Summary
The DNA replication checkpoint, also known as the intra-S or S-phase checkpoint, plays a central role in ensuring the accuracy of DNA replication. When replication is impeded by DNA damage or other conditions, this checkpoint delays cell cycle progression and coordinates resumption of replication with DNA repair pathways. One of its critical functions is to stabilize stalled replication forks in a replication-competent state, presumably by maintaining proper assembly of replisome components and preserving DNA structures. Here we describe a series of assays used to study the replication checkpoint. These assays allow us to investigate the specific functions of proteins involved in the replication checkpoint in fission yeast.
Keywords: The DNA replication checkpoint, S-phase stress response, Cds1, Chk1, Rad3, Cds1 kinase assays, Pulsed-field gel electrophoresis, Rad22-YFP DNA repair foci
1. Introduction
Environmental toxins or drugs can cause DNA damage and lead to an arrest of DNA replication forks (1–3). Arrested forks are among the most serious of threats to genomic integrity because they can collapse, break, or rearrange (4). To suppress these genome-destabilizing events, all eukaryotic cells are equipped with a DNA replication stress response pathway, termed the DNA replication checkpoint or the S-phase checkpoint (1–3). In humans, defects in this checkpoint cause genetic instability, leading to a strong predisposition to cancer (3, 5–11).
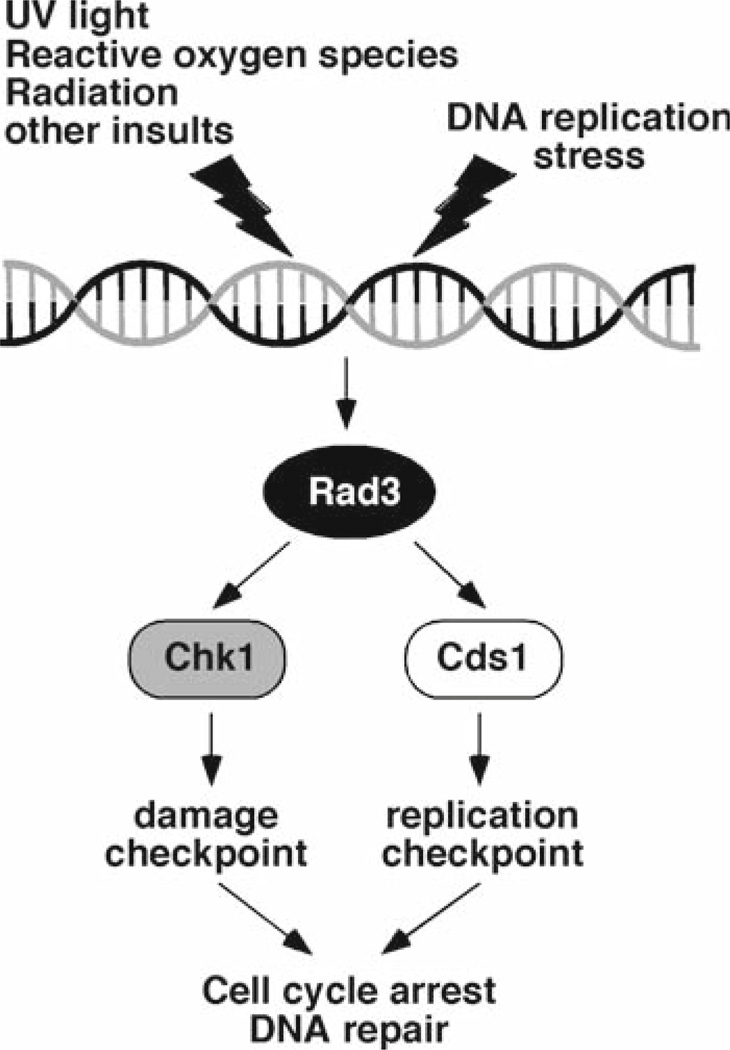
The replication checkpoint is activated when replication forks are arrested by DNA damage, protein complexes bound to chromatin, starvation of deoxyribonucleotides, or other conditions. This checkpoint arrests cell cycle progression, specifically preventing the onset of mitosis when DNA is not fully replicated, while at the same time regulating other less well-understood processes that are required for recovery from fork arrest (1–3). Central to this system are protein kinases such as human ATM and ATR (1–3, 5, 12). In humans, major ATM/ATR downstream targets include p53, Chk1, and Chk2. Both Chk1 and Chk2 arrest the cell cycle by phosphorylating Cdc25, which inhibits its phosphatase activity and in some cases promotes its degradation, thereby preventing it from activating Cdc2 (Cdk1), a kinase essential for mitotic onset. The checkpoint proteins are also thought to facilitate DNA repair and recombination pathways (3, 5, 6). In the model organism Schizosaccharomyces pombe, the ATM/ATR homolog Rad3 also controls downstream effector kinases Cds1 (Chk2 homolog) and Chk1 (Fig. 1). Cds1 and Chk1 define redundant pathways of checkpoint activation in response to fork arrest, although Cds1 acts as the main kinase for activation of the replication checkpoint (Fig. 1) (1, 3, 13–15). Thus, the mechanisms of checkpoint responses appear to be highly conserved throughout evolution.
Fig. 1.
S. pombe checkpoint pathway. DNA damage or replication stress activates the Rad3-dependent checkpoint pathway. Rad3 sends a checkpoint signal to downstream checkpoint effectors, Chk1 and Cds1 to arrest the cell cycle and facilitate DNA repair pathways.
