Abstract
Benign metastasizing leiomyoma, a rare condition of controversial origin, is characterized by the occurrence of extrauterine smooth muscle tumors primarily affecting the lungs of women with a history of uterine leiomyomas. Numerous genetic studies of uterine leiomyoma with rearrangements of the HMGA2 and HMGA1 loci defined in prominent subgroups have been conducted. In contrast, cytogenetic and molecular descriptions of benign metastasizing leiomyoma are few, and, in particular, this entity has not been previously subjected to single nucleotide polymorphism (SNP) array analysis. In this study, conventional karyotypic, and/or molecular cytogenetic, and SNP array characterization of a pleuropulmonary benign mestasizing leiomyoma and a synchronous deep soft tissue leiomyoma of the thigh, which arose in a 56-year-old female with a remote history of uterine leiomyomata, revealed rearrangement of the HMGA1 (6p21) locus and nearly identical genomic profiles, including loss of chromosome 7 material in both lesions. These findings suggest that both the deep soft tissue and pleuropulmonary lesions were derived from the same abnormal clone and are genetically related to uterine leiomyomata.
Keywords: Benign metastasizing leiomyoma, cytogenetics, HMGA1, fluorescence in situ hybridization (FISH)
Benign metastasizing leiomyoma (BML) is a rare, benign smooth muscle tumor first described by Steiner in 1939 (1). This tumor primarily occurs in premenopausal women with a history of uterine leiomyomata and previous myomectomy or hysterectomy. Occasionally, the uterine lesions are discovered synchronously. The lung is the most common site of involvement, but other distant extra-uterine sites reported include abdomino-pelvic lymph nodes, heart, skeletal muscle, breast, brain, and bone, among others (2–8).
Several theories regarding the pathogenesis of BML have been postulated. Some authors have proposed it arises de novo (5,9). Others assert that distant organ or lymph node involvement occurs because uterine leiomyoma fragments gain venous access or enter dilated lymphatic channels following surgical manipulation (8,10–13). Metaplastic transformation of coelomic epithelium due to hormonal stimulation has also been put forward as an explanation for BML (8,14,15). The possibility also exists that pre-existing or concurrent uterine smooth muscle tumors are actually leiomyosarcomas (11,12).
Conventional cytogenetic studies have provided valuable insight regarding the histopathogenesis of numerous mesenchymal neoplasms. To our knowledge, only five pulmonary BMLs have been previously characterized karyotypically (16). In the current study, a chromosomally aberrant clone detected in a pulmonary BML from a 56-year-old female was subsequently shown to share similar genomic imbalances with a synchronous deep soft tissue leiomyoma using molecular cytogenetic (fluorescence in situ hybridization [FISH] with chromosome specific probes) and whole genome single nucleotide polymorphism (SNP) array analysis.
Case history
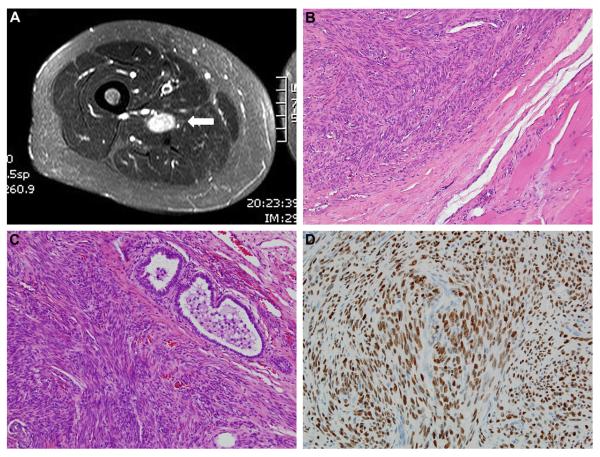
A 56-year-old female presented with worsening and refractory hypertension of six months’ duration. Her past medical history was significant for total abdominal hysterectomy performed 20 years prior. Radiographic studies (renal ultrasound and abdominal CT angiogram) demonstrated evidence of fibromuscular dysplasia of the right renal artery, a 3 cm mass near the spleen, and bilateral enlarged, heterogeneous adnexa. Moreover, plain radiographs and chest CT revealed multiple bilateral lung nodules. Consequently, the patient underwent evaluation for malignancy of unknown primary, which included a PET scan that showed a moderately fludeoxyglucose (FDG) avid soft tissue mass in the right adductor magnus muscle (Figure 1A). The lung nodules and perisplenic mass were non-avid.
Figure 1.
