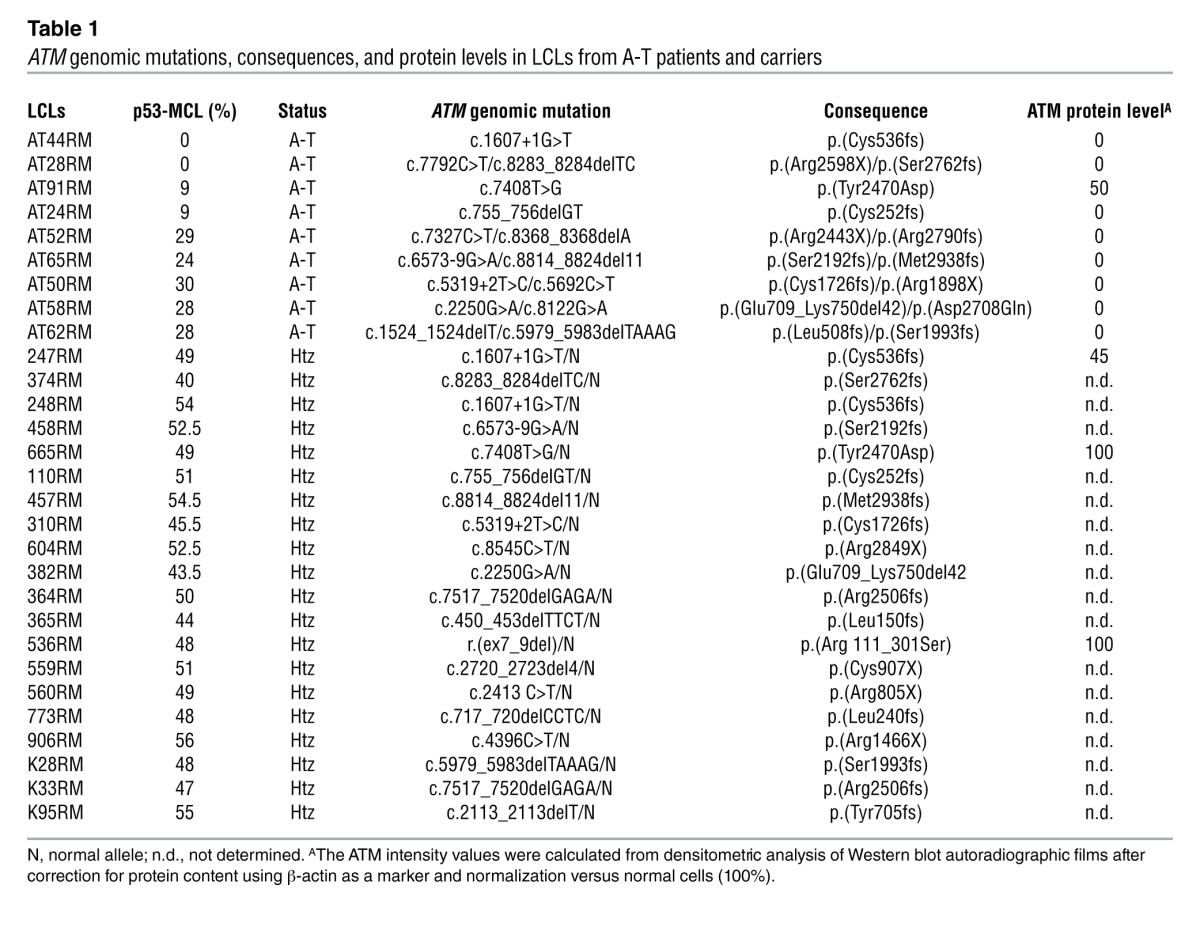
Centrosomes duplicate at each cell cycle and are the major organizers of the bipolar mitotic spindle. A number of different proteins reside at the centrosome permanently, while some transiently localize to the centrosome during the cell cycle or in response to damage (27). Since we previously demonstrated that p53 moves to centrosomes at each mitosis in an ATM-dependent manner (22–24), we blindly measured the percentage of mitotic cells with p53 centrosomal localization (p53 mitotic centrosomal localization, or p53-MCL) in EBV-derived LCLs obtained from 4 A-T patients, 1 of their parents (the obligate A-T carriers), and from 4 wild-type ATM (wtATM) relatives (families 1–4). To this end, a double indirect immunofluorescence (IF) analysis was performed with anti-p53 and anti–γ-tubulin Abs (see Methods for details). We observed 3 completely different behaviors: wtATM donors had p53-MCL in more than 75% of their mitotic LCLs; A-T patients had an opposite behavior, with less than 30% p53-MCL; and A-T carriers expressed an intermediate phenotype, with approximately 50% p53-MCL in their LCLs (Figure 1, B and C). Because of this initial clear-cut readout, we asked whether this p53 behavior could be used to determine mutant ATM zygosity. Thus, we blindly measured, in 3 different laboratories, p53-MCL in 30 LCLs obtained from 10 A-T homozygotes and 20 heterozygotes carrying different types of ATM mutations (e.g., deletions, insertions, and missense mutations) that result in different levels of ATM protein expression (e.g., from absence to an approximate physiological amount) (Table 1). Eleven LCLs from healthy donors with wtATM assessed by direct sequencing were analyzed for comparison. As shown in Figure 1D, the percentage of mitotic cells with p53-MCL, assessed by double indirect IF, can easily and significantly (P < 0.0001) discriminate among the 3 genotypes independently of the type of ATM mutations.
