We wish to draw colleagues' attention to the detection in European poultry of H5N1 highly pathogenic notifiable avian influenza A (HPNAI) viruses from clade (genetic grouping) 2.3.2 affecting domestic poultry and wild birds in Romania and Bulgaria, respectively. Previous European occurrences of viruses related to the recent HPAI H5N1 epizootic in both wild birds and poultry in Europe have involved exclusively viruses from clade 2.2 (Alexander, 2007; Brown, 2009). This represents the most westerly spread of clade 2.3.2 viruses which have shown an apparently expanding range of geographical dispersal since mid-2009 following confirmation of infections in wild waterfowl species in Mongolia and Eastern Russia. Infection in a feral pigeon (Colombia livia) in Moscow during October of that year was the most westerly detection of HPNAI H5N1clade 2.3.2 virus until now (OIE, 2009). Poultry outbreaks with this genotype were reported in Nepal during early February 2010 (OIE, 2010a). Nucleotide sequencing at the Veterinary Laboratories Agency (VLA) in the UK of original sample material from two outbreaks revealed a close relationship with HPNAI H5N1 clade 2.3.2 viruses.
On 13 March 2010, official notification was made to the competent veterinary authority in Romania of the suspicion of AI infection based on gross pathology findings at post-mortem examination of two hens from two backyard flocks with a population of 47 hens from Letea, a small locality in the Danube delta, county of Tulcea. HPNAI virus of H5N1 subtype was confirmed on 15 March 2010 by RT-PCR using standard protocols (details available on request). The outbreak involved a cluster of six backyard flocks within a 400 metre radius (Defra, 2010; OIE, 2010b). On 29 March, a second outbreak of disease was confirmed affecting all 80 hens from one backyard flock in Plauru village, Tulcea County, located 55 km east of the first cluster (OIE, 2010c). H5N1 HPNAI was detected by RT-PCR (details available on request) following the discovery of 28 dead and 52 sick hens by a veterinarian on 24 March.
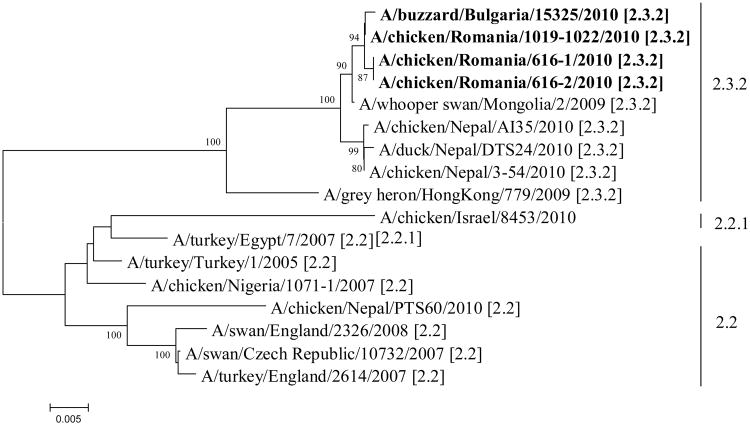
In a separate incident on 29 March 2010, The National Veterinary Service of Bulgaria notified the OIE of the detection of an H5N1 HPNAI virus from a pooled sample of lungs, trachea, liver, caecal tonsils and gizzard collected from a dead Common Buzzard (Buteo buteo). The bird had been found on 15 March 2010 in the St. Constantine and Helena resort on the Black Sea coast and submitted to the regional laboratory for AI in Varna as part of an ongoing Influenza surveillance and research collaboration with the Center of Excellence for Influenza Research and Surveillance at St. Jude Children's Research Hospital, USA (OIE, 2010d). During necropsy of the buzzard, no lesions on the internal organs were observed and in particular, no signs of HPNAI infection. Samples from the positive buzzard were sent to the National Reference Laboratory for AI in Sofia and the H5N1 subtype was confirmed. Clinical material from the poultry outbreaks in Romania and the Common Buzzard from Bulgaria was forwarded to the EU Reference Laboratory for Avian Influenza and Newcastle Disease, VLA-Weybridge for further virological investigation and confirmation that they were high pathogenicity isolates. Sequencing of the amplified haemagglutinin (HA) gene from the Romanian isolates (A/chicken/Romania/616-1/2010, A/chicken/Romania/616-2/2010 and A/chicken/Romania/1019-1022/2010) revealed a cleavage site (motif PQRERRRKRGLF) consistent with viruses of high pathogenicity belonging to clade 2.3.2 of the contemporary Eurasian H5N1 lineage. These findings were in accordance with those obtained by the National Reference Laboratory in Romania. The HA gene sequence of the Romanian isolates had a 99.3% similarity with the H5N1 HPNAI clade 2.3.2 virus isolates from Nepal, while the Bulgarian isolate, A/buzzard/Bulgaria/15325/2010, revealed a 99.9% similarity to the Romanian isolates (Figure 1).
