Summary
Due to photobleaching and phototoxicity induced by high-intensity excitation light the number of fluorescence images that can be obtained in live cells is always limited. This limitation becomes particularly prominent in multi-dimensional recordings when multiple Z-planes are captured at every time point. Here we present a simple technique, termed Predictive-Focus Illumination (PFI), which helps to minimize cells’ exposure to light by decreasing the number of Z-planes that need to be captured in live-cell 3-D time lapse recordings. PFI utilizes computer tracking to predict positions of objects-of-interest (OOIs) and restricts image acquisition to small dynamic Z-regions centered on each OOI. Importantly, PFI does not require hardware modifications and it can be easily implemented on standard wide-field and spinning-disk confocal microscopes.
Keywords: light microscopy, digital imaging, software, live-cell microscopy, photodamage
Introduction
Live-cell fluorescence microscopy is an essential tool for visualizing the dynamics of individual cells, organelles, or macromolecular assemblies inside the cell. Unfortunately, the number of images that can be recorded during an experiment is limited by photobleaching (deterioration of fluorophore) and phototoxicity (deleterious reaction of live cells to high-intensity light). Both phenomena primarily arise due to the production of reactive oxygen species (Dixit & Cyr, 2003) and are commonly referred to as “photodamage” (Rieder & Cole, 2000, Khodjakov & Rieder, 2006, Stephens & Allan, 2003).
Photodamage becomes particularly restrictive in high-resolution multi-dimensional recordings where multiple Z-planes need to be collected at every time point. In conventional microscopy excitation light passes through the entire thickness of the specimen irrespective of the exact position of the focal plane. Thus, cells are irradiated with the same amount of light during each exposure. As a result, photodamage effects induced by a single Z-series are similar to those induced by the same number of ‘in-focus’ images.
Several approaches for decreasing photodamage in 3-D imaging have been proposed, most notably Selective Plane Illumination Microscopy (SPIM) (Huisken et al., 2004) and Controlled Light Exposure Microscopy (CLEM) (Hoebe et al., 2007). SPIM is based on a radically different excitation-light path in which only one focal plane is illuminated by a light sheet oriented roughly orthogonal to the optical axis of the microscope (Huisken et al., 2004). In CLEM, which has been implemented only in the conventional point-scanning confocal microscopy, excitation light is rapidly (within dwell time) attenuated for those pixels that show a slow initial accumulation of the signal (Hoebe et al., 2007). While both SPIM and CLEM offer clear advantages, implementation of these techniques requires sophisticated and expensive hardware modifications. A simpler approach to reducing phototoxicity in live cell recordings is likely to become popular among cell biologists, particularly if such an approach does not require hardware upgrades and is compatible with microscopy modalities currently used in live-cell microscopy.
Here we present a simple technique, termed Predictive-Focus Illumination (PFI), which can be enabled on typical wide-field (WF) and spinning-disk (SDC) confocal systems by utilizing open-source software. Although PFI is in principle applicable to other types of microscopy such as laser-scanning confocal (LSCM) or two-photon excitation (TPE), we focused our work specifically on WF and SDC because high sensitivity and a reasonable level of photodamage in these modalities make them quite popular among cell biologists (Wang et al., 2005, Murray et al., 2007). Further, most WF and SDC microscopy workstations are readily supported by μManager, a java-based open-source microscopy software (Edelstein et al., 2010) which makes PFI implementable by the end user in the environment of a cell biology laboratory.