Benign metastasizing leiomyoma. (A) MRI of well-circumscribed thigh mass (white arrow). (B) Thigh mass exhibiting bland, relatively cellular, smooth muscle proliferation without atypia, necrosis, or mitotic activity. (C) Similar histopathologic appearance for the pleuropulmonary lesion. (D) Strong, diffuse nuclear staining for estrogen receptor in the pleuropulmonary lesion.
The soft tissue mass in the thigh was excised. Grossly, the thigh mass was a firm, tan, well-circumscribed nodule, 2.2 cm in greatest dimension, with a white-pink whorled surface on cut section. Histopathologically, the lesion was composed of relatively well-organized bundles of smooth muscle cells without significant cytologic atypia, lesional necrosis, or identifiable mitotic activity (Figure 1B). The lesional cells were diffusely and strongly positive for α-smooth muscle actin, and the diagnosis of soft tissue leiomyoma was made.
Since this was considered an incidental finding and not the source of diffuse pulmonary metastasis, a pleuropulmonary and a subpleural nodule were subsequently biopsied. The gross and microscopic appearances of the lung nodules were identical to that of the soft tissue leiomyoma (Figure 1C). In addition to α-smooth muscle actin, the lesional cells in the lung nodules were also positive for estrogen receptor-α (Figure 1D) and progesterone receptor. Estrogen receptor-α and progesterone receptor immunohistochemical staining were retrospectively performed on the thigh nodule, and were both positive.
Histopathologic re-review of the hysterectomy specimen from 1990 confirmed the presence of multiple leiomyomata, the largest of which was 7 cm. While focal ischemic-type necrosis was noted, no cytologic atypia or coagulative tumor necrosis was identified. No mitotic activity was observed in over 50 high-power fields. The blocks from this original specimen had been discarded years prior to the patient’s current presentation.
Materials and methods
Conventional cytogenetic analysis
Representative fresh, lesional tissue from a right middle lobe lung nodule was received from Vanderbilt University Medical Center (VUMC) for conventional cytogenetic analysis. The tumor sample was disaggregated mechanically and enzymatically with scissors and collagenase, and the sample was cultured in RPMI-1640 media supplemented with 20% fetal bovine serum and antibiotics for 4–6 days as previously described (17). Metaphase chromosomes were banded with Giemsa trypsin, and karyotypes were described according to the International System for Cytogenetic Nomenclature 2009 (18).
Molecular cytogenetic analysis
In an effort to assess the presence or absence of shared genomic imbalances between the thigh and lung lesions, FISH studies were performed using a chromosome 7 specific centromere probe (CEP 7; Abbott Molecular, Des Plaines, IL) and a 22q13 BAC probe cocktail (RP11-63H6, RP4-742C19, RP11-7L8, RP11-111A3; Invitrogen, Grand Island, NY) on 4 μm sections from formalin-fixed paraffinembedded (FFPE) tissue, as per slight modification of the manufacturer’s instructions. In addition, based on the karyotypic observance of a 6p21 rearrangement, FISH was also performed using a HMGA1 break-apart probe as previously described on both thigh and lung lesions (19).
Specifically, tissue sections were deparaffinized in Hemo-D three times at room temperature for 5 minutes each, followed by dehydration in 95% ethanol twice for 1 minute each, and airdried. Tissue sections were then pretreated in 0.2N HCl at room temperature for 20 minutes, rinsed in distilled water at room temperature for 3 minutes, incubated in Pretreatment Solution (Abbott Molecular) at 80°C for 30 minutes, rinsed in distilled water again at room temperature for 3 minutes, followed by digestion in protease solution (25 mg Protease I in 50 ml of Protease buffer, Abbott Molecular) at 37°C for 15 minutes, and finally post-fixed with 1% formaldehyde in 1× phosphate buffered saline (PBS)/MgCl2 for 10 minutes. The slides were then dehydrated in an ethanol series (70%, 85%, and 100%) at room temperature for 2 minutes each and air-dried. Following pretreatment, the tissue and probes were co-denatured at 80°C for 5 minutes and incubated overnight at 37°C using the HYBrite system (Abbott Molecular). Posthybridization washing was performed in 2× SSC/0.1% NP-40 at 72°C for 2 minutes, followed by 2× saline sodium citrate (SSC)/0.1% NP-40 at room temperature for 1 minute. The slides were then counterstained with 4,6-diamidino-2-phenylindole (DAPI) II (Abbott Molecular).
Hybridization signals were assessed in 200 interphase nuclei with strong and well-delineated signals by two different individuals (D.H. and J.M.B.). Normal uterine myometrium served as a negative control.