Table 1.
ATM genomic mutations, consequences, and protein levels in LCLs from A-T patients and carriers
Inhibition of p53 centrosomal localization is A-T specific.
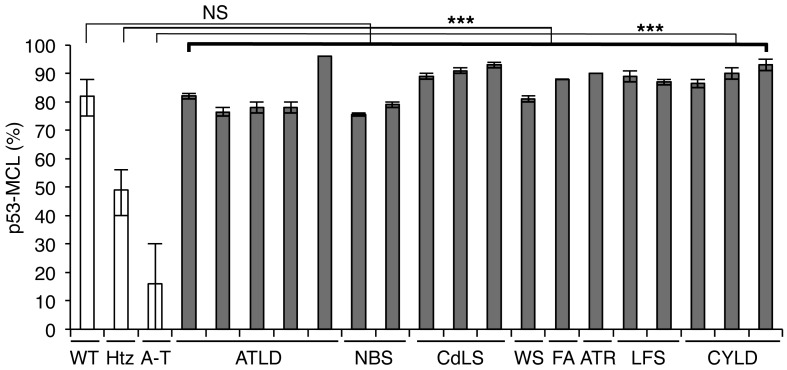
ATM is a key sensor of the DNA double-strand breaks that critically contribute to the DNA damage response by activating p53 as well as a large series of regulators of cell-cycle checkpoints, DNA repair machinery, replication forks, and telomeres (4–7). Mutations in some of these tATM substrates, such as MRE11, NBS1, SMC1A, and WRN have been shown to cause different syndromes (i.e., A-T–like disorder, Nijmegen breakage syndrome, Cornelia de Lange syndrome, and Werner syndrome, respectively) that can share some clinical symptoms and/or cellular characteristics with A-T (4, 21, 28, 29). Thus, we evaluated whether p53-MCL can also be affected in these syndromes by analyzing LCLs from different patients. As shown in Figure 2, the percentage of mitotic cells with p53-MCL was comparable to that of wtATM-carrying cells in all LCLs, supporting an ATM-specific role for the centrosomal localization of p53. This result is consistent with the DNA damage independence of ATM-mediated p53 localization at the centrosome during mitosis (23, 24). To further assess the specificity of p53-MCL in A-T, we analyzed LCLs from 1 patient affected by radiosensitive Fanconi anemia group A; 1 patient with the Seckel syndrome variant caused by mutation of ATR, the ATM- and Rad3-related protein that phosphorylates p53 at the same Ser15 residue phosphorylated by ATM; 2 Li-Fraumeni syndrome patients carrying 2 different p53 mutations; and 3 patients with cylindromatosis, an unrelated syndrome caused by mutation in the CYLD oncosuppresor gene. Also, in these cases (Figure 2), the percentage of mitotic cells with p53-MCL was comparable to that of healthy donors. Thus, these results support high A-T specificity in p53-MCL evaluation, indicating that this specificity can be used as a test to identify A-T homozygotes and heterozygotes.
Figure 2. p53-MCL in LCLs derived from patients with different syndromes.
LCLs derived from patients affected by syndromes other than A-T showed a percentage of p53-MCL comparable to that of wtATM-carrying donors. White columns indicate the data shown in Figure 1D; gray columns indicate the percentage of p53-MCL for each LCL analyzed by 2 different operators who examined 100 metaphases per sample in quadruplicate. Data are expressed as mean ± SD. ***P < 0.0001; two-tailed Student’s t test. NS, not significant; ATLD, A-T–like disorder; NBS, Nijmegen breakage syndrome; CdLS, Cornelia de Lange syndrome; WS, Werner syndrome; FA, Fanconi anemia group A; ATR, Seckel syndrome; LFS, Li-Fraumeni syndrome; CYLD, cylindromatosis.
p53-MCL detection in PBMCs is a highly accurate assay.
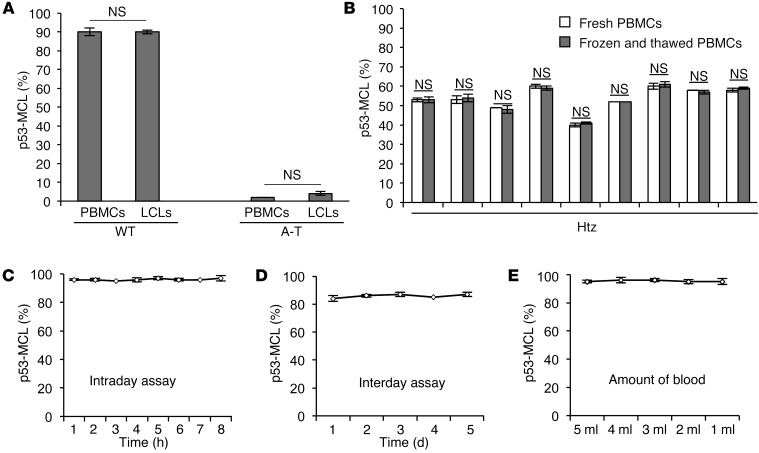
We then asked whether the p53-MCL assay could be performed on PBMCs. Freshly isolated PBMCs from 1 A-T patient, 1 A-T carrier, and 1 healthy donor were stimulated to proliferate by incubation with phytohemagglutinin (PHA). Since p53 moves to the centrosome in a microtubule-dependent manner (22), we could not rely on the use of colchicine or other inhibitors of microtubule polymerization. Thus, the percentage of p53-MCL was assayed at different time points after PHA stimulation (Table 2). No significant difference was observed in the donors at the different times. However, the 60-hour PHA-stimulation time point showed the largest quantity of mitotic cells in the 3 samples, hence, all subsequent experiments were performed using 60-hour PHA stimulation. When double indirect p53 and anti–γ-tubulin IF was performed, no difference in the percentage of cells with p53-MCL was observed between LCLs and PBMCs derived from 1 A-T patient and 1 healthy donor (Figure 3A), or in PBMCs derived from 9 A-T carriers and analyzed as fresh cell populations, or after frozen in DMSO and then thawed (Figure 3B). These findings indicate that p53-MCL is not modified by EBV-mediated immortalization or cell manipulation. Of relevance, these 2 characteristics allow for testing on stored PBMCs, obviating the need for laborious and time-consuming stabilization of LCLs. In addition, we found that the assay could be efficiently performed by using PBMCs isolated from 2-day-old samples of whole blood maintained at room temperature and containing heparin, EGTA, or lithium chloride as anticoagulants (data not shown).