Principles of Predictive-Focus Illumination (PFI)
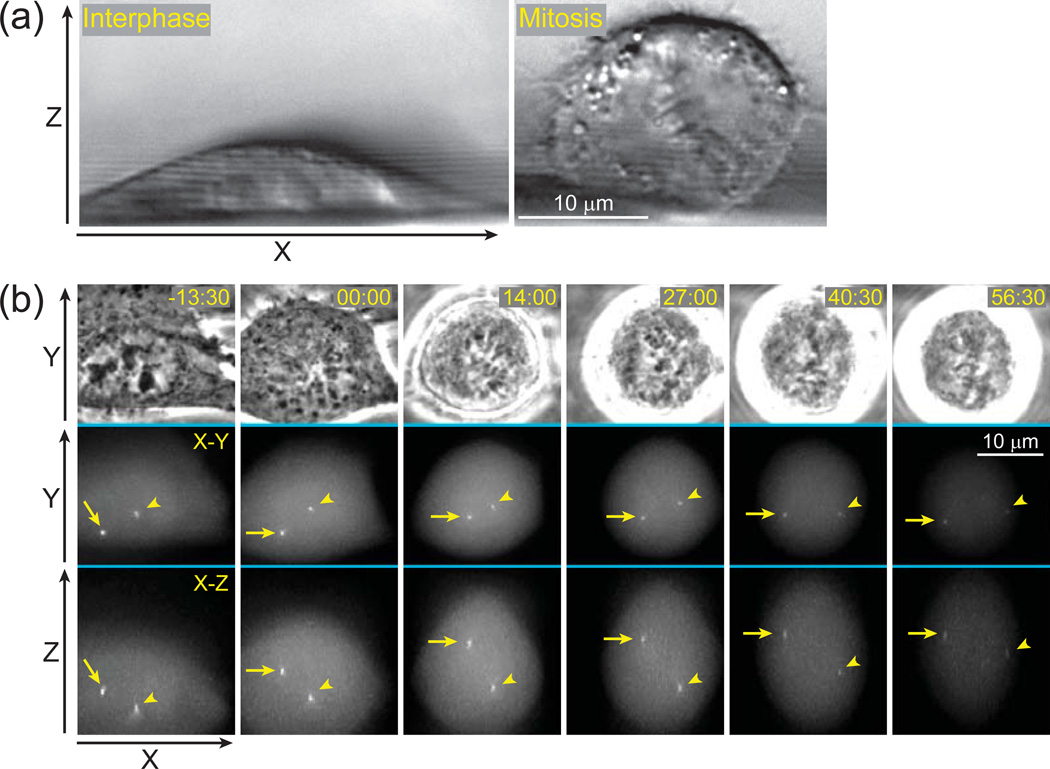
In the conventional approach to four-dimensional time-lapse recordings, the number of focal planes collected at every time point is determined by the range of movements that the object can potentially exhibit during the course of recording. For example, in applications that deal with the behavior of motile intracellular organelles the entire volume of the cell must be scanned to ensure that an in-focus image of the Object of Interest (OOI) is captured. This can be achieved with just a few Z-planes for a thin interphase cell. However, when the cell enters mitosis it rounds up and as a result a much wider Z-range is required to cover the depth of the cell (Fig.1a). Thus, if the cell undergoes transitions between interphase and mitosis during the experiment, the Z-range of the entire time-lapse recording is determined by the thickness of the rounded mitotic cell even though during interphase this range is exceedingly wide and most of the recorded Z-planes contain no useful information on the OOIs. Yet, recording these “empty” Z-planes exposes cells to unnecessary irradiation which in turn increases both photobleaching and phototoxicity. Another example of the inefficiency in conventional imaging comes from the experiments where movements of a small object are followed in a relatively thick specimen such as movements of cells in 3-D matrices or movements of individual organelles in relatively thick cells (Fig.1b). In these cases most of the recorded images contain nothing but out-of-focus blur. We sought to circumvent the problem of unnecessary exposure in 3-D time-lapse recordings by augmenting image acquisition with a feedback loop that detects the OOIs and predicts their positions at the next time-point. These predictions are then used to restrict the Z-range of acquisition to a small number of focal planes centered on each OOI. The principle layout of this approach which we term Predictive Focus Illumination (PFI) is illustrated in (Fig. 2a).
Figure 1.
Conventional approach to 3-D time-lapse imaging results in unnecessary exposure to light. (a) Shape of a typical animal cell changes during the cell cycle. Cultured cells grown on solid substrates are significantly thinner during interphase (left panel) than during mitosis (right panel). Differential Interference Contrast (DIC) images depicting a side-view of human cells (RPE-1) grown on a 120-µm fluorocarbon string mounted on a coverslip. (b) An example of 3-D time-lapse GFP-fluorescence recording (selected frames) in which movements of centrosomes (arrow and arrowhead) are followed in a mitotic human cell (RPE-1). Each time-point is presented as a selected phase-contrast image (top row) and two maximal-intensity projections, one along Z (middle) and one along Y axis (bottom). Although only a few focal planes are required to capture in-focus images of the centrosomes (yellow boxes), a much larger number of focal planes have to be collected (61 planes spanning ~24 µm per time-point) due to gradual rounding of the cell and extensive movements of the centrosomes along Z-axis. These unnecessary focal planes expose cells to additional irradiation resulting in photobleaching (notice gradually decreasing intensity) and phototoxicity (the cell arrests in mitosis). Time in minutes : seconds. Time 00:00 corresponds to nuclear envelope breakdown (onset of mitosis).