Figure 1.
Phylogenetic analysis of the HA gene of seventeen HPAI H5N1 isolates. The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches for values greater than 70% (Felsenstein, 1985). The evolutionary distances were computed using the Tamura-Nei method (Tamura and Nei, 1993). There were a total of 1610 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 (Tamura et al., 2007).
Most of the reported H5N1 HPNAI detections in wild birds during the latter half of 2009 were associated with viruses belonging to clade 2.3.2. This represents an evolving and changing epidemiological dynamic, whereby virus from a clade other than clade 2.2 has apparently spread to wild birds, with potential maintenance and spread through such populations. There are, therefore, some similarities with the first emergence and spread of H5N1 HPNAI clade 2.2 viruses but careful monitoring is required to determine whether the clade 2.3.2 genotype is replacing clade 2.2, or both genotypes can co-exist. Host population dynamics and abnormal migratory movements are also key components in assessing risk to European poultry and there are relevant parallels to the situation in 2005/6 (Defra, 2010; Hesterberg et al., 2009). Further characterization of the clade 2.3.2 H5N1 HPNAI viruses is underway along with a thorough review of the existing molecular diagnostic tools to confirm their fitness-for-purpose.
A fuller description of these investigations will be provided in due course. Early detection of disease in Romania and Bulgaria was achieved through established programmes of AI surveillance in poultry and wild birds operating in these Member States. Influenza A viruses are clearly circulating in wild birds and poultry, and while it is not possible to reliably predict the scale of threat posed by the apparent ongoing westward spread of clade 2.3.2 HPNAI H5N1 viruses, ongoing vigilance for clinical signs of disease as part of existing passive surveillance frameworks for AI, and the prompt reporting of suspect cases in poultry is advised.
Acknowledgments
The authors thank Dr. Dennis Alexander of the VLA for his constructive review of the manuscript. This work was funded, in part, by the National Institute of Allergy and Infectious Diseases, the National Institutes of Health, under contract number: HHSN266200700005C and by the EU Reference Laboratory for Avian Influenza (contract number: EU9302).
References
- Alexander D. An overview of the epidemiology of avian influenza. Vaccine. 2007;25:5637–5644. doi: 10.1016/j.vaccine.2006.10.051. [DOI] [PubMed] [Google Scholar]
- Brown I. Summary of avian influenza activity in Europe, Asia, and Africa, 2006-2009. Avian Dis. 2009;54:187–193. doi: 10.1637/8949-053109-Reg.1. [DOI] [PubMed] [Google Scholar]
- Defra [assessed 6 July 2010];Preliminary Outbreak Assessment. 2010 Available at: http://www.defra.gov.uk/foodfarm/farmanimal/diseases/atoz/ai/latest-situation/index.htm.
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Hesterberg U, Harris K, Stroud D, Guberti V, Busani L, Pittman M, Piazza V, Cook A, Brown I. Avian influenza surveillance in wild birds in the European Union in 2006. Influenza Other Respi Viruses. 2009;3:1–14. doi: 10.1111/j.1750-2659.2008.00058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OIE. Immediate notification Highly pathogenic avian influenza. Russia: 2009. Weekly Disease Information, Vol. 22 - NO. 45, 5 November 2009. http//www.oie.int/wahis/public.php?page=weekly_report_index&admin=0. [Google Scholar]
- OIE. Immediate notification Highly pathogenic avian influenza. Nepal: 2010a. Weekly Disease Information, vol. 22 - No. 45, 5 February 2010. http://www.oie.int/wahis/public.php?page=weekly_report_index&admin=0. [Google Scholar]
- OIE. Immediate notification Highly pathogenic avian influenza. Romania: 2010b. Weekly Disease Information, Vol. 23 - No. 11, 16 March 2010. http: http://www.oie.int/wahis/public.php?page=weekly_report_index&admin=0. [Google Scholar]
- OIE. Follow-up report No 1 Highly pathogenic avian influenza. Romania: 2010c. Weekly Disease Information, Vol. 23 - No. 13, 30 March 2010. http://www.oie.int/wahis/public.php?page=weekly_report_index&admin=0. [Google Scholar]
- OIE. Immediate notification Highly pathogenic avian influenza. Bulgaria: 2010d. Weekly Disease Information, Vol. 23 - No. 13, 1 April 2010. http://www.oie.int/wahis/public.php?page=weekly_report_index&admin=0. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol and Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol and Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol and Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]