Abstract
Cellular iron homeostasis is maintained by the coordinate posttranscriptional regulation of genes responsible for iron uptake, release, use, and storage through the actions of the iron regulatory proteins IRP1 and IRP2. However, the manner in which iron levels are sensed to affect IRP2 activity is poorly understood. We found that an E3 ubiquitin ligase complex containing the FBXL5 protein targets IRP2 for proteasomal degradation. The stability of FBXL5 itself was regulated, accumulating under iron- and oxygen-replete conditions and degraded upon iron depletion. FBXL5 contains an iron- and oxygen-binding hemerythrin domain that acted as a ligand-dependent regulatory switch mediating FBXL5's differential stability. These observations suggest a mechanistic link between iron sensing via the FBXL5 hemerythrin domain, IRP2 regulation, and cellular responses to maintain mammalian iron homeostasis.
Although iron is an essential cofactor for many proteins, its chemical properties also promote side reactions that damage macromolecules. Failure to maintain proper iron homeostasis can lead to a variety of diseases including anemia and iron overload disorders (1–4). When cellular iron bio-availability is low, the iron regulatory proteins IRP1 and IRP2 coordinate the posttranscriptional regulation of many genes affecting cellular and systemic iron homeostasis by binding to iron response elements (IREs) found within their mRNAs (5, 6). For example, when IRPs bind to the IRE located within the 5′ untranslated region (UTR) of the ferritin mRNA, initiation of protein synthesis is repressed (1, 2). Conversely, IRP binding to IREs within the transferrin receptor 1 (TfR1) 3′UTR stabilizes the mRNA and increases its expression (7). When cellular iron bioavailability is high, IRP1 assembles an iron-sulfur cluster and loses its affinity for IREs (6), whereas IRP2 is preferentially degraded by the proteasome (8, 9). However, the underlying mechanism(s) by which cells directly sense iron (as well as oxygen) bio-availability and relate those changes to differences in IRP2 stability remain controversial (10–15).
To address these questions, we performed a small interfering RNA (siRNA) screen to identify an E3 ubiquitin ligase that regulates IRP2 stability (16). A clonal human embryonic kidney (HEK) 293 cell line stably transfected with a plasmid constitutively expressing N-terminal hemaggluti-nin (HA)–tagged and C-terminal FLAG-tagged IRP2 accumulated high levels of HA-IRP2-FLAG in cells depleted of iron upon treatment with the chelator deferoxamine mesylate (DFO). As observed for endogenous IRP2, low levels of HA-IRP2-FLAG accumulated in cells incubated in the presence of excess iron [ferric ammonium citrate (FAC); fig. S1B]. As an alternative to immunoblots, HA-IRP2-FLAG accumulation levels could be assessed using a high-throughput luminescent proximity assay (AlphaScreen; fig. S1A).
Inappropriate accumulation of HA-IRP2-FLAG under iron-replete conditions, consistent with reduced E3 ligase activity, was best observed with siRNAs targeting expression of the F-box–containing FBXL5 protein (17). Compared with a nontargeting (NT) control siRNA, knockdown of FBXL5 expression by either of two independent siRNAs (FBXL5 1 or 2) led to complete stabilization of HA-IRP2-FLAG under iron-replete conditions (Fig. 1A). Similar results were observed for endogenous IRP2 in HEK 293 cells (Fig. 1A), HeLa cells (fig. S2A), nontumorigenic HBEC-30 cells (fig. S2B), and a clonal cell line containing a tetracycline (Tet)–inducible FBXL5 short hairpin RNA (shRNA) (see below). F-box–containing proteins typically assemble within SCF E3 ubiquitin ligase complexes containing SKP1, Cullin 1 (CUL1), and RBX1 (18), each of which were also identified in the unbiased siRNA screen and whose knockdown partially stabilized IRP2 (Fig. 1A).
Fig. 1.