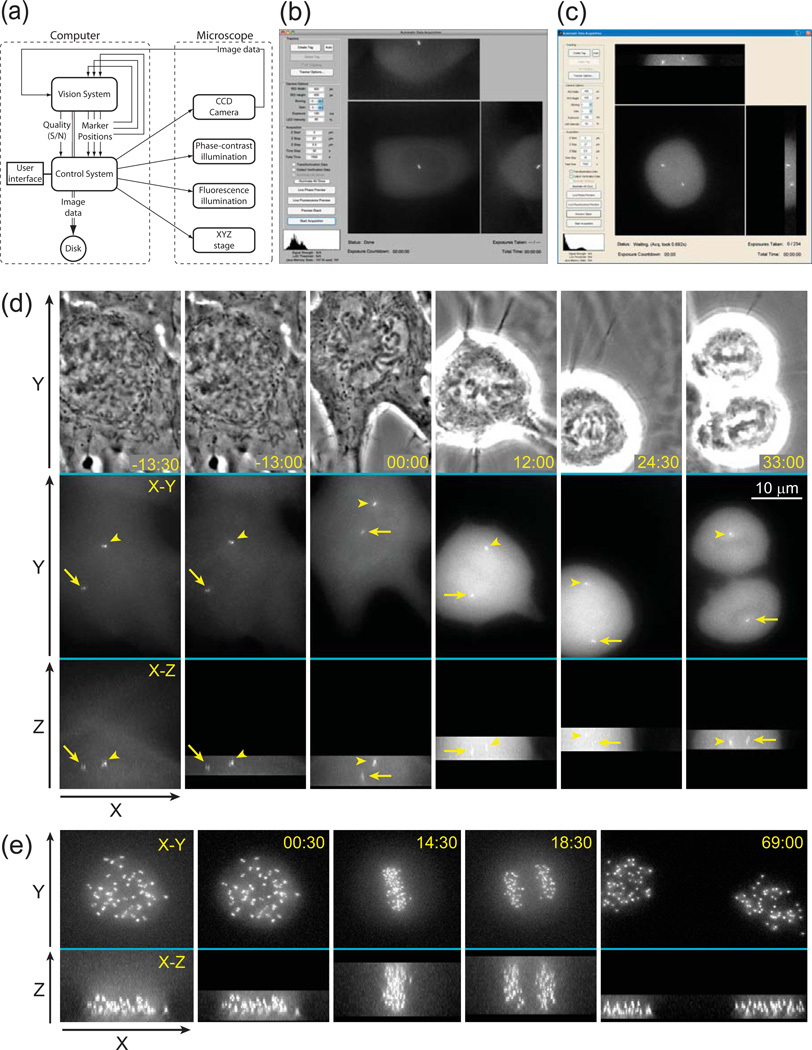
Figure 2.
Principle layout and implementation of the Predictive-Focus Illumination (PFI) system. (a) Bloc-diagram of the PFI algorithm. (b) Computer screenshot of the PFI system after the initial preview. A data of 55 Z-planes has been collected and the user is prompted to manually mark OOIs or to execute their automatic detection. In this example the system runs under Mac OS X. (c) Computer screenshot of the PFI system during the acquisition loop. Only the focal planes adjacent to the positions of OOIs were illuminated. In this example the system runs under MS Windows 7. (d) Example of PFI recording. The cell line, light intensity, and acquisition parameters are the same as in Fig.1b; however, in this case only the focal planes adjacent to the positions of centrosomes are recorded in the PFI mode. (e) Example of PFI recording with a large number of OOIs. In this example 46 pairs of kinetochores were followed in a human RPE-1 cell. Notice that during anaphase kinetochores are spread throughout the entire Z-range (frame 18:30). However, during earlier and later stages of mitosis a significant number of exposures is avoided. Also notice that cells successfully progresses through mitosis with minimal photobleaching. Time in minutes : seconds. Time 00:00 corresponds to nuclear envelope breakdown.