Recent studies have identified a group of proteins that are involved in the activation of the replication checkpoint kinase Cds1 and the stabilization of replication forks. In fission yeast, Mrc1, a mediator of the replication checkpoint, is essential for Cds1 activation in a manner dependent on Rad3 (16, 17). Swi1 forms a replication fork protection complex with Swi3 and is required for proper activation of Cds1 and replication fork stabilization (18, 19). Hsk1-Dfp1, the Cdc7-Dbf4-related kinase, functions in conjunction with the Swi1–Swi3 complex and is also important for activation of Cds1 and fork stabilization (20, 21). Furthermore, Ctf18, a component of an alternative replication factor C complex, has shown to be involved in these mechanisms. Ctf18 and Swi1–Swi3 function in separate and redundant pathways required for the replication checkpoint and sister chromatid cohesion (22). Taken together, these facts suggest that a complicated network of proteins is involved in checkpoint signaling and fork stabilization, ensuring accuracy in copying the genome. In this chapter, we will describe a collection of experiments that we have used to investigate the replication checkpoint in S. pombe. These experiments include assays to evaluate sensitivities of cells to S-phase stressing agents, epistasis analysis involving checkpoint defective mutants, Cds1 kinase assay, pulsed-field gel electrophoresis (PFGE) of chromosomes, and visualization of DNA damage during S-phase.
2. Materials
2.1. Serial Dilution Growth Assays
YES (yeast extract and supplements medium). 5 g/L yeast extract, 30 g/L glucose, 187.5 mg/L leucine, 187.5 mg/L histidine, 187.5 mg/L adenine, and 100 mg/L uracil.
1 M hydroxyurea (HU): dissolved in water and stored at −20°C (Sigma-Aldrich, St. Louis, MO).
Methylmethane sulfonate (MMS) (straight solution) (Sigma-Aldrich).
10 mM camptothecin (CPT) (Sigma-Aldrich): dissolved in DMSO and stored at −20°C.
Hemacytometer.
10-cm petri dishes.
100% ethanol.
Replica plater for 96-well plate (8 × 6 Array, 48-pin) (R2383, Sigma-Aldrich).
8-channel pipetter.
UV crosslinker (we used a Stratalinker from Stratagene, La Jolla, CA).
2.2. Cds1 Kinase Assay
1 M HU.
STOP buffer. 150 mM NaCl, 50 mM NaF, 10 mM EDTA, 1 mM NaN3.
Anti-Cds1 antibody (kindly provided by Dr. Teresa Wang, Stanford University).
Protein A-Sepharose.
Lysis Buffer. 50 mM Tris–HCl, pH 7.5, 80 mM β-glycerol phosphate, 250 mM NaCl, 15 mM nitrophenylphosphate, 50 mM NaF, 5 mM EDTA, 1 mM DTT, and 0.1% NP-40 supplemented with protease inhibitor cocktail (complete EDTA-free protease inhibitor cocktail from Roche, Basel, Switzerland) and p-4-amidoinophenyl-methane sulfonyl fluoride hydrochloride monohydrate (p APMSF) (Sigma-Aldrich): prepare fresh before use.
0.5-mm glass beads.
FastPrep cell disruptor (Qbiogene, Irvine, CA).
Protein Assay Dye Reagent Concentrate (BioRad, Hercules, CA).
2× Kinase Buffer. 20 mM HEPES, pH 7.5, 150 mM KCl, 10 mM MgCl2, 1 mM EDTA, 2 mM DTT.
1× Kinase Buffer. 10 mM HEPES, pH 7.5, 75 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT.
γ-32P-ATP (3,000 Ci/mmol).
10 mM ATP.
10 mg/mL MBP (Myelin Basic Protein, a Cds1 substrate): dissolved in water and stored at −20°C (Sigma-Aldrich).
2× SDS-PAGE sample buffer. 100 mM Tris–HCl, pH 8.0, 4% SDS, 4 mM EDTA, 20% glycerol, 0.005% bromophenol blue, and 10% mercaptoethanol.
PhosphorImager system (e.g., Storm 840 from GE Healthcare).
Liquid scintillation counter.
2.3. Pulsed-Field Gel Electrophoresis
CSE buffer: 20 mM citric acid, 20 mM Na2 HPO4, adjusted to pH 5.6, 1.2 M sorbitol, 40 mM EDTA.
1 M HU.
Zymolyase 100T (Seikagaku, Tokyo, Japan).
TSE buffer. 10 mM Tris–HCl, pH 7.5, 0.9 M sorbitol, 45 mM EDTA, pH 8.0.
Low Melt Agarose (BioRad).
Plug molds for CHEF gel system (BioRad).
Tris–EDTA–SDS buffer. 0.25 M EDTA, pH 8.0, 50 mM Tris–HCl, pH 7.5, 1% SDS.
NDS buffer. 10 mM Tris base, 0.5 M EDTA, adjusted to pH 9.5, 1% lauryl sarcosine.
20 mg/mL Proteinase K (Invitrogen, Carlsbad, CA): dissolved in water and stored at −20°C.
0.5 M EDTA, pH 8.0.
TE buffer. 10 mM Tris–HCl, pH 8.0, 1 mM EDTA, pH 8.0.
Megabase agarose (BioRad).
1× TAE buffer. 40 mM Tris–Acetate, pH 8.3, 1 mM EDTA
50 mg/mL Ethidium bromide.
Hemacytometer.
Pulsed-field gel electrophoresis system (we used a CHEFDR II system from BioRad).
2.4. Rad22-YFP Foci Detection
Glass slides.
Glass coverslips.
Hemacytometer.
Fluorescence microscope.
S. pombe rad22-YFP strain (will be available from National BioResource Project, Japan)
3. Methods
3.1. Serial Dilution Growth Assays Used to Determine the Effects of Fork Stalling Agents
Stalled replication forks activate the replication checkpoint. Therefore, many checkpoint mutants show sensitivity to hydroxyurea (HU), which inhibits ribonucleotide reductase, thereby depleting the dNTP pool available for DNA synthesis and leading to stalled replication forks (1–3). Some mutants also show sensitivity to methylmethane sulfonate (MMS, which promotes alkylation of DNA templates, causing stalled replication forks), ultraviolet (UV, which causes the formation of cyclobutane dimers and other lesions leading to an arrest of replisome progression), or camptothecin (CPT, which induces replication fork breakage by trapping topoisomerase I-DNA complexes) (23–26). In S. pombe, mutations in Cds1, a master kinase for the replication checkpoint, render cells highly sensitive to HU (24). There is a group of proteins that are required for activation of Cds1 in fission yeast. These include Mrc1, Swi1–Swi3, and Ctf18, whose mutations also render cells sensitive to HU (16, 18, 19, 22). In addition, cells with mutations in proteins involved in fork stabilization, including Swi1–Swi3 and Ctf18, display sensitivity to MMS and CPT (22, 27). In this section, we describe the use of serial dilution growth assays, colloquially known as “spot assays,” to evaluate sensitivity of S. pombe mutant cells to S-phase stressing agents. This approach serves as a first step toward understanding the possible role of a gene of interest in the activation of Cds1 and/or fork stabilization.