IRP2 is stabilized under iron-replete conditions after siRNA-mediated suppression of SCFFBXL5. (A) IRP2 accumulation was assessed using the AlphaScreen assay (top) or by immunoblot analysis of endogenous IRP2 (bottom). Knockdown efficiency of the SKP1, CUL1, and RBX1 siRNAs is shown in fig. S2D. (B) IRP1 stabilization under iron-replete conditions after siRNA-mediated suppression of FBXL5, as measured by immunoblot analysis of HEK 293 cell lines stably transfected with either a myc-tagged wild-type (WT) or 3C>3S IRP1 Tet-inducible expression construct. (C) Measurement of RNA binding activity from lysates after siRNA-mediated suppression of FBXL5 in cells expressing HA-IRP2-FLAG. Because human IRP1-IRE and IRP2-IRE complexes migrate similarly, antibodies were added to supershift individual complexes. (D) Relative TfR1 and FBXL5 mRNA accumulation levels measured by qRT-PCR. Assays were performed in triplicate with data represented as the mean ± SE with P values determined using Student's unpaired t test (***P < 0.001).
Despite a greater than 60% amino acid identity, IRP1 and IRP2 are regulated differently. Such differences are not attributable to the 73–amino acid insert unique to IRP2 (19), as deletion of this region does not preclude iron-dependent IRP2 degradation or sensitivity to FBXL5 knockdown (fig. S2C). Moreover, when IRP1 is not bound to an iron-sulfur cluster, it can be targeted for proteasomal degradation under iron-replete conditions, similar to IRP2 (20, 21). Although little change in IRP1 levels was observed in FAC-treated HEK 293 cell lines stably expressing myc-tagged wild-type (WT) IRP1 after transfection with three independent FBXL5 siRNAs, a myc-tagged IRP1 (3C > 3S) lacking three cysteines required for Fe-S cluster assembly (20) responded in an analogous manner to endogenous IRP2 (Fig. 1B). Together, these data suggest that FBXL5 can recognize both apo-IRP1 and IRP2, likely through a common element.
We performed electrophoretic mobility shift assays to demonstrate that this accumulated IRP2 upon FBXL5 knockdown is functional. Knockdown of FBXL5 expression under iron-replete conditions increased total IRE binding activity (IRP1/2) to levels similar to those observed under iron-deficient conditions (Fig. 1C). Although IRP1's RNA binding activity is sensitive to cellular iron availability, FBXL5 knockdown had little effect on IRP1's IRE binding activity, which suggests that it does not simply reduce cellular iron uptake or availability. In contrast, the increased IRP2-FLAG levels upon FBXL5 knockdown in FAC-treated cells elevated total IRP2 RNA binding activity (Fig. 1C). Furthermore, in Tet-inducible FBXL5 shRNA cells, loss of FBXL5 expression (+Tet) resulted in a factor of ∼2.5 increase in TfR1 mRNA accumulation under iron-replete conditions (Fig. 1D) consistent with the increased IRP2 levels (see below). Thus, reduced FBXL5 expression results in inappropriate accumulation of IRP2 under iron-replete conditions and increased IRP2 IRE binding activity with concomitant misregulation of IRP target genes.