Table 2.
Time-course study of p53-MCL among wild-type, heterozygous, and homozygous carriers
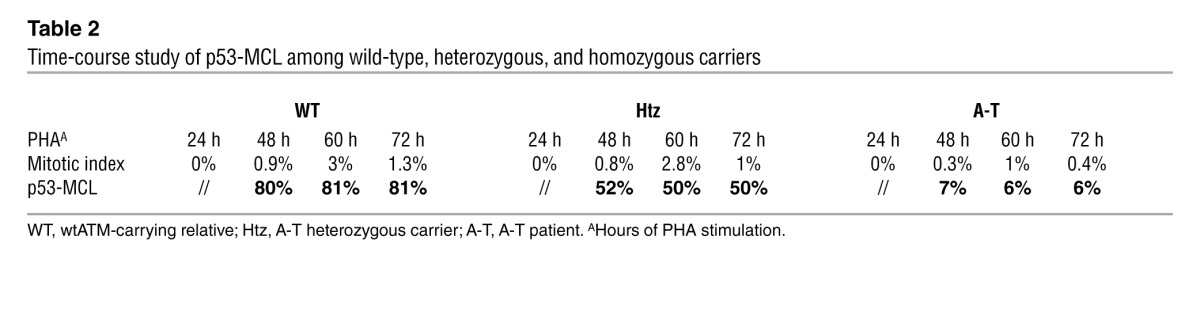
Figure 3. Assessment of p53-MCL test precision.
(A) Summary data of p53-MCL percentages in PBMCs and LCLs derived from 1 A-T patient and 1 healthy donor. No significant difference was observed between the 2 groups. Two-tailed Student’s t test. (B) Summary data of p53-MCL percentages in fresh PBMCs and frozen and thawed PBMCs. No significant difference was observed after freezing and thawing PBMCs from 9 different A-T carriers. (C) Intraday assay variability testing was performed on a single blood sample from 1 healthy donor as follows: PBMCs were PHA-stimulated for 60 hours and used to make 8 different coverslip preparations, each of which was immunostained at the indicated times after fixation. No significant difference was observed between the preparations. (D) Interday assay variability was performed by collecting and assessing PBMCs from the same healthy donor on 5 different days. No significant difference was observed among the samples. (E) To verify the minimal amount of blood required to perform the test, we collected 15 ml of blood from a single healthy donor and made 5 aliquots with different amounts of blood (from 1 to 5 ml). The test performed on PBMCs purified from the 5 different samples showed no significant differences, demonstrating that the assay can be performed with as little as 1 ml of whole blood. A 2-tailed Student’s t test was performed. All samples were analyzed by examining 100 metaphases per sample in quadruplicate. Data are expressed as mean ± SD. NS, not significant.
Next, we evaluated the test’s precision by assessing its intraday and interday variability. In the first case, PBMCs were isolated on day 0 from a healthy donor, stimulated with PHA for 60 hours, and used to make 8 different coverslip preparations that were immediately fixed. Double indirect IF was performed on 1 coverslip every hour, 8 times. For the interday variability, blood samples and the relative PBMCs from a single healthy donor were collected and assessed as described, on 5 different days, with a 2- to 3-month interval between the 5 days. No significant variations were observed among the samples for both intraday (coefficient of variation [CV] = 0.6%) (Figure 3C) and interday (CV = 1.4%) (Figure 3D) assays. Finally, we verified the minimal amount of blood required to perform the test. As shown in Figure 3E, as little as 1 ml of whole blood was necessary, indicating that the assay can also be performed for diagnosis in young children.
p53-MCL assessed in PBMCs identifies BC-associated ATM variants.