To evaluate real-life advantages of PFI we implemented this technique on a typical 3-D wide-field fluorescence microscopy workstation consisting of a research-grade inverted microscope (Nikon TE-300) with an electronically controlled X-Y stage (Ludl) and a piezo-controlled objective lens (PI). Standard XF100 filter cube (Omega Optical) was used for GFP fluorescence and illumination density was kept at ~0.2 W/cm2. Images were recorded on a Photometrics Coolsnap HQ CCD camera. A Java-based open-source microscopy environment, μManager (Edelstein et al., 2010), was combined with MATLAB (Mathworks) to control the microscope during the development stage. However, MATLAB code can be compiled so that the user-oriented final software would require only μManager and thus be fully compatible with the range of microscopy hardware supported by this free program.
In our implementation of PFI the user is prompted to define the value of Z-step and the range of Z-scanning sufficient to cover potential movements of the object. At the first time point all focal planes within this range are recorded (Fig. 2b). This first volume is then used to define the OOIs that can be selected manually by the user or auto-detected. Coordinates of the OOIs are used to define the Z-range of image acquisition at the next time-point. New coordinates of each OOI are then automatically extracted and used to restrict the subsequent Z-range to just a few planes centered on the predicted Z-positions of the OOIs (Fig. 2c).
OOI detection and tracking algorithm
PFI requires that OOIs are detected and their movements are tracked in 3-D. Detection and tracking must be achieved during relatively short intervals between the consecutive time-points, which implies that the algorithms utilized in the PFI system should not be computationally expensive. Further, due to significant variability in the shape and size of different biological objects, different algorithms would be advantageous in particular applications.
For PFI development we decided to utilize a relatively simple and not computationally-expensive algorithm where small globular organelles such as centrosomes and kinetochores, or tips of dynamic microtubules are modeled as small bright spheres blurred by the point spread function (PSF) of the optical system. Visualizing the behavior of this type of organelles is an important application among cell biologist interested in cell division, chromosome segregation, or in cytoskeletal dynamics (Jaqaman et al., 2010, Stumpff et al., 2008, Matov et al., 2010, Thoma et al., 2010, Magidson et al., 2011, Kitajima et al., 2011).
Rather than modeling the particular PSF for a given microscope, our algorithm treats each OOI as a multivariate Gaussian brightness function, which we call a “blob”, owing to its appearance. This model is quite effective in detecting centrioles and kinetochores in the image plane, centered on the brightest point of the organelle. Due to significant anisotropy of the 3D sampling, we do not attempt to model the OOIs as 3D blobs, but use a 2D Gaussian model for individual slices of a stack.
The resulting blob detection filter is a Laplacian of Gaussian (LoG) scaled so that the central positive lobe of the LoG impulse response matches the central cross-section of an OOI image. Applying this technique to various imaging modalities requires nothing more than an adjustment of the LoG scale parameters (constants in the software) which makes our algorithm equally applicable to WF and SDC microscopy.
The algorithm is designed to simultaneously detect and track blobs frame by frame in four-dimensional image data (3-D volumes over time). User-provided input at the initial stage of recording results in the first set of blob-location guesses. At each subsequent time point new location estimates are derived from the previous time point’s image by pairing up responses between adjacent time points using a maximum a posteriori (MAP) decision rule based on a 3-D Laplacian probability distribution model of OOI motion over the time interval, and the current set of blobs.
Algorithm inputs include 1) Image (I); 2) Initial Tag Position Guesses (xk, yk, zk); 3) Tag Sigma - Laplacian of Gaussian (LoG) Standard Deviation (σt); 4) Distance Sigma (σd); and 5) Re-Detection Sigma (σr).
Preprocessing
First, each slice the image is convolved with a 2-D LoG filter designed to detect 2-D elliptical blobs, LoG(I) = h * I where the impulse response is:
The convolution produces a strong response for bright ellipsoids surrounded by darker background in the image data. The responses to noise spikes are significantly weaker while larger areas of homogeneous fluorescence on average provide no response (output = 0). This allows us to only retain and tag for tracking consideration the strongest 1% of the output pixels which reduces computation time in the matching process (this threshold can be relaxed to track OOIs of various sizes and geometries). Locations and associated response values for the strong objects are denoted as Xi, Yi, Zi, Ri. Due to anisotropy in the image data Zi needs to be rescaled to match the scale of Xi, Yi.
Peak Selection via Energy Minimization
To identify which of the responses corresponds to the most probable new location of the previous frame’s OOI we rely on the lowest energy metric that characterizes the separation between the previous frames tag and a given response value Xi, Yi, Zi, Ri.
For a single tag k, the best match is found from: argmin(i){Energy(i,k,)}.