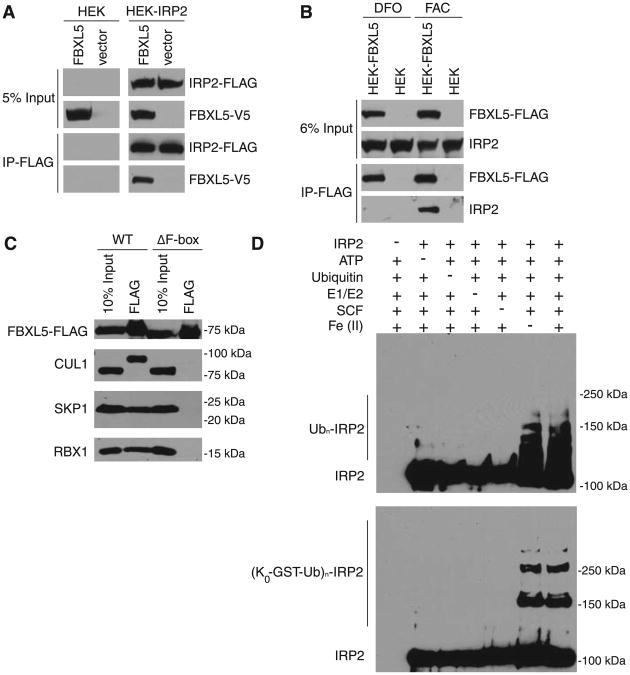
A physical interaction between FBXL5 and IRP2 was observed as antibodies to FLAG coimmunoprecipitated FBXL5-V5 with IRP2-FLAG (Fig. 2A) and FLAG-tagged FBXL5 coimmunoprecipitated endogenous IRP2 (Fig. 2B). To investigate whether this interaction reflected an E3 ligase–substrate relationship, we prepared re-combinant SCFFBXL5. Affinity-purified FLAG-tagged WT FBXL5, but not FBXL5 lacking the F-box domain, assembled into a stoichiometric (fig. S3, A and B) SCFFBXL5 complex (Fig. 2C). Purified SCFFBXL5, but not the ΔF-box FBXL5 variant (fig. S3C), was able to ubiquitinate re-combinant IRP2 in vitro (Fig. 2D).
Fig. 2.
IRP2 is a substrate for SCFFBXL5. (A) FBXL5-V5 coimmunoprecipitates with HA-IRP2-FLAG. (B) Coimmunoprecipitation of endogenous IRP2 and FBXL5-FLAG from HEK 293T cells incubated with either DFO or FAC. (C) Immunoblot analysis of FLAG immunoprecipitates from insect cells coexpressing SKP1, CUL1, and RBX1 with either full-length FLAG-FBXL5 (WT) or FLAG-FBXL5 lacking the F-box domain (ΔF-box). (D) Recombinant SCFFBXL5 ubiquitinates IRP2 in vitro when supplemented with purified E1 and E2 enzymes, adenosine triphosphate (ATP), and WT ubiquitin (top) or glutathione S-transferase (GST)–tagged ubiquitin lacking lysines (K0-GST-Ub) and thus unable to form polyubiquitin chains (bottom).
To reveal the mechanism by which SCFFBXL5 targets IRP2 for degradation in both an iron- and oxygen-dependent manner (2, 6), we conducted an immunoblot analysis, which revealed that both FBXL5-FLAG (Fig. 3A) and endogenous FBXL5 (Fig. 3C) protein levels were low when iron is limiting but increased by more than an order of magnitude under iron-replete conditions. This difference was attenuated by addition of the proteasome inhibitor MG132 (Fig. 3A). Interestingly, FBXL5-FLAG protein levels were inversely regulated with respect to IRP2 (Fig. 3B). Because FBXL5 mRNA levels did not change significantly as a function of iron (Fig. 1D) and iron-dependent regulation of exogenous FBXL5-FLAG did not require elements from the FBXL5 promoter or UTRs, these data indicate that FBXL5 is posttranslationally targeted for pro-teasomal degradation in an iron-dependent manner. FBXL5-FLAG levels from iron-replete cells were also substantially lower when incubated under low-O2 conditions (Fig. 3D). Thus, both iron- and O2-dependent regulation of IRP2 may be mediated by reciprocal effects on FBXL5's stability.
Fig. 3.
FBXL5 protein accumulation is dependent on iron and oxygen availability. (A) Immunoblot analysis of stably transfected FBXL5-FLAG protein accumulation under iron-deplete (DFO) or iron-replete (FAC) conditions. (B) Time course of IRP2 and FBXL5 responses to changes in iron availability. HEK FBXL5-FLAG cells were incubated 16 hours with either FAC or DFO, then switched to low-or high-iron conditions, respectively. Quantitation is provided in table S1. (C) Endogenous IRP2 and FBXL5 levels were assessed by immunoblot analysis in WT cells (293 TRex) or in a clonal cell line stably expressing a Tet-inducible FBXL5 shRNA. (D) Accumulation of FBXL5 under iron-replete conditions is attenuated (by a factor of 2.5) in cells incubated under low-oxygen conditions (1% O2) for 16 hours. (E and F) Residues 1 to 161 mediate iron- and oxygen-dependent regulation of FBXL5 accumulation. Assessment of FBXL5 protein accumulation under iron-deplete (DFO) or iron-replete (FAC) conditions by immunoblot analysis of transiently transfected FBXL5 constructs. Quantitation is provided in table S2.