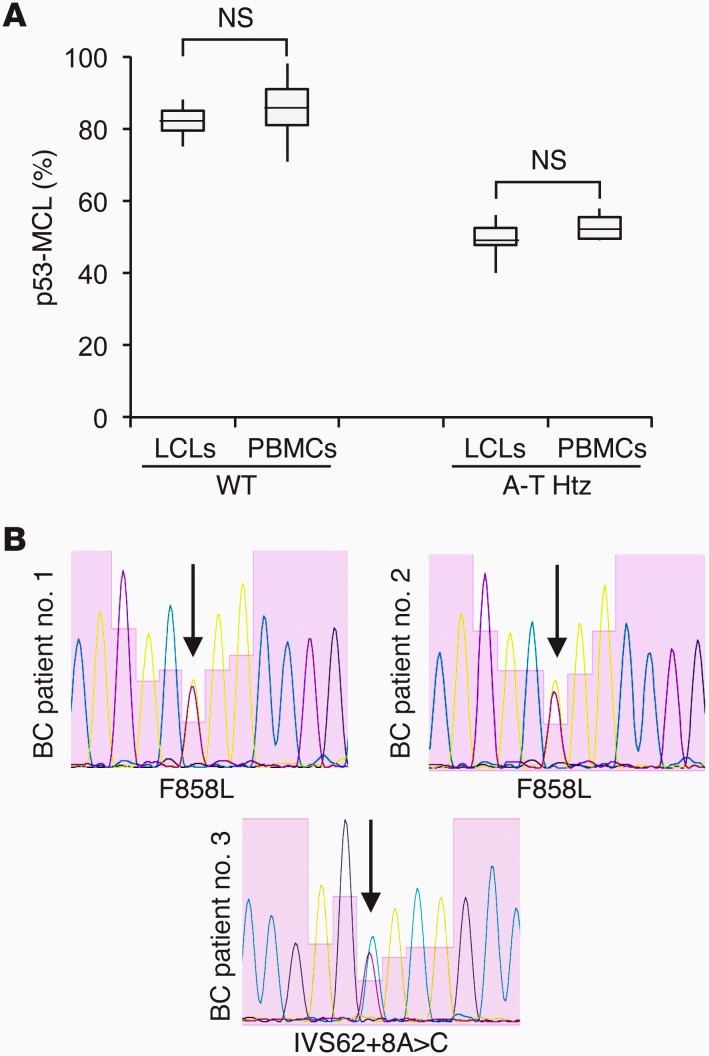
ATM is considered an intermediate-risk BC susceptibility gene (13, 30, 31). Thus, to begin challenging our PBMC test for p53-MCL on unknown targets, a group of 80 BC patients (Table 3) and 100 healthy donors were assessed for p53-MCL. All healthy donors showed a percentage of mitotic cells with p53-MCL in the range identified for the wtATM-carrying donors (Figure 4A). In contrast, 7 of 80 BC patients showed a p53-MCL comparable to that of LCLs from the A-T heterozygotes (Figure 4A). This result is in agreement with the expected cancer increase in A-T carriers compared with the general population (8.75% vs. 1.69%–3.43%; i.e., the theoretical frequency of A-T carriers in the Italian population; Italian A-T Registry). Of interest — though at this point the numbers are too low for statistical evaluation — 6 of 7 patients had an early-onset tumor appearance (37–42 years of age), and their BCs were hormone receptor–positive and HER2-negative.
Table 3.
Baseline characteristics of BC patients
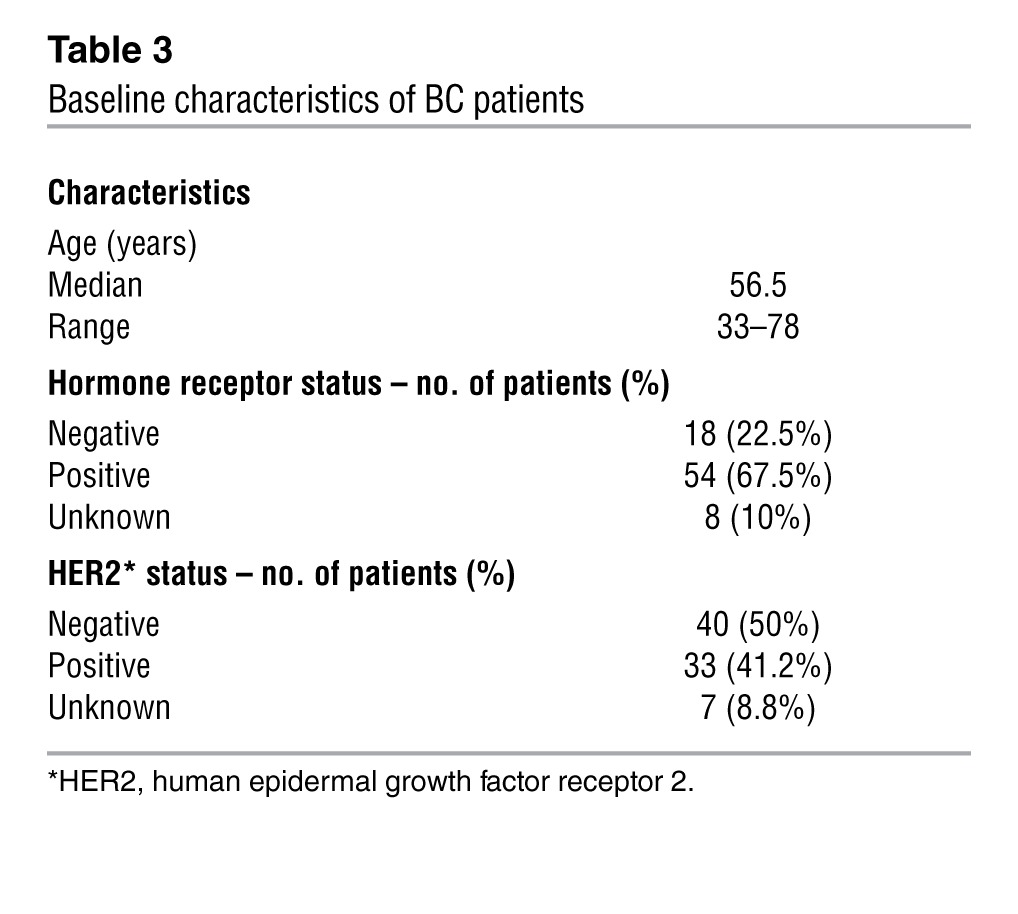
Figure 4. ATM in BC patients.