After finding the strongest matching response for a given tag j, the tag must be excluded from subsequent detections to prevent that a single response peak matches multiple tags. To do this, peak responses (Ri) are attenuated before attempting to match the next tag j.
Ra is the set of all tag responses. The response of the previously matched peak i is set to zero and responses immediately adjacent to the matched peak are attenuated. The tag location is subsequently refined to sub pixel accuracy using a Gaussian fit of the blob.
Estimating Accuracy and Error Detection
The quality of our tracking results is ensured by several quantitative measures that are taken at each time point. Signal strength is calculated in a manner similar to signal to noise ratio, using the LoG amplitude of each detected tag as a signal value and the average level of the LoG image the noise value.
Low values of this metric indicate that tracking results are suspicious. In practice we found that values lower than 8 for this parameter signify that tag detection failed. The tracking results are also checked by examination of the Euclidean distances between the positions of the same OOI in two consecutive time points. If an OOI appears to have move farther than the specified maximum position change, or the detection quality falls below 8, the software alerts the user who then has an option to manually re-identify the OOI.
Validation of PFI advantages
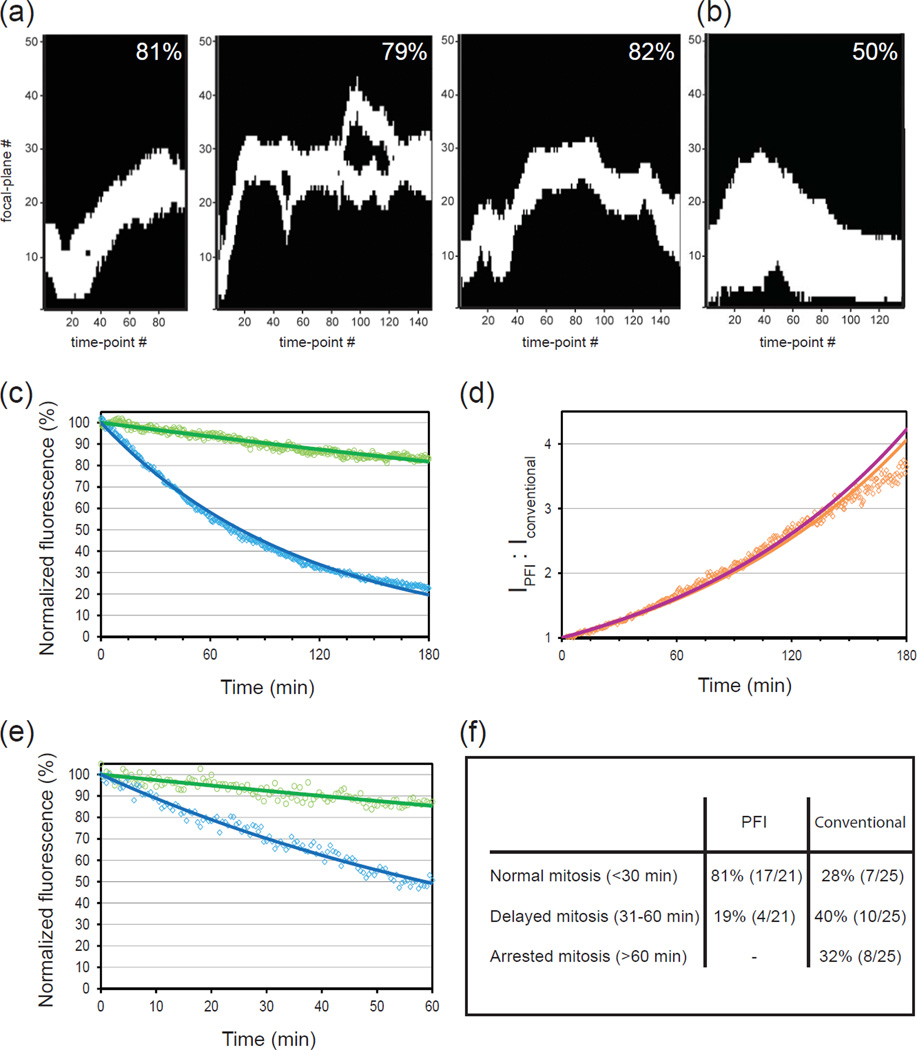
As evident from the examples of PFI recordings (Fig. 2d–e) this approach results in a significant decrease in the number of exposures required in typical 3-D time-lapse recordings. The exact advantage depends on the number of OOIs as well as the size and geometry of the cell. PFI is most beneficial for the experiments that involve tracking a few small objects that move extensively in large 3-D space. For experiments that involve such popular application as tracking centrosome movements (Piel et al., 2000, Loncarek et al., 2008) during mitosis, PFI can decrease the number of required exposures up to 5-fold (Fig. 3a) which is a remarkable achievement. Obviously, PFI becomes less advantageous in experiments with a larger number of evenly distributed OOIs at every time point. In practice, we found PFI to be advantageous even in relatively complex situations such as recordings of kinetochore movements during mitotic spindle formation. In our tests, PFI decreases the number of exposures approximately 2-fold in normal human RPE-1 cells with 92 labeled kinetochores (Fig. 3b).
