Abstract
Motivation: A common question arises at the beginning of every experiment where RNA-Seq is used to detect differential gene expression between two conditions: How many reads should we sequence?
Results: Scotty is an interactive web-based application that assists biologists to design an experiment with an appropriate sample size and read depth to satisfy the user-defined experimental objectives. This design can be based on data available from either pilot samples or publicly available datasets.
Availability: Scotty can be freely accessed on the web at http://euler.bc.edu/marthlab/scotty/scotty.php
Contact: gabor.marth@bc.edu
Supplementary information: Supplementary data are is available at Bioinformatics online.
1 INTRODUCTION
An experiment’s power to detect differences in expression is based on its ability to distinguish true biological differences between conditions from the variability that occurs in repeated measurements of the same condition. In an RNA-Seq experiment, this variability stems from three sources: biological variance, technical measurement imprecision and Poisson variance stemming from the inherent nature of counting experiments (Supplementary Section S1). These sources of variability lead to uncertainty about the gene’s true average expression in each condition, limiting the resolution of the differences that can be detected as statistically significant.
The uncertainty that is caused by biological and non-Poisson technical variance can be countered by increasing the number of biological replicates of each condition. Biological replicates are used because their measurements are subject to both. In contrast, Poisson uncertainty is reduced to the same degree if a fixed number of reads is used to either add more replicates or sequence the existing replicates more deeply (Supplementary Section S2, Fig. S1). Given a fixed number of reads, the most power will be achieved if these reads are used to sequence the highest number of biological replicates possible. However, biological material and library construction increase costs each time an additional replicate is added. Further, measuring a large fraction of the genes with low read counts can produce a dataset that is biased against identifying differentially expressed genes with low read counts because these genes will be measured with higher noise.
We devised a simple web-based tool, Scotty, which allows users to optimize the replicate number and read depth to maximize the statistical power achieved, while excluding configurations that require too many replicates, are too expensive, do not have sufficient power or result in datasets where large subsets of genes are measured with a high measurement bias. Scotty is similar in function to existing programs that are available for microarray experiments (Seo et al., 2006), but is specifically adapted to RNA-Seq.
2 WORKFLOW AND IMPLEMENTATION
The general workflow for a biologist using Scotty is shown in Supplementary Figure S2. First, Scotty uses prototype data to quantify the rate at which new RNAs are measured and the degree of variability between replicates. Because these attributes are determined by both biology and experimental noise, they will be most accurately estimated from pilot data generated using the same techniques that will be used in the actual experiment, preferably by the same laboratory (Section 4). However, Scotty also enables users to run power analyses using pre-loaded publicly available datasets as prototypes. Pilot data will ideally consist of read count data from two pairs of replicates: one each from the control and test condition (Supplementary Section S3).
To model power, Scotty fits the observed data to theoretical distributions, which were selected based on empirical observations. Scotty first assesses how many reads are required to measure a specified number of genes or transcripts as described in Supplementary Section S4. Scotty then estimates how much variance is present between replicates of the same condition, which largely determines how many replicates are required (Supplementary Section S5). Scotty then recommends to the user the optimal experimental configuration by testing a matrix of experimental designs. The possible designs are constrained by user-defined parameters specifying the maximum number of replicates and reads per replicate. For each replicate count, 10 different read depths are tested for power, cost and a bias metric. Statistical power is calculated using a t-test (Chow et al., 2002; Harrison and Brady, 2004) for reasons described in Supplementary Section S6.
Under default settings, Scotty calculates the percentage of genes or transcripts with a 2× fold change in the test condition relative to the control that will be detected at P < 0.01. A power difference metric is used to determine how many measurements will be significantly affected by Poisson noise. It defines a maximum power for each expression level as the percentage that would be expected to be detected if there were no Poisson noise. As read counts increase, power asymptotically approaches this maximum power. Under the default settings, the power difference metric is the percentage of measurements that are measured with <50% of maximum power.
To test how well Scotty’s results can be generalized to other methods, we used simulation experiments to compare the power of a t-test with the power of the DESeq, a statistical package whose approach shares information between genes to provide a better estimate of variance when it is poorly measured owing to low replicate number (Anders and Huber, 2010). We found that while DESeq has substantially higher power to detect differentially expressed genes when there are two replicates present the power of the two methods was similar when there were three or more replicates (Supplementary Section S6).