(A) Comparison of p53-MCL percentages between LCLs derived from wtATM donors (n = 11 from Figure 1D); PBMCs derived from healthy donors (n = 100); LCLs from obligate A-T carriers (n = 20 from Figure 1D); and PBMCs derived from BC patients scored as A-T carriers based on the p53-MCL test (n = 7). Box-and-whisker plot: horizontal lines within the box represent median values, and the bottom and top of the box show the lower and upper quartiles. Two-tailed Student’s t test. (B) Electropherograms of ATM exon 19 (BC patient no. 1 and BC patient no. 2), and intron 62 (BC patient no. 3) from BC patients scored as A-T carriers based on the p53-MCL test.
We sequenced the ATM gene in 3 BC patients (the only available ones, see Methods for details) and in 10 patients who scored as wtATM-carrying donors based on their p53-MCL test. In the latter cases, direct sequencing confirmed the wild-type status of the ATM gene. In contrast, the 3 cases identified as A-T heterozygous based on their p53-MCL test showed monoallelic ATM mutations (Figure 4B): 1 had the intronic c.8786+8A>C variant, while the other 2 had the c.2572T>C (p.F858L) missense mutation. Of relevance, these ATM modifications were not described in A-T patients, but have been linked to cancer predisposition, indicating that the p53-MCL test can functionally recognize at least some of the ATM cancer predisposition variants (32, 33).
Discussion
We provide here a preliminary setup and validation of a new functional assay for evaluating mutations in the human ATM gene by measuring the percentage of mitotic cells with p53 localization at the centrosome. At variance with existing tests, p53-MCL unambiguously distinguishes between A-T homozygotes and heterozygotes in a very highly sensitive and specific manner. Indeed, the results obtained by analyzing all the wtATM-carrying donors and ATM mutation carriers were stable enough within each genotype to allow for significant discrimination, without the need for reference samples. In addition, none of the LCLs derived from patients with genetic diseases causing ataxia (i.e., A-T–like disorder), or showing radiosensitivity (i.e., Nijmegen breakage syndrome, Werner syndrome, or Fanconi anemia group A) have reduced p53-MCL. Similar data were also obtained by analyzing ATR-defective and p53-mutated LCLs (Seckel syndrome and Li-Fraumeni syndrome) or other disorders (Cornelia de Lange syndrome and cylindromatosis). Although the definitive validation of this assay requires larger numbers, our data indicate that the p53-MCL test has a sensitivity and specificity not attained by other existing A-T functional assays.
From clinical and epidemiological points of view, screening for mutations by full gene sequencing is still too expensive and difficult in most countries (34). Thus, young children developing ataxia experience a long diagnostic process before their PBMCs are immortalized or their ATM gene is sequenced. The issue is even more challenging for the identification of heterozygous carriers. Since the discovery of IR hypersensitivity in A-T cells in vitro in 1975 (35), and the development of the colony survival assay (15, 16), which remains one of the most important laboratory tests for A-T diagnosis, several assays have been developed in an attempt to improve A-T identification. Unfortunately, none of these tests, with the exception of ATM sequencing, has yet obtained unambiguous results. In our opinion, this is due to two inherent reasons. The first is linked to ATM’s role in the DNA damage response, the best-characterized ATM function, on which most A-T tests are based. ATM works within a network of several sensor and effector proteins whose mutations can cause cell phenotypes to partially overlap those of the A-T cells, thereby reducing the test’s specificity. The second reason is linked to the quantitative differences induced by the diverse amounts of wild-type protein present in A-T carriers compared with that found in wild-type homozygotes and A-T homozygotes. Because of the huge number of mutations and the complexity of the pathways analyzed in the different assays, these quantitative differences consist of a continuum of partially overlapping readouts that are unable to yield unambiguous results (13, 15–21). These problems have a limited impact on the identification of A-T homozygotes, but constitute a formidable obstacle to carrier prediction. To our surprise, at variance with these assays, the p53-MCL assay does not measure a continuous quantitative variation (e.g., radiosensitivity), but rather a “binary” outcome. At the single-cell level, p53 does or does not localize at the centrosome, and we did not find any detectable quantitative difference, such as lower p53 quantities, at the centrosome. At the cell-population level, the number of cells showing one or the other phenotype allowed us to clearly discriminate the genotype.
From a biological point of view, it is possible that more precise and quantitative assays than double indirect IF will allow us to discover more subtle differences in the amount of p53 at the centrosomes. However, in anaphase and telophase, the last steps of mitosis, p53 was at the centrosome in all of the cells we analyzed, independently of the genotype (data not shown). This suggests the existence of compensatory mechanisms for the final phase of cell division and supports the concept that p53-MCL is an ATM-dependent qualitative characteristic.
