Abstract
Most retroviruses require translational recoding of a viral messenger RNA stop codon to maintain a precise ratio of structural (Gag) and enzymatic (Pol) proteins during virus assembly1,2. Pol is expressed exclusively as a Gag–Pol fusion either by ribosomal frameshifting or by read-through of the gag stop codon3. Both of these mechanisms occur infrequently and only affect 5–10% of translating ribosomes, allowing the virus to maintain the critical Gag to Gag–Pol ratio4–8. Although it is understood that the frequency of the recoding event is regulated by cis RNA motifs, no mechanistic explanation is currently available for how the critical protein ratio is maintained. Here we present the NMR structure of the murine leukaemia virus recoding signal and show that a protonation-dependent switch occurs to induce the active conformation. The equilibrium is such that at physiological pH the active, read-through permissive conformation is populated at approximately 6%: a level that correlates with in vivo protein quantities. The RNA functions by a highly sensitive, chemo-mechanical coupling tuned to ensure an optimal read-through frequency. Similar observations for a frameshifting signal indicate that this novel equilibrium-based mechanism may have a general role in translational recoding.
Genetic recoding either by frameshifting or by read-through requires cis-acting elements downstream of the recoding site in the mRNA. In murine leukaemia virus (MLV), a 63-nucleotide read-through signal folds as a pseudoknotted7–9 structure (MLV-PK) that can independently direct recoding of stop codons5,6,10, even in heterologous RNA (see Supplementary Text and Supplementary Fig. 1a). Secondary structure mapping indicates that MLV-PK adopts a classic hairpin-type fold9 consisting of two stems, S1 and S2, connected by a single base loop L1 and an 18-nucleotide loop L2 (Supplementary Fig. 1a). The 8-base spacer between the UAG stop codon and the first base pair of stem S1 have been proposed to be unstructured7,8,11. Although a majority of RNA pseudoknots contain specific tertiary interactions between stem and loop elements12, RNA melting data monitored by changes in UV absorption. indicated that these interactions in MLV-PK have an unexpected and pronounced dependence on pH (Supplementary Fig. 2). Assignment of the NMR data (Supplementary Figs 3 and 4) unambiguously confirmed the presence of a pH-dependent equilibrium between two inter-converting conformations. Marked chemical shift perturbations are observed as the minor conformation becomes increasingly populated at lower pH values: this allowed us to select reporter residues, A17 and A14, by which to evaluate the equilibrium (Fig. 1b). As the N1 position of A17 becomes protonated, typical adjacent C2 chemical shift perturbations occur that report on the pKa for this protonation event. Concurrent with protonation at A17, the C2 position of the A14 residue in the minor groove of stem S1 experiences a downfield chemical shift change (Fig. 1b) and we observe the appearance of nuclear Overhauser enhancements (NOEs) from the A14 H2 resonance to ribose resonances of G49 and U50 in loop L2. Together these data indicate that A14 enters a more electronegative environment upon the approach of loop L2 and hence reports on formation of S1–L2 tertiary interactions. Interestingly, curve fitting for both the A17 and A14 resonances yielded a pKa value of 6.23 and a pfold value of 6.20, respectively, implying that the two processes of protonation and tertiary structure formation may be coupled (pfold = pH where 50% of MLV-PK have a tertiary structure) (Fig. 1c and Supplementary Fig. 5). We calculate that at physiological pH (7.4), a distribution would result in which ~6% of the MLV-PK population would have a tertiary structure whereas the remaining ~94% would lack the tertiary contacts. As this distribution correlates both with our in vivo observations (Supplementary. 6a) and with the previously observed in vivo levels of Gag–Pol and Gag4–8, respectively, we asked if an equilibrium (PKactive ⇌ PKinactive) determines the Gag:Gag–Pol ratio.