Figure 3.
Quantification of PFI advantages over conventional illumination. (a) Three typical illumination profiles for recordings of centrosome behavior during mitosis (left panel corresponds to the cell presented in Fig. 2c). In these profiles white areas represent focal planes that were captured at each time point. Black areas represent focal planes that would additionally be captured in conventional-illumination recordings. The decrease in total light exposure achieved by PFI is shown for each recording. Notice, that when the two centrosomes reside in significantly different focal planes PFI software records two separate Z-ranges each centered on a single centrosome (middle panel). (b) Illumination profile for PFI recording with a large number of OOI (corresponds to the cell presented in Fig.1e). Notice that PFI decreases total light exposure two-fold. (c) In-vitro comparison of photobleaching in PFI (green) vs. conventional (blue) recordings (average of 5 recordings). Each curve represents changes in the intensity of fluorescein beads mounted on the coverslip. Because the beads remain stationary during the experiment the difference in total exposure time at each time point is 7/61 (the number of Z-planes collected in PFI vs. conventional modes). (d) Level of signal preservation achieved by PFI in vitro. Magenta line represents a theoretical prediction based on the 7/61 decrease in light exposure. Orange dots are observed experimental values; orange line represents an exponential fit of experimental data. Notice a close match between theoretical predictions and the experiment. (e) In-vivo comparison of photobleaching in conventional vs. PFI time-lapse recordings of centrin-GFP-labeled centrosomes (average of 5 recordings) in human RPE-1 cells during interphase. (d) Duration of mitosis in RPE-1 cells when the behavior of centrosomes was followed via PFI vs. conventional illumination.
Since the current implementation of PFI employs a simple blob-detection algorithm, its ability to follow cells with a large number of objects seems unexpected. Indeed, precise tracking of kinetochores requires complex off-line algorithms (Thomann et al., 2003, Jaqaman et al., 2010). In contrast, our software does not identify each and every kinetochore – the goal of OOI-detection in PFI is to rapidly define the range of Z-scanning for the next time point. As long as the outmost peripheral OOIs are detected, the Z-range will be set properly which result in a significant decrease of total light exposure.
The several-fold decrease in total light exposure enabled by PFI is expected to alleviate photodamage effects in fluorescence microscopy. To numerically assess the advantages of PFI we first compared photobleaching curves of 100-nm beads (Molecular Probes) mounted on the coverslip and excited in conventional vs. PFI modes. The excitation/emission peaks of the beads are similar to that of GFP. This allowed us to use the same imaging conditions as in our live-cell experiments, except that the intensity of the excitation light was increased 4-fold to compensate for the beads’ photostability. Expectedly, photobleaching of PFI-imaged beads was significantly reduced (Fig.3c). Because, signal intensity decreases exponentially due to photobleaching, the advantage of PFI becomes particularly prominent during long-term experiments. In a typical WF system with minimal confocal effects, the ratio between the residual intensity of fluorophore excited via PFI vs. conventional microscopy should increase exponentially over time, and we find that experimental data closely match theoretical predictions (Fig.3d).
A similar level of photobleaching decrease was observed in live-cell recordings of human RPE-1 cells with centrin-GFP-labeled centrosomes (Fig.3e). It is noteworthy that in contrast to in-vitro experiments, the signal intensity of GFP-tagged organelles is potentially affected not only by photobleaching but also by the physiological factors such as new protein synthesis and the level dynamic exchange between different compartments within the cell. Previous characterization of centrin-GFP behavior suggests that during interphase the intensity of individual centrosomes remains relatively constant over a period of ~1 hr, although centrosome-associated centrin undergoes a constant exchange with the soluble cytoplasmic pool (La Terra et al., 2005, Magidson et al., 2007). We find that while the brightness of centrosomes in conventional recordings falls down ~2-fold, over the course of 60-min time-lapse recordings (30-sec. intervals, 61 Z-planes per time point); PFI-illuminated centrosomes retain ~85% of their original intensity. Thus, the extent of photobleaching under PFI is ~3-fold less than under conventional illumination (Fig. 3e).