We serendipitously discovered this ATM-dependent behavior of p53, though we still do not understand why half of the A-T carriers’ cells behave like wild-type cells and the other half like A-T cells. We found no differences in gender, type of ATM mutations, amounts of residual ATM protein, or p53 polymorphisms at codon 72. Limiting dilution cloning of heterozygous LCLs yielded only clones with a percentage of mitotic cells with p53-MCL, comparable to that of healthy donors. This indicates that the 50% distribution is not linked to each cell division, and that among the A-T heterozygous cells, those able to localize p53 at their centrosomes have a cloning advantage (data not shown). It will be interesting to compare the single-cell clones with the parental-cell populations in order to understand the molecular cause of the peculiar distribution found in A-T carriers.
We took advantage of these observations and set up a p53-MCL assay on PBMCs. We found that as little as 1 ml of whole blood is sufficient to calculate the percentage of mitotic cells with p53-MCL with a high degree of precision. In addition, when we applied our test to search for unknown A-T carriers among BC patients, in whom A-T–carrier frequency should be 2- to 4-fold higher than in the general population, we identified 7 putative A-T heterozygotes (i.e., the expected increment) and 2 cancer-predisposition ATM variants. The role of monoallelic ATM mutations in BC susceptibility has been a subject of debate. Some studies have indicated that ATM mutations causing A-T in biallelic carriers (i.e., protein truncations or splice junction variants) confer a risk for BC, others have reported little evidence of an associated risk, and still other studies have shown that a subset of rare, evolutionarily unlikely missense substitutions are important (13, 30–33). These studies indicate that useful screenings for ATM mutations linked to BC susceptibility require full gene sequencing to identify the different classes of ATM variants and to further discriminate between deleterious missense mutations and neutral ATM polymorphisms. Of interest, our initial evaluation of 80 BC patients showed that p53-MCL can functionally recognize at least some of the ATM cancer predisposition variants. Future studies will show whether and to what extent p53-MCL can discriminate between ATM-neutral polymorphisms and deleterious missense mutations.
In conclusion, we have established a simple, fast, minimally invasive, reliable, and inexpensive test to determine mutant ATM zygosity, opening the possibility of performing large-scale screening in the general population for different clinical aspects, such as early diagnosis, genetic counseling, cancer predisposition, susceptibility to IR, and selection for specific targeted therapies.
Methods
Cells and culture conditions.
EBV-immortalized LCLs and freshly isolated PBMCs were cultured in RPMI-1640 GlutaMAX supplemented with 15% heat-inactivated fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (all from Invitrogen). PBMCs were isolated from donors’ heparinized blood samples by Lympholyte-H (Cedarlane) density gradient centrifugation. Briefly, blood was layered over an equal volume of Lympholyte-H (Cedarlane) and centrifuged at 800g for 20 minutes at room temperature. The PBMC layer was removed and washed twice at 200g for 10 minutes in RPMI-1640 GlutaMAX (Invitrogen). GlutaMAX (Invitrogen) PBMCs were seeded at a concentration of 1 × 106/ml in T12.5-cm2 tissue culture BD Falcon flasks (BD Biosciences) and incubated at 37°C in a 5% CO2 atmosphere. PBMCs were stimulated to proliferate by incubation with 5 μg/ml PHA (Sigma-Aldrich) for 60 hours.
IF assay.
Proliferating cells (i.e., LCLs or PHA-stimulated PBMCs) were washed, spread onto glass coverslips by centrifugation, and fixed in 3.7% formaldehyde for 10 minutes at room temperature. Next, cells were permeabilized by a first incubation in 0.25% Triton X-100 in PBS (Lonza) for 5 minutes at room temperature, and a second incubation in 100% methanol for 10 minutes at –20°C. Fixed and permeabilized cells were preincubated in 0.25% Triton X-100 (Lonza) with 3% BSA in PBS for 30 minutes at 37°C in a humidified chamber, and then incubated for 2 hours with anti-p53 and anti–γ-tubulin Abs. Mouse monoclonal anti-p53 clone DO-7 (M 7001; Dako) and rabbit anti–γ-tubulin (T3559; Sigma-Aldrich) were used. Ab specificity for the identification of protein centrosomal localization was previously described (22–24). Ab dilutions in PBS with 1% BSA were empirically established; usually, the anti-p53 Ab was diluted at 1:300 and the anti–γ-tubulin at 1:2,000. After 3 washes in 0.05% Tween 20 in PBS, cells were incubated with rhodamine-conjugated anti-rabbit and FITC-conjugated anti-mouse Abs (Jackson ImmunoResearch) for 30 minutes at 37°C. After 3 washes, cells were counterstained with 0.2 μg/ml DAPI for 5 minutes at room temperature to visualize DNA, and then mounted on glass slides in Vectashield (Vector Laboratories). Cells were examined under an Olympus BX53 microscope equipped with epifluorescence and photographs were taken (×100 objective) using a cooled camera device (ProgRes MF). Percentages of p53-MCL were measured by counting 100 cells in metaphase and analyzing 2 coverslips for each sample. To assess the variability among operators, percentages of p53-MCL were blindly measured on the same preparations by at least 2 operators in 2 different laboratories. Confocal analysis was performed with a Zeiss ApoTome System.
ATM gene sequencing.
Genomic DNA was extracted from PBMCs by Quick-gDNA MiniPrep (Zymo Research) according to the manufacturer’s instructions. Sixty-two ATM exons were amplified using AmpliTaq Gold (Applied Biosystems), as described (36), and subjected to direct sequencing at the Genechron Laboratory (Rome, Italy). Of the 7 BC patients scored as positive for A-T heterozygosity by the p53-MCL test, only 3 samples were sequenced because 2 patients died before we had time to collect DNA samples and the other 2 no longer allowed us to obtain blood samples. ATM gene sequencing in the LCLs was performed as described (36).