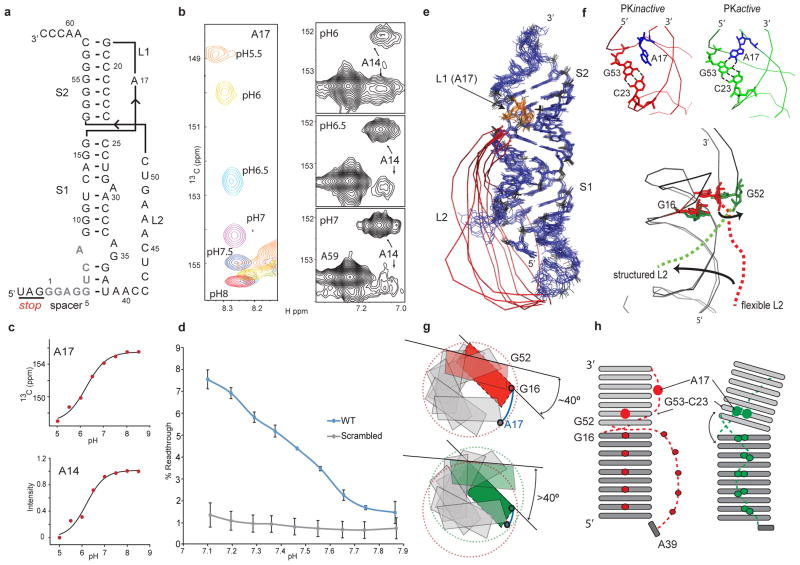
Figure 1. MLV-PK conformational-equilibrium-dependent read-through.
a, NMR-derived secondary structure of MLV-PK. Spacer residues (grey) G5–A8 originally predicted to be unstructured are involved in stem S1 formation and consequently loop L2 is shortened. Residue numbers are labeled according to their position in the in vitro construct b, Adenosine C2 chemical shift changes in residues A17 and A14 during pH titrations (Supplementary Fig. 5). c, Nonlinear data fits26 for residues A17 (left) and A14 (right) to determine the pKa and pfold respectively (Supplementary Fig. 5). d, The effects of the translation pH on MLV-PK read-through activity in vitro. Error bars indicate standard error (n = 3). Decreasing the pH from 7.8 to 7.1 leads to a 500% increase in read-through levels. At pH 7.0, our NMR data indicate that ~8% of MLV-PK is in the folded conformation, which correlates very closely to the in vitro read-through level of ~7.5% at pH 7.1. e, NMR structure ensemble of PKinactive with residue A17 in orange and the flexible loop L2 in red. A 180° rotated view is shown in Supplementary Fig. 10a. f, Close-up views of the stem S2 and loop L1 major groove in both forms of MLV-PK and the A17+-C23:G53 base-triple in PKactive. Dotted lines indicate hydrogen-bond formation. g, Overlay of the PKinactive (red) and PKactive (green) structures showing the global transition upon PKactive formation. h, Schematic of the secondary structure model (bottom), a view from the helical axis (top). A17 protonation in MLV-PK overwinds the helical twist, which consequently leads to an interhelical bend and releases the steric hindrance at the S1–S2 junction.
To test functionally this hypothesis, we developed a dual luciferase reporter13 in vitro translation system and modulated the response by varying the assay pH (Fig. 1d). A control experiment using a scrambled MLV-PK sequence showed that translational read-through by ribosomes is not affected in the pH range tested. These data show that at pH 7.4 ~5.5% of ribosomal recoding occurs—a level in agreement with our measured value of conformational exchange from the NMR experiments. Notably, however, the activity of MLV-PK was found to be strictly dependent upon the pH of the translation system, and exhibits enhanced read-through activity at lower pH (Fig. 1d). Thus, these results indicate that formation of tertiary structure in MLV-PK leads Figto read-through of the stop codon and, furthermore, that enriching the population with the tertiary fold enhances read-through-stimulating activity. Mutational analysis to test the importance of protonation at A17 uncovered additional protonation sites, which demonstrate that MLV-PK is a complex multiple-proton sensor, and that protonation of A17 is one of two or more chemical triggers that govern the equilibrium (see Supplementary Text and Supplementary Figs 6b and 7).