SNP array analysis
DNA was extracted from representative unstained, formalin-fixed paraffin-embedded tissue sections as described previously (20). Samples were processed with the 250K Nsp Assay Kits (Affymetrix, Santa Clara, CA). Briefly, 1 μg of genomic DNA was digested with Nsp restriction enzyme, ligated to the adaptors, and amplified by polymerase chain reaction (PCR) using a universal primer. After purification of PCR products with SNPClean magnetic beads (Agencourt Biosciences, Beverly, MA), amplicons were quantified, fragmented, labeled, and hybridized to 250K Nsp arrays. Following washing and staining, the arrays were scanned to generate .CEL files for downstream analysis. Data acquired from the Affymetrix GeneChip Operating System v4.0 (GCOS) was analyzed using Affymetrix Gene-Chip Genotyping Analysis Software (GTYPE) 4.1. Copy number analysis was performed with Copy Number Analyzer for Affymetrix GeneChip arrays (CNAG 3.0) (21–23).
Results
Conventional cytogenetic analysis
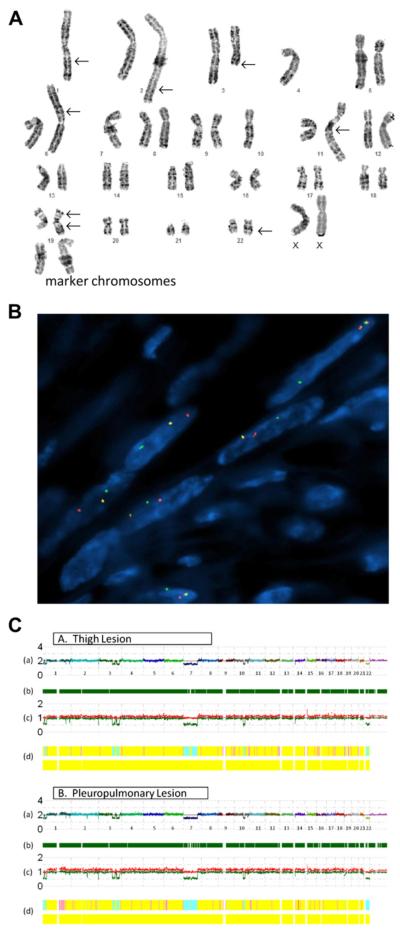
Traditional karyotyping of the pleuropulmonary nodule revealed the following abnormal composite clone in 12 cells: 42–44,XX,-1,add(1)(q25),add(2)(q37),del(3)(q13),-4,add(6) (p21),-7,-10,add(11)(q25),add(19)(p13),der(19)add(19)(p13) add(19)(q13.2),add(22)(q12),+2-5mar (Figure 2A). Eight cells were karyotypically normal.
Figure 2.
A) Conventional karyotype of the pleuropulmonary benign metastasizing leiomyoma revealing a hypodiploid clone characterized by loss of chromosomes 1, 4, 7, and 10; gain of 2–5 marker chromosomes; additional material of unknown origin on 1q, 2q, 6p, 11q, 19p, and 22q; a deletion of 3q; and a derivative 19 with additional unknown material on both long and short arms. (B) FISH analysis with a HMGA1 break-apart probe demonstrates split red and green signals indicative of a rearrangement of this locus. (C) SNP array whole genome view of the thigh and pleuropulmonary lesions (A and B, respectively) showing nearly identical genomic profiles, with the exception of an additional loss of 2q22.1 in the pleuropulmonary lesion but not the thigh lesion; (A) estimated copy number as log2 ratio–chromosomes are color coded and sequential along the x-axis; (B) dark green bars represent heterozygous reads; (C) allele-based analysis (high and low alleles are shown in red and green, respectively); and (D) hidden Markov model estimates for copy number (top bar: pink = 3, yellow = 2, aqua = 1) and LOH likelihood (bottom bar: negative for the presence of LOH).
Molecular cytogenetic analysis
Loss of one CEP7 probe signal and one 22q13 BAC cocktail probe signal were detected in 59% and 53% of the tumor cells of the thigh lesion, respectively, and in 55% and 51% of the interphase cells from the karyotyped lung nodule, respectively. A rearrangement of the HMGA1 (6p21) locus was identified in 65% and 74% of the tumor cells from the deep soft tissue and pleuropulmonary leiomyomata, respectively (Figure 2B).