To characterize PFI advantages in decreasing phototoxicity we compared the duration of mitosis in cells followed via conventional vs. PFI time-lapse recordings. High-intensity light as well as other stressful conditions is known to delay mitotic progression and can even irreversibly arrest cells in mitosis (Mikhailov et al., 2002, Mikhailov et al., 2005). Thus, reproducible and rapid progression through cell division serves as a good indicator of low phototoxicity. Recordings of RPE-1 cells with centrin-GFP-labeled centrosomes were initiated during G2 and cells were followed under identical environmental conditions in conventional vs. PFI modes. The great majority of cells followed by PFI completed mitosis in less than 30 min (time from nuclear envelope breakdown to anaphase onset) while the rest of the cells experienced a minor delay. In contrast, conventional illumination with the same excitation light intensity and exposure time delayed mitotic progression or completely arrested mitosis in the majority of cells (Fig. 3d).
As an added benefit, under many circumstances PFI enables higher temporal resolution recordings. OOI detection requires only ~2.5 sec on a typical personal computer (e.g., an Intel i7 2.67 GHz CPU, 3 GB RAM, Windows-7 operating system). This time is significantly shorter than the time saved at each time point due to a 50–80% decrease in the number of recorded which ultimately shortens the acquisition cycle.
The fewer images recorded in PFI contain essentially the same amount of information pertinent to the OOIs as in conventional recordings. Because PFI simply restricts the range of Z-scanning to the focal planes immediately adjacent to each OOI, PFI datasets are fully quantitative. The intensity of each object as well as background intensities in the areas adjacent to each OOI remains intact. Further, because PFI datasets contain optical planes adjacent to the OOI, these datasets can be deconvolved using standard algorithms. These features along with seamless compatibility between PFI and popular live-cell imaging modalities such as wide-field and spinning-disk confocal compare PFI favorably with other techniques based on adaptive attenuation of the excitation light. However, in contrast to conventional recordings, PFI datasets contain no information of the “bystander objects” that were not deemed as OOIs. Portions of the cell that were originally of no interest could be missing entirely from the final image data.
Future improvement of PFI under development include adaptive intensity control of the LED light source to achieve minimum lighting for imaging, adaptive Z stepping instead of fixed Z stepping, and a more accurate tracking algorithm for the complete OOI motion history.
Conclusions
PFI offers a simple but effective means for alleviating photodamage in live-cell 3-D time-lapse microscopy. In sharp contrast to other techniques aimed at the same goal (Hoebe et al., 2007, Caarls et al., 2011) PFI is compatible with wide-field and spinning-disk confocal systems that are currently most popular in live-cell microscopy. While the exact benefits of PFI depend on the geometry of the sample, this approach decreases light exposure 2–5 fold in time-lapse recordings that involve G2/M transitions in typical mammalian cells. Thus, although not universal, PFI has potentials to gain popularity with a large group of cell biologists who study mechanisms of spindle assembly and chromosome movements. With the development of more sophisticated detection/tracking algorithms the range of potential PFI applications can be expanded to include, for example, cell migration, nuclear positioning, and Golgi/vesicular traffic.
Acknowledgments
This work was supported in part by the National Science Foundation (NSF) Smart Lighting Engineering Research Center (EEC-0812056) and by the School of Engineering at Rensselaer Polytechnic Institute. Research on mitosis in the Khodjakov lab is supported by the National Institutes of Health grant GM59363 (to A.K.). MatLab code to enable PFI is available for download from www.rpi.edu/exactURL/provided/upon/acceptance/.