Although conformational heterogeneity precludes us from assigning all protonation sites and their effect on the global structure of PKactive, we can examine the role of A17+ by obtaining a partial structure of PKactive (see Methods). Comparison with the complete PKinactive structure allows us to propose a mechanism for the conformational transitions that are required for read-through activity (see Supplementary Figs 8, 9, 10 and 11 and Supplementary Table 1). In both conformations a G–U wobble and three distinct A–U Watson–Crick base pairs are observed in the NMR data (Supplementary Fig. 8). Whereas two canonical A–U base pairs are readily assigned to stem S1 (U12–A30 and A14–U27), the third pair can only be formed between bases U6 and A37. Thus, in contrast to previous chemical probing studies9, our NMR assignments unambiguously prove that the helical stem S1 is extended by additional base pairing between residues G5–A8 in the spacer and loop L2 residues A34–U38 (Fig. 1a, e and Supplementary Fig. 1). The extended stem S1 helix also contains a 1 × 2 internal bulge14,15 formed by bases A8, A34 and G35 (see also Supplementary Fig. 9). Additionally, our structures consistently show residue A29 in stem S1 to be looped out towards the major groove and not involved in tertiary interactions even in PKactive (Supplementary Fig. 10). A direct consequence of the 12-base-pair (including the internal A8–G35 stack) extended stem S1 is a shortened 4-nucleotide spacer and a shortened loop L2 that has no long-range NOEs to stem S1 and hence is flexible in PKinactive (Fig. 1e).
In the PKinactive structure (Fig. 1e and Supplementary Table 1 and Supplementary Fig. 10), the extended stem S1 coaxially stacks on stem S2 to form a collinear, quasi-continuous helix as evidenced by typical stacking NOEs between G16 and G52 across the helical junction (Supplementary Fig. 4). Collinear stacking of stems S1 and S2 is possible because S2 contains seven base pairs, which leaves an ~8-Å phosphorus–phosphorus distance for loop L1 across the S2 major groove. This distance can be spanned by the single nucleotide A17 without introducing any major twist or bend at the helical junction16. However, this coaxial stacking leads to steric hindrance at the junction, which, combined with the shortened nature of loop L2 prevents S1–L2 interactions. In PKactive, protonation leads to changes that include a loss of stacking interactions between G16 and G52 at the helical junction, a marked chemical shift change for residues at the S1–L2 turn (Supplementary Fig. 11) and the appearance of NOEs from G52 H2′/3′, G52 H8 and G53 H8 to the protonated A17+ H2 (Supplementary Fig. 12). These data indicate that in PKactive A17+ forms an A17+–C23:G53 base triple in the major groove (Fig. 1f). This repositioning of A17+ decreases the length of loop L1 below the required minimal distance for coaxial stacking and, accordingly, an inter-helical bend is introduced that relieves the steric hindrance at the junction and allows for S1–L2 tertiary interactions (Figs 1f–h).
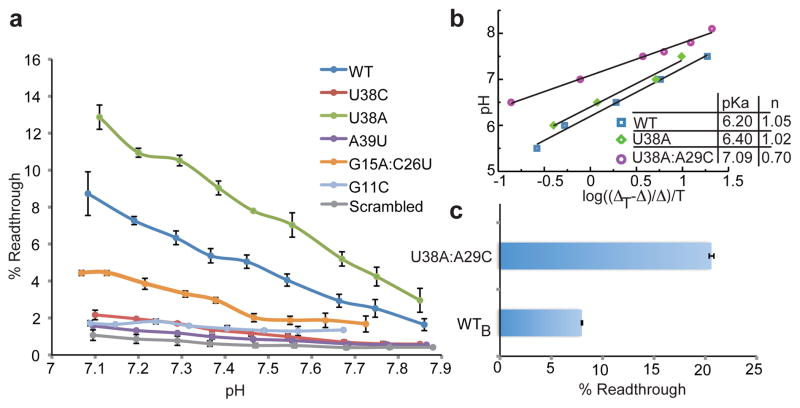
Because our data show that MLV-PK contains critical motifs that may regulate the structural transition (A17, A29, the helical junction and the S1–L2 turn), we proposed that manipulating these sites to favour tertiary contacts may modulate the equilibrium between PKactive and PKinactive. Given that adenosine-stacking interactions are known to increase thermodynamic stability at turns17, we asked if a U38A mutant would allow for increased A38-to-A39 stacking at the S1–L2 turn and explain the previously observed increase in read-through levels8. In addition, we used an A29C mutant to test if the looped-out A29 sterically hinders S1–L2 triplex formation. Read-through levels in vivo are significantly stimulated in both U38A (~80% increase) and A29C (~20%) (Supplementary Fig. 6c). Remarkably, the pKa for A17 in the U38A mutant is increased to 6.40 (Fig. 2a, b) and inter-helical bending occurs at higher pH (data not shown). Consequently, this construct has an increased response to pH change (Fig. 2a). These data indicate that improved adenosine stacking in U38A facilities S1–L2 folding, which is tightly coupled to the protonation event at the distal A17 site via the helical junction. Next, we asked if the two hyperactive mutants could have an additive effect in a U38A:A29C double mutant. Notably, a synergistic effect was observed for the double mutant, affording a ~260% increase in read-through levels in vivo, which correlates with a significantly elevated pKa of 7.09 for A17 (Fig. 2b, c and Supplementary Fig. 13). Furthermore, we could also design mutants that disrupt the structural integrity of the stem S1 and the S1–L2 turn and shift the equilibrium towards the PKinactive form: the pH response is thus attenuated (G15A:C26U) or abrogated (G11C, U6A, A39U) (Fig. 2a and Supplementary Figs 6c and 11b).