SNP array analysis
SNP array results of both the lung and thigh lesions were concordant with the corresponding conventional cytogenetic results, showing a loss of one copy number of 1p36.33-1p36.23, 1p36.13-1p35.2, 2q37.3, 3q23, 3q25.1, 3q25.32-3q26.33, 7p22.3-7q31.33, 10q22.3-10q23.33, and 22q11.1-22q13.33 (Figure 2C). The pleuropulmonary BML had an additional loss of 2q22.1.
Discussion
The nature of BML has been debated. A histologically benign lesion, it has been hypothesized to arise either: 1) de novo; 2) subsequent to mechanical instrumentation leading to vascular/lymphatic dissemination; 3) via metastasis from a low-grade uterine leiomyosarcoma; 4) as a passive deposit from intravenous leiomyomatosis; or 5) as a hormonally induced metaplastic transformation. Occurring predominantly in women having undergone gynecological surgery for uterine leiomyomata, the lesion’s pathogenesis has been an enigma. Genetic studies could be useful in providing mechanistic insight.
Only a solitary cytogenetic study of BML has been previously published. Nucci et al. (16) analyzed five pulmonary BML specimens from five different patients. All cases showed 19q and 22q terminal deletions; loss of 1p and 13q material was detected in three and two lesions, respectively; and two lesions exhibited rearrangements of 6p21. Similar cytogenetic changes involving 19q, 22q, and 6p21 were identified in the case reported herein, which further supported the non-random association of these anomalies with BML. A previous metaphase-based comparative genomic hybridization study of a case of BML revealed no genomic imbalances (24). X-chromosome inactivation analysis of the lung and uterine lesions from this same patient confirmed the presence of a shared (identical) inactivation pattern. Patton et al. (10) assessed the variable length of the polymorphic CAG repeat sequence within the human androgen receptor gene on pulmonary and uterine lesions from two informative patients and found identical patterns of androgen receptor allelic inactivation, which indicated that the lesions were clonal.
Roughly 40% of primary uterine leiomyomata are characterized by non-random structural chromosomal abnormalities (25). Of these karyotypically abnormal uterine leiomyomata, at least 20% demonstrate 12q15 rearrangements involving the HMGA2 locus sometimes accompanied by interstitial deletions of chromosome 7 (commonly involving q22-32) (25). Alternatively, 7q deletions may occur as the sole anomaly. Fewer than 10% of cytogenetically aberrant leiomyomas harbor rearrangements of 6p21 (HMGA1 locus) either as the sole anomaly or accompanied by other clonal changes (25). Interestingly, a karyotypic abnormality of 6p21 accompanied by loss of 7q material was identified in the current lung lesion with rearrangement of HMGA1 confirmed by FISH in interphase cells of both the thigh and lung lesions. Notably, HMGA1 rearrangements have not been identified in leiomyosarcoma. These findings would suggest the hypothesis proposing BML as metastatic disease from a low-grade uterine leiomyosarcoma is less likely.
In addition to FISH, a SNP array approach was employed to compare the genomic profile of the deep soft tissue thigh mass with the cytogenetically characterized pleuropulmonary BML, since only FFPE tissue was available. Remarkably, the genomic profiles of the thigh and lung lesions were nearly identical, including partial loss of chromosomes or chromosomal arms 1p, 2q, 3q, 7, 10q, and 22q in addition to the aforementioned HMGA1 rearrangement. The strikingly similar genetic profiles of these anatomically distinct lesions suggests a common origin from the same abnormal clone. For the pleuropulmonary BML, near-complete concordance could also be demonstrated between the SNP array and conventional karyotypic findings. Loss of material involving chromosomes 4, 6, 11, and 19 noted by standard karyotyping but not by SNP array analysis might be explained by the presence of this chromosomal material within marker chromosomes.
The initial clinical impression was that the smooth muscle tumor of the thigh was an incidental deep soft tissue leiomyoma. This lesion was well circumscribed grossly and histologically—traits emblematic of either a primary deep soft tissue leiomyoma or “benign metastasis.” Aside from the pleuropulmonary lesion, the thigh lesion also expressed estrogen and progesterone receptors—findings compatible with hormonally driven uterine smooth muscle derivation. Features diagnostic of leiomyosarcoma, such as coagulative tumor cell necrosis, abundant mitoses, atypical mitoses, and cytologic atypia, were absent in this case. These observations coupled with the genetic findings suggest the thigh lesion represents a BML.