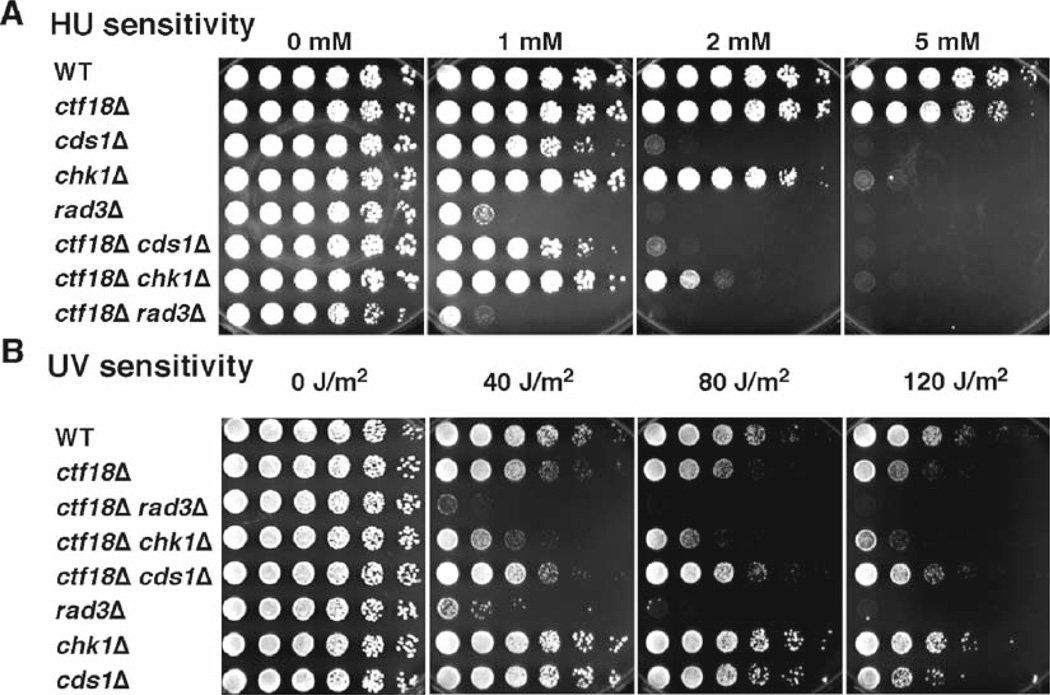
Cds1 and Chk1 define redundant pathways of checkpoint activation in response to fork arrest (1, 3, 13–15). Therefore, cds1sΔ chk1Δ double mutant cells display extreme sensitivity to HU and UV, both of which cause stalled replication forks. Because both Cds1 and Chk1 pathways are controlled by the Rad3 kinase (Fig. 1), cds1Δ chk1Δ and rad3Δ show similar sensitivities to HU or UV (1, 3, 13–15). Therefore, epistasis analysis with checkpoint mutants in HU and UV survival assays determines whether a gene of interest functions in the Cds1 or Chk1 pathway. For example, we have previously shown that the Ctf18 protein is involved in the Cds1-dependent checkpoint pathway (22). As shown in Fig. 2, ctf18Δ chk1Δ cells are more sensitive to HU and UV than either single mutant, while ctf18Δcds1Δ and ctf18Δ rad3 double mutant cells show HU and UV sensitivity similar to either single mutant. These results suggest that Ctf18 is involved in the Cds1-dependent replication checkpoint.
Fig. 2.
Ctf18 is involved in the replication checkpoint enforced by the Cds1 kinase. Synergistic interaction of ctf18Δ and chk1Δ in HU (A) and UV (B) survival assays shows that Ctf18 is required for survival of replication fork arrest. For HU sensitivity assays, fivefold serial dilution of cells was incubated on YES agar medium supplemented with the indicated amounts of HU for 2–4 days at 32°C. For UV survival assays, fivefold serial dilution of cells was plated on YES agar medium and exposed to the indicated doses of UV. Agar plates were then incubated for 2–3 days at 32°C. ctf18Δ showed a strong synergistic interaction with chk1Δ, but not with cds1Δ and rad3Δ, suggesting that Ctf18 is involved in the Cds1 pathway.
Inoculate S. pombe cells into 5 mL of YES, and grow cells overnight until the OD600 reaches ~1.0. If cells are overgrown (OD600 of more than 2.0), dilute cells into YES at OD600 of 0.4, and grow cells for 4 h.
Prepare 1/10 dilution of culture, and use 10 µL of diluted cell suspension to measure cell density in a hemacytometer (see Fig. 2 in Chapter “Chromatin Immunoprecipitation of Replication Factors Moving with the Replication Fork”).
Adjust cell density to 2.0 × 107 cells/mL in a 1.5-mL microcentrifuge tube. If the cell density of a culture is less than 2.0 × 107, then centrifuge the culture and suspend cells in an appropriate amount of YES to obtain the cell density of 2.0 × 107 cells/mL.
Prepare two 10-cm petri dishes, one containing sterilized water and the other one containing 100% ethanol.
Sterilize the pins of a 48-pin-replica plater with 100% ethanol and the flame of a Bunsen burner. Allow the plater to cool to room temperature.
Using an 8-channel pipetter, add 200 µL of sterilized water from the 10-cm petri dish to columns #2 to #6 of a sterilized 96-well plate (see Fig. 3).
Add 250 µL of cell suspension (2.0 × 107 cell/mL) to column #1.