To identify the region of FBXL5 that confers such regulation, we transfected a series of V5-tagged FBXL5 deletion mutants (fig. S4) into HEK 293T cells followed by incubation with DFO or FAC. As observed for the full-length protein, all but one deletion construct showed preferential accumulation under conditions of excess iron. In contrast, the FBXL5 ΔN-term protein accumulated at constitutively high levels under both conditions (Fig. 3E). Moreover, expression of FBXL5 residues 1 to 161 demonstrated that this region was sufficient to recapitulate iron-dependent degradation (Fig. 3F).
Residues 1 to 161 of the human FBXL5 protein are predicted to contain five α helices encompassing several conserved histidine and glutamic acid residues (figs. S5 and S6A), similar to hemerythrin-like four-helix up and down bundles with an additional C-terminal helix packed against the core (fig. S6B). Although not previously reported in mammalian proteins, hemerythrin domains have been frequently reported to contain μ-oxo diiron centers (22) that reversibly bind oxygen (fig. S6C) and often function as O2-transport proteins, O2 sensors, or metal storage depots in marine invertebrates and bacteria (23). Recombinantly expressed (fig. S6D) FBXL5 hemerythrin domain (WT) copurified with iron, whereas mutation of one of the predicted bridging carboxylates (E61A) was sufficient to abolish iron binding (Fig. 4A). UV-visible absorbance spectroscopy of the WT domain also showed a broad absorbance peak around 330 nm, characteristic of oxidized μ-oxo diiron centers in hemerythrins (24), that decreased upon reduction with dithionite to remove oxygen (Fig. 4B). Circular dichroism spectrometry showed that the WT domain is largely α-helical with minima at 207 and 222 nm (25). In contrast, the spectrum of the E61A mutant is indicative of an unstructured protein (Fig. 4C). Thermal denaturation experiments (Fig. 4D) revealed that treatment of the hemerythrin domain with dithionite and the iron chelator o-phenanthroline lowers both the melting temperature and cooperativity of unfolding, which indicates that these reagents destabilize the tertiary fold of the domain (25). Thus, the FBXL5 N terminus folds into an α helix–rich structure capable of binding both iron and oxygen.
Fig. 4.
FBXL5 contains an iron- and oxygen-binding hemerythrin domain that regulates its stability. (A) Iron content of recombinant WT or variant (E61A) FBXL5 hemerythrin domains. Assays were performed in triplicate with data represented as the mean ± SE, with P values determined using Student's unpaired t test (***P < 0.001). (B) UV chromatogram of the FBXL5 hemerythrin domain before and after dithionite addition. (C) Circular dichroism spectra of purified FBXL5 hemerythrin domains. (D) Measurementofmean molar residual ellipticity at 222 nm as a function of thermal de-naturation of the WT domain treated with dithionite and iron chelator (o-Phen). (E) Luciferase activity (upper panels) and protein accumulation levels (lower panels; quantitation provided in table S3) in HEK 293T cells transiently transfected with fusion protein expression constructs. E.V., empty vector. Assays were performed in triplicate with data represented as the mean ± SE. (F) Immunoblot analysis of protein accumulation from transiently transfected FBXL5 expression constructs.