ATM protein levels by Western blotting.
Levels of ATM protein expression were determined by Western blotting using specific Abs and β-actin (A2066; Sigma-Aldrich) for normalization. The cells were washed with PBS buffer plus 0.1 mM Na3VO4, pelleted and lysed in Laemmli buffer (0.125 M Tris-HCl pH 6.8, 5% SDS). Total cell extracts were size fractionated using the NuPAGE Novex system (Invitrogen).
Statistics.
We determined statistical significance of differences between 2 groups by 2-tailed Student’s t test. P values less than 0.01 were considered significant.
Study approval.
Collection and analyses of PBMCs from healthy donors, A-T carriers, and A-T patients were approved by the Ethical Committee of S. Andrea Hospital Sapienza Rome University. Those from BC patients were approved by the Ethical Committee of the Regina Elena National Cancer Institute. Donors provided written informed consent.
Acknowledgments
We are grateful to D. Delia (Italian National Cancer Institute, Milan, Italy), M.F. Lavin (Queensland Institute of Medical Research, Brisbane, Australia), A. Musio (National Research Council, Pisa, Italy), F. Rosselli (Gustave Roussy, Villejuif, France), and A.M. Taylor (University of Birmingham, United Kingdom) for their gift of LCLs. We thank S. Bacchetti, A. Musio, and C. Rinaldo for their critical review of the manuscript. We are particularly grateful to M. Crescenzi for helpful discussions, suggestions, and critical evaluation of the manuscript. This work was supported by grants from the Ministero della Salute and the Associazione Italiana per la Ricerca sul Cancro (to S. Soddu).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(3):1335–1342. doi:10.1172/JCI67289.
References
- 1.Gatti RA, Boder E, Vinters HV, Sparkes RS, Norman A, Lange K. Ataxia-telangiectasia: an interdisciplinary approach to pathogenesis. Medicine (Baltimore). 1991;70(2):99–117. [PubMed] [Google Scholar]
- 2.Su Y, Swift M. Mortality rates among carriers of ataxia-telangiectasia mutant alleles. Ann Intern Med. 2000;133(10):770–778. doi: 10.7326/0003-4819-133-10-200011210-00009. [DOI] [PubMed] [Google Scholar]
- 3.Perlman S, Becker-Catania S, Gatti RA. Ataxia-telangiectasia: diagnosis and treatment. Semin Pediatr Neurol. 2003;10(3):173–182. doi: 10.1016/S1071-9091(03)00026-3. [DOI] [PubMed] [Google Scholar]
- 4.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9(10):759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 5.Derheimer FA, Kastan MB. Multiple roles of ATM in monitoring and maintaining DNA integrity. FEBS Lett. 2010;584(17):3675–3681. doi: 10.1016/j.febslet.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bensimon A, Aebersold R, Shiloh Y. Beyond ATM: the protein kinase landscape of the DNA damage response. FEBS Lett. 2011;585(11):1625–1639. doi: 10.1016/j.febslet.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 7.McKinnon PJ. ATM and the molecular pathogenesis of ataxia telangiectasia. Annu Rev Pathol. 2012;7:303–321. doi: 10.1146/annurev-pathol-011811-132509. [DOI] [PubMed] [Google Scholar]
- 8.Uziel T, et al. Genomic organization of the ATM gene. Genomics. 1996;33(2):317–320. doi: 10.1006/geno.1996.0201. [DOI] [PubMed] [Google Scholar]
- 9.Concannon P, Gatti RA. Diversity of ATM gene mutations detected in patients with ataxia-telangiectasia. Hum Mutat. 1997;10(2):100–107. doi: 10.1002/(SICI)1098-1004(1997)10:2<100::AID-HUMU2>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 10.Mitui M, et al. Independent mutational events are rare in the ATM gene: haplotype prescreening enhances mutation detection rate. Hum Mutat. 2003;22(1):43–50. doi: 10.1002/humu.10232. [DOI] [PubMed] [Google Scholar]
- 11.Swift M, Morrell D, Cromartie E, Chamberlin AR, Skolnick MH, Bishop DT. The incidence and gene frequency of ataxia-telangiectasia in the United States. Am J Hum Genet. 1986;39(5):573–583. [PMC free article] [PubMed] [Google Scholar]
- 12.Chessa L, Lisa A, Fiorani O, Zei G. Ataxia-telangiectasia in Italy: genetic analysis. Int J Radiat Biol. 1994;66(6 suppl):S31–S33. doi: 10.1080/09553009414551831. [DOI] [PubMed] [Google Scholar]
- 13.Tavtigian SV, et al. Rare, evolutionarily unlikely missense substitutions in ATM confer increased risk of breast cancer. Am J Hum Genet. 2009;85(4):427–446. doi: 10.1016/j.ajhg.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson L, et al. Diagnosis of ataxia-telangiectasia by T-lymphocyte cloning assay. Lancet. 1985;2(8466):1242. doi: 10.1016/s0140-6736(85)90766-4. [DOI] [PubMed] [Google Scholar]
- 15.Huo YK, et al. Radiosensitivity of ataxia-telangiectasia, X-linked agammaglobulinemia, and related syndromes using a modified colony survival assay. Cancer Res. 1994;54(10):2544–2547. [PubMed] [Google Scholar]