Mix the cell suspension by pipetting up and down using an 8-channel pipetter set to 50 µL, and transfer 50 µL of cell suspension to column #2 to make fivefold dilution. Repeat fivefold serial dilutions until column #6 (see Note 1 and Fig. 3).
Place the replica plater in cell suspensions on the 96-well plate to transfer cell suspension to pins.
Touch the replica plater to a YES agar plate or a YES agar plate containing a drug. Repeat the transfer of cells from the 96-well plate to another YES plate containing a different drug. It is important to wash pins of the replica plater with water and then with ethanol at each replication to avoid crosscontamination of different drugs. For UV sensitivity assays, replica plate cells to YES agar medium, and expose the YES plates to short-wavelength (254-nm) UV in a Stratalinker (see Note 2).
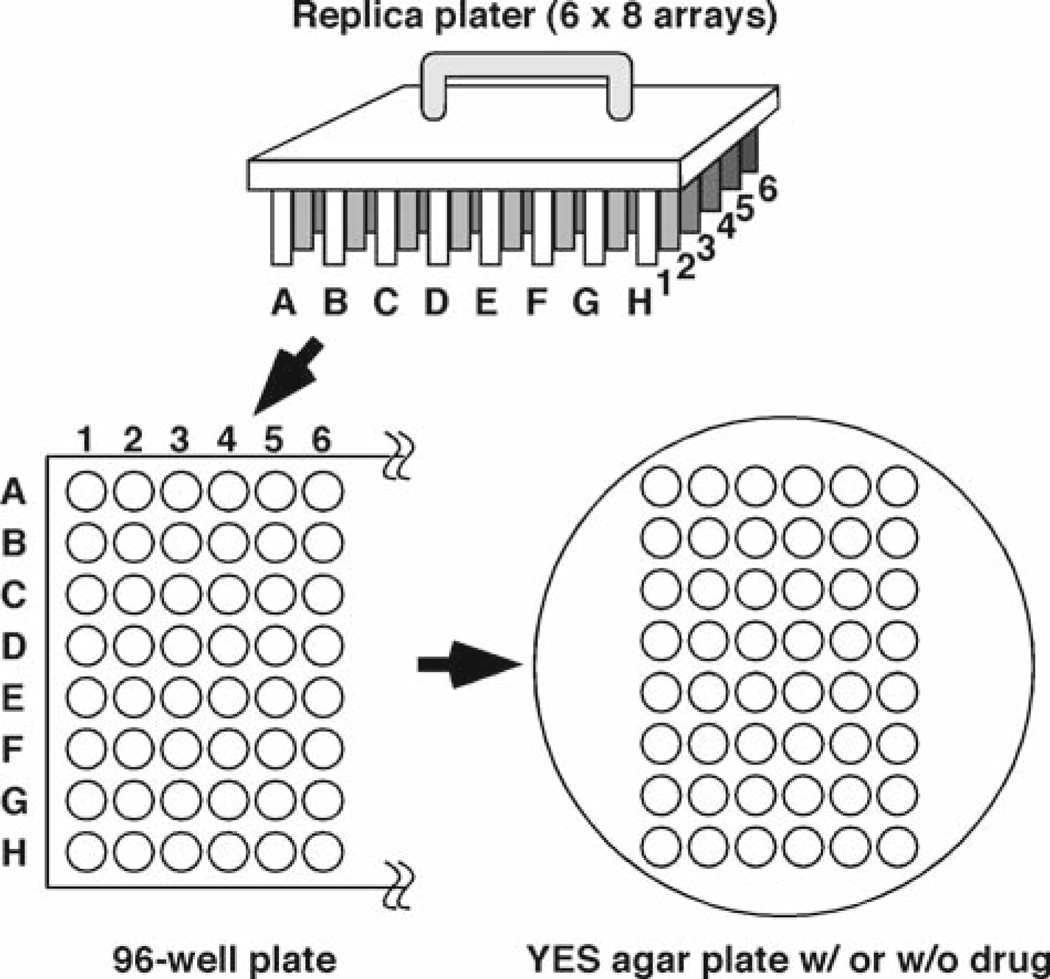
Allow agar plates to absorb cell suspension. Incubate agar plates at appropriate temperatures to allow cell growth. Compare cell growth of different cell lines after several days of incubation. Document using a digital camera or a scanner linked to a computer. Growth of eight strains can be simultaneously compared on a single plate (rows A through H, see Fig. 3).
Fig. 3.
Using a 48-pin replica plater, serial dilution of cells can be transferred from a 96-well plate to a YES agar medium. Fivefold serial dilutions of cells are added from column #1 through #6. Eight strains (rows A through H) can be tested simultaneously.