In conclusion, this unique case represents a novel approach to the study of BML. In the past, the genetic distinction between leiomyoma and leiomyosarcoma of the uterus was well defined, with recurrent anomalies such as HMGA2 (12q15) and HMGA1 (6p21) rearrangements and loss of 7q in subsets of the former, and lack of these changes but much more complex and non-recurring aberrations that reflected a high level of genomic instability in the latter. The findings of the current study confirm the presence of non-random genetic changes characterizing the entity known as “benign metastasizing leiomyoma,” which have in common alterations witnessed as recurrent in uterine leiomyoma. Interestingly, the cytogenetic/loss of heterozygosity (LOH) pattern of 1p loss, which is often accompanied by 19q and/or 22q loss, represents a distinct but uncommon uterine leiomyoma subgroup characterized by increased cellularity with transcriptional profiles similar to those of uterine leiomyosarcomas (26). The identification of 6p21 (HMGA1 locus) rearrangements in four of seven BMLs studied to date (including two in the current study) suggests these lesions are genetically related to uterine leiomyomata, and that this genetic aberration may also be associated with a subset of leiomyomata with possible metastatic potential when accompanied by 1p, 19q, and/or 22q loss. Finally, application of SNP array and FISH studies for assessment of synchronous lesions arising in distinct anatomic locations (i.e., deep soft tissue and pleuropulmonary) in this patient revealed essentially identical genetic profiles further supporting a shared derivation instead of an independent tumor origin for these lesions.
Acknowledgments
The authors thank Marilu Nelson, BS, MS for her valuable technical assistance. This study was supported in part by a grant from the National Institutes of Health U-10-CA98543-091 (J.A.B.).
References
- 1.Steiner PE. Metastasizing fibroleiomyoma of the uterus: report of a case and review of the literature. Am J Pathol. 1939;15:89–110. 7. [PMC free article] [PubMed] [Google Scholar]
- 2.Jo JH, Lee JH, Kim DC, et al. A case of benign metastasizing leiomyoma with multiple metastasis to the soft tissue, skeletal muscle, lung, and breast. Korean J Intern Med. 2006;21:199–201. doi: 10.3904/kjim.2006.21.3.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang MW, Kang SK, Yu JH, et al. Benign metastasizing leiomyoma: metastasis to the ribs and vertebra. Ann Thorac Surg. 2001;91:924–926. doi: 10.1016/j.athoracsur.2010.08.030. [DOI] [PubMed] [Google Scholar]
- 4.Galvin SD, Wademan B, Chu J, et al. Benign metastasizing leiomyoma: a rare metastatic lesion in the right ventricle. Ann Thorac Surg. 2010;89:279–281. doi: 10.1016/j.athoracsur.2009.06.050. [DOI] [PubMed] [Google Scholar]
- 5.Cho KR, Woodruff JD, Epstein JI. Leiomyoma of the uterus with multiple extrauterine smooth muscle tumors: a case report suggesting multifocal origin. Hum Pathol. 1989;20:80–83. doi: 10.1016/0046-8177(89)90207-4. [DOI] [PubMed] [Google Scholar]
- 6.Thomas EO, Gordon J, Smith-Thomas S. Diffuse uterine leiomyomatosis with uterine rupture and benign metastatic lesions of the bone. Obstet Gynecol. 2007;109:528–530. doi: 10.1097/01.AOG.0000237314.07944.32. [DOI] [PubMed] [Google Scholar]
- 7.Senay S, Kaya U, Cagil H, et al. Surgical management of intravenous leiomyomatosis with cardiac extension. Do we need total circulatory arrest? Thorac Cardiovasc Surg. 2007;55:322–323. doi: 10.1055/s-2007-964953. [DOI] [PubMed] [Google Scholar]
- 8.Abell MR, Littler ER. Benign metastasizing uterine leiomyoma. Multiple lymph nodal metastases. Cancer. 1975;36:2206–2213. doi: 10.1002/cncr.2820360938. [DOI] [PubMed] [Google Scholar]
- 9.Horiuchi K, Yabe H, Mukai M, et al. Multiple smooth muscle tumors arising in deep soft tissue of lower limbs with uterine leiomyomas. Am J of Surg Path. 1998;22:897–901. doi: 10.1097/00000478-199807000-00012. [DOI] [PubMed] [Google Scholar]