Figure 2. Functional data for modulation of the equilibrium.
a, The effects of the translation pH on read-through activity of MLV-PK S1–L2 turn mutants in vitro. The data show the positive effect of increased stacking in U38A, which is abrogated in U38C and correlates with in vivo activity (Supplementary Fig. 6c). Error bars indicate standard error (n = 3). b, Hill plots27 (R ≥ 0.99) to determine the pKa values of A17 in the indicated MLV-PK constructs. ΔT is the total chemical shift difference for the C2 carbon of A17 in PKinactive and PKactive and Δ is the difference between the C2 chemical shift at a given pH and the maximum chemical shift. c, In vivo read-through levels for the MLV-PK wild type (WTB) and the U38A:A29C mutant.
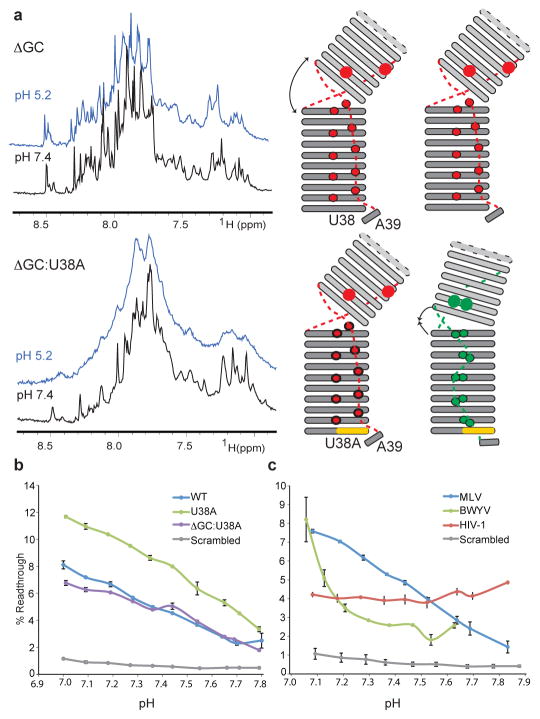
Lastly, we decided to engineer an artificial inter-helical bend by deleting one base pair in S2 (ΔGC) to test the importance of the mechanical response. Noticeably, our NMR data for ΔGC show that although the junction is unstacked at high pH, its ability to form the PKactive conformer is abrogated (Fig. 3a and Supplementary Fig. 14). This construct is not pH responsive, indicating that the correct register for the S2–L1 triple-helix formation does not form. We were able to rescue this mutant by using a ΔGC:U38A double mutant (Fig. 3b). Because U38A stabilizes the S1–L2 turn, this construct readjusts the inter-helical bend by bringing loop L2 in register with stem S1. This restores the pH response and conclusively proves that long-range effects occur between the junction and the S1–L2 turn. Although it was not possible to perform the in vitro translation experiment on the ΔGC mutant owing to the associated introduction of an in-frame U38AA stop codon, we could test the ΔGC:U38A construct (Fig. 3b). In agreement with our NMR data this construct rescues the pH-modulated recoding activity, albeit with a lower efficiency compared to U38A (Fig. 3b). These experiments prove that the mechanical and chemical responses in MLV-PK are indeed coupled, and confirm our equilibrium-based RNA conformational switch model. Lastly, similar pH-dependent, frameshift activity by the well-characterized frameshifting signal from beet western yellows virus (BWYV)18,19 pseudoknot (Fig. 3d) suggests that fine-tuning a proton-driven structural equilibrium may be a common mechanism for regulating recoding of gene expression.