3.2. Cds1 Kinase Assay
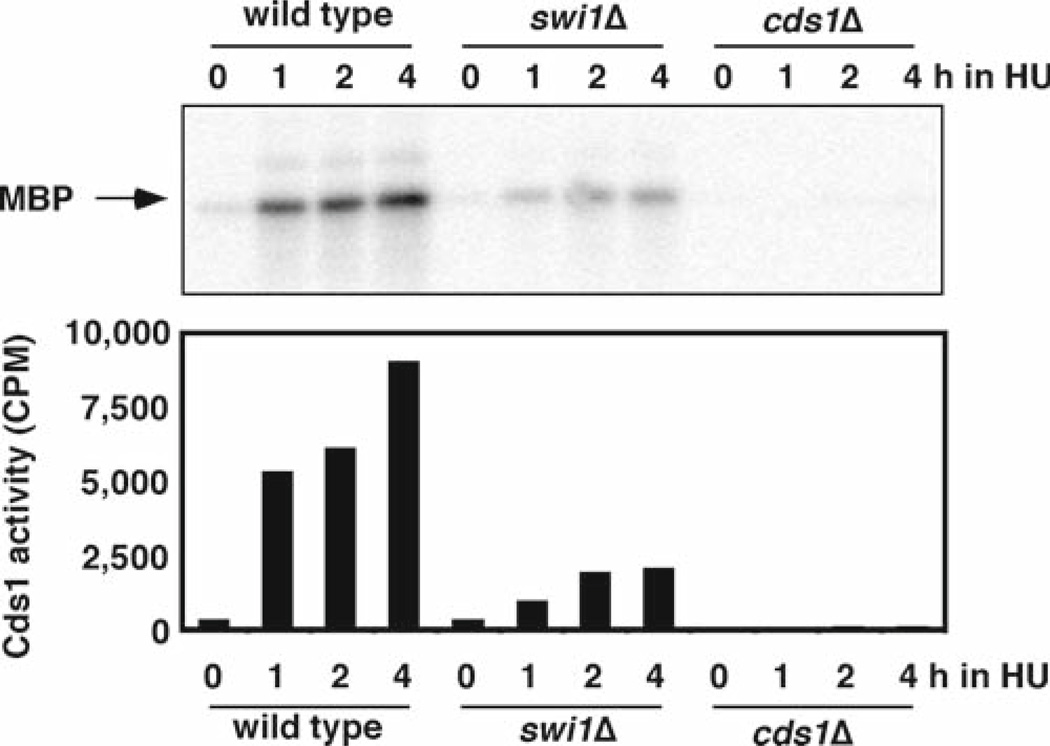
If a gene of interest is implicated in the Cds1-dependent replication checkpoint pathway, it is obviously important to test whether the mutant cells show a decreased level of Cds1 activity. In this section we describe a Cds1 kinase assay using myelin basic protein (MBP) as a substrate. We will treat cells with HU to activate Cds1. Figure 4 shows an example of Cds1 kinase assays using swi1Δ and cds1Δ mutants.
Fig. 4.
Cds1 activation is strongly reduced in swi1Δ cells. Cells of the indicated genotypes were incubated in YES liquid medium supplemented with 12 mM HU for 0, 1, 2, and 4 h at 30°C. Kinase activity of immunoprecipitated Cds1 was measured using myelin basic protein (MBP) as a substrate. The radiolabeled MBP was detected after gel electrophoresis (upper panel). The radioactivity levels (counts per minute, CPM) of MBP were then determined in a liquid scintillation counter (lower panel).
3.2. 1.Preparation of Cells
Inoculate S. pombe cells into 5 mL of YES, and grow cells overnight. Next day, dilute cells into 100 mL of YES and grow overnight until the OD600 reaches ~1.0. If cells are overgrown (OD of more than 2.0), dilute cells into YES at OD600 of 0.4, and grow cells for 4 h.
Dilute cells at OD600 of 0.4 in 200 mL of YES and grow cells.
Add 2.4 mL of HU (final concentration 12 mM) and continue to grow cells.
Transfer 50 mL of cell culture to a centrifuge tube at indicated times (0, 1, 2, and 4 h after the addition of HU). Centrifuge at 1,000 × g for 3 min at 4°C to collect cells.
Resuspend cells in 10 mL of STOP buffer and centrifuge again.
Resuspend cells in 1 mL of STOP buffer, transfer to a 1.5-mL screw top tube (see Note 3), aspirate buffer, and immediately freeze cell pellet at −80°C.
3.2.2. Immunoprecipitation of Cds1
Prepare anti-Cds1 antibody-bound protein A beads a day before immunoprecipitation (see Note 4): Mix 20 µL bed volume of protein A sepharose (prewashed with Lysis buffer), 20 µL of Lysis buffer, and 1 µL of anti-Cds1 antibody for one sample. Rotate overnight at 4°C.
Suspend cell pellet with 200 µL of Lysis buffer.
Add glass beads until they reach the surface of the buffer.
Break cells using a FastPrep cell disruptor at 4°C (output 6.0, 20 s, two cycles, 2 min interval between cycles) (see Note 5).
To recover cell lysate, pierce the bottom of the tube with a heated needle, and place tube in a new 1.5-mL microcentrifuge tube. Centrifuge at 800 × g using a microcentrifuge for 30 s to collect cell lysate in the new tube.
Discard the tube containing glass beads, add 400 µL of Lysis Buffer to the cell lysate, and mix well.
Centrifuge at 16,000 × g for 5 min at 4°C, transfer supernatant to a new 1.5-mL microcentrifuge tube.
Centrifuge again at 16,000 × g for 10 min at 4°C, transfer supernatant to a new 1.5-mL microcentrifuge tube.
Measure protein concentration using BioRad Protein Assay Dye Reagent Concentrate and adjust concentrations to the lowest concentrated sample.
Add 40 µL of Cds1-antibody-bound protein A beads (50% slurry) prepared at step 1 to each sample.
Rotate samples for 1–2 h at 4°C.
Wash beads three times with 500 µL of ice-cold Lysis Buffer
Wash beads three times with 500 µL of ice-cold 1× Kinase Buffer
3.2.3. Kinase Reaction
Prepare Kinase Reaction Cocktail. Mix 10 µL of 2× Kinase Buffer, 2 µL of γ-32P-ATP (5 µCi), 0.2 µL of 10 mM ATP, 0.5 µL of 10 mg/mL MBP, and 7.3 µL of H2O for one kinase reaction.
Add 20 µL of Kinase Reaction Cocktail to antibody-bound protein A beads prepared earlier.
Incubate for 15 min at 30°C. Mix every 1–3 min to avoid precipitation of beads.
Stop the kinase reaction by adding 25 µL of 2× SDS Sample Buffer
Boil samples for 5 min, and store samples at −20°C.
3.2.4. Detection of Cds1 Kinase Activity
To visualize MBP, run a 15% SDS-PAGE gel using 10 µL of samples prepared earlier.
After SDS-PAGE, stain the gel with Coomassie Brilliant Blue, and dry the gel.
Wrap the dried gel with plastic wrap and detect radioactivity incorporated in MBP with a phosphorImager.
After imaging, cut out MBP bands. The radioactivity levels (cpm) of MBP bands should be determined in a liquid scintillation counter.