- 16.Sun X, et al. Early diagnosis of ataxia-telangiectasia using radiosensitivity testing. J Pediatr. 2002;140(6):724–731. doi: 10.1067/mpd.2002.123879. [DOI] [PubMed] [Google Scholar]
- 17.Chun HH, et al. Improved diagnostic testing for ataxia-telangiectasia by immunoblotting of nuclear lysates for ATM protein expression. Mol Genet Metab. 2003;80(4):437–443. doi: 10.1016/j.ymgme.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Du L, Lai CH, Concannon P, Gatti RA. Rapid screen for truncating ATM mutations by PTT-ELISA. Mutat Res. 2008;640(1–2):139–144. doi: 10.1016/j.mrfmmm.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Porcedda P, et al. A rapid flow cytometry test based on histone H2AX phosphorylation for the sensitive and specific diagnosis of ataxia telangiectasia. Cytometry A. 2008;73(6):508–516. doi: 10.1002/cyto.a.20566. [DOI] [PubMed] [Google Scholar]
- 20.Honda M, Takagi M, Chessa L, Morio T, Mizuatni S. Rapid diagnosis of ataxia-telangiectasia by flow cytometric monitoring of DNA damage-dependent ATM phosphorylation. Leukemia. 2009;23(2):409–414. doi: 10.1038/leu.2008.195. [DOI] [PubMed] [Google Scholar]
- 21.Nahas SA, Butch AW, Du L, Gatti RA. Rapid flow cytometry-based structural maintenance of chromosomes 1 (SMC1) phosphorylation assay for identification of ataxia-telangiectasia homozygotes and heterozygotes. Clin Chem. 2009;55(3):463–472. doi: 10.1373/clinchem.2008.107128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ciciarello M, et al. p53 displacement from centrosomes and p53-mediated G1 arrest following transient inhibition of the mitotic spindle. J Biol Chem. 2001;276(22):19205–19213. doi: 10.1074/jbc.M009528200. [DOI] [PubMed] [Google Scholar]
- 23.Tritarelli A, et al. p53 localization at centrosomes during mitosis and postmitotic checkpoint are ATM-dependent and require serine 15 phosphorylation. Mol Biol Cell. 2004;15(8):3751–3757. doi: 10.1091/mbc.E03-12-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oricchio E, Saladino C, Iacovelli S, Soddu S, Cundari E. ATM is activated by default in mitosis, localizes at centrosomes and monitors mitotic spindle integrity. Cell Cycle. 2006;5(1):88–92. doi: 10.4161/cc.5.1.2269. [DOI] [PubMed] [Google Scholar]
- 25.Doxsey S, Zimmerman W, Mikule K. Centrosome control of the cell cycle. Trends Cell Biol. 2005;15(6):303–311. doi: 10.1016/j.tcb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Fukasawa K. Oncogenes and tumour suppressors take on centrosomes. Nat Rev Cancer. 2007;7(12):911–924. doi: 10.1038/nrc2249. [DOI] [PubMed] [Google Scholar]
- 27.Fukasawa K. p53, cyclin-dependent kinase and abnormal amplification of centrosomes. Biochim Biophys Acta. 2008;1786(1):15–23. doi: 10.1016/j.bbcan.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ammazzalorso F, Pirzio LM, Bignami M, Franchitto A, Pichierri P. ATR and ATM differently regulate WRN to prevent DSBs at stalled replication forks and promote replication fork recovery. EMBO J. 2010;29(18):3156–3169. doi: 10.1038/emboj.2010.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Revenkova E, et al. Cornelia de Lange syndrome mutations in SMC1A or SMC3 affect binding to DNA. Hum Mol Genet. 2009;18(3):418–427. doi: 10.1093/hmg/ddn369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson D, et al. Cancer risks and mortality in heterozygous ATM mutation carriers. J Natl Cancer Inst. 2005;97(11):813–822. doi: 10.1093/jnci/dji141. [DOI] [PubMed] [Google Scholar]
- 31.Renwick A, et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat genet. 2006;38(8):873–875. doi: 10.1038/ng1837. [DOI] [PubMed] [Google Scholar]
- 32.Stredrick DL, et al. The ATM missense mutation p.Ser49Cys (c.146C>G) and the risk of breast cancer. Hum Mutat. 2006;27(6):538–544. doi: 10.1002/humu.20323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meier M, et al. Relation between genetic variants of the ataxia telangiectasia-mutated (ATM) gene, drug resistance, clinical outcome and predisposition to childhood T-lineage acute lymphoblastic leukaemia. Leukemia. 2005;19(11):1887–1895. doi: 10.1038/sj.leu.2403943. [DOI] [PubMed] [Google Scholar]
- 34.Narod SA. BRCA mutations in the management of breast cancer: the state of the art. Nat Rev Clin Oncol. 2010;7(12):702–707. doi: 10.1038/nrclinonc.2010.166. [DOI] [PubMed] [Google Scholar]
- 35.Taylor AM, Harnden DG, Arlett CF, Harcourt SA, Lehmann AR, Stevens S. Ataxia telangiectasia: a human mutation with abnormal radiation sensitivity. Nature. 1975;258(5534):427–429. doi: 10.1038/258427a0. [DOI] [PubMed] [Google Scholar]
- 36.Magliozzi M, et al. DHPLC screening of ATM gene in Italian patients affected by ataxia-telangiectasia: fourteen novel ATM mutations. Dis Markers. 2006;22(4):257–264. doi: 10.1155/2006/740493. [DOI] [PMC free article] [PubMed] [Google Scholar]