REFERENCES
- Caarls W, Rieger B, De Vries AH, Arndt-Jovin DJ, Jovin TM. Minimizing light exposure with the programmable array microscope. Journal of Microscopy. 2011;241:101–110. doi: 10.1111/j.1365-2818.2010.03413.x. [DOI] [PubMed] [Google Scholar]
- Dixit R, Cyr R. Cell damage and reactive oxygen species production induced by fluorescence microscopy: effect on mitosis and guidelines for non-invasive fluorescence microscopy. Plant Journal. 2003;36:280–290. doi: 10.1046/j.1365-313x.2003.01868.x. [DOI] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using æManager. Current Protocols in Molecular Biology. 2010;92:14.20.11–14.20.17. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebe RA, Van Oven CH, Gadella TWJ, Dhonukshe PB, Van Noorden CJF, Manders EMM. Controlled light-exposure microscopy reduces photobleaching and phototoxicity in fluorescence live-cell imaging. Nature Biotechnology. 2007;25:249–253. doi: 10.1038/nbt1278. [DOI] [PubMed] [Google Scholar]
- Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- Jaqaman K, King EM, Amaro AC, Winter JR, Dorn JF, Elliott HL, McClelland SE, Porter IM, Posch M, Toso A, Danuser G, McAinsh AD, Meraldi P, Swedlow JR. Kinetochore alignment within the metaphase plate is regulated by centromere stiffness and microtubule depolymerases. J. Cell Biol. 2010;188:665–667. doi: 10.1083/jcb.200909005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL. Imaging the division process in living tissue culture cells. Methods. 2006;38:2–16. doi: 10.1016/j.ymeth.2005.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Ohsugi M, Ellenberg J. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell. 2011;146:568–581. doi: 10.1016/j.cell.2011.07.031. [DOI] [PubMed] [Google Scholar]
- La Terra S, English CN, Hergert P, McEwen BF, Sluder G, Khodjakov A. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J. Cell Biol. 2005;168:713–720. doi: 10.1083/jcb.200411126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J, Hergert P, Magidson V, Khodjakov A. Control of daughter centriole formation by the pericentriolar material. Nature Cell Biology. 2008;10:322–328. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V, Loncarek J, Hergert P, Rieder CL, Khodjakov A. Laser microsurgery in the GFP era: A cell biologist's perspective. In: Berns MW, Greulich KO, editors. Laser Manipulations of Cells and Tissues. Elsevier; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V, O'Connell CB, Loncarek J, Paul R, Mogilner A, Khodjakov A. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell. 2011;146:555–567. doi: 10.1016/j.cell.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matov A, Applegate K, Kumar P, Thoma C, Krek W, Danuser G, Wittmann T. Analysis of microtubule dynamic instability using a plus-end growth marker. Nat Methods. 2010;7:761–768. doi: 10.1038/nmeth.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailov A, Cole RW, Rieder CL. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Current biology : CB. 2002;12:1797–1806. doi: 10.1016/s0960-9822(02)01226-5. [DOI] [PubMed] [Google Scholar]
- Mikhailov A, Shinohara M, Rieder CL. The p38-mediated stress-activated checkpoint. A rapid response system for delaying progression through antephase and entry into mitosis. Cell Cycle. 2005;4:57–62. doi: 10.4161/cc.4.1.1357. [DOI] [PubMed] [Google Scholar]
- Murray JM, Appleton PL, Swedlow JR, Waters JC. Evaluating performance in three-dimensional fluorescence microscopy. J Microsc. 2007;228:390–405. doi: 10.1111/j.1365-2818.2007.01861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 2000;149:317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Cole R. Microscopy-induced radiation damage, microtubules, and progression through the terminal stage of G 2 (Prophase) in verterate somatic cells. Cold Spring Harbor Symposia on Quantitative Biology. 2000;65:369–376. doi: 10.1101/sqb.2000.65.369. [DOI] [PubMed] [Google Scholar]
- Stephens DJ, Allan VJ. Light microscopy techniques for live cell imaging. Science. 2003;300:82–86. doi: 10.1126/science.1082160. [DOI] [PubMed] [Google Scholar]
- Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Developmental Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma CR, Matov A, Gutbrodt KL, Hoerner CR, Smole Z, Krek W, Danuser G. Quantitative image analysis identifies pVHL as a key regulator of microtubule dynamic instability. J Cell Biol. 2010;190:991–1003. doi: 10.1083/jcb.201006059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomann D, Dorn J, Sorger PK, Danuser G. Automatic fluorescent tag detection in 3D with super-resolution: application to the analysis of chromosome movement. Journal of Microscopy. 2003;211:230–248. doi: 10.1046/j.1365-2818.2002.01066.x. [DOI] [PubMed] [Google Scholar]
- Wang E, Babbey CM, Dunn KW. Performance comparison between the high-speed Yokogawa spinning disc confocal system and single-point scanning confocal systems. J Microsc. 2005;218:148–159. doi: 10.1111/j.1365-2818.2005.01473.x. [DOI] [PubMed] [Google Scholar]