3.3. Pulsed-Field Gel Electrophoresis
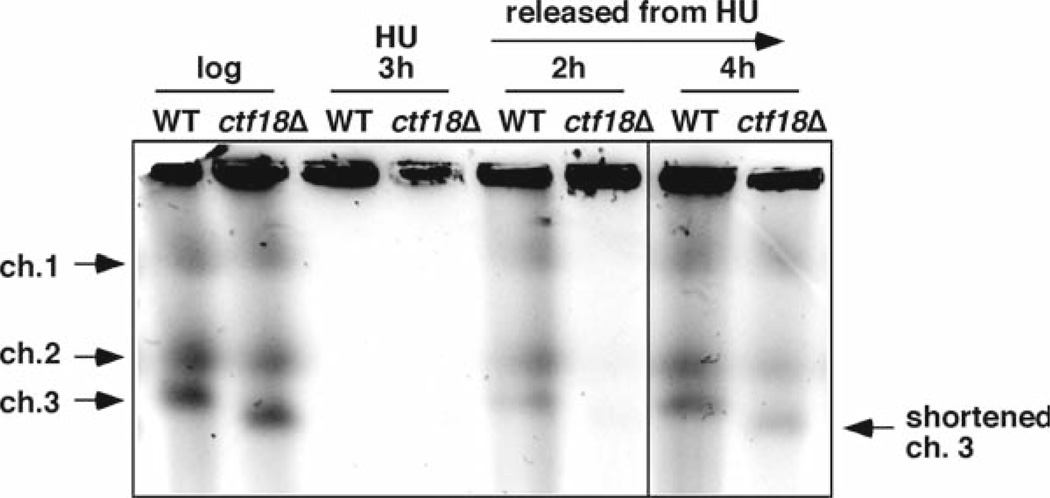
One of most important functions of the replication checkpoint is to stabilize replication forks by maintaining proper assembly of replisome components and preserving DNA structures when problems are encountered (28–32). Therefore, it is also important to determine whether a protein involved in Cds1 activation is also required for replication fork stabilization. The replication fork stalls in the presence of HU; however, wild-type cells can recover from fork arrest because forks are maintained in a state competent for resumption of DNA synthesis. In contrast, if replication forks are not stably maintained in the absence of the proper replication checkpoint, forks may collapse or rearrange, resulting in a defect in replication recovery after fork abnormality. To evaluate fork stability in S. pombe mutants, we utilize pulsed-field gel electrophoresis (PFGE). Figure 5 shows an example of PFGE analysis of ctf18Δ cells treated with HU.
Prepare a midlog phase S. pombe cell culture (OD600 = 0.4) in 300 mL YES.
Monitor cell density using a hemacytometer, and collect a sample of 2.5 × 108 cells by centrifugation (1,000 × g, 3 min, 4°C) for a log-phase sample. Wash cells with CSE buffer once, and store cell pellet at −80°C, and proceed to step 7.
Add HU to the culture to a final concentration of 12 mM, and grow cells for additional 3 h in a 30°C shaker.
Monitor cell density using a hemacytometer, and collect a sample of 2.5 × 108 cells by centrifugation (1,000 × g, 3 min, 4°C) for an HU-treated sample. Wash cells with CSE buffer once, and store cell pellet at −80°C, and proceed to step 7.
Wash remaining cells twice with fresh YES medium, and return the culture to the 30°C shaker.
In 1, 2, and 4 h, monitor cell density using a hemacytometer, and collect a sample of 2.5 × 108 cells by centrifugation (1,000 × g, 3 min, 4°C). Wash cells with CSE buffer once, store cell pellet at −80°C, and proceed to step 7.
Suspend cells from the collected samples in 1 mL of CSE containing 1 mg/mL of Zymolyase 100T, and incubate at 37°C for 2 h.
Pellet cells by centrifugation (1,000 × g, 3 min, 4°C).
Resuspend cells at a concentration of 8 × 108 cells/mL in 300 µL TSE.
Warm the cell suspension to 42°C.
Add 300 µl of 1.1% low melting temperature agarose in TSE.
Dispense aliquots into plug molds (five aliquots per sample), and allow plugs to solidify for 30 min at 4°C.
Transfer plugs into a centrifuge tube containing 3 mL of Tris–EDTA–SDS, and incubate at 55°C for 90 min.
Replace the buffer with 3 mL of NDS supplemented with 1 mg/mL Proteinase K (Invitrogen, Carlsbad, CA), and incubate plugs at 55°C for 24 h (see Note 6).
Replace the buffer with 3 mL of fresh NDS supplemented with 1 mg/mL Proteinase K, and incubate plugs at 55°C for 24 h (see Note 6).
To analyze chromosome DNA embedded in plugs, equilibrate plugs in 5 mL TE three times for 30 min each (see Note 7).
Run on 0.8% Megabase agarose gel (BioRad, Hercules, CA) in 1× TAE using a CHEF-DR II system (BioRad, Hercules, CA) at the following settings: block 1, 2 v/cm, initial, and final switch time of 1,800 s, 14°C, pump speed 70, for 72 h.
Stain gels with 0.5 µg/mL ethidium bromide in H2O for 30 min, then destain with water for 1–2 h.
Visualize chromosomes using a UV transilluminator.
Fig. 5.
Ctf18 is required for the efficient resumption of replication following fork damage. Chromosome samples from either wild-type or ctf18Δ cells were examined by PFGE. Cells were grown until midlog phase and then incubated in the presence of 12 mM HU for 3 h at 30°C. Cells were then washed and released into fresh medium. Chromosomal DNA samples were prepared at the indicated times. ctf18Δ cells showed a delay in recovery of DNA replication after fork arrest. ctf18Δ cells also displayed a short chromosome III probably due to replication and/or recombination defects.