Figure 3. Compensatory effects of the S1–L2 turn on the engineered inter-helical bend and equilibrium-based frameshifting levels.
a, One-dimensional NMR data at pH 7.4 (black) and pH 5.2 (blue) for the indicated MLV-PK constructs (left) to probe the long-range conformational transition. See Supplementary Fig. 2b for comparison with wild-type MLV-PK. Corresponding schematic topology models (right) indicate the degree of inter-helical bending and compensation by the S1–L2 turn. Broadening of the line widths are indicative of tertiary structure formation, which is abrogated in GC but compensated in the GC:U38A mutant. b, The effects of the translation pH on the in vitro read-through activity of the above MLV-PK constructs. Error bars indicate standard error (n = 3). c, Comparison of the effects of the translation pH on MLV-PK read-through and BWYV and human immunodeficiency virus-1 (HIV-1) frameshift activity in vitro. Error bars indicate standard error (n = 3). In HIV-1, the recoding signal is understood to be a stem-loop28 in which no protonation sites are predicted. In our pH-modulated assay the HIV-1 construct is not dependent on protonation for activity.
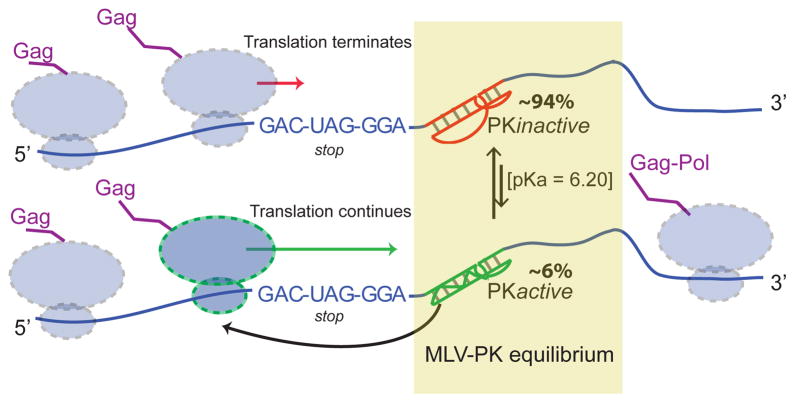
RNA is capable of a wide range of structural transitions ranging from subtle rearrangements to global folding that determine function20. MLV-PK is a highly responsive, multi-proton sensor that couples a chemical and mechanical response to induce a conformational transition. We present a novel equilibrium-based mechanism for regulation of the frequency of ribosomal recoding, wherein the only determinant required is an RNA conformational switch (Fig. 4). Previous explanations for how the recoding frequency is maintained have invoked mechanisms such as ribosomal mechanical stress, thermodynamic and kinetic stability of the recoding signal, ribosomal pausing and ribosomal helicase unwinding21,22. These processes will have a role in recoding but, according to our proposed mechanism, they would occur only if the translating ribosome encounters the active conformation of the RNA. Further support for our mechanism comes from the slow exchange lifetime of MLV-PK on the NMR time scale (>100 ms), which is slower than the known rate of ribosomal decoding in vivo23. This ensures that the pseudoknot tertiary fold will be maintained while a translating ribosome attempts to decode the Gag: Gag–Pol boundary.
Figure 4. Model for equilibrium-based mechanism.
Gene expression via translational recoding is regulated by a dynamic equilibrium (PKactive ⇌ PKinactive) between an active, read-through permissive conformation and an inactive, non-permissive conformation the distribution of which determines the Gag:Gag–Pol ratio. Ribosomes that encounter the PKactive conformation continue translating through the stop codon. Structural and functional data indicate that PKactive interacts with the ribosome before the stop codon is in the decoding centre owing to the shortened nature of the linker.
METHODS SUMMARY
Detailed methods for RNA sample preparation, biophysical characterization12,24, structure determination24,25 and translational recoding assays13 can be found in Methods.
METHODS
RNA sample preparation
DNA sequences were designed to include the T7 promoter, an insert sequence corresponding to nucleotides 1–63 of MLV-PK, a SmaI linearization site and restriction sites for BamHI and EcoRI for insertion of the amplified product into pUC19. RNA samples for biophysical experiments were transcribed and purified as described24. Samples were prepared in various buffers as required for the experiments (see later).