- 10.Patton KT, Cheng L, Papavero V, et al. Benign metastasizing leiomyoma: clonality, telomere length, and clinicopathologic analysis. Mod Pathol. 2006;19:130–140. doi: 10.1038/modpathol.3800504. [DOI] [PubMed] [Google Scholar]
- 11.Weiss SW, Goldblum JR, editors. Enzinger and Weiss’s soft tissue tumors. 5th Edition Mosby Inc; Philadelphia, PA: 2008. pp. 530–1146. [Google Scholar]
- 12.Nuovo GJ, Schmittgen TD. Benign metastasizing leiomyoma of the lung: clinicopathologic, immunohistochemical, and micro-RNA analyses. Diagn Mol Pathol. 2008;17:145–150. doi: 10.1097/PDM.0b013e31815aca19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koh DM, Burn PR, King DM. Benign metastasizing uterine leiomyoma with intracaval leiomyomatosis. Br J Radiol. 2000;73:435–437. doi: 10.1259/bjr.73.868.10844871. [DOI] [PubMed] [Google Scholar]
- 14.Hoynck van Papendrecht HP, Gratama S. Leiomyomatosis peritonealis disseminata. Eur J Obstet Gynecol Reprod Biol. 1983;14:251–259. doi: 10.1016/0028-2243(83)90268-x. [DOI] [PubMed] [Google Scholar]
- 15.Awonuga AO, Shavell VI, Imudia AN, et al. Pathogenesis of benign metastasizing leiomyoma: a review. Obstet Gynecol Surv. 2010;65:189–195. doi: 10.1097/OGX.0b013e3181d60f93. [DOI] [PubMed] [Google Scholar]
- 16.Nucci MR, Drapkin R, Dal Cin P, et al. Distinctive cytogenetic profile in benign metastasizing leiomyoma: pathogenetic implications. Am J Surg Pathol. 2007;31:737–743. doi: 10.1097/01.pas.0000213414.15633.4e. [DOI] [PubMed] [Google Scholar]
- 17.Bridge JA, Neff JR, Mouron BJ. Giant cell tumor of bone: chromosomal analysis of 48 specimens and review of the literature. Cancer Genet Cytogenet. 1992;58:2–13. doi: 10.1016/0165-4608(92)90125-r. [DOI] [PubMed] [Google Scholar]
- 18.Shaffer LG, Slovak MC, Campbell LJ, editors. ISCN 2009: an international system for human cytogenetic nomenclature. Karger; Basel: 2009. [Google Scholar]
- 19.Medieros F, Araujo AR, Erickson-Johnson MR, et al. HMGA1 and HMGA2 rearrangements in mass-forming endometriosis. Genes Chromosomes Cancer. 2010;49:630–634. doi: 10.1002/gcc.20772. [DOI] [PubMed] [Google Scholar]
- 20.Lyons-Weiler M, Hagenkord J, Sciulli C, et al. Optimization of the Affymetrix GeneChip mapping 10k 2.0 assay for routine clinical use on formalin-fixed paraffin-embedded tissues. Diagn Mol Pathol. 2008;7:3–13. doi: 10.1097/PDM.0b013e31815aca30. [DOI] [PubMed] [Google Scholar]
- 21.Nannya Y, Sanada M, Nakazaki K, et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65:6071–6079. doi: 10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto G, Nannya Y, Kato M, et al. Highly sensitive method for genomewide detection of allelic composition in nonpaired, primary tumor specimens by use of affymetrix single-nucleotidepolymorphism genotyping microarrays. Am J Hum Genet. 2007;81:114–126. doi: 10.1086/518809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monzon FA, Hagenkord J, Lyons-Weiler M, et al. Whole genome SNP arrays as a potential diagnostic tool for the detection of characteristic chromosomal aberrations in renal epithelial tumors. Mod Pathol. 2008;21:599–608. doi: 10.1038/modpathol.2008.20. [DOI] [PubMed] [Google Scholar]
- 24.Teitze L, Günther K, Hörbe A, et al. Benign metastasizing leiomyoma: a cytogenetically balanced but clonal disease. Hum Pathol. 2000;31:126–128. doi: 10.1016/s0046-8177(00)80210-5. [DOI] [PubMed] [Google Scholar]
- 25.Hodge JC, Morton CC. Genetic heterogeneity among uterine leiomyomata: insights into malignant progression. Hum Mol Gen. 2007;16:R7–R13. doi: 10.1093/hmg/ddm043. [DOI] [PubMed] [Google Scholar]
- 26.Christacos NC, Quade BJ, Dal Cin P, et al. Uterine leiomyomata with deletions of 1p represent a distinct cytogenetic subgroup associated with unusual histologic features. Genes Chromosomes Cancer. 2006;45:304–312. doi: 10.1002/gcc.20291. [DOI] [PubMed] [Google Scholar]