3.4. Detection of Rad22-YFP DNA Repair Foci
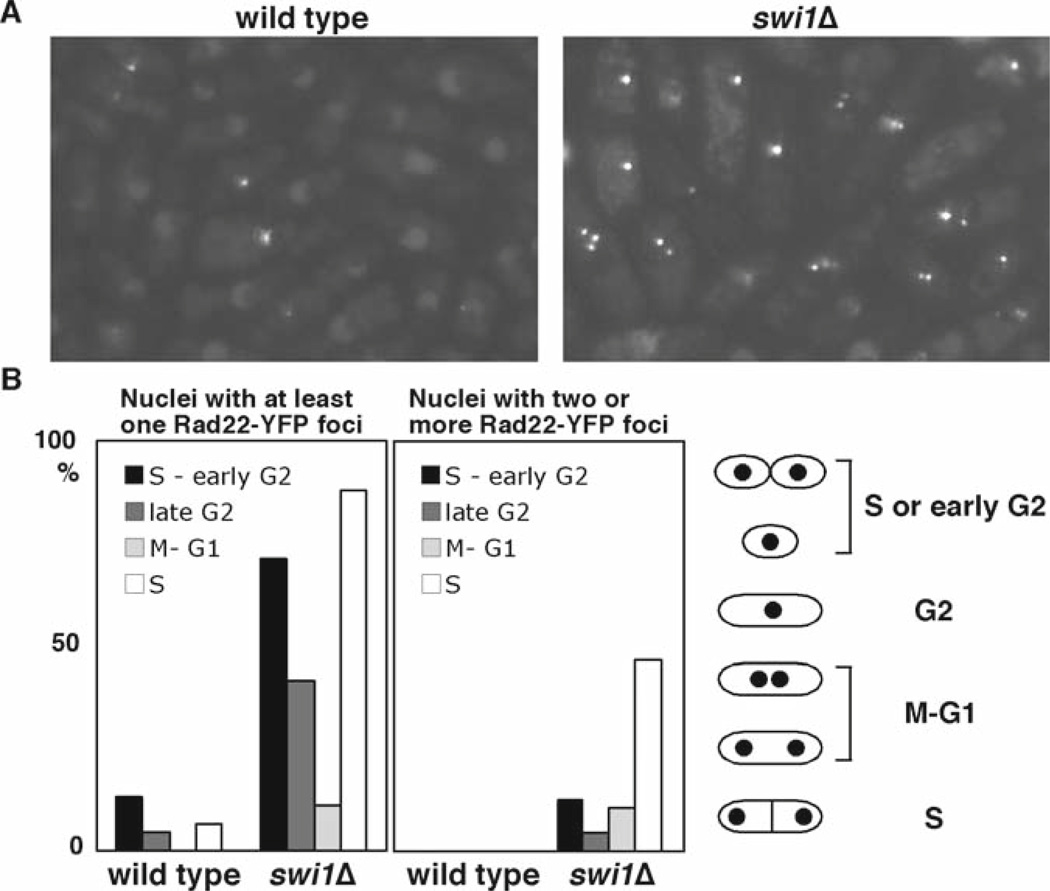
Some factors involved in the replication checkpoint are also involved in replication fork stabilization even in the absence of genotoxic stress (18–20, 22). When cells experience collapsed replication forks, cells show an increase in DNA damage because of an accumulation of abnormal DNA structures at the replication forks (18, 19). To visualize DNA damage in live cells, we utilize cells expressing Rad22-YFP fusion protein from its endogenous promoter at its genomic locus. Rad22 is a homolog of budding yeast Rad52 and is shown to bind single-stranded DNA (ssDNA) during homologous recombination at double-strand breaks and other sites that have exposed ssDNA segments, leading to the formation of Rad22-YFP DNA repair foci at the site of DNA damage during S-phase (18, 33, 34). Figure 6 shows an example of spontaneous Rad22-YFP foci accumulated in swi1 cells.
Inoculate cells expressing Rad22-YFP in 5 mL YES and grow cells at 25°C until midlog phase (see Note 8).
Collect cells by centrifugation (see Note 9) and keep cell pellet (with small amount of YES) on ice.
Place 2 µL of cell suspension (from the bottom of tube) on a glass slide. Cover the cell suspension with a 22 mm × 22 mm coverslip.
Observe cells with a fluorescence microscope, and capture Rad22-YFP fluorescence images. In total, more than 200 cells should be monitored.
Estimate the cell cycle position of cells containing Rad22- YFP foci by analyzing the cell length, number, and position of nuclei, and the presence of a division plate (see Fig. 2 in Chapter “Chromatin Immunoprecipitation of Replication Factors Moving with the Replication Fork”).
Fig. 6.
swi1Δ cells experience replication abnormality. (a) Rad22-YFP foci formation was significantly elevated in swi1Δ cells. Cells of the indicated genotype expressing genomic Rad22-YFP were grown in YES medium at 25°C until midlog phase. (b) Quantification of Rad22-YFP foci according to cell cycle stages. S and early G2 cells had the most Rad22-YFP foci. The percentages of nuclei that have at least one focus or harbor two or more foci are shown.
Acknowledgments
We thank Adam Leman and Jordan Rapp for helpful discussion. This work was supported by a Leukemia Research Foundation grant (E.N.), Drexel University College of Medicine start-up funds (E.N.), and NIH grant GM59447 (P.R.).
Footnotes
We use water to dilute cells on a 96-well plate. Water has more surface tension compared to YES medium, allowing a better transfer of cells from a 96-well plate to a 48-pin replica plater.
The cell suspension should be absorbed into the agar medium before UV irradiation. After UV irradiation, the plates should be incubated in dark in an incubator to avoid possible photoreactivation repair although this pathway may not exist in S. pombe.
It is important to use tubes that fit properly into the FastPrep cell disruptor.
If the anti-Cds1 antibody is not available, use strains that are engineered to express tagged Cds1. In this case, an antibody against or an affinity column for the tag should be used. The cds1–2HA6His strain is available from National BioResource Project, Japan.
It is important to monitor cell disruption under a microscope. More than 90% of cells should be disrupted.
To activate Proteinase K, samples containing Proteinase K should be preincubated for 30 min at 37°C.
If plugs are not used for electrophoresis immediately, store the plugs in 5 mL 0.5 M EDTA at 4°C. Equilibrate plugs again as described in Subheading 3.3.16 before electrophoresis.
We grow cells at 25°C to obtain stronger yellow fluorescent protein (YFP) signals.
We increase cell density by centrifugation. This allows us to monitor many cells in one image.