UV melting analysis
RNA samples for the wild-type, U38A and A39U MLV-PK constructs were annealed at 95 °C under dilute conditions (1–2 μM) in UV buffer (20 mM cacodylate, 1 M NaCl). Absorbance versus temperature melting curves for the MLV-PK RNA samples were acquired at wavelengths of 260 nm and 280 nm. The heating rate was fixed at 0.5 °C min−1 with a Beckman DU800 single beam spectrophotometer equipped with a Peltier heating device. The pH experiments were performed at pH 5.5 and 7.5 and absorbance data were converted to first derivative versus temperature plots and analysed as described previously12.
In vivo translation assay
Read-through was monitored using the p2luc dual luciferase reporter system13. The complete MLV-PK, BWYV and HIV-1 recoding sequence (MLV-PK, GACCCTAGATGACTAGGGAGGTCAGGGTCAGGAGCCCCCCCCTGAACCCAGGAAAC CCTCAAAGTCGGGGGGCAACCCGTC; BWYV, CAATTCATCGGGAAACTAAGTGCGCGGCACCGTCCGCGGAACAAACGGAAG; HIV-1, GAGACAGGCTAATTTTTTAGGGAAGATCTGGCCTTCCCACAAGGGAAGGCCAGGAA TTTTCTTCAGAGCAGACCATAGCC) were inserted between the Renilla (Rluc) and firefly (Fluc) luciferase genes using either the SalI and BamHI or the BamHI and SacI restriction sites. To normalize for transfection and translation efficiency, control constructs were transfected in parallel to the sample plasmid. The control plasmid was identical to the MLV-PK construct except for a single U to C mutation in the stop codon and represented a read-through efficiency of 100% at the mutated codon. The control constructs for BWYV and HIV-1 contained the same sequences in the −1 reading frame and the slippery sites were abrogated: (BWYV, CAATTCATCCGGGAAGCTAAGTGCGCGGCACCGTCCGCGGAACAAACGGAAG; HIV-1, GAGACAGGCTAACTTCTTAAGGGAAGATCTGGCCTTCCCACAAGGGAAGGCCAGGG AATTTTCTTCAGAGCAGACCATAGCC). Dual luciferase assays were performed in 96-well format. 293A cells (Invitrogen) were plated at a density of 2 × 104 cells per well in 125 μl DMEM plus penicillin and streptomycin and 10% FBS and grown overnight at 37 °C in 5% CO2. Transfections were performed with Fugene6 (Roche) and transfection mixes were made according to the manufacturer’s instructions (50 ng reporter, 0.150 μl Fugene6, serum-free medium in a 17.5 μl final volume). Transfections were performed in triplicate and 5 μl of the transfection mix was added to each well. Twenty-four hours post-transfection, the supernatant was decanted and 25 μl 1× PLB (passive lysis buffer; Promega) was added to each well. Lysates were incubated at ambient temperature with agitation for 10 min then subjected to one freeze–thaw cycle (−80 °C). Dual luciferase measurements were performed with 20 μl of the lysate using a dual luciferase reporter assay system (Promega). The luciferase values from the control were used to normalize those from the sample, allowing us to report read-through normalized to 100%.
In vitro translation assay
Dual luciferase plasmids containing the MLV-PK recoding sequence inserted between the SalI and BamHI sites were linearized with HpaI and recovered by ethanol precipitation. The linearized template (1 μg) was used to synthesize capped RNA using the mMESSAGE mMACHINE kit (Ambion) and the manufacturer’s protocol (2 h incubation). The capped RNA was polyadenylated for 1 h using the poly(A) tailing kit (Ambion) and the mRNA was purified using the MEGAClear kit (Ambion). RNA integrity was confirmed by running 1 μg on a denaturing glyoxal gel. The pH of the rabbit reticulocyte lysate (Ambion) was altered by the addition of dilute HCl and NaOH, the concentration of which was empirically determined. 5.75 μl of HCl (9.35, 6.32, 3.33, 0.0 mM) or NaOH (0.53, 1.30, 2.04, 2.78, 3.51, 4.23, 4.93, 5.63, 6.34 mM) was dispensed into 0.2 ml PCR tubes. Translation mixes (prepared in triplicate) were combined on ice and contained 1.25 μl 20× translation buffer (Ambion), 0.5 μl 50× (2.5 mM) methionine, 17 μl lysate and 0.5 μl (0.125 μg) reporter mRNA. The translation mix was added to the dilute acid or base, briefly vortexed, and incubated at 30 °C for 1.5 h. The reaction was quenched on ice for 10 min, 5 μl was removed for the dual luciferase assay and the pH of the remaining reaction was measured at 30 °C with an InLab Micro pH probe (Mettler-Toledo).