3 PERFORMANCE AND OUTPUT
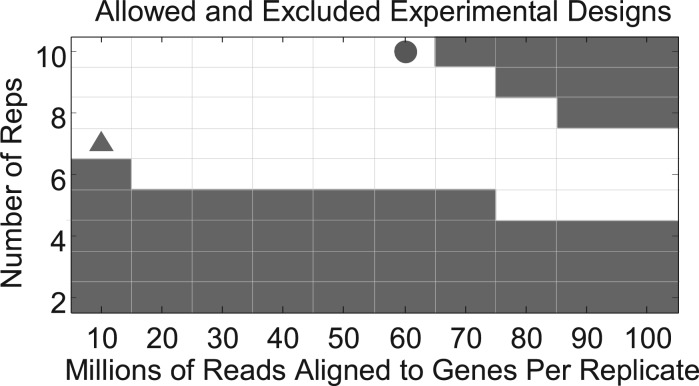
Scotty’s primary output is a matrix showing the power of the experimental configurations that are permitted under the user constraints and pinpoints the cheapest and the most powerful design choices (Fig. 1). Other figures show the cost and measurement bias of each design. Basic quality metrics for pilot data are also included. To test the accuracy of Scotty’s power estimates, we simulated datasets having a known number of differentially expressed genes sequenced to different depths. We found that two replicates with 10 million reads each were sufficient to predict power in configurations of up to 10 replicates and 100 million reads with >0.99 correlation with the number of differentially expressed genes that could be identified in the data (Supplementary Fig. S3).
Fig. 1.
An example output from the Scotty application. This figure shows the user which of the tested experimental configurations do (white) and do not (shaded) conform to the user-defined constraints. Scotty then indicates the optimal configuration based on cost (filled triangle) and power (filled circle)
4 PILOT DATA VERSUS EXISTING DATASETS
We set out to examine if publicly available datasets are sufficient for designing a new experiment, or whether pilot data are required.
We analyzed four publicly available human liver RNA-Seq datasets (Supplementary Section S7, Fig. S4) and asked how well one such dataset predicts the rate of gene discovery (saturation) in another. We find substantial variability among saturation curves (less between experiments from the same lab, more across groups) and predictions may over- or under-estimate the number of genes quantified at a given read depth by up to 55%. Furthermore, there was less variability between individuals within the same dataset than across experiments, suggesting that the primary source of differences is experimental artifact rather than biological variability. Similarly, we find large differences in how well biological replicates reproduce within each experiment (Supplementary Fig. S5). These observations suggest that pilot data generated by the same laboratory will provide a more accurate prediction of power than publicly available experiments.
5 DISCUSSION
While general guidance exists for biologists on designing RNA-Seq experiments (Fang and Cui, 2011), Scotty is, to our knowledge, the first interactive tool aiding RNA-Seq experimental design. As the accuracy of Scotty’s projections is determined by the degree of similarity between the pilot and the main experiment, pilot data should be collected under conditions closely resembling the experimental conditions.
ACKNOWLEDGEMENTS
We thank our system administrator Tony Schreiner for substantial help in deploying Scotty on the web and Kourosh Zarringhalam, Patricia Smith, and Igor Lasic for helpful suggestions.
Funding: R01 HG004719 from NHGRI to G.T.M.
Conflict of Interest: none declared.
REFERENCES
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow SC, et al. A note on sample size calculation for mean comparisons based on noncentral t-statistics. J. Biopharm. Stat. 2002;12:441–456. doi: 10.1081/BIP-120016229. [DOI] [PubMed] [Google Scholar]
- Fang Z, Cui X. Design and validation issues in RNA-seq experiments. Brief Bioinform. 2011;12:280–287. doi: 10.1093/bib/bbr004. [DOI] [PubMed] [Google Scholar]
- Harrison D, Brady A. Sample size and power calculations using the noncentral t-distribution. Stata J. 2004;4:142–153. [Google Scholar]
- Seo J, et al. An interactive power analysis tool for microarray hypothesis testing and generation. Bioinformatics. 2006;22:808–814. doi: 10.1093/bioinformatics/btk052. [DOI] [PubMed] [Google Scholar]