References
- 1.Boddy MN, Russell P. DNA replication checkpoint. Curr. Biol. 2001;11:R953–R956. doi: 10.1016/s0960-9822(01)00572-3. [DOI] [PubMed] [Google Scholar]
- 2.Osborn AJ, Elledge SJ, Zou L. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 2002;12:509–516. doi: 10.1016/s0962-8924(02)02380-2. [DOI] [PubMed] [Google Scholar]
- 3.Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Toward maintaining the genome: DNA damage and replication checkpoints. Annu Rev Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- 4.McGlynn P, Lloyd RG. Recombinational repair and restart of damaged replication forks. Nat Rev Mol Cell Biol. 2002;3:859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- 5.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 6.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 7.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 8.Kolodner RD, Putnam CD, Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- 9.McGowan CH. Checking in on Cds1 (Chk2): A checkpoint kinase and tumor suppressor. Bioessays. 2002;24:502–511. doi: 10.1002/bies.10101. [DOI] [PubMed] [Google Scholar]
- 10.Paulovich AG, Toczyski DP, Hartwell LH. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 11.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 12.Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297:547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- 13.O’Connell MJ, Walworth NC, Carr AM. The G2-phase DNA-damage checkpoint. Trends Cell Biol. 2000;10:296–303. doi: 10.1016/s0962-8924(00)01773-6. [DOI] [PubMed] [Google Scholar]
- 14.Rhind N, Russell P. Checkpoints: it takes more than time to heal some wounds. Curr Biol. 2000;10:R908–R911. doi: 10.1016/s0960-9822(00)00849-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhind N, Russell P. Chk1 and Cds1: linchpins of the DNA damage and replication checkpoint pathways. J Cell Sci. 2000;113(Pt 22):3889–3896. doi: 10.1242/jcs.113.22.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka K, Russell P. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat Cell Biol. 2001;3:966–972. doi: 10.1038/ncb1101-966. [DOI] [PubMed] [Google Scholar]
- 17.Zhao H, Tanaka K, Noguchi E, Noguchi C, Russell P. Replication checkpoint protein Mrc1 is regulated by Rad3 and Tel1 in fission yeast. Mol Cell Biol. 2003;23:8395–8403. doi: 10.1128/MCB.23.22.8395-8403.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noguchi E, Noguchi C, Du LL, Russell P. Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol Cell Biol. 2003;23:7861–7874. doi: 10.1128/MCB.23.21.7861-7874.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noguchi E, Noguchi C, McDonald WH, Yates JR, III, Russell P. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol Cell Biol. 2004;24:8342–8355. doi: 10.1128/MCB.24.19.8342-8355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsumoto S, Ogino K, Noguchi E, Russell P, Masai H. Hsk1-Dfp1/Him1, the Cdc7-Dbf4 kinase in Schizosaccharomyces pombe, associates with Swi1, a component of the replication fork protection complex. J Biol Chem. 2005;280:42536–42542. doi: 10.1074/jbc.M510575200. [DOI] [PubMed] [Google Scholar]
- 21.Takeda T, Ogino K, Tatebayashi K, Ikeda H, Arai K, Masai H. Regulation of initiation of S phase, replication checkpoint signaling, and maintenance of mitotic chromosome structures during S phase by Hsk1 kinase in the fission yeast. Mol Biol Cell. 2001;12:1257–1274. doi: 10.1091/mbc.12.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ansbach AB, Noguchi C, Klansek IW, Heidlebaugh M, Nakamura TM, Noguchi E. RFCCtf18 and the Swi1–Swi3 complex function in separate and redundant pathways required for the stabilization of replication forks to facilitate sister chromatid cohesion in Schizosaccharomyces pombe. Mol Biol Cell. 2008;19:595–607. doi: 10.1091/mbc.E07-06-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capasso H, Palermo C, Wan S, Rao H, John UP, O’Connell MJ, Walworth NC. Phosphorylation activates Chk1 and is required for checkpoint-mediated cell cycle arrest. J Cell Sci. 2002;115:4555–4564. doi: 10.1242/jcs.00133. [DOI] [PubMed] [Google Scholar]
- 24.Lindsay HD, Griffiths DJ, Edwards RJ, Christensen PU, Murray JM, Osman F, Walworth N, Carr AM. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walworth N, Davey S, Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature. 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- 26.Wan S, Capasso H, Walworth NC. The topoisomerase I poison camptothecin generates a Chk1-dependent DNA damage checkpoint signal in fission yeast. Yeast. 1999;15:821–828. doi: 10.1002/(SICI)1097-0061(199907)15:10A<821::AID-YEA422>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 27.Sommariva E, Pellny TK, Karahan N, Kumar S, Huberman JA, Dalgaard JZ. Schizosaccharomyces pombe, Swi1, Swi3, and Hsk1 are components of a novel S-phase response pathway to alkylation damage. Mol Cell Biol. 2005;25:2770–2784. doi: 10.1128/MCB.25.7.2770-2784.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 29.Paciotti V, Clerici M, Scotti M, Lucchini G, Longhese MP. Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol Cell Biol. 2001;21:3913–3925. doi: 10.1128/MCB.21.12.3913-3925.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- 31.Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- 32.Tercero JA, Longhese MP, Diffley JF. A central role for DNA replication forks in checkpoint activation and response. Mol Cell. 2003;11:1323–1336. doi: 10.1016/s1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- 33.Kim WJ, Lee S, Park MS, Jang YK, Kim JB, Park SD. Rad22 protein, a rad52 homologue in Schizosaccharomyces pombe, binds to DNA double-strand breaks. J Biol Chem. 2000;275:35607–35611. doi: 10.1074/jbc.M007060200. [DOI] [PubMed] [Google Scholar]
- 34.Ostermann K, Lorentz A, Schmidt H. The fission yeast rad22 gene, having a function in mating-type switching and repair of DNA damages, encodes a protein homolog to Rad52 of Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:5940–5944. doi: 10.1093/nar/21.25.5940. [DOI] [PMC free article] [PubMed] [Google Scholar]