NMR data acquisition, resonance assignment and structure calculations
For NMR experiments RNA samples were resuspended in NMR buffer (10 mM Tris-HCl at pH 6.5 and pH 7.5 for PKactive and PKinactive, respectively, and 10 mM NaCl). NMR data were acquired using Bruker 700 MHz and 900 MHz spectrometers equipped with cryoprobes. Spectra were recorded at 298 K and 308 K with the exception of data for the imino region for which data were also recorded at 278 K. Assignments for non-exchangeable 1H and 13C signals were obtained from two-dimensional NOESY, two-dimensional HMQC and three-dimensional HMQC-NOESY data sets recorded with unlabelled and nucleotide-specific (ACCN-MLV-PK, GCCN-MLV-PK) selectively labelled samples and two-dimensional NOESY samples obtained for nucleotide-specifically deuterated samples (GUH-MLV-PK and ACH-MLV-PK)24. Structures were calculated as described24 using manually assigned restraints in CYANA25. The statistics table for the PKinactive structure ensemble is included in Supplementary Table 1. The changes that occur upon A17 protonation in stem S2, loop L1 and the helical junction were used to generate a model for this region of PKactive. The NOEs were confirmed with various MLV-PK mutants (U6A, A17U, A17C) (see Supplementary Figs 11 and 12). Molecular images were generated with PyMOL (http://www.pymol.org).
Supplementary Material
Acknowledgments
We thank J. Atkins for the p2luc plasmid, the New York Structural Biology Center for NMR time and A. Hawkins, S. Leiman and J. Gullinger for their contributions. B.H.-L. and S.P.G. would also like to thank the late D. Wolf for his inspiration and many helpful discussions. S.P.G. acknowledges grant R37 CA30488 from the NCI/NIH and is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Author Contributions: V.M.D’S., B.H.-L., M.A.D. and S.P.G. conceived of and designed the experiments. V.M.D’S., M.A.D., C.S., N.S. and B.H.-L. did the structural analysis, B.H.-L. did the recoding assays, and M.A.D. and J.M.N. did the in vivo assay for the double mutant. M.A.D. and V.M.D’S. wrote the manuscript with assistance from B.H.-L.
Author Information: Coordinates and restraints for the final ensemble of ten structures of PKinactive have been deposited in the Protein Data Bank under accession code 2LC8. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Felsenstein KM, Goff SP. Expression of the gag-pol fusion protein of Moloney murine leukemia virus without gag protein does not induce virion formation or proteolytic processing. J Virol. 1988;62:2179–2182. doi: 10.1128/jvi.62.6.2179-2182.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shehu-Xhilaga M, Crowe SM, Mak J. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J Virol. 2001;75:1834–1841. doi: 10.1128/JVI.75.4.1834-1841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baranov PV, Gesteland RF, Atkins JF. Recoding: translational bifurcations in gene expression. Gene. 2002;286:187–201. doi: 10.1016/s0378-1119(02)00423-7. [DOI] [PubMed] [Google Scholar]
- 4.Philipson L, et al. Translation of MuLV and MSV RNAs in nuclease-treated reticulocyte extracts: enhancement of the gag-pol polypeptide with yeast suppressor tRNA. Cell. 1978;13:189–199. doi: 10.1016/0092-8674(78)90149-6. [DOI] [PubMed] [Google Scholar]
- 5.Yoshinaka Y, Katoh I, Copeland TD, Oroszlan S. Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc Natl Acad Sci U S A. 1985;82:1618–1622. doi: 10.1073/pnas.82.6.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wills NM, Gesteland RF, Atkins JF. Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc Natl Acad Sci USA. 1991;88:6991–6995. doi: 10.1073/pnas.88.16.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wills NM, Gesteland RF, Atkins JF. Pseudoknot-dependent read-through of retroviral gag termination codons: importance of sequences in the spacer and loop 2. EMBO J. 1994;13:4137–4144. doi: 10.1002/j.1460-2075.1994.tb06731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alam SL, Wills NM, Ingram JA, Atkins JF, Gesteland RF. Structural studies of the RNA pseudoknot required for readthrough of the gag-termination codon of murine leukemia virus. J Mol Biol. 1999;288:837–852. doi: 10.1006/jmbi.1999.2713. [DOI] [PubMed] [Google Scholar]