To determine whether the FBXL5 hemerythrin (Hr) domain could regulate a heterologous protein, we transfected HEK 293T cells with constructs expressing the domain fused to either the N terminus (Hr-Luc) or C terminus (Luc-Hr) of firefly luciferase. Unlike unmodified luciferase (Luc), both Hr-Luc (∼2.5×) and Luc-Hr (∼2.0×) luciferase activities were higher in lysates from FAC-treated cells and correlated with changes in fusion protein accumulation (Fig. 4E). Incubation under low-oxygen conditions also reduced fusion protein luciferase activity and accumulation under iron-replete conditions (Fig. 4E).
Because deletion of the hemerythrin domain resulted in constitutive FBXL5 accumulation (Δ1-161; Fig. 4F) and fusion of the domain conferred iron- and O2-dependent regulation of stability in the context of a heterologous protein (Fig. 4E), a recognition motif for an E3 ubiquitin ligase (degron) likely resides within this domain. Several F-box proteins mediate their own stability through autoubiquitination (26), which may contribute to the partial stabilization of the ΔF-box variant (Fig. 3E). However, knockdown of FBXL5 expression has no effect on the iron-dependent accumulation of the hemerythrin domain itself (fig. S7), indicating that its degron is likely recognized by an as yet unidentified E3 ligase. In the folded iron- and oxygen-bound state, this degron may be efficiently sequestered from the E3 ligase within the hemerythrin fold, promoting FBXL5 accumulation and subsequent IRP2 degradation.
If the hemerythrin domain is unable to assemble a diiron center (e.g., iron-deficient conditions), the domain may unfold to reveal the degron. Degradation of FBXL5 would then preclude the assembly of the SCF complex allowing IRP2 to accumulate and bind IREs. Indeed, prior studies have implicated the involvement of such a labile protein in IRP2 degradation (8). Consistent with this model, mutation of iron-binding ligands prevented FBXL5 accumulation (Fig. 4F), suggesting an iron-insensitive, and unfolded, hemerythrin domain in which the degron is constitutively exposed. In contrast, ΔN-term FBXL5 overexpression partially circumvented iron-dependent IRP2 regulation, as observed by a reduction of IRP2 accumulation under iron-deplete conditions (fig. S8). However, the inability of ΔN-term FBXL5 overexpression to fully block IRP2 accumulation in DFO-treated cells may indicate the presence of additional mechanism(s) by which iron and oxygen regulate SCFFBXL5. Underscoring this possibility is the observation that FBXL5 and IRP2 failed to coimmunoprecipitate in lysates from DFO-treated cells, which suggests that iron availability may also affect substrate recognition (Fig. 2B). We did not observe a further increase in ubiquitination activity when additional iron was added to recombinant SCFFBXL5 in vitro (Fig. 2D), which suggests two possibilities: Either there was sufficient iron present during purification, or the minimal reconstitution assay presented here did not fully recapitulate all aspects of SCFFBXL5 regulation.
Like several F-box–containing proteins reported to act as ligand-regulated effector proteins in plants (27), FBXL5 is an example of a ligand-regulated F-box protein in metazoans. Our observations suggest a direct mechanistic link between sensing iron and oxygen availability via FBXL5's hemerythrin domain, IRP2 induction, and the regulation of mammalian iron homeostasis.
Supplementary Material
Acknowledgments
We thank R. Eisenstein for IRP1 reagents and helpful discussions; Z. Chen for SCF expression plasmids; W. Gao for K0-GST-Ubiquitin and helpful discussions; M. Brown and J. Goldstein for E2 expression constructs; K. Gardner and F. Correa for assistance with circular dichroism; the UTSW HTS facility for assistance with siRNA screening; and L. Wang, G. Pineda, S. Laxman, X. Du, and J. Wang for helpful discussions. R.K.B. is the Michael L. Rosenberg Scholar in Medical Research and was supported by the Burroughs Wellcome Fund, the Robert A. Welch Foundation, the Texas Advanced Research Program, and NIH grant CA115962. This investigation was conducted in a facility constructed with support from the Research Facilities Improvement Program (grant C06 RR 15437-01) from the NIH National Center for Research Resources.