- 9.Brierley I, Pennell S, Gilbert RJ. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat Rev Microbiol. 2007;5:598–610. doi: 10.1038/nrmicro1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honigman A, Wolf D, Yaish S, Falk H, Panet A. cis Acting RNA sequences control the gag-pol translation readthrough in murine leukemia virus. Virology. 1991;183:313–319. doi: 10.1016/0042-6822(91)90144-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng YX, Yuan H, Rein A, Levin JG. Bipartite signal for read-through suppression in murine leukemia virus mRNA: an eight-nucleotide purine-rich sequence immediately downstream of the gag termination codon followed by an RNA pseudoknot. J Virol. 1992;66:5127–5132. doi: 10.1128/jvi.66.8.5127-5132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nixon PL, Giedroc DP. Energetics of a strongly pH dependent RNA tertiary structure in a frameshifting pseudoknot. J Mol Biol. 2000;296:659–671. doi: 10.1006/jmbi.1999.3464. [DOI] [PubMed] [Google Scholar]
- 13.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 14.Schroeder S, Kim J, Turner DH. G.A and U.U mismatches can stabilize RNA internal loops of three nucleotides. Biochemistry. 1996;35:16105–16109. doi: 10.1021/bi961789m. [DOI] [PubMed] [Google Scholar]
- 15.Badhwar J, Karri S, Cass CK, Wunderlich EL, Znosko BM. Thermodynamic characterization of RNA duplexes containing naturally occurring 1 × 2 nucleotide internal loops. Biochemistry. 2007;46:14715–14724. doi: 10.1021/bi701024w. [DOI] [PubMed] [Google Scholar]
- 16.Michiels PJ, et al. Solution structure of the pseudoknot of SRV-1 RNA, involved in ribosomal frameshifting. J Mol Biol. 2001;310:1109–1123. doi: 10.1006/jmbi.2001.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clanton-Arrowood K, McGurk J, Schroeder SJ. 3′ terminal nucleotides determine thermodynamic stabilities of mismatches at the ends of RNA helices. Biochemistry. 2008;47:13418–13427. doi: 10.1021/bi801594k. [DOI] [PubMed] [Google Scholar]
- 18.Cornish PV, Hennig M, Giedroc DP. A loop 2 cytidine-stem 1 minor groove interaction as a positive determinant for pseudoknot-stimulated -1 ribosomal frameshifting. Proc Natl Acad Sci U S A. 2005;102:12694–12699. doi: 10.1073/pnas.0506166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atkins JF, Gesteland RF. Recoding: expansion of decoding rules enriches gene expression. Springer; 2010. [Google Scholar]
- 20.Al-Hashimi HM, Walter NG. RNA dynamics: it is about time. Curr Opin Struct Biol. 2008;18:321–329. doi: 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giedroc DP, Cornish PV. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 2009;139:193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agirrezabala X, Frank J. Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Q Rev Biophys. 2009;42:159–200. doi: 10.1017/S0033583509990060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhlman B, Luisi DL, Young P, Raleigh DP. pKa values and the pH dependent stability of the N-terminal domain of L9 as probes of electrostatic interactions in the denatured state. Differentiation between local and nonlocal interactions. Biochemistry. 38:4896–4903. doi: 10.1021/bi982931h. [DOI] [PubMed] [Google Scholar]
- 25.Legault P, Pardi A. In-situ probing of adenine protonation in RNA by 13C NMR. J Am Chem Soc. 1994;116:8390–8391. [Google Scholar]
- 26.D’Souza V, Dey A, Habib D, Summers MF. NMR structure of the 101-nucleotide core encapsidation signal of the Moloney murine leukemia virus. J Mol Biol. 2004;337:427–442. doi: 10.1016/j.jmb.2004.01.037. [DOI] [PubMed] [Google Scholar]
- 27.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 28.Staple DW, Butcher SE. Solution structure and thermodynamic investigation of the HIV-1 frameshift inducing element. J Mol Biol. 2005;349:1011–1023. doi: 10.1016/j.jmb.2005.03.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.