Footnotes
Supporting Online Material: www.sciencemag.org/cgi/content/full/1176326/DC1
Materials and Methods
References
References and Notes
- 1.Muckenthaler MU, Galy B, Hentze MW. Annu Rev Nutr. 2008;28:197. doi: 10.1146/annurev.nutr.28.061807.155521. [DOI] [PubMed] [Google Scholar]
- 2.Wallander ML, Leibold EA, Eisenstein RS. Biochim Biophys Acta. 2006;1763:668. doi: 10.1016/j.bbamcr.2006.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beutler E. Annu Rev Med. 2006;57:331. doi: 10.1146/annurev.med.57.121304.131310. [DOI] [PubMed] [Google Scholar]
- 4.Wrighting DM, Andrews NC. Curr Top Dev Biol. 2008;82:141. doi: 10.1016/S0070-2153(07)00006-3. [DOI] [PubMed] [Google Scholar]
- 5.Hentze MW, Muckenthaler MU, Andrews NC. Cell. 2004;117:285. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 6.Rouault TA. Nat Chem Biol. 2006;2:406. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- 7.Koeller DM. Proc Natl Acad Sci USA. 1989;86:3574. doi: 10.1073/pnas.86.10.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo B, Phillips JD, Yu Y, Leibold EA. J Biol Chem. 1995;270:21645. doi: 10.1074/jbc.270.37.21645. [DOI] [PubMed] [Google Scholar]
- 9.Samaniego F, Chin J, Iwai K, Rouault TA, Klausner RD. J Biol Chem. 1994;269:30904. [PubMed] [Google Scholar]
- 10.Iwai K. Proc Natl Acad Sci USA. 1998;95:4924. doi: 10.1073/pnas.95.9.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamanaka K. Nat Cell Biol. 2003;5:336. doi: 10.1038/ncb952. [DOI] [PubMed] [Google Scholar]
- 12.Bourdon E. Blood Cells Mol Dis. 2003;31:247. doi: 10.1016/s1079-9796(03)00161-x. [DOI] [PubMed] [Google Scholar]
- 13.Hanson ES, Rawlins ML, Leibold EA. J Biol Chem. 2003;278:40337. doi: 10.1074/jbc.M302798200. [DOI] [PubMed] [Google Scholar]
- 14.Wang J. Mol Cell Biol. 2004;24:954. doi: 10.1128/MCB.24.3.954-965.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zumbrennen KB, Hanson ES, Leibold EA. Biochim Biophys Acta. 2008;1783:246. doi: 10.1016/j.bbamcr.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.See supporting material on Science Online.
- 17.Zhang N. Biochem Biophys Res Commun. 2007;359:34. doi: 10.1016/j.bbrc.2007.05.068. [DOI] [PubMed] [Google Scholar]
- 18.Willems AR, Schwab M, Tyers M. Biochim Biophys Acta. 2004;1695:133. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 19.Iwai K, Klausner RD, Rouault TA. EMBO J. 1995;14:5350. doi: 10.1002/j.1460-2075.1995.tb00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clarke SL. EMBO J. 2006;25:544. doi: 10.1038/sj.emboj.7600954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fillebeen C, Chahine D, Caltagirone A, Segal P, Pantopoulos K. Mol Cell Biol. 2003;23:6973. doi: 10.1128/MCB.23.19.6973-6981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stenkamp RE. Chem Rev. 1994;94:715. [Google Scholar]
- 23.French CE, Bell JM, Ward FB. FEMS Microbiol Lett. 2008;279:131. doi: 10.1111/j.1574-6968.2007.01011.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhang JH, Kurtz DM, Jr, Xia YM, Debrunner PG. Biochemistry. 1991;30:583. doi: 10.1021/bi00216a037. [DOI] [PubMed] [Google Scholar]
- 25.Greenfield NJ. Nat Protocols. 2007;1:2876. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galan JM, Peter M. Proc Natl Acad Sci USA. 1999;96:9124. doi: 10.1073/pnas.96.16.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Somers DE, Fujiwara S. Trends Plant Sci. 2009;14:206. doi: 10.1016/j.tplants